Abstract
CDKN2A is an evolutionarily conserved gene encoding proteins implicated in tumor suppression, ocular development, aging, and metabolic diseases. Like the human form, mouse Cdkn2a encodes two distinct proteins – p16lnk4a, which blocks cyclin-dependent kinase activity, and p19Arf, which is best known as a positive regulator of the p53 tumor suppressor – and their functions have been well-studied in genetically engineered mouse models. Relatively little is known about how expression of the two transcripts is controlled in normal development and in certain disease states. To better understand their coordinate and transcript-specific expression in situ, we used a transposase-aided approach to generate a new BAC transgenic mouse model in which the first exons encoding Arf and Ink4a are replaced by fluorescent reporters. We show that mouse embryo fibroblasts (MEFs) generated from the transgenic lines faithfully display induction of each transgenic reporter in cell culture models, and we demonstrate the expected expression of the Arf reporter in the normal testis, one of the few places where that promoter is normally expressed. Interestingly, the TGFβ-2-dependent induction of the Arf reporter in the eye - a process essential for normal eye development - does not occur. Our findings illustrate the value of BAC transgenesis in mapping key regulatory elements in the mouse by revealing the genomic DNA required for Cdkn2a induction in cultured cells and the developing testis, and the apparent lack of elements driving expression in the developing eye.
Keywords: Arf, Ink4a, cis-regulation, BAC transgenic mice, eye development
Introduction
CDKN2A, an evolutionarily conserved genetic locus in mammals, encodes two functionally distinct proteins, p16INK4a and p14ARF (p19Arf in mouse) that were first recognized to block cancer [1–3]. They link two critical tumor suppressor pathways: p16INK4a functionally activates the protein product of the RB1 gene that, among other things, imposes a G1 phase cell cycle arrest [4]. The p14ARF protein primarily acts in the nucleus to indirectly activate p53 [5]. It also harbors p53-independent activities, including repressing Pdgfrb expression in the developing mouse eye [6]. Human CDKN2A is induced in response to senescence and many oncogenic signals [7,8], and its deletion is one of the most frequent events in human cancer [9].
While mouse orthologs of RB1 and, in some genetic backgrounds, TP53 can play essential roles in cellular differentiation and normal development [10–12], genetically engineered mice lacking Cdkn2a, or Ink4a or Arf individually, were initially felt to be normal, except for cancer susceptibility [13–15]. It is now clear that mice lacking Arf are blind due to the formation of a dense, retrolental fibrovascular plaque in late stages of eye development, a defect mimicking persistent hyperplastic primary vitreous (PHPV) [16]. The animals also display more subtle abnormalities in spermatogenesis [17]. Both findings are consistent with the temporally- and spatially-restricted expression of the Arf promoter; the expression in the primary vitreous depends on Tgfb2 [18,19], but factors driving expression in the testis are not known.
A broader role for CDKN2A in human disease is suggested by the fact that polymorphisms in a gene-poor region lying upstream of CDKN2A and the flanking CDKN2B confer risk for coronary artery disease (CAD) and Type 2 diabetes mellitus [20,21]. Knocking out the orthologous gene-poor region in the mouse reproduced elements of the human phenotype in that the mutant animals were overweight and displayed increased mortality when fed a high-fat, high-cholesterol diet [22]. That phenotype correlated with decreased expression of both Cdkn2a and Cdkn2b in the heart [22] and in the eye [23], Absence of this so-called CAD risk interval in mouse embryo fibroblasts (MEFs) impairs Arf induction by TGFβ [23]. How the expression of these genes is influenced by metabolic stress, such as from a high-fat/high-cholesterol diet, and the nature of the CDKN2A expressing cells is not known.
Most studies of human INK4A and ARF expression have focused on the promoter regions [4,24], although the aforementioned findings suggest the existence of more distant cis regulatory elements. There is also much evidence for the coordinate control of these two transcripts [4,25–27], but their expression can be uncoupled, as in the developing eye and the testis [28,29]. Furthermore, most of the above-mentioned studies utilized cultured cells with relatively little translation of these findings in vivo. For example, the context- and cell type-dependent capacity of oncogenic RAS to induce the transcripts was only realized by analyses of different tumor types in vivo [30].
Certain mouse models have been developed to study Arf and Ink4a expression: GFP or β-galactosidase for Arf [18,31] and luciferase for Ink4a [32]. Each model brings certain advantages and disadvantages regarding the ability to quantify expression and to resolve expression in closely apposed cells; none of them allow the expression of both promoters to be tracked at the same time. We sought to develop a new model by replacing the first coding exon of Arf and Ink4a with different fluorescent reporters that would enable two-color fluorescence detection at single cell resolution. We chose to leverage the power of Tol2 transposon-aided production of transgenic mice using a bacterial artificial chromosome (BAC) so that expression could be studied without disrupting one of the native transcripts encoded at CDKN2A.
Materials and Methods
Plasmids and BAC recombineering
Plasmids were generous gifts from the following: mouse BAC12397 (pBeloBAC11-based C57BL/6 genomic DNA) from Dr. Charles J. Sherr (St. Jude Children’s Research Hospital); pSIM18 mobile recombineering plasmid from Dr. Donald Court (NCI at Frederick); iTol2-Kanamycin from Dr. Maximiliano L. Suster (University of Bergen); and pLD53-SC.A.EB from Dr. Nathaniel Heintz (Rockefeller University). To generate pLD53-based shuttle vectors, PCR-amplified fragments of ~500bp flanking regions of exon1α/1β and nhrGFPII/dTomato coding sequences with overlapping homology sequences were subcloned into the pLD53-SC.A.EB backbone using the Gibson Assembly Kit (New England Biolabs) according to manufacturer’s recommendations. Correctly assembled shuttle vectors were identified by restriction digest and confirmed by Sanger sequencing.
To generate the dual-reporter construct, BAC-carrying bacteria was first transformed with pSIM18 recombineering plasmid. PCR-amplified iTol2-kanamycin cassette with flanking 50bp arms homologous to the BAC backbone vector was introduced as linear dsDNA, and positive clones were identified by antibiotic resistance and confirmed through restriction digestion. Three rounds of overnight culturing at 37°C was carried out to eliminate the pSIM18 plasmid. pLD53-p16-nhrGFPII and pLD53-p19-dTomato shuttle vectors were sequentially introduced to iTol2-modified BAC to replace Cdkn2a exons 1α and 1β, respectively. pLD53 shuttle co-integrated BAC clones were identified by ampicillin resistance, followed by overnight culturing on plates containing 5% sucrose. Candidate sucrose-resistant clones were then screened by PCR and confirmed by restriction digestion to identify correctly modified constructs. Sequences for all primers used can be found in Table S1.
Animals
Dual-fluorescent BAC transgenic mice were generated in the Transgenic Core at UT Southwestern Medical Center. In brief, the final BAC transgene plasmid was purified using NucleoBond BAC 100 (Clontech) and resuspended in injection buffer (10mM Tris-HCl, 0.1mM EDTA, 100mM NaCl, and 1x polyamines) at 3 or 5 ng/μl. Transposase mRNA was also resuspended in injection buffer at 10 or 30 ng/μl. Following standard procedures described previously [33], BAC cDNA and transposase mRNA were co-injected into the pronucleus of C57BL/6 fertilized oocytes. Injected oocytes were transferred into the oviduct of the pseudopregnant female mice. Potential F0 founder and future progeny were screened by PCR using NaOH-extracted tail DNA and primers specific for reporter cassettes listed in Table S1. Lines with documented germline transgene transmission were maintained by breeding hemizygous transgenic animals to C57BL/6 wildtype animals (The Jackson Laboratory). chr4Δ70kb/Δ70kb mice [22] were obtained from Mouse Models of Human Cancer Consortium Repository (MMHCC) and maintained on a mixed C57BL/6 × 129/Sv genetic background. ArfGfp/+ mice [31], obtained from Dr. Charles Sherr, were also maintained on a mixed C57BL/6 × 129/Sv genetic background.
Cell culture and treatment
The 10T1/2 mouse pericyte-like cell line was obtained from American Tissue Type Culture Collection (ATCC) and cultured in DMEM medium with 10% FBS supplemented with Pen/Step. BAC DNA was transiently transfected into 10T1/2 cells using Lipofectamine2000 (Thermo Fisher Scientific) according to the manufacturer’s recommendations. Primary MEFs were isolated at E13.5 or E14.5 and cultivated as previously described [34]. MEFs were serially propagated according to a “3T3” protocol by plating 3×105 cells/6cm dish every 3 days as previously described [35]. To quantify the reporter expression in MEFs, cells were harvested and resuspended in 1x DPBS and analyzed with FACSCanto flow cytometer (BD Biosciences) for nhrGFPII and dTomato reporter expression and analyzed using FlowJo software.
Tissue isolation and histology studies
Tissues and embryos were isolated and fixed in 4% PFA overnight at 4°C. After brief PBS washes, tissues were equilibrated in 30% sucrose and embedded in O.C.T. medium over dry ice. Frozen sections (10μm) were washed with PBS, permeabilized with 0.1% Triton-X/PBS, and blocked with 10% serum in 0.1% Triton-X/PBS for immunohistochemistry, or directly stained using TO-PRO-3 and mounted in 50% glycerol/PBS for imaging. Confocal fluorescent images were acquired using a Zeiss LSM510 META inverted microscope (UTSW O’Brien Cell Biology and Imaging Core).
Quantitative RT-PCR
Total RNA was extracted from cultured cells and cDNA was prepared as previously described [23]. For RNA extraction from testis, dissected tissues were flash frozen in liquid nitrogen and homogenized in QIAzol (Qiagen) using a hand-held mini tissue homogenizer. Total RNA was isolated using the miRNeasy mini kit (Qiagen) with on-column DNaseI (Qiagen) treatment to remove genomic DNA. Quantitative mRNA analysis was performed using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) and the CFX96 Touch System (Bio-Rad) for real-time PCR detection. Sequences for qRT-PCR primers are listed in Table S1.
Statistical analysis
Quantitative data are presented as column bar graphs with mean ± SD from at least three representative experiments. Statistical significance (P < 0.05) between two populations was determined by Student’s t-test.
Results and Discussion
Generation of dual fluorescent BAC transgenic reporter mice to monitor Cdkn2a expression
To study the context-specific regulation of Arf and Ink4a in primary MEFs and in vivo, we designed and generated a dual-fluorescent Cdkn2a BAC reporter that could be utilized in transposon-aided BAC transgenesis (Fig. 1a). The original BAC vector contained ~150kb of mouse genomic DNA, including Cdkn2a/b and flanking DNA (BAC12397, Fig. 1a). Cdkn2a exons 1α and 1β were replaced with cDNA coding for a nuclear-localized GFP variant (nhrGFPII) and dTomato, respectively, through a two-step, RecA-based recombineering approach (Fig. S1a) [36]. Note that only the fluorescent reporters would be translated from the Ink4a (p16-nhrGFPII) and Arf promoters (p19-dTomato) (Fig. S2).
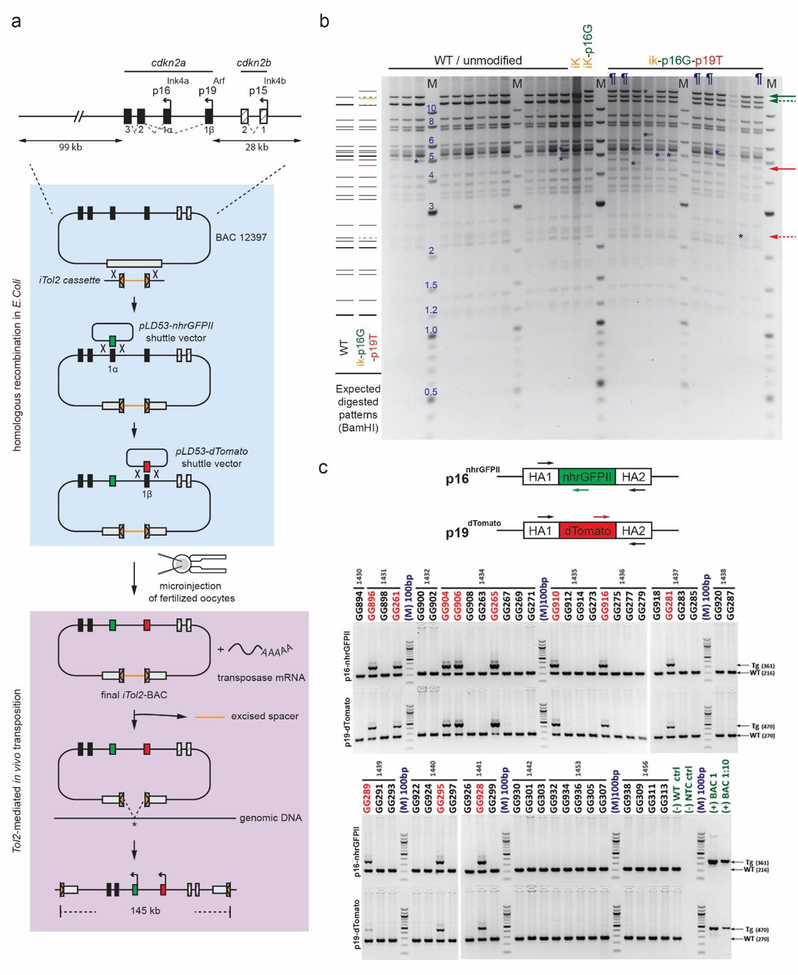
Figure 1.
Design, generation, and verification of the Cdkn2a reporter BAC construct. a Schematic diagram displays mouse Cdkn2a gene locus (top), BAC modification steps by homologous recombination (middle), and transposase-aided integration (bottom). The pBeloBAC11 vector backbone is indicated by the gray box in middle and bottom panels. b Photo of representative, ethidium bromide-stained agarose gel of unmodified (WT) and modified BAC cDNA following BamHI digestion. Expected restriction fragments (left) and fragment sizes (M; middle) are displayed in kilobases. Yellow, green, and red lines and arrows show new fragments expected after iTol2, p16-nhrGFPII, and p19-dTomato insertion, respectively. Dashed arrows identify loss of unmodified wildtype bands; solid arrows indicate expected transgene insertion bands. Definitions - iK: iTol2-inserted BAC; ik-p16G: iTol2 and p16-nhrGFPII-inserted BAC; ik-p16G-p19T: iTol2, p16-nhrGFPII and p19-dTomato-inserted BAC; ¶: confirmed BACik-p16G-p19T clones; *, unexpected restriction fragment. c Photo of representative, ethidium bromide-stained agarose gel showing PCR products resulting from amplification of genomic DNA from individual BAC transgenic founders. Schematic diagram (top panel) shows location of PCR primers to amplify either wildtype (WT) or transgenic (TG) DNA for p16-nhrGFPII and p19-dTomato. Arrows (right side) indicate expected PCR product sizes. Molecular weight (M) is indicated in nucleotide base pairs (bp). Definition - HA: Homology Box A represents the sequence flanking transgene insertion site.
To increase the efficiency of BAC transposition, transposable elements from the medeka Tol2 transposon [37] were inserted into the BAC backbone vector through the plasmid-based lambda phage Red/ET recombineering system (Fig. 1a) [38]. Modifications in the final BAC clone (BACik-p16G p19T) were verified using PCR amplification and restriction-digested fingerprinting (Fig. 1b and S1b). Functionality of the final BACik-p16G-p19T clone was tested by transfecting it into mouse 10T1/2 fibroblasts in vitro, and p16-nhrGFPII and p19-dTomato expression was detectable in individual cells 72 hours later (Fig. 2a and additional data not shown).
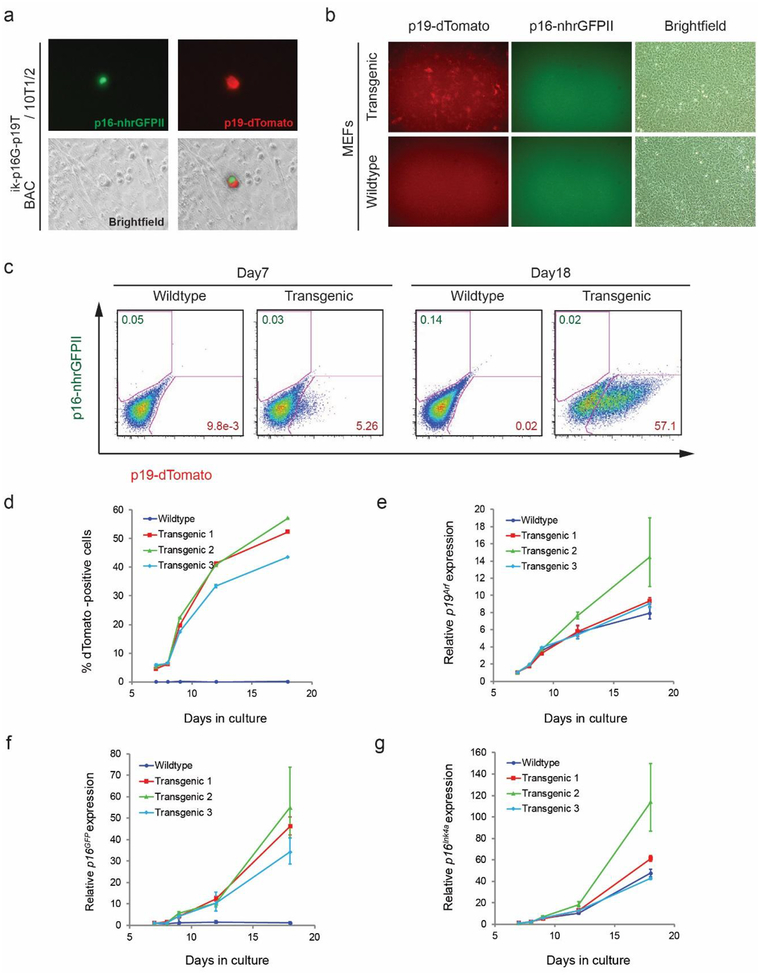
Figure 2.
Induction of the Cdkn2a reporter with serial passage of transgenic mouse embryonic fibroblasts. a Fluorescence photomicrograph of 10T1/2 fibroblasts transfected with of BACik-p16G-p19T reporter shows isolated cell expressing both green (p16-nhrGFPII) and red (p19-dTomato) reporters. b, c Fluorescence photomicrograph (b) and flow cytometry plots (c) of MEFs derived from wildtype and BACik-p16G-p19T transgenic mice at passage number 5 (b) or following 7 or 18 days in culture (c). Numerous dTomato positive cells were detected in transgenic cells, but nhrGFPII protein was not visible, even though mRNA was induced (f, g). d Quantitative analysis of dTomato-positive MEFs from three different transgenic lines or a single wildtype line. Cells were quantified by flow cytometry with serial passage for the indicated number of days. e-g Quantitative analysis of relative mRNA expression of endogenous mRNA for Arf (e), p16-nhrGFPII (f), and Ink4a mRNA (g) in wildtype or transgenic MEFs upon serial passage.
The purified BACik-p16G-p19T plasmid was injected in the UT Southwestern Transgenic Core facility into the pronucleus of fertilized mouse oocytes, with and without co-injection of Tol2 transposase mRNA (Fig. 1a). Out of the 50 adults animals screened, eleven F0 founders were identified by PCR genotyping (Fig. 1c). We note that founder animals were only successfully generated when transposase mRNA was co-injected, and increasing the concentration of BAC DNA and including transposase mRNA seemed to facilitate transgenesis, which is consistent with previous studies (Table S2) [39,40]. Though Tol2-mediated transgenesis is reported to preferentially drive single-copy integration [39], PCR genotyping showed evidence for a multicopy concatamer in at least one of the lines (Figure S3). This fact likely accounted for our inability to map the BAC integration site using PCR-based approaches [41]. Germline transmission was confirmed in 9 of the 11 potential founders (Table S3), and those lines were studied further.
In vitro confirmation of Arf reporter induction in serially-passed MEFs
We examined the expression of p19-dTomato and p16-nhrGFPII using primary MEFs derived from transgenic lines and cultivated ex vivo. The so-called “culture shock” observed in MEFs is one well-established method to study dynamic induction of p19Arf and p16lnk4a [13]. As expected in early passage MEFs, few p19-dTomato-positive cells were observed by florescence microscopy, consistent with prior analyses of p19Arf [13,18]. However, more than half of the cells showed expression of the p19-dTomato at passage 5 (day 18), a finding that was confirmed using flow cytometry (Fig. 2b, c). The relative fraction of dTomato-positive cells closely paralleled relative Arf mRNA expression measured by real-time qRT-PCR (r2= 0.8339, p<0.0001) (Fig. 2d, e). Real-time qRT-PCR showed that nhrGFPII mRNA also increased in parallel to native Ink4a following kinetics previously reported in cultured MEFs (Fig. 2f, g) [32]. We conclude that induction of mRNA encoding both the p19-dTomato and p16-nhrGFPII reporters in cultured cells shows that key cis regulatory elements reside within the BAC construct.
In contrast to the Arf reporter, though, neither direct fluorescence microscopy nor flow cytometry revealed the green fluorescence signal from the p16-nhrGFPII in serially passed MEFs isolated from 3 different transgenic lines, despite robust induction of the mRNA (Fig. 2b, c). Sequencing of genomic DNA derived from the transgenic mouse showed that lack of fluorescence was not due to a deleterious mutation in the nhrGFPII altering the coding sequence or disrupting translation initiation (Fig. S2 and sequencing results not shown). We speculate that another post-transcriptional effect may account for inability to detect the green fluorescence signal. For example, the dTomato cassette included a woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) [42], which can boost gene expression, but that element was not included in the nhrGFPII cassette (Fig. S2). Exactly how much that influences nhrGFPII protein expression and detection is not yet clear.
Expression of the Arf reporter in postnatal testis but not in the developing eye
As mentioned earlier, temporally- and spatially-restricted expression of Arf during embryonic development or in normal adult tissues is found in the testis [16,28,31], and this provided an excellent opportunity to validate the p19-dTomato reporter. We detected p19-dTomato expression in whole mounted testis by direct fluorescence and localized the expressing cells to outer elements of the seminiferous tubules in the transgenic testis from postnatal day (P) 12 through P21, consistent with previous reports using GFP and β-galactosidase reporters in Arf Gfp/Gfp and Arf lacZ/lacZ mice (Fig. 3a and data not shown) [17,31]. This expression pattern was identified in eight of the nine transgenic animals (Fig. 3b), indicating that the expression is not likely to be influenced by random BAC integration.
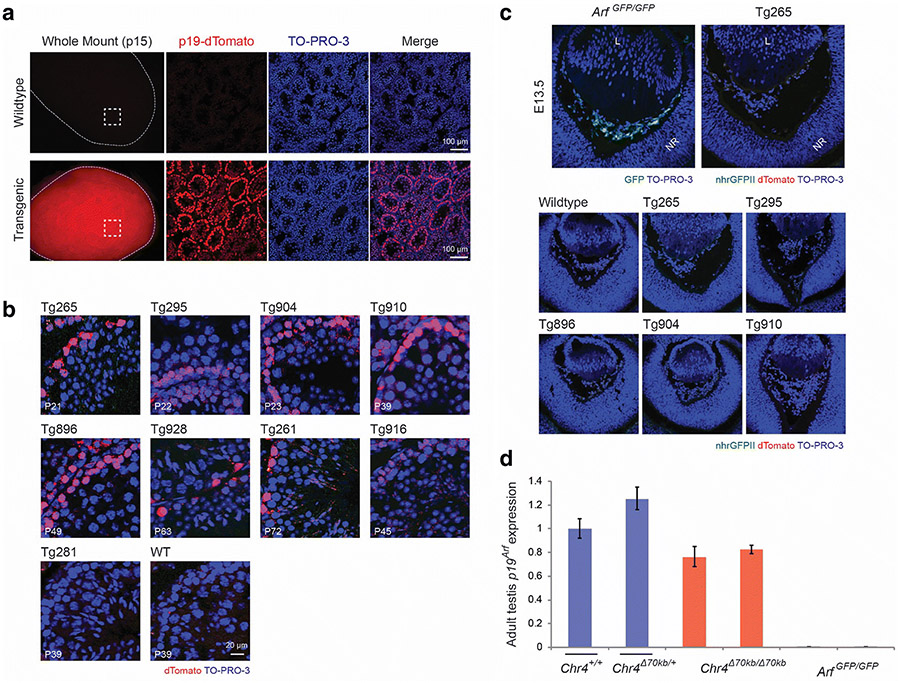
Figure 3.
Detectable expression of the p19-dTomato reporter in the testis but not in newborn mouse eye in BACik-p16G-p19T transgenic animals. a, b Representative fluorescence photomicrographs of and mount (left column) and cryostat sections of the mouse testis taken from wild type and transgenic mice at postnatal day (P) 15 (a) or as indicated (b). The Tg265 transgenic line is displayed in (a); transgenic lines in (b) are indicated. TO-PRO-3 staining highlights nuclei. c Representative fluorescence photomicrographs of cryostat sections show the developing eye in wildtype, Arf GFP/GFP, or BAC transgenic lines (Tg) in mouse embryos at embryonic day (E) 13.5. Note that Gfp expression from the native Arf promoter is readily detected in Arf GFP/GFP animals, but dTomato expression from the p19-dTomato BAC reporter is not. TO-PRO-3 staining highlights nuclei. Definitions – L: lens; NR: neuroretina. d Quantitative analysis of relative Arf mRNA expression, measured by quantitative RT-PCR, using RNA extracted from postnatal testes from wildtype (Chr4 +/+) and transgenic animals as indicated. Arf mRNA expression is normalized to Gapdh and presented as average relative to wildtype animals. Error bars denote standard deviation.
Arf mRNA and p19Arf protein is also expressed in the developing mouse eye from E13.5, when the hyaloid vessels in the primary vitreous first form, through P5, when they begin to regress. This expression pattern was first identified by RT-PCR [16], and it was verified by two different reporters as well as immunofluorescence staining for p19Arf [6,18]. In the current experiments, we again showed GFP expression in the primary vitreous of ArfGFP/GFP mouse eyes at E13.5, but surprisingly, p19-dTomato was not detectable in eyes from any of five transgenic lines shown to express the reporter in the testis (Fig. 3b, c). This finding demonstrates that cis regulatory elements present in the ~150kb BAC transgene differentially control Arf promoter activity in different anatomic sites.
The differential expression of the p19-dTomato reporter could be due to the absence in the BAC of a distant cis enhancer required for Arf induction in the eye. The aforementioned gene-poor segment orthologous to the human CAD risk interval could serve as that distant enhancer because it is essential for Arf expression in the Chr4 Δ70kb/Δ70kb eye [23] but was not included in the BACik-p16G-p19T construct. If true, expression of native Arf in the testis should similarly be independent of that distant cis element. Indeed, qRT-PCR for Arf mRNA in wildtype and Chr4 Δ70kb/Δ70kb animals showed this to be the case (Fig. 3d). Therefore, the random integration of the BAC transgene in five different lines likely uncoupled that putative cis enhancer from the reporter, dampening its expression in the eye.
It should be emphasized that Arf expression in the mouse eye depends on TGFβ [18], and deletion of the distant regulatory element in Chr4 Δ70kb/Δ70kb MEFs specifically impairs TGFβ-dependent induction of this gene in cultured MEFs [23]. This fact is consistent with our recent finding that TGFβ augments histone 3 lysine 27 acetylation (H3K27ac) in three peaks lying within the CAD risk interval in HeLa cells, and those peaks are essential for induction of human ARF [43]. We are in the process of using computational approaches to identify syntenic regions in the mouse genome and using other functional strategies to more finely define the TGFβ-dependent cis enhancers in the mouse [44]. Regrettably, the importance of those elements was not clear when the BACik-p16G-p19T construct was designed, and we have not yet identified a publicly available BAC that spans this entire sequence.
There are other possible explanations for our findings. For example, position effects from random integration could still play a role in silencing the dTomato reporter in the eye. The chance for this to happen in multiple independent lines in a way that does not interfere with dTomato expression in the testis and cultured fibroblasts is very remote. Alternatively, key regulatory elements in the BAC could have been disrupted in generating the construct and mouse lines. Yet, PCR analyses of DNA derived from transgenic mouse showed that a) both ends of the BAC were integrated (Fig. S3), and b) none of 16 regions across the BAC harbored obvious insertions or deletions that might have disrupted a specific enhancer (Fig. S3). Remaking the transgenic mouse with a BAC transgenic construct that includes the candidate cis enhancer element represents an attractive approach to provide definitive proof of these distant, TGFβ-dependent enhancers.
We are cognizant of the fact that one prior report suggested that a BAC similar to the one we used seemed to contain the needed regulatory elements: BAC-based transgenic delivery of mouse Arf cDNA was reported to be sufficient to rescue the PHPV-like developmental eye disease in a single BAC transgenic line [45]. In contrast to our findings, that report showed that induction of the transgenic Arf cDNA did not follow the same kinetics as the native Arf transcript in cultured MEFs [45], and the expected Arf expression was not documented at E13.5. We can reconcile our findings and this previous report in two ways. First, integration site-specific effects may enable baseline – but not TGFβ-induced – Arf expression in the developing eye at a sufficient level to mollify the ocular developmental defects without actually mirroring the normal expression pattern. Alternatively, we note that the BAC construct in our studies contained approximately ~65 kb of additional DNA 3’ to the Cdkn2a gene. In principle, that DNA could contain cis repressor elements that must be counterbalanced by the aforementioned distant enhancers to yield the appropriate temporal and spatially-restricted expression. That latter hypothesis is readily tested using BAC-based transgenesis in the mouse.
Although we did not meet our primary goal of generating a dual-reporter mouse to track the expression of two distinct transcripts, our work adds to the literature of BAC-based transgenic mouse production. The relative ease of homologous recombination-based generation makes the generation of complex transgenic relatively straightforward, as others have described [36,46]. Our findings confirm a previous report that incorporation of transposable elements into the BAC and co-injection of transposase mRNA into pronuclei can facilitate the generation of transgenic animals [39]. As we move forward, we envision testing whether the deletion or addition of candidate regulatory elements represses testicular or restores ocular expression. Finally, the growing use of CRISPR/Cas9 to edit the mammalian genome will enable the generation of reporters knocked into a native locus [47,48]; however, the inactivation of the native gene product in the process can result in a phenotype and possibly alter the expression of the promoter. Indeed, Arf expressing cells are more easily visualized of Arf lacZ/lacZ and Arf Gfp/Gfp animals than in heterozygous animals retaining native p19Arf expression [6,18]. In contrast, a BAC transgenic mouse reporter enables the study of the Arf promoter without the potentially confounding effect of haploinsufficiency of an endogenous gene.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge technical assistance provided by S. Singleterry, and many helpful comments by other members of the Skapek laboratory. We also acknowledge support from the National Institutes of Health National Eye Institute (R01 EY019942), from the Cancer Prevention and Research Institute of Texas (RP120685-P2), and from the UTSW Harold C. Simmons Comprehensive Cancer Center from the National Cancer Institute (CA142543).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest
We have no conflict of interest related to this report.
References
- 1.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS, Johnson BE, Skolnick MH (1994) A cell cycle regulator potentially involved in genesis of many tumor types. Science 264:436–440. 10.1126/science.8153634 [DOI] [PubMed] [Google Scholar]
- 2.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA (1994) Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368:753–756. 10.1038/368753a0 [DOI] [PubMed] [Google Scholar]
- 3.Quelle DE, Zindy F, Ashmun RA, Sherr CJ (1995) Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993–1000. 10.1016/0092-8674(95)90214-7 [DOI] [PubMed] [Google Scholar]
- 4.Gil J, Peters G (2006) Regulation of the INK4b–ARF–INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7:667–677. 10.1038/nrm1987 [DOI] [PubMed] [Google Scholar]
- 5.Somasundaram K, EI-Deiry WS (2000) Tumor suppressor p53: regulation and function. Front Biosci 5:D424–437. 10.2741/Somasund [DOI] [PubMed] [Google Scholar]
- 6.Silva RL, Thornton JD, Martin AC, Rehg JE, Bertwistle D, Zindy F, Skapek SX (2005) Arf-dependent regulation of Pdgf signaling in perivascular cells in the developing mouse eye. EMBO J 24:2803–2814. 10.1038/sj.emboj.7600751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bringold F, Serrano M (2000) Tumor suppressors and oncogenes in cellular senescence. Exp Gerontol 35:317–329. 10.1016/S0531-5565(00)00083-8 [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ (2012) Ink4-Arf locus in cancer and aging. Wiley Interdiscip Rev Dev Biol 1:731–741. 10.1002/wdev.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim WY, Sharpless NE (2006) The regulation of INK4/ARF in cancer and aging. Cell 127:265–275. 10.1016/j.cell.2006.10.003 [DOI] [PubMed] [Google Scholar]
- 10.Almog N, Rotter V (1997) Involvement of p53 in cell differentiation and development. Biochim Biophys Acta 1333:F1–27. 10.1016/S0304-419X(97)00012-7 [DOI] [PubMed] [Google Scholar]
- 11.Lipinski MM, Jacks T (1999) The retinoblastoma gene family in differentiation and development. Oncogene 18:7873–7882. 10.1038/sj.onc.1203244 [DOI] [PubMed] [Google Scholar]
- 12.Stiewe T (2007) The p53 family in differentiation and tumorigenesis. Nat Rev Cancer 7:165–167. 10.1038/nrc2072 [DOI] [PubMed] [Google Scholar]
- 13.Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ (1997) Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19 ARF. Cell 91:649–659. 10.1016/S0092-8674(00)80452-3 [DOI] [PubMed] [Google Scholar]
- 14.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho RA (1996) Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27–37. 10.1016/S0092-8674(00)81079-X [DOI] [PubMed] [Google Scholar]
- 15.Sharpless NE, Bardeesy N, Lee K-H, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA (2001) Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86–91. 10.1038/35092592 [DOI] [PubMed] [Google Scholar]
- 16.McKeller RN, Fowler JL, Cunningham JJ, Warner N, Smeyne RJ, Zindy F, Skapek SX (2002) The Arf tumor suppressor gene promotes hyaloid vascular regression during mouse eye development. Proc Natl Acad Sci U S A 99:3848–3853. 10.1073/pnas.052484199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churchman ML, Roig I, Jasin M, Keeney S, Sherr CJ (2011) Expression of Arf tumor suppressor in spermatogonia facilitates meiotic progression in male germ cells. PLoS Genet 7:e1002157 10.1371/journal.pgen.1002157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman-Anderson NE, Zheng Y, McCalla-Martin AC, Treanor LM, Zhao YD, Garfin PM, He T-C, Mary MN, Thornton JD, Anderson C, Gibbons M, Saab R, Baumer SH, Cunningham JM, Skapek SX (2009) Expression of the Arf tumor suppressor gene is controlled by TGFβ2 during development. Development 136:2081–2089. 10.1242/dev.033548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Zhao YD, Gibbons M, Abramova T, Chu PY, Ash JD, Cunningham JM, Skapek SX (2010) Tgfβ signaling directly induces Arf promoter remodeling by a mechanism involving Smads 2/3 and p38 MAPK. J Biol Chem 285:35654–35664. 10.1074/jbc.M110.128959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC (2007) A common allele on chromosome 9 associated with coronary heart disease. Science 316:1488–1491. 10.1126/science.1142447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hannou SA, Wouters K, Paumelle R, Staels B (2015) Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trends Endocrinol Metab 26:176–184. 10.1016/j.tem.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 22.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA (2010) Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature 464:409–412. 10.1038/nature08801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Y, Devitt C, Liu J, Mei J, Skapek SX (2013) A distant, cis-acting enhancer drives induction of Arf by Tgfβ in the developing eye. Dev Biol 380:49–57. 10.1016/j.ydbio.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esteller M (2002) CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 21:5427–5440. 10.1038/sj.onc.1205600 [DOI] [PubMed] [Google Scholar]
- 25.Bernard D, Martinez-Leal JF, Rizzo S, Martinez D, Hudson D, Visakorpi T, Peters G, Carnero A, Beach D, Gil J (2005) CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene 24:5543–5551. 10.1038/sj.onc.1208735 [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Gu X, Su I-h, Bottino R, Contreras JL, Tarakhovsky A, Kim SK(2009) Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev 23:975–985. 10.1101/gad.1742509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164–168. 10.1038/16476 [DOI] [PubMed] [Google Scholar]
- 28.Martin AC, Thornton JD, Liu J, Wang X, Zuo J, Jablonski MM, Chaum E, Zindy F, Skapek SX (2004) Pathogenesis of persistent hyperplastic primary vitreous in mice lacking the Arf tumor suppressor gene. Invest Ophthalmol Vis Sci 45:3387–3396. 10.1167/iovs.04-0349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zindy F, Quelle DE, Roussel MF, Sherr CJ (1997) Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15:203–211. 10.1038/sj.onc.1201178 [DOI] [PubMed] [Google Scholar]
- 30.Young NP, Jacks T (2010) Tissue-specific p19Arf regulation dictates the response to oncogenic K-ras. Proc Natl Acad Sci U S A 107:10184–10189. 10.1073/pnas.1004796107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, Cleveland JL, Roussel MF, Sherr CJ (2003) Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci U S A 100:15930–15935. 10.1073/pnas.2536808100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burd CE, Sorrentino JA, Clark KS, Darr DB, Krishnamurthy J, Deal AM, Bardeesy N, Castrillon DH, Beach DH, Sharpless NE (2013) Monitoring tumorigenesis and senescence in vivo with a p16INK4a-luciferase model. Cell 152:340–351. 10.1016/j.cell.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy A, Gertsenstein M, Vintersten K, Behringer R (2003) Manipulating the mouse embryo: a laboratory manual, 3rd edn Cold Spring Harbor Laboratory Press, NY. [Google Scholar]
- 34.Sharpless NE (2006) Chapter 28 - Preparation and immortalization of primary murine cells In: Celis JE (ed) Cell Biology, 3rd edn Academic Press, Burlington, pp 223–228. 10.1016/B978-012164730-8/50029-0 [DOI] [Google Scholar]
- 35.Todaro GJ, Green H (1963) Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 17:299–313. 10.1083/jcb.17.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong S, Yang XW, Li C, Heintz N (2002) Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6kγ origin of replication. Genome Res 12:1992–1998. 10.1101/gr.476202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urasaki A, Morvan G, Kawakami K (2006) Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174:639–649. 10.1534/genetics.106.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datta S, Costantino N, Court DL (2006) A set of recombineering plasmids for gram-negative bacteria. Gene 379:109–115. 10.1016/j.gene.2006.04.018 [DOI] [PubMed] [Google Scholar]
- 39.Suster ML, Sumiyama K, Kawakami K (2009) Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics 10:477 10.1186/1471-2164-10-477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Keuren ML, Gavrilina GB, Filipiak WE, Zeidler MG, Saunders TL (2009) Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res 18:769–785. https://doi:10.1007/s11248-009-9271-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suster ML, Abe G, Schouw A, Kawakami K (2011) Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc 6:1998–2021. https://doi:10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- 42.Zufferey R, Donello JE, Trono D, Hope TJ (1999) Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol 73:2886–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y-T, Xu L, Bennett L, Hooks JC, Liu J, Zhou Q, Liem P, Zheng Y, Skapek SX (2019) Identification of de novo enhancers activated by TGFβ to drive expression of CDKN2A and B in HeLa cells. Mol Cancer Res. https://doi:10.1158/1541-7786.Mcr-19-0289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kristensen DM, Wolf YI, Mushegian AR, Koonin EV (2011) Computational methods for Gene Orthology inference. Brief Bioinform 12:379–391. https://doi:10.1093/bib/bbr030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matheu A, Pantoja C, Efeyan A, Criado LM, Martín-Caballero J, Flores JM, Klatt P, Serrano M (2004) Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev 18:2736–2746. 10.1101/gad.310304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narayanan K, Chen Q (2011) Bacterial artificial chromosome mutagenesis using recombineering. J Biomed Biotechnol 2011:971296 10.1155/2011/971296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H, Wang H, Jaenisch R (2014) Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat Protoc 9:1956–1968. 10.1038/nprot.2014.134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.