Abstract
Mutational activation of the epidermal growth factor receptor (EGFR) is a major player in the pathogenesis of non-small cell lung cancer (NSCLC). NSCLC patients with constitutively active EGFR mutations (mEGFR) eventually develop drug resistance against EGFR tyrosine-kinase inhibitors (TKIs); therefore, better understandings of key components of mEGFR signaling are required. Here, we initially observed aberrantly high expression of protein kinase Cα (PKCα) in lung adenocarcinomas, especially those with mEGFR, and proceeded to examine the role of PKCα in the regulation of the signaling pathways downstream of mutant EGFR (mtEGFR). The results showed that NSCLC cell lines with constitutively active EGFR mutations tend to have very or moderately high PKCα levels. Furthermore, PKCα was constitutively activated in HCC827 and H4006 cells which have an EGFR deletion mutation in exon 19. Interestingly, mtEGFR was not required for the induction of PKCα at protein and message levels, suggesting that the increased levels of PKCα are due to independent selection. Whereas, mtEGFR activity was required for robust activation of PKCα. Loss of functions studies revealed that the NSCLC cells rely heavily on PKCα for the activation of the mTORC1 signaling pathway. Unexpectedly, the results demonstrated that PKCα was required for activation of Akt upstream of mTOR but only in cells with the mtEGFR and with the increased expression of PKCα. Functionally, inhibition of PKCα in HCC827 led to caspase-3-dependent apoptosis and a significant decrease in cell survival in response to cellular stress induced by serum starvation. In summary, the results identified important roles of PKCα in regulating mTORC1 activity in lung cancer cells, whereby a primary switching occurs from PKCα-independent to PKCα-dependent signaling in the presence of mEGFR. The results present PKCα as a potential synergistic target of personalized treatment for NSCLC with constitutively active mutant forms of EGFR and constitutively active PKCα.
Keywords: signaling, oncogenes, kinases
INTRODUCTION
Lung cancer represents the leading cause of cancer-related deaths all over the world (1, 2). The treatment options for lung cancer are extremely limited due to the fact that most cases are usually diagnosed at late stages of the disease. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for 85% of the diagnosed cases, and is divided into three main types: adenocarcinoma, large cell carcinoma, and squamous cell carcinoma (3, 4). NSCLC can be associated with activating mutations in several oncogenes such as KRAS and epidermal growth factor (EGFR) or due to inactivation of some tumor suppressors such as PTEN, p53, Pb, and p16 (5–7). Mutations in EGFR are mostly in the form of exon 19 deletions affecting the LREA motif (delE746–750) or an exon 21 substitution of arginine for leucine at position 858 (L858R). These mutations lead to constitutive activation of EGFR leading to induction of its downstream pro-survival signaling cascades (8), and they occur in 10 up to 50% (in Asia-Pacific populations) of NSCLC patients with EGFR mutations (9, 10). NSCLC patients with either delE746–750 or L858R EGFR mutations have been shown to be initially sensitive to tyrosine kinase inhibitor (TKI) molecules such as erlotinib and gefitinib. However, they ultimately losoe their sensitivity due to emergence of resistance (11, 12).
The protein kinase C (PKC) family of serine-threonine kinases comprises ten members that are classified according to their structural domains and activators into three sub-families: diacylglycerol (DAG)-responsive/Ca+2 dependent (conventional; cPKC α, βI, βII, and γ), Ca+2-independent (novel; nPKCδ, ε, η, and θ), and atypical (aPKCζ and ι) that are independent of calcium and DAG (13, 14). Previous studies have demonstrated the roles of PKC family members in cancer progression and metastasis. PKCα has been shown to mediate cell proliferation, invasion, and anti-apoptosis in different cancer types, such as bladder (15), breast (16, 17), colon (18), gastric (19), glioma (20), melanoma (21), liver (22), and lung cancers (23–26). More importantly, PKCα plays anti-apoptotic and metastatic roles in NSCLC cells (24, 25, 27). PKCα has also been implicated in resistance to doxorubicin in lung cancer which was linked to the phosphorylation of the antiapototic protein RLIP76 at its threonine-297 residue (23). Moreover, miR-203-mediated downregulation of PKCα in A549 cells was associated with increased apoptosis (27). In regard to its role in metastasis, the activation of PKCα following its interaction with the substrate, discs large homology 1 (DLG1), was shown to induce migration of NSCLC cells (24). Moreover, increased PKCα activity was observed in lung metastatic nodules from diethylnitrosamine (DEN)-induced hepatocellular carcinoma in rats (25). Recently, PKCα was shown to mediate erlotinib resistance and epithelial-to-mesenchymal transition (EMT) in H1650 NSCLC cells (28). These findings suggest that PKCα could be a therapeutic target in NSCLC patients. In patients with NSCLC, PKCα has been shown to be highly expressed with higher levels being found in adenocarcinoma than squamous cell carcinoma (29). Therefore, in the present study, we aimed to investigate the possible regulation of PKCα by persistent EGFR activity driven by exon 19 deletion (delE746–750) and the role of PKCα in cell survival. The results point to a key acquired role of PKCα in reconfiguring signaling downstream of mutant EGFR. These results and their implications for the biology and therapeutics of NSCLC are discussed.
RESULTS
PKCα is highly expressed in NSCLC cell lines with EGFR mutation
It was reported that PKCα is highly expressed in NSCLC and preferentially expressed in adenocarcinoma compared with squamous carcinoma of the lung (29). In order to further understand the regulation of PKCα in NSCLC, the expression of PKCα was analyzed by examining Cancer Cell Line Encyclopedia (CCLE) database (30) available on cBioPortal website (http://www.cbioportal.org/study?id=cellline_ccle_broad). Six NSCLC cell lines with EGFR mutations (HCC827, HCC2279, HCC4006, HCC2935 with delE746-A750; H3255 with L858R; H1975 with L858R + T790M) and 8 NSCLC cell lines with wild-type EGFR (H1666, H1299, H1781, H292, HCC78, HCC95, H661, and H1437) were selected for comparisons of PKC expression levels. As can be seen, the mean Z score for PKCα (but not PKCβ or PKCγ) is significantly higher (1.37 ± 0.42) in NSCLC cell lines with EGFR mutations than cells with wild type EGFR (−0.54 ± 0.14; Fig. 1A–C). To corroborate the data, we randomly checked the protein level and mRNA level of PKCα in some NSCLC cells analyzed in the CCLE. Indeed, in the H292 cell line that harbors a wild-type EGFR, we could not detect PKCα protein by western blotting (Fig. 1D). Moreover, the mRNA level was also quite low; however, both protein and mRNA levels of PKCα were significantly elevated by 15-fold in HCC827 harboring E746-A750 deletion in exon 19 of EGFR compared to H292 cells with wild type EGFR (Fig. 1D). It was also observed that H4006, H3255, and H1975 expressed high PKCα protein levels (data not shown). To further explore the correlation between PKCα and the status of EGFR mutation in NSCLC, we screened a tumor tissue array. The results showed that 18 out of 40 patients’ tissue samples were positive for mtEGFR (deletion E746-A750) (45%) with high expression of PKCα. In addition, PKCα was highly expressed in 10 samples with no staining for the deletion mutant of EGFR, suggesting that some of the PKCα positive tumors may harbor additional mutations in EGFR or other oncogenes. However, the optical density of PKCα staining was significantly higher in samples with mtEGFR than those with negative staining for mtEGFR (Fig. 2A&B). By examining the lung carcinoma TCGA datasets, we found that there is a significant upregulation in PKCα mRNA expression in patients with EGFR mutations (82 patients) compared to those without EGFR mutations (936 patients; Supplementary Fig. 1). Collectively, these results indicate that NSCLC cell lines and patients harboring activating EGFR mutation tend to have high PKCα expression.
FIGURE 1. PKCα is highly expressed in NSCLC cell lines with EGFR mutations.
Mean Z scores from CCLE for PKCα (A), PKCβ (B) and PKCγ (C) were compared between 6 NSCLC cell lines with mtEGFR and 8 NSCLC cells with wild-type EGFR; (D) H292 and HCC827 cells were seeded in tissue culture plate over night, and then were harvested to examine the protein and mRNA levels of mtEGFR, PKCα by western blotting and real-time PCR.*** P<0.001.
FIGURE 2. PKCα is highly expressed in NSCLC patients with mtEGFR.
(A) Non-small cell lung carcinoma TMA, which includes 40 cases of non-small cell lung carcinoma and normal adjacent lung tissues in duplicate, were stained with either mtEGFR (E746-A750del Specific) or PKCα antibody following the standard immunohistochemistry procedure; (B) Mean optical density (OD) for PKCα staining were calcualted by ImageJ and compared between patients with positive and negative mtEGFR staining. ** p<0.01.
Activating mutations in EGFR lead to stimulation of PKCα in NSCLC cells
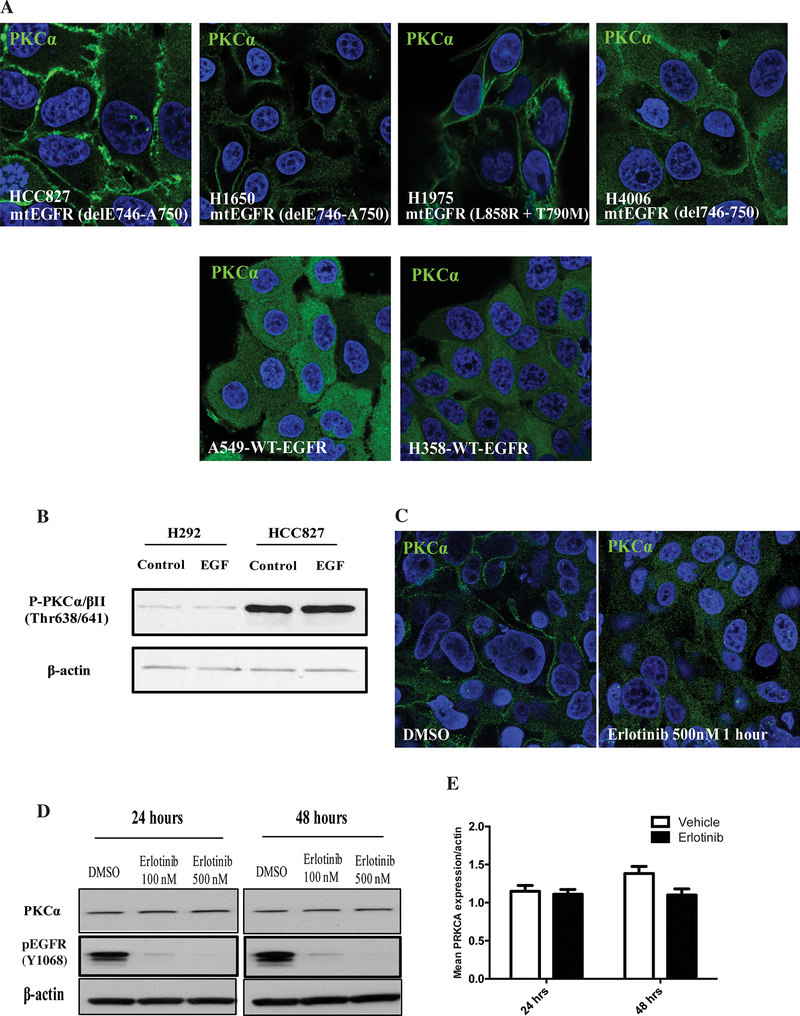
Since we observed the connection between PKCα and mtEGFR in NSCLC cell lines, it became important to determine the activity of PKCα in these cells. Thus, the localization of PKCα in these cells was assessed under serum starvation. As shown in Fig. 3A, in HCC827, H1650, H1975 and H4006, which contain activating EGFR mutation, PKCα was mostly located on the plasma membrane, indicating that PKCα was ‘constitutively’ active in the cells even under serum starvation. On the other hand, in A549 and H358 that have wild type EGFR, PKCα was mostly cytoplasmic (Fig. 3A). To further understand the status of PKCα in HCC827 cells, we evaluated the phosphorylation of PKCα/βII at Thr638/Thr641 in H292 and HCC827 since the phosphorylation of PKCα/βII at Thr638/Thr641 is important for their catalytic activity and represents an active form of PKCs. As shown in Figure 3B, the phosphorylation of PKCα/βII at Thr638/Thr641 in HCC827 cells was considerably higher than that in H292 cells, which suggests a robust PKCα catalytic activity in HCC827. Moreover, addition of EGF to HCC827 cells did not increase the phosphorylation of PKCα, consistent with the high ‘intrinsic’ activity of mtEGFR. Taken together, these results therefore support a correlation between mtEGFR and PKCα activation.
FIGURE 3. PKCα is activated in NSCLC cells.
(A) HCC827, H1650, H1975, H4006, A549 and H358 cells were serum starved for 5 hours, and then immunofluorescence was performed to visualize endogenous PKCα; (B) H292 and HCC827 cells were starved for 5 hours and then stimulated with 10ng/mL of EGF for 5 min as indicated; (C) HCC827 cells were starved and treated with DMSO or 500nM erlotinib for 1 hour followed by immunofluorescence to visualize endogenous PKCα; (D and E) HCC827 cells were starved and treated with DMSO, 100nM erlotinib, or 500nM erlotinib for 24 hours or 48 hours, followed by western blotting to analyze PKCα and phosphorylated EGFR protein level (D) or by real-time PCR to analyze PKCα mRNA level (E).
To determine whether mtEGFR is responsible for the activation of PKCα, we tested the effects of the EGFR specific inhibitor erlotinib on the localization of PKCα. As can be seen, the inhibitor was able to alter the localization of PKCα from the plasma membrane to the cytoplasm at a concentration of 500nM for 1 hour (Fig. 3C), thereby establishing a mechanistic link between activity of mtEGFR and activation of PKCα. On the other hand, and quite interestingly, prolonged treatment with erlotinib did not exert any effects on PKCα protein and mRNA expression, but decreased phosphorylation of PKCα (Fig. 3D&E, Supplementary Fig. 2), revealing that the increased expression of PKCα is not dependent on basal activity of mtEGFR, but that the activation of PKCα is. This result suggests that the increased expression of PKCα must represent a separate selection pressure in the cells with mtEGFR.
It then became important to determine whether EGFR mutation is sufficient to induce the sustained activation of PKC. Accordingly, H292 cells were co-transfected with wild type GFP- PKCα and empty vector, wild type EGFR or mtEGFR (containing E746- A750 deletion in exon 19). As can be seen, PKCα remained in the cytoplasm in cells transfected with PKCα and empty vector or wild type EGFR, but PKCα became mostly located on the plasma membrane when co-transfected with mtEGFR (Fig. 4A). Similarly, in H292 stably expressing PKCα and mtEGFR, PKCα is mostly located on the plasma membrane (Supplementary Fig. 3). Taken together, these results demonstrate that mtEGFR is necessary and sufficient to activate PKCα in the context of lung cancer cells with mtEGFR and high levels of PKCα.
FIGURE 4. EGFR mutation is sufficient to induce the sustained activation of PKC.
(A) H292 cells were co-transfected with PKCα-GFP and empty vector or PKCα-GFP and mtEGFR. After 24 hours, cells were serum starved for 5 hours, followed by immunofluorescence to visualize the location of PKCα-GFP, wild-type EGFR-GFP, or mtEGFR-GFP. Quantitation of GFP-positive cells with membrane localization is shown next to the images; (B) HCC827 cells were starved and treated with 5μM U73122 for 1 hour, and the localization of endogenous PKCα was analyzed by immunofluorescence; (C) HCC827 cells were starved and treated with DMSO or 500nM erlotinib for 1 hour and then the phospho-PLCγ Tyrosine 783 and PLCγ 1 and β-actin were analyzed by western blotting; (D) HCC827 cells were starved and treated with DMSO or 500nM erlotinib for 1 hour and then the phospho-PLCγ Tyrosine 783 was analyzed by immunofluorescence.
Since EGFR couples to PKC through the activation of phospholipase C (PLC)γ via phosphorylation on tyrosine 783, we next examined the effects of the PLCγ inhibitor U73122 on the localization of PKCα. As shown in Figure 4B, treatment with U73122 for 1 hour induced the relocation of PKCα from the plasma membrane to the cytoplasm. Similarly, downregulation of PLCγ1 by two different siRNAs also induced relocation of PKCα from the plasma membrane to the cytoplasm in HCC827 and in EGF-stimulated H292 stably expressing PKCα (Supplementary Fig. 3). Moreover, erlotinib inhibited the basal phosphorylation of PLCγ1 in HCC827 cells (Fig. 4C&D). Taken together, these results demonstrate that mtEGFR constitutively activates PKCα and drives the PKCα isoenzyme to the plasma membrane through PLCγ1 and this effect requires persistent activity of mtEGFR.
Activation of mTORC1 in NSCLC cell lines with mtEGFR requires PKCα and PLCγ
Since the data above suggest that mtEGFR continuously activates PKCα in HCC827 cells, it became important to determine the consequences of the sustained activation of PKCα in the NSCLC cell lines. Our laboratory previously reported that sustained/persistent activation of classical PKC results in the activation of mTORC1 (31). Therefore, mTORC1 signaling was examined in the NSCLC cell lines with mtEGFR. First, we examined the effect of the classical PKC inhibitors, Go6976 and enzastaurin, on p70S6K in HCC827 cells. As can been seen, at concentrations as low as 50nM, Go6976 was able to inhibit the basal phosphorylation of p70S6K with no effect on tyrosine phosphorylation of EGFR (Fig. 5A). Moreover, the more specific classical PKC inhibitor enzastaurin also inhibited the phosphorylation of p70S6K with no effect on tyrosine phosphorylation of EGFR (Fig. 5A). We likewise observed a similar inhibitory effect of these inhibitors in H4006 (Supplementary Fig. 2). In addition, we used PKCα siRNA to knock down PKCα in HCC827 and observed a decrease in the phosphorylation of p70S6K with or without EGF treatment (Fig. 5B). Since we showed that PLCγ is necessary for the activation of PKCα in HCC827, we then tested the effect of U73122 on the phosphorylation of p70S6K in HCC827 cells. The results showed that treatment with U73122 effectively reduced the phosphorylation of p70S6K with no effect on tyrosine phosphorylation of EGFR (Fig. 5C). Similar effect on p70S6K was observed after knocking down of PLCγ1 by using siRNA (Supplementary Fig. 3). Collectively, these data clearly demonstrate that the activation of mTORC1 requires a signaling pathway from the mtEGFR transduced by PLCγ and PKCα.
FIGURE 5. Activation of mTORC1 in NSCLC cell lines with mtEGFR requires PKCα, and PLCγ.
(A) HCC827 cells were starved and then treated with various concentrations of Go6976 or enzastaurin as indicated, following with western blotting to detect phosphorylation of S6K and phosphorylation of EGFR; (B) HCC827 cells were transfected with control siRNA or PKCα siRNA for 72 hours, and then serum starved for 5 hours following with 10ng/ml EGF for 5 min; (C) HCC827 cells were starved and then treated with different concentration of U73122 for 1 hour, followed by western blotting to detect pEGFR, pS6K, S6K and actin.
PKCα is critical for the activation of PI3K/Akt prompted by mtEGFR in HCC827
In glioma cells, it was shown that high levels of EGFR couple preferentially to PKCα which couples to mTOR in an Akt-independent manner (20). We therefore evaluated the roles of PKCα in the activation of Akt. The results showed that treatment with Go6976 and enzastaurin reduced the phosphorylation of Akt at Ser473 in HCC827 cells (Fig. 6A). The results also showed that PKCα knockdown via siRNA or CRISPR repressed the phosphorylation of Akt (Fig. 6B&C). As the phosphorylation of Akt at Thr308 was shown to be correlated with its activity in NSCLC (32), we evaluated the effect of enzastaurin and Go6976 on the phosphorylation of Akt at Thr308. As shown in Figure 6D, both inhibitors effectively inhibited the phosphorylation at Thr308. Similarly, deletion of PKCα via CRISPR also reduced the phosphorylation of Akt on T308 (Fig. 6D). Taken together, these results indicate that cPKC is critical for Akt activation in HCC827, unlike the results in glioma.
FIGURE 6. PKCα is involved in the activation of PI3K/Akt in HCC827 cells with mtEGFR.
(A) HCC827 cells were starved for 5 hours and then treated with Go6976 or enzastaurin at different concentration as indicated, followed by western blotting to detect P-AKT (S473) and actin; (B) HCC827 cells transfected with control or PKCα siRNA were starved for 5 hours, and then protein levels were checked by western blotting; (C) Vector and PKCα CRISPR HCC827 cells were serum starved for 5 hours followed by immunoblotting; (D) HCC827 cells were starved for 5 hours, followed by Go6976 or enzastaurin treatment for 1 hour; (E) HCC827 cells were starved and then treated with MK-2206 and KU-0063794 for 1 hour, and then phosphorylation and total protein level of targeted proteins were analyzed by western blotting; (F) HCC827 cells were starved for 5 hours and then treated with 5μM U73122 for 1 hour; (G) HCC827 cells were transfected with control, Gab1, or ErbB3 siRNA for 72 hours and then starved for 5 hours followed by western blotting to detect phosphorylated or total protein levels of targeted proteins; (H) HCC827 cells were starved for 5 hours and then treated with DMSO, Go6976 or erlotinib, followed by immunoprecipitation of mtEGFR to analyze the interaction of Gab1 and mtEGFR.
Next, we evaluated if Akt is essential for mTORC1 activity in HCC827 by using an Akt inhibitor, MK-2206. As shown in Figure 6E, treatment with MK-2206 abolished phosphorylation of p70S6K, the same as the treatment with highly specific dual-mTOR inhibitor of mTORC1 and mTORC2, KU-0063794. Notably, the inhibitors did not modulate phosphorylation of the mtEGFR on Y1068, indicating lack of effects on initial status of EGFR activation. These results, also unlike those in glioma cells, indicate that Akt is required for activation of mTOR in mt-EGFR cell lines.
To consolidate these data, we checked the effect of U73122 on the phosphorylation of Akt at Ser473, as we showed that PLCγ plays a vital role in the regulation of PKCα activity prompted by mtEGFR in HCC827. As shown in Figure 6F, U73122 treatment significantly inhibited the phosphorylation of Akt at Ser473. Collectively, these data indicate that the activation of Akt in HCC827 requires the coordination of mtEGFR/ PLCγ/ PKCα signaling pathway.
Finally, it has been shown that the Gab adaptor proteins and ErbB3 play critical roles in activation of the PI3K pathway by interacting with mtEGFR in EGFR-mutant NSCLC cell lines. Therefore, to evaluate the role of PKCα in the activation of PI3K/AKT by mtEGFR, HCC827 cells were transfected with control, Gab1, or ErbB3 siRNAs. As shown in Figure 6G and Supplementary Figure 4, downregulation of Gab1 by two different siRNAs reduced the phosphorylation of p70S6K. On the other hand, ErbB3 knockdown had no effect on the phosphorylation of p70S6K (Fig. 6G). Next, we assessed whether PKC plays a role in the interaction of Gab1 and mtEGFR. Notably, both Go6976 and erlotinib decreased the interaction between Gab1 and mtEGFR (Fig. 6H). Moreover, Gab1 and mtEGFR interaction was less in PKCα CRISPR cells (Supplementary Fig.4). Together, these data implicate an important role of PKCα in the activation of PI3K by mtEGFR through the Gab1 pathway.
PKCα gain of function is sufficient to activate the Akt/mTOR pathway
Since the results strongly suggest that PKCα activates mTORC1 in response to mtEGFR, we investigated if this regulation of mTORC1 is exclusive to mtEGFR but not WT EGFR. Indeed, as shown in Figure 7A, mTOR was activated constitutively in serum free conditions in HCC827 cells, and this high level of activation did not respond to additional EGF and was cPKC-dependent. On the other hand, mTOR in H292 cells, with WT EGFR, was activated by EGF and this was largely PKC-independent.
FIGURE 7. PKCα gain of function is sufficient to activate the Akt/mTOR pathway.
(A) H292 and HCC827 cells were starved for 5 hours, and then pretreated with Go6976 for 1 hour followed by treatment with 10ng/mL EGF for 5 min; (B) H292 cells were transiently transfected with vector or PKCα-GFP, and then cells were starved for 5 hours followed by treatment with 10ng/mL EGF for 5 min; (C) H292 cells were stably transfected with vector, mtEGFR or PKCα, and then cells were analyzed by western blotting for phosphorylated or total targeted protein.
Next, we examined if PKCα is sufficient to impart activation of mTOR in serum free conditions in lung cancers with WT EGFR. As can be seen, transient overexpression of PKCα in H292 cells, which have WT EGFR and very low levels of endogenous PKCα, resulted in significantly increased basal phosphorylation of S6K and increased response to EGF (Fig. 7B). In cells with stable overexpression of PKCα, there was also persistent induction of phosphorylation of S6K that was not seen with overexpression of mtEGFR (Fig. 7C). Moreover, overexpression of PKCα appeared to selectively enhance phosphorylation of Akt on T308 and not S473. Also, overexpression of PKCα did not increase the phosphorylation of ERK1,2. In contrast, overexpression of mtEGFR significantly increased the activation of ERK1,2 but had no effects on the phosphorylation of Akt or S6K. Altogether these results reveal that expression of mtEGFR is not sufficient to drive activation of mTOR. On the other hand, enhanced expression of PKCα is sufficient to activate the Akt/mTOR pathway, and this is further driven by EGF. These results help define the independent contribution of elevated expression of PKCα in lung cancer cells with mEGFR as it becomes required for allowing activation of Akt and mTOR.
mtEGFR/PKCα/mTOR signaling pathway regulates cell survival in HCC827
To further explore the significance of this gain of function of PKCα, we examined several oncogenic properties in H292 and HCC827 cells. We found that cancer cells with mtEGFR grow much better in serum free conditions compared with H292 cells with wild type EGFR (Fig. 8A). The growth advantage of HCC827 was reversed by treatment with erlotinib and became similar to the growth observed in H292 (Fig. 8B), suggesting that this advantage is contributed by the mtEGFR. Next, we assessed the effects of inhibition of cPKC. Interestingly, the inhibition of cPKC specifically abolished this advantage for mtEGFR, and a similar albeit reduced effect was observed with inhibition of mTOR (Fig. 8C). Moreover, loss of PKCα by CRISPR repressed the growth advantage of HCC827 cells under serum starvation (Fig. 8D). Interestingly, stable co-expression of PKCα and mtEGFR in H292 cells significantly improved growth under serum starvation more than expressing mtEGFR alone (Fig. 8E). Taken together, these results begin to demonstrate that mtEGFR/PKC signaling pathway plays an important role in cell survival. To gain further insight into the cell survival advantage of HCC827 under serum starvation, we examined the apoptosis in HCC827 cells. The results showed that inhibition of PKCα induced autophagy and caspase-3-dependent apoptosis, as did inhibition of mtEGFR (Fig. 9A). Moreover, PKCα CRISPR cells showed higher cleaved caspase-3 levels in response to erlotinib treatment (Fig. 9B). Taken together, these results demonstrate that mtEGFR/PKC signaling pathway plays an important role in apoptosis and autophagy and loss of PKCα could sensitize cells with mtEGFR to erlotinib.
FIGURE 8. mtEGFR/PKCα/mTOR signaling pathway regulates cell survival in HCC827.
(A) H292 and HCC827 cells were grown in medium with serum or without serum as indicated. MTT OD value was read at 1, 2, and 3 days after the feeding; (B) H292 cells and HCC827 cells were grown in medium without serum plus DMSO or 100nM erlotinib for 48 hours; (C) H292 and HCC827 cells were cultured in medium without serum plus DMSO, 100nM Go6976, or 100nM rapamycin for 48 hours; (D) Vector and PKCα CRISPR HCC827 cells were grown under serum free conditions, and MTT OD values were measured after 24, 48, and 72 hours; (E) Control H292 cells and those stably expressing PKCα and or mtEGFR were grown under serum free conditions and MTT OD values were measured after 24, 48, and 72 hours.* p<0.05; ** p<0.01.
FIGURE 9. mtEGFR/PKCα signaling regulates apoptosis and autophagy in HCC827 cells.
HCC827 cells were grown in serum-free medium plus DMSO, 100nM Go6976, or 100nM erlotinib for 48 hours. The LC3-I, LC3-II, pro-caspase 3 and cleaved caspase 3 were detected by Western blotting; (B) Vector and PKCα CRISPR HCC827 cells were treated with vehicle or 100 nM erlotinib for 24 hours followed by immunoblotting for caspase-3 and cleaved caspase-3.
DISCUSSION
To date, research has established the role of EGFR mutation and its subsequent persistent activation in the development of NSCLC, while PKCα has also been shown to play a part in the progression of NSCLC as well as the development of drug resistance. Here, we explored the possible regulation of PKCα by persistent EGFR activation driven by exon 19 deletion (delE746–750). The results clearly demonstrate that PKCα is highly expressed in cells harboring mutant EGFR and in tissues from NSCLC patients with activating EGFR mutation. Moreover, the results showed that active EGFR is required and sufficient to activate PKCα in these NSCLC cells via a PLCγ-dependent mechanism. Strikingly, results demonstrate that PKCα plays a role in rewiring the PI3K/AKT/mTORC1 signaling downstream of mtEGFR through the Gab1 pathway. Biologically, PKC-dependent signaling pathways downstream of mtEGFR are important for promoting cell survival and reducing apoptosis. Collectively, the results reveal a previously unknown role for PKCα in modulating PI3K/AKT/mTORC1 signaling in the context of NSCLC with EGFR mutation.
The major conclusion from this study relates to the interactions of mEGFR and PKCα. First, the results show that the mutation of EGFR correlates with high expression of PKCα. The results of this study show that this induction is not directly dependent on the activity of EGFR since inhibition of EGFR by erlotinib had no effect on mRNA and protein expression of PKCα. Thus, we propose that cells with activating EGFR mutation select for overexpression of PKCα which then imparts a growth advantage to those cells. This is clearly illustrated and corroborated by the gain-of-function studies that showed that expression of mtEGFR on its own is unable to turn on either Akt or mTOR and that this response requires additional expression of PKCα to activate these critical pathways and to stimulate cell growth under serum starvation. Second, the results show that the persistent activity of mtEGFR causes continuous activation of PKCα, as demonstrated by increased phosphorylation of PKCα/βII and membrane localization of PKCα under serum starvation in cells with activating EGFR mutation. Moreover, overexpression of mtEGFR was sufficient to induce the sustained activation of transiently or stably overexpressed PKC in NSCLC cells with wild-type EGFR. Thus, mtEGFR is both necessary and sufficient for activation of PKCα in NSCLC lung cancer cells.
A major revelation from these studies is that mEGFR utilizes PKCα to rewire downstream signaling. The results show that in cells with mtEGFR, PKCα is required for activation of Akt/mTOR pathway while on the other hand, EGF-induced mTOR activation is PKCα-independent in cells with wild-type EGFR. The data show that PKCα is necessary and sufficient to activate Akt/mTOR pathway in cells with increased levels of PKCα (and mtEGFR) as demonstrated by increased phosphorylation of Akt and S6K by overexpression of PKCα in cells with wild-type EGFR. This is supported by inhibitor results as well as with knock down.
Functionally, we implicate PKCα in important biology of NSCLC cells with mtEGFR. The results showed that NSCLC cells with activating EGFR mutations have a growth advantage in serum-free conditions compared with cells with wild-type EGFR. Importantly, this growth advantage is dependent on the activities of both EGFR and PKC that was supported by the growth advantage of wild-type EGFR cells stably expressing mtEGFR and PKC. Similarly, PKCα plays a role in apoptosis and autophagy in response to erlotinib and Go6976 and sensitizing cells with mtEGFR to erlotinib. Taken together, these findings clearly demonstrate a pro-survival role played by PKCα in the biology of NSCLC in the context of mtEGFR. Previous studies have revealed significant roles played by PKCs in cancer progression. However, depending on the isoform and context, a specific PKC could function either as a tumor suppressor or a tumor promoter. For example, PKCβII inhibition with enzastaurin reduced proliferation and migration of A549 cells in a PLD/mTOR- dependent manner (31), and inhibition of PKCβ has been considered as a therapeutic approach for lung cancer (33, 34). Similarly, PKCε has recently been shown to be required for progression and metastasis of NSCLC cells (35), and previous studies have demonstrated the important role played by PKCα in modulating NSCLC invasion (24, 26). On the other hand, PKCα has recently been shown to act as a tumor suppressor of KRAS-mediated lung cancer (36). In addition, PKC has recently been shown to be a tumor suppressor in colon cancer (37). It should be noted though that the expression of PKCα appears to be low in KRAS mutated NSCLC. These findings together with the results presented herein suggest distinct roles of PKCα in NSCLC depending on the mutation of either KRAS or EGFR oncogene.
The stimulation of survival and growth associated with EGFR mutations involves signaling through PI3K/Akt pathway. Activation of Akt is achieved through phosphorylation of threonine 308 (Thr308) and serine 473 (Ser473) residues. The phosphorylation of Akt at its Thr308 residue mostly occurs by action of PDK1 while phosphorylation of Ser473 occurs by action of mTORC2 (38–40). Activated Akt then acts on several downstream targets, such as mTORC1 and p70S6 to promote cell growth and survival (41). Akt activity has been shown to be a useful biomarker for cancer prognosis and response to therapy. However, phosphorylation of Akt at Thr308 has been demonstrated to be a more reliable biomarker for Akt activity in vivo (39), and higher phosphorylation levels of Akt at Thr308, but not Ser473, correlates with poor survival in NSCLC (42) and acute myeloid leukemia (43). These findings suggest that the degree of Akt phosphorylation at Thr308 could be a useful indicator of Akt activity. That was recently confirmed in tissue samples from NSCLC patients, where Akt phosphorylation at Thr308 was shown to correlate with the phosphorylation of several substrates downstream of Akt (32). Here we found that inhibition or downregulation of PKCα was associated with less phosphorylation of Akt at both residues. Interestingly, PKCα overexpression NSCLC with wild-type EGFR selectively induced the phosphorylation of Akt Thr308 and had little effect on Ser473. These results indicate that PKCα is critical for the Akt activity in NSCLC cells with mtEGFR. As such, Akt phosphorylation at T308 might be relevant as a biomarker of PKC activity in NSCLC tissues with mutant EGFR. The current results also reveal that PKCα-dependent activation of PI3K/Akt downstream of mtEGFR involves Gab1, an adaptor protein that has been implicated in PI3K signaling in response to EGF following its tyrosine phosphorylation (44, 45). Gab1 can also induce PI3K by amplifying EGFR signaling by a positive feedback loop as previously described (44). Recently, Gab1 has also been shown to be activated by EGFR mutation in NSCLC cells (46).
In conclusion, our results demonstrate a previously unidentified role of PKCα in mediating PI3K/Akt/mTORC1 signaling and biology downstream of activated EGFR in NSCLC cells. Therefore, targeting PKCα and/or EGFR could be a promising therapeutic strategy for NSCLC patients who are harboring activating EGFR mutations.
MATERIALS AND METHODS
Materials
Dulbecco’s Modified Eagle Medium (DMEM) with high glucose was from Invitrogen. The HCC827, H292, H4006, 293T cell lines were purchased from American Type Culture Collection. Enzastaurin and erlotinib were from LC Laboratories (Woburn, MA). Go6976 and U0126 were purchased from EMD Millipore (Billerica, MA). Anti-phospho-p70S6K (Thr389), S6K, mTOR, phospho- Erk½ (Thr202/Tyr204), Erk½, (E747-A750del Specific) EGFR, EGFR, P-Akt (Ser473), P-Akt (Thr308), Akt, PKCα and ErbB3 antibodies were from Cell Signaling Technology (Danvers, MA), and anti-actin antibody was purchased from Sigma-Aldrich (St. Louis, MO). Anti-EEA1 antibody was from BD Biosciences (San Jose, CA). siRNAs for PKCα and Gab1 were from Qiagen (Valencia, CA) and those for PLCγ1 were from ambion (life technologies). Dual mTOR inhibitor (KU-0063794) and Akt inhibitor (MK-2206 2HCl) were purchased from Selleckchem. All other reagents were from Sigma-Aldrich (St. Louis, MO).
Cell Culture and Cell Counting
HCC827, H4006, H1975, and H292 cells were grown in RPMI supplemented with 10% (v/v) fetal bovine serum. For cell counting, cells were trypsinized from culture dishes, and 10μl of cell suspension was mixed with Trypan blue in a 1:1 ratio (v/v) for 1 minute and then cell number was counted using Countess® automated cell counter (Invitrogen).
Immunohistochemistry
Human tissue arrays were obtained from Folio (Catalogue number: FOLIO-800). Deparaffinization of tissue slides was performed at 60°C for 1 hr. After 10 min of cooling, tissues were hydrated in two separate baths of xylene for 5 minute each, followed by a bath in 2% hydrogen peroxide/methanol solution for 30 minutes and finally incubated in 3 different concentrations of ethanol solutions (100%, 95%, 70%). After neutralizing in water, antigen retrieval was performed in citrate buffer (pH 6) at 100–120°C for 10 minutes. Sections were cooled at 4°C for 30 min and subsequently blocked using 5% BSA/Tris buffer solution/ 0.1% Tween (TBST) for 30 min at room temperature. Blocked tissues were incubated with unconjugated primary antibody overnight: anti-EGFR (E746-A750del Specific) or anti-PKCα (Cell Signaling), both diluted 1:50 in block solution. Tissues were washed for 5 min 3x with TBST and incubated with conjugated secondary antibody (Biocare MACH 3 Rabbit HRP). Sections were then washed and counterstained with nuclear stain and mounted in Vectashield mounting medium (Vector Labs). Microscopy was performed using a Zeiss microscope. Optical density (OD) for PKCα staining were calculated by ImageJ software version 2 (https://imagej.nih.gov/ij/).
Plasmid Constructs and Transient Transfection
All plasmids were generated by standard protocols. Wild type GFP-PKCα and GFP-EGFR were described previously (47–49). The GFP-mtEGFR construct was made by using pEGFP-N1 vector and GFP-EGFR as a template for the QuickChange II Site-Direct Mutagenesis Kit. Cells were plated on 6- well plates at a density 5×104 cells/well. After 24 hours, X-tremeGENE 9 was used for transient transfection following the manufacturer’s protocol. After transfection, cells were grown in normal medium with 10% FBS for 24 to 48 hours and then starved with medium without serum for 5 hours, followed by different treatments.
Generation of H292 Cells Stably Expressing PKCα and/or or mtEGFR
pENTR™ Directional TOPO® Cloning Kit (Life technologies, NY, USA) was used for cloning of PKCα and mtEGFR into pENTR/D-TOPO entry vector. Human PKCα and mtEGFR were amplified from GFP-PKCα and GFP-mtEGFR plasmid constructs using the following primers: PKCα Fwd: CACCATGGCTGACGTTTTCCCG; PKCα Rvs: TACTGCACTCTGTAAGATGGG; mtEGFR Fwd: CACCATGGGACCCTCCGGGACG; mtEGFR Rvs: TCATGCTCCAATAAATTCACTGC. PCR products were then purified and cloned into pENTR/D-TOPO and transformed into One Shot® Competent E. coli (Life technologies) and plasmids were isolated form selected colonies and sequenced. PKCα and mtEGFR were cloned into pLenti6.3/TO/V5-DEST (Addgene) and pLenti CMV/TO Puro DEST (Addgene), respectively using clonase reaction following the Gateway® LR Clonase® II enzyme mix’s protocol (Life technologies) for 2 hours at 25°C. Two μl from clonase reaction were transformed into STBL3 cells (life technologies) and plated into ampicillin LB plates followed by plasmid isolation and sequencing.
For generating lentiviral particles, 293T cells were seeded into 100mm dishes at a density of 2.5×106 cells in 10ml DMEM with 10% FBS on the day before transfection. For each transfection, 13.5 μl X-tremeGENE 9 was diluted into 150μl optimum/dish. Next, 1.5μl from each of the following plasmids was added: PKCα/mtEGFR pLentiviral vector, viral envelope plasmid (pCMV-VSV-G; Addgene), packaging plasmid (pCMV-dR8.2 dvpr; Addgene). As a control, pLenti CMV/TO Puro and pLenti CMV Blast Empty vectors (Addgene) were used.
The mixture was then incubated for 15 min at room temperature and then added to the cells. One day later, medium was aspirated and replaced with 10ml DMEM. Two days later, medium was harvested, filtered through 48μm PVDF filters, aliquoted, and stored at −80°C.
H292 cells were seeded into 35mm dishes (150,000 cells in 2ml RPMI). Next day, media were aspirated then replaced with 1ml RPMI with 16μg/ml polyprene followed by addition of 1ml of PKCα, mtEGFR, or control lentiviral particles. Medium was changed after 24 hrs with fresh RPMI. One day later, cells were expanded into 100mm dishes in 10ml RPMI with selective antibiotic (2μg/ml puromycin for mtEGFR and 50μg/ml blasticidin for PKCα). Cells were selected for 10 days followed by confirmation of protein overexpression with western blotting. For PKCα and mtEGFR co-expression experiment, H292 cells were first infected with PKCα lentivirus and were selected with blasticidin for 10 days followed by infection with mtEGFR lentivirus and cells were then selected with puromycin for 7–10 days.
siRNA Transfection
siRNAs were transfected into NSCLC cells with Lipofectamine RNAiMAX following the manufacturer’s recommendation. Transfected cells were grown in medium with 10% FBS for 48 to 72 hours then followed by specific treatments as indicated.
Generation of PKCα Knockout Cells Using Crispr/Cas9 Technology
To knockout PKCα from HCC827 cells, we used the Lenticrispr v2.0 system (50). The LentiCRISPR v2 plasmid was a gift from Feng Zhang (Addgene plasmid # 52961). Briefly, guide RNAs targeting the PKCα gene were designed using the ChopChop algorithm (http://chopchop.cbu.uib.no/) and cloned into the plasmid as described previously (51). To generate lentivirus, 2μg plasmids were co-transfected with 2μg VSV-G and 2μg dVPR into 293T cells (2500K/100mm dish). Virus-containing media were harvested and filtered (0.22μM PVDF membrane) after 72h. HCC827 cells (100K/6-well) were infected with 1ml virus-containing media in the presence of polybrene (8μg/ml). After 48h, cells were split into 100mm dishes containing selective media (2μg/ml puromycin). After 7 days of selection, puromycin was removed and cells were maintained in normal growth medium.
Immunofluorescence
Cells were plated on 35mm confocal dishes (MatTek) at a density of 3 to 5×105 cells/dish. After 24 to 48 hours, cells were then starved with medium without serum for 5 hours, followed by treatment. Fixation and permeabilization were conducted as described previously (52). After incubation with 2% human serum for 1 hour, cells were stained with primary antibodies and secondary antibodies as described before (52). All images were taken by Leica TCS SP8, and pictures are representative of a least three fields examined from three independent experiments.
MTT Assay
Cells (5×104) were plated in 6 well plates, and after 24 hours, they were treated as indicated. MTT (Thiazolyl Blue Tetrazolium Bromide: 5mg/ml in PBS, 400μl) and fresh medium was added to each well, and following brief agitation, cells were incubated at 37°C for 3–4 hours. Media were aspirated, and 2ml MTT solvent was added. Following brief agitation to ensure full dissolution, optical density was read at 595nm.
Immunoprecipitation
HCC827 were serum starved for 5 hours and then treated with vehicle, Go6976, or erlotinib for 1 hour. After washing with ice-cold PBS, cells were collected and lysed in lysis buffer (50mM Tris-HCl, pH 7.4, 150mM NaCl, 1mM EDTA, and 1% Triton X-100) with protease and phosphatase inhibitor cocktails (Sigma-Aldrich) on ice for 20 min. Lysates were cleared by centrifugation, and protein concentration was determined by Bicinchoninic Acid Assay (BCA). Lysates containing 500μg - 1mg protein were pre-cleared with protein G magnetic beads (EMD Millipore) for 1h at 4°C, and subsequent immunoprecipitations were carried out using anti-(E747-A750del Specific) EGFR antibody by end-to-end rotation overnight at 4°C following the manufacturer’s protocol. The next day, the pre-formed antibody-antigen complex was added to the protein G magnetic beads and incubated for another 1h at 4°C. Antigen-antibody-bead complexes were separated by using the magnetic stand and washed three times with 1X PBS containing 0.1% Tween. After boiling with sample loading buffer, proteins were resolved on 4–20% TGX gels (BioRad Laboratories), and immunoblotting was carried out as described above.
Statistical Analysis
Statistical significance was calculated with student’s t-test or by two-way ANOVA with Bonferroni Post-test where appropriate. A p-level of below 0.05 was considered to be statistically significant.
Supplementary Material
SUPPLEMENTARY FIGURE 1.
The expression of PKCα was examined in adenocarcinoma and squamous cell carcinoma patients with (82 samples) or without (936 samples) EGFR mutations from TCGA database. Expression data were downloaded (http://gdac.broadinstitute.org/), log2 transformed and plotted.
SUPPLEMENTARY FIGURE 2.
H4006 cells were serum starved and then treated with various concentrations of Go6976 (A) or enzastaurin (B) as indicated, following with western blotting to detect phosphorylation of S6K, EGFR, ERK, and AKT; (C) HCC827 were treated with different doses of erlotinib for 48 hours followed by detection of pPKCα and βII, pEGFR, and total PKCα by western blotting; (D) HCC827 cells were transfected with 2 siRNA against PKCα followed by detection phosphorylation of AKT and S6K by western blotting.
SUPPLEMENTARY FIGURE 3.
(A) H292 cells stably expressing PKC α and/or mtEGFR were serum starved for 6 hours followed by immunofluorescence to visualize PKCα localization; (B) HCC827 were transfected with 2 different PLCγ1 siRNAs for 48 hours, the cells were then serum starved for 5 hours followed by immunofluorescence to visualize PKCα localization and immunoblotting to examine the effect on phosphorylation of S6K; (C) H292 stably expressing PKCα were transfected with PLCγ1 siRNA for 48 hours, then serum starved overnight followed by stimulation with EGF 100 ng/ml for 1 hour and immunofluorescence for PKCα localization.
SUPPLEMENTARY FIGURE 4.
(A) HCC827 cells were transfected with two different Gab1 siRNAs for 48 hours and then starved for 5 hours followed by western blotting to detect phosphorylated and total S6K; (B) Vector and PKCα HCC827 CRISPR cells were starved for 5 hours followed by immunoprecipitation of mtEGFR to analyze the interaction of Gab1 and mtEGFR.
Acknowledgements
This work was supported by NCI grant CA97132.
ABBREVIATIONS USED
- cPKC
Classical protein kinase C
- EGFR
Epidermal growth factor receptor
- ERK
Extracellular related kinase
- mEGFR
EGFR mutations
- mtEGFR
mutant EGFR
- mTOR
Mammalian target of rapamycin
- mTORC1
mTOR Complex 1
- NSCLC
Non-small cell lung cancer
- PI3K
phosphoinositide 3-kinase
- S6K
p70 Ribosomal S6 Kinase
Footnotes
Competing Interests
Authors declare no conflict of interest in relation to the work described.
REFERENCES
- 1.Kanne JP. Screening for lung cancer: what have we learned? American Journal of Roentgenology. 2014;202(3):530–5. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136(5):E359–E86. [DOI] [PubMed] [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA, editors. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship Mayo Clinic Proceedings; 2008: Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shames DS, Wistuba II. The evolving genomic classification of lung cancer. The Journal of pathology. 2014;232(2):121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science (New York, NY). 1991;253(5015):49–53. [DOI] [PubMed] [Google Scholar]
- 6.Reissmann PT, Koga H, Takahashi R, Figlin RA, Holmes EC, Piantadosi S, et al. Inactivation of the retinoblastoma susceptibility gene in non-small-cell lung cancer. The Lung Cancer Study Group. Oncogene. 1993;8(7):1913–9. [PubMed] [Google Scholar]
- 7.Jin G, Kim MJ, Jeon H-S, Choi JE, Kim DS, Lee EB, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung cancer. 2010;69(3):279–83. [DOI] [PubMed] [Google Scholar]
- 8.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science (New York, NY). 2004;305(5687):1163–7. [DOI] [PubMed] [Google Scholar]
- 9.Gazdar A Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene. 2009;28:S24–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper WA, Lam DC, O’Toole SA, Minna JD. Molecular biology of lung cancer. Journal of thoracic disease. 2013;5 Suppl 5:S479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonicelli A, Cafarotti S, Indini A, Galli A, Russo A, Cesario A, et al. EGFR-targeted therapy for non-small cell lung cancer: focus on EGFR oncogenic mutation. Int J Med Sci. 2013;10(3):320–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steins M, Thomas M, Geissler M. Erlotinib. Recent results in cancer research Fortschritte der Krebsforschung Progres dans les recherches sur le cancer. 2014;201:109–23. [DOI] [PubMed] [Google Scholar]
- 13.Steinberg SF. Structural basis of protein kinase C isoform function. Physiological reviews. 2008;88(4):1341–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nature Reviews Cancer. 2007;7(4):281–94. [DOI] [PubMed] [Google Scholar]
- 15.Kong C, Zhu Y, Liu D, Yu M, Li S, Li Z, et al. Role of protein kinase C-alpha in superficial bladder carcinoma recurrence. Urology. 2005;65(6):1228–32. [DOI] [PubMed] [Google Scholar]
- 16.Gupta A, Galoforo S, Berns C, Martinez A, Corry P, Guan KL, et al. Elevated levels of ERK2 in human breast carcinoma MCF‐7 cells transfected with protein kinase Cα. Cell proliferation. 1996;29(12):655–63. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Han J, Lee SK, Choi M-Y, Kim J, Lee J, et al. Berberine suppresses the TPA-induced MMP-1 and MMP-9 expressions through the inhibition of PKC-α in breast cancer cells. Journal of Surgical Research. 2012;176(1):e21–e9. [DOI] [PubMed] [Google Scholar]
- 18.Oster H, Leitges M. Protein kinase C α but not PKCζ suppresses intestinal tumor formation in ApcMin/+ mice. Cancer Research. 2006;66(14):6955–63. [DOI] [PubMed] [Google Scholar]
- 19.Jiang X-H, Tu S-P, Cui J-T, Lin MC, Xia HH, Wong WM, et al. Antisense targeting protein kinase C α and β1 inhibits gastric carcinogenesis. Cancer Research. 2004;64(16):5787–94. [DOI] [PubMed] [Google Scholar]
- 20.Fan Q-W, Cheng C, Knight ZA, Haas-Kogan D, Stokoe D, James CD, et al. EGFR signals to mTOR through PKC and independently of Akt in glioma. Science signaling. 2009;2(55):ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith SD, Enge M, Bao W, Thullberg M, Costa TD, Olofsson H, et al. Protein kinase Calpha (PKCalpha) regulates p53 localization and melanoma cell survival downstream of integrin alphav in three-dimensional collagen and in vivo. The Journal of biological chemistry. 2012;287(35):29336–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh Y-H, Wu T-T, Huang C-Y, Hsieh Y-S, Hwang J-M, Liu s. p38 mitogen-activated protein kinase pathway is involved in protein kinase Cα–regulated invasion in human hepatocellular carcinoma cells. Cancer research. 2007;67(9):4320–7. [DOI] [PubMed] [Google Scholar]
- 23.Singhal SS, Yadav S, Singhal J, Drake K, Awasthi YC, Awasthi S. The role of PKCα and RLIP76 in transport‐mediated doxorubicin‐resistance in lung cancer. FEBS letters. 2005;579(21):4635–41. [DOI] [PubMed] [Google Scholar]
- 24.O’Neill AK, Gallegos LL, Justilien V, Garcia EL, Leitges M, Fields AP, et al. Protein kinase Cα promotes cell migration through a PDZ-dependent interaction with its novel substrate discs large homolog 1 (DLG1). Journal of Biological Chemistry. 2011;286(50):43559–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Porta CA, Tessitore L, Comolli R. Changes in protein kinase C alpha, delta and in nuclear beta isoform expression in tumour and lung metastatic nodules induced by diethylnitrosamine in the rat. Carcinogenesis. 1997;18(4):715–9. [DOI] [PubMed] [Google Scholar]
- 26.Cheng CY, Hsieh HL, Sun CC, Lin CC, Luo SF, Yang CM. IL‐1β induces urokinse‐plasminogen activator expression and cell migration through PKCα, JNK½, and NF‐κB in A549 cells. Journal of cellular physiology. 2009;219(1):183–93. [DOI] [PubMed] [Google Scholar]
- 27.Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, et al. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PloS one. 2013;8(9):e73985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abera MB, Kazanietz MG. Protein kinase Cα mediates erlotinib resistance in lung cancer cells. Molecular pharmacology. 2015;87(5):832–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahn M, Su C, Li S, Chedid M, Hanna KR, Graff JR, et al. Expression Levels of Protein Kinase C-α in Non–Small-Cell Lung Cancer. Clinical lung cancer. 2004;6(3):184–9. [DOI] [PubMed] [Google Scholar]
- 30.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Osta M, Liu M, Adada M, Senkal CE, Idkowiak-Baldys J, Obeid LM, et al. Sustained PKCβII activity confers oncogenic properties in a phospholipase D-and mTOR-dependent manner. The FASEB Journal. 2014;28(1):495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent E, Elder D, Thomas E, Phillips L, Morgan C, Pawade J, et al. Akt phosphorylation on Thr308 but not on Ser473 correlates with Akt protein kinase activity in human non-small cell lung cancer. British journal of cancer. 2011;104(11):1755–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vansteenkiste J, Ramlau R, Von Pawel J, San Antonio B, Eschbach C, Szczesna A, et al. A phase II randomized study of cisplatin-pemetrexed plus either enzastaurin or placebo in chemonaive patients with advanced non-small cell lung cancer. Oncology. 2012;82(1):25–9. [DOI] [PubMed] [Google Scholar]
- 34.Willey CD, Xiao D, Tu T, Kim KW, Moretti L, Niermann KJ, et al. Enzastaurin (LY317615), a protein kinase C beta selective inhibitor, enhances antiangiogenic effect of radiation. International Journal of Radiation Oncology* Biology* Physics. 2010;77(5):1518–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caino MC, Lopez-Haber C, Kim J, Mochly-Rosen D, Kazanietz MG. Proteins kinase Cϵ is required for non-small cell lung carcinoma growth and regulates the expression of apoptotic genes. Oncogene. 2012;31(20):2593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill K, Erdogan E, Khoor A, Walsh M, Leitges M, Murray NR, et al. Protein kinase Cα suppresses Kras-mediated lung tumor formation through activation of a p38 MAPK-TGFβ signaling axis. Oncogene. 2014;33(16):2134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antal CE, Hudson AM, Kang E, Zanca C, Wirth C, Stephenson NL, et al. Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. 2015(1097–4172 (Electronic)). [DOI] [PMC free article] [PubMed]
- 38.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Current biology. 1997;7(4):261–9. [DOI] [PubMed] [Google Scholar]
- 39.Vanhaesebroeck B, Alessi DR. The PI3K–PDK1 connection: more than just a road to PKB. Biochemical Journal. 2000;346(3):561–76. [PMC free article] [PubMed] [Google Scholar]
- 40.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science (New York, NY). 2005;307(5712):1098–101. [DOI] [PubMed] [Google Scholar]
- 41.Hanada M, Feng J, Hemmings BA. Structure, regulation and function of PKB/AKT—a major therapeutic target. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2004;1697(1):3–16. [DOI] [PubMed] [Google Scholar]
- 42.Tsurutani J, Fukuoka J, Tsurutani H, Shih JH, Hewitt SM, Travis WD, et al. Evaluation of two phosphorylation sites improves the prognostic significance of Akt activation in non–small-cell lung cancer tumors. Journal of clinical oncology. 2006;24(2):306–14. [DOI] [PubMed] [Google Scholar]
- 43.Gallay N, Dos Santos C, Cuzin L, Bousquet M, Gouy VS, Chaussade C, et al. The level of AKT phosphorylation on threonine 308 but not on serine 473 is associated with high-risk cytogenetics and predicts poor overall survival in acute myeloid leukaemia. Leukemia. 2009;23(6):1029–38. [DOI] [PubMed] [Google Scholar]
- 44.Rodrigues GA, Falasca M, Zhang Z, Ong SH, Schlessinger J. A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Molecular and cellular biology. 2000;20(4):1448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattoon DR, Lamothe B, Lax I, Schlessinger J. The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC biology. 2004;2(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song X, Fan P-D, Bantikassegn A, Guha U, Threadgill DW, Varmus H, et al. ERBB3-independent activation of the PI3K pathway in EGFR-mutant lung adenocarcinomas. Cancer research. 2015;75(6):1035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng X, Hannun YA. An essential role for autophosphorylation in the dissociation of activated protein kinase C from the plasma membrane. J Biol Chem. 1998;273(41):26870–4. [DOI] [PubMed] [Google Scholar]
- 48.Idkowiak-Baldys J, Becker KP, Kitatani K, Hannun YA. Dynamic sequestration of the recycling compartment by classical protein kinase C. J Biol Chem. 2006;281(31):22321–31. [DOI] [PubMed] [Google Scholar]
- 49.Liu M, Idkowiak-Baldys J, Roddy PL, Baldys A, Raymond J, Clarke CJ, et al. Sustained activation of protein kinase C induces delayed phosphorylation and regulates the fate of epidermal growth factor receptor. PLoS One. 2013;8(11):e80721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pulkoski-Gross MJ, Jenkins ML, Truman J-P, Salama MF, Clarke CJ, Burke JE, et al. An intrinsic lipid-binding interface controls sphingosine kinase 1 function. Journal of Lipid Research. 2018;59(3):462–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Idkowiak-Baldys J, Baldys A, Raymond JR, Hannun YA. Sustained receptor stimulation leads to sequestration of recycling endosomes in a classical protein kinase C- and phospholipase D-dependent manner. J Biol Chem. 2009;284(33):22322–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY FIGURE 1.
The expression of PKCα was examined in adenocarcinoma and squamous cell carcinoma patients with (82 samples) or without (936 samples) EGFR mutations from TCGA database. Expression data were downloaded (http://gdac.broadinstitute.org/), log2 transformed and plotted.
SUPPLEMENTARY FIGURE 2.
H4006 cells were serum starved and then treated with various concentrations of Go6976 (A) or enzastaurin (B) as indicated, following with western blotting to detect phosphorylation of S6K, EGFR, ERK, and AKT; (C) HCC827 were treated with different doses of erlotinib for 48 hours followed by detection of pPKCα and βII, pEGFR, and total PKCα by western blotting; (D) HCC827 cells were transfected with 2 siRNA against PKCα followed by detection phosphorylation of AKT and S6K by western blotting.
SUPPLEMENTARY FIGURE 3.
(A) H292 cells stably expressing PKC α and/or mtEGFR were serum starved for 6 hours followed by immunofluorescence to visualize PKCα localization; (B) HCC827 were transfected with 2 different PLCγ1 siRNAs for 48 hours, the cells were then serum starved for 5 hours followed by immunofluorescence to visualize PKCα localization and immunoblotting to examine the effect on phosphorylation of S6K; (C) H292 stably expressing PKCα were transfected with PLCγ1 siRNA for 48 hours, then serum starved overnight followed by stimulation with EGF 100 ng/ml for 1 hour and immunofluorescence for PKCα localization.
SUPPLEMENTARY FIGURE 4.
(A) HCC827 cells were transfected with two different Gab1 siRNAs for 48 hours and then starved for 5 hours followed by western blotting to detect phosphorylated and total S6K; (B) Vector and PKCα HCC827 CRISPR cells were starved for 5 hours followed by immunoprecipitation of mtEGFR to analyze the interaction of Gab1 and mtEGFR.