Abstract
Crassulacean acid metabolism (CAM) has evolved as a water-saving strategy, and its engineering into crops offers an opportunity to improve their water use efficiency. This requires a comprehensive understanding of the regulation of the CAM pathway. Here, we use the facultative CAM species Talinum triangulare as a model in which CAM can be induced rapidly by exogenous abscisic acid. RNA sequencing and metabolite measurements were employed to analyse the changes underlying CAM induction and identify potential CAM regulators. Non-negative matrix factorization followed by k-means clustering identified an early CAM-specific cluster and a late one, which was specific for the early light phase. Enrichment analysis revealed abscisic acid metabolism, WRKY-regulated transcription, sugar and nutrient transport, and protein degradation in these clusters. Activation of the CAM pathway was supported by up-regulation of phosphoenolpyruvate carboxylase, cytosolic and chloroplastic malic enzymes, and several transport proteins, as well as by increased end-of-night titratable acidity and malate accumulation. The transcription factors HSFA2, NF-YA9, and JMJ27 were identified as candidate regulators of CAM induction. With this study we promote the model species T. triangulare, in which CAM can be induced in a controlled way, enabling further deciphering of CAM regulation.
Keywords: Abscisic acid, crassulacean acid metabolism, metabolome, Talinum triangulare, time course, transcriptome
Exogenous abscisic acid leads to fast transcriptome reprogramming, which translates into altered metabolome composition. This includes nocturnal acidification, indicating induction of the CAM pathway.
Introduction
Crassulacean acid metabolism (CAM) has independently evolved as a carbon-concentrating and water-saving strategy in approximately 6% of vascular plants (Winter et al., 2008; Silvera et al., 2010). CAM species rely on the temporal separation of primary carbon assimilation and its subsequent incorporation into carbohydrates through the Calvin–Benson–Bassham cycle (CBBC). Stomata open towards the end of the light period, enabling CO2 uptake and its assimilation in the form of HCO3– by the concerted actions of phospohoenolpyruvate carboxylase (PPC) and malate dehydrogenase (MDH). Malate is the main organic acid stored in the vacuole, but accumulation of citrate and isocitrate has also been observed (Lüttge, 1988; Medina et al., 1993; Herppich et al., 1995; Chen et al., 2002; Winter and Holtum, 2014). During the light period, stomata are closed and, depending on the type of CAM species, one of three decarboxylating enzymes releases nocturnally fixed CO2 from malic acid, thus providing Rubisco with its substrate (Holtum et al., 2005). The diurnal cycling of acidification and organic acid degradation is tightly interconnected with carbohydrate metabolism. Soluble sugars (fructose, glucose, or sucrose) or polysaccharides [starch, (poly)fructans] are degraded in the dark to provide carbon skeletons in the form of PEP for nocturnal carboxylation. The sugar/polysaccharide pool is then regenerated in the light via gluconeogenesis (Borland and Taybi, 2004; Holtum et al., 2005; Borland et al., 2016; Taybi et al., 2017). To mitigate futile cycling, temporal control of enzyme activities is essential and occurs at both transcriptional and post-transcriptional levels, including diurnally regulated expression of PHOSPHOENOLPYRUVATE CARBOXYLASE (PPC) and PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE (PPCK) (Nimmo et al., 1999; Cushman et al., 2008).
Despite the strict temporal regulation, CAM remains a highly plastic adaptation that has been defined in different modes, including obligate and facultative CAM, CAM-idling, and CAM-cycling (Lüttge, 2004). In particular, it has been proposed that facultative CAM could serve as a model to allow the identification of the minimal gene set required for a fully functional CAM pathway and to shed light on the regulation of these genes, thus allowing CAM engineering into C3 crops (Borland et al., 2014; Hartwell et al., 2016). In facultative CAM species, CAM is induced in response to environmental stresses (e.g. drought) and abscisic acid (ABA) has been identified as an important signal in CAM induction (Chu et al., 1990; Dai et al., 1994; Taybi and Cushman, 2002). Characteristics of CAM, such as nocturnal acidification, accumulation of PPC transcript, and increased extractable activities of central CAM enzyme, have been induced by exogenous ABA in Portulacaria afra (Ting, 1981), Mesembryanthemum crystallinum (Holtum and Winter, 1982; Chu et al., 1990; Dai et al., 1994), or Kalanchoë blossfeldiana (Taybi et al., 1995). In Talinum triangulare, increased amounts of transcripts encoding proteins of both CAM and ABA signalling pathways were detected in response to water withdrawal, returning to non-stress levels upon rewatering (Brilhaus et al., 2016).
In the plant, ABA is perceived by soluble receptors of the PYRABACTIN RESISTANCE/PYRABACTIN RESISTANCE-LIKE/REGULATORY COMPONENTS OF ABA RECEPTOR group, which upon ABA binding physically interact with clade A PP2C phosphatases (PP2CA), preventing dephosphorylation of SNF1-RELATED PROTEIN KINASEs 2 (SnRK2) (Park et al., 2009). Autophosphorylated SnRK2s subsequently modulate the activity of their downstream targets, ranging from other kinases to anion channels and numerous transcription factors (TFs), many of them recognizing ABA-responsive elements in the promoter sequences of target genes (Yoshida et al., 2010; Antoni et al., 2011).
Despite detailed understanding of ABA signalling and accumulating evidence of the ABA inducibility of CAM, a study further elucidating the role of ABA in CAM induction is lacking. We therefore conducted this study with the following aims: (i) to discover how rapidly the CAM pathway can be activated by exogenous ABA; (ii) to determine whether it is possible to distinguish drought-induced and ABA-induced facultative CAM; and (iii) to identify the CAM regulators downstream of ABA signalling.
Materials and methods
Plant material and growth conditions
Talinum triangulare plants used in this study originated from two subsequent, controlled self-pollination events, increasing the homogeneity of the plant material. Seeds were germinated in multiplication substrate (Floraton 3, Floragard) and 4-week-old seedlings were transferred to pots with D 400 soil and Cocopor (Stender). Two weeks before the treatment, the plants were placed into a controlled-environment plant chamber (MobyLux GroBanks, CLF Plant Climatics) with the following growth conditions: 12 h light/12 h dark at 25 °C/23 °C. The light intensity at the leaf level was 150–200 µmol s–1 m–2. To avoid unintended induction of CAM by drought, all plants were watered as needed, with the same amount of water per pot.
Treatments and harvest of leaf material
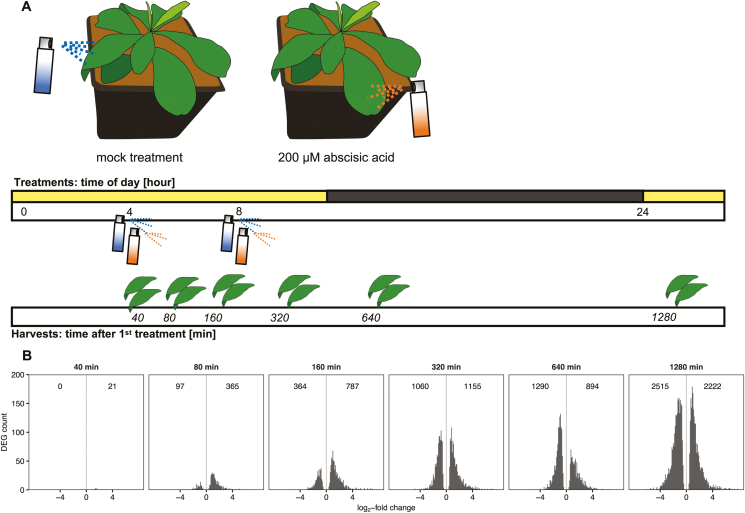
To determine a suitable ABA concentration, a range from 10 to 600 µM was tested (see Supplementary Fig. S1 at JXB online) prior to the work presented here. Using 2-month-old plants, two mature and fully illuminated leaves of each plant were treated either with (+/–) ABA [Sigma-Aldrich; 200 µM solution in 0.095% (v/v) methanol] or mock solution (0.095% (v/v) methanol). Both solutions contained 0.02% (v/v) Tween-20 (PanReac AppliChem). The first treatment took place after 4 hours in the light and the second treatment followed 4 hours later (Fig. 1). Treated leaves were harvested and snap-frozen in liquid nitrogen 40, 80, 160, 320, 640, and 1280 min after the first treatment. Two biological replicates originating from two independent plants were harvested per time point. The treated leaves from the same plant were pooled prior to all analyses.
Fig. 1.
Early response of Talinum triangulare to exogenous abscisic acid. (A) Experimental design. Prior to the treatments, T. triangulare plants were adapted to a 12 h/12 h light/dark, 25 °C/23 °C cycle. At 2 months of age, two mature leaves per plant were sprayed with 200 µM ABA or mock solution [0.095% (v/v) methanol]. The first treatment was applied 4 hours into the light period and was followed by a second treatment 4 hours later. Treated leaves were harvested and snap-frozen 40, 80, 160, 320, 640, and 1280 min after the first treatment. (B) Number of significantly (q≤0.01) down- and up-regulated genes (differentially expressed genes; DEGs) in ABA-treated leaves compared with mock-treated leaves at each time point.
Titratable acidity
Approximately 30 mg of ground leaf material was incubated in 500 µl 50% (v/v) methanol at 90 °C while shaking. After centrifugation at 13,000 g for 5 min at room temperature, 450 µl of the supernatant was collected and the remaining pellet was extracted once more. Both fractions were pooled and stored at –20 °C until use. Titratable acidity was measured using bromothymol blue (Carl Roth) as a pH indicator: 60 µl of extract was mixed with 40 µl distilled H2O and 4 µl bromothymol blue (1 mg/ml stock). Using the microinjector of a Synergy H1 (BioTek) plate reader, samples were titrated with 1 mM KOH in 5 µl steps. After each step, absorbance at 445 nm and 615 nm was recorded, and the ratio of the absorbances at 615/445 nm was calculated. Based on the 615/445 ratio of buffer standards (pH 4.6–7.5), the pH of analysed extracts was determined. From the volume of KOH required to titrate extracts to pH 7.0, the titratable acidity was calculated. Two extractions were performed from each sample, and each extract was titrated in three replicates.
Protein and starch measurements
Approximately 25 mg of ground leaf material was used for extraction in 80% (v/v) ethanol with 10 mM 2-(N-morpholino)ethanesulfonic acid (pH 6). Extraction was performed three times with 500 µl extraction buffer while shaking for 1 hour at 90 °C. After each extraction step, samples were centrifuged at 12,000 g for 10 min and the supernatant was collected and pooled with previous fractions. From each sample, two extracts were prepared and stored at –20 °C until analysed. The protein concentration was measured using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
Pellets obtained during protein extraction were gelatinized with 300 µl 0.2 M NaOH at 90 °C for 40 minutes. After adjustment to pH 5–6, 200 µl hydrolysis buffer was added [20 mM acetate buffer, pH 4.8; 0.5 U α-amylase (10102814001, Roche), and 4 U amyloglucosidase (A7095, Roche)] and the mixture was incubated overnight at 37 °C. Glucose resulting from starch degradation was measured enzymatically: 10 µl of digest was added to 100 mM HEPES-NaOH (pH 7.5) with 10 mM MgCl2, 1 mM NADP+, 2 mM ATP, 0.1 U glucose-6-phosphate dehydrogenase (G6PDHII-RO, Roche), and 4 U hexokinase (11426362001, Roche). The final reaction volume was 200 µl and the increase of absorbance at 340 nm was monitored.
Metabolite profiling
Approximately 50 mg of ground leaf material was used to obtain extracts for metabolite profiling by gas chromatography-mass spectrometry (GC-MS) as described by Fiehn et al. (2000). Data analysis was performed using MassHunter Software, version B.07.00 (Agilent). For relative quantification, all metabolite peak areas were normalized to the peak area of the internal standard ribitol added prior to extraction and the amount of leaf material. From each sample, two extracts were prepared and analysed.
Amino acids were measured by liquid chromatography-mass spectrometry (LC-MS). Extracts were prepared in 80% (v/v) ethanol (Fisher Scientific) as described by Di Martino et al. (2003), and data analysis was conducted using MassHunter Software, version B.07.00 (Agilent). From each sample, two extracts were prepared and analysed. Metabolites analysed by both methods are listed in Supplementary Dataset S3.
Total RNA extraction, preparation of Illumina libraries, and sequencing
Total RNA was extracted from ground leaf tissue using the Monarch® Total RNA Miniprep Kit (New England Biolabs Inc.) following the manufacturer’s instructions. Contaminating DNA was digested with DNase I (New England Biolabs Inc.). The integrity of input RNA was analysed on a 2100 Bioanalyzer (Agilent). Libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit (Illumina) following the Illumina TruSeq Stranded mRNA Sample Preparation Guide #15031047 Rev. E, with the following adaptations: 300 ng total RNA as the basis for library preparation, adapter index dilution to 50% with RNase-free water, and an additional purification step with AMPure XP (Beckman Coulter).
The concentration of the resulting libraries was estimated using a NanoDrop 8000 (Thermo Fisher Scientific) and quality control was performed with a High Sensitivity NGS Fragment Analysis Kit (1 bp–6000 bp) DNF-474 (Advanced Analytical Technologies, Inc.). Based on quantification with a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific), libraries were adjusted to 2 nM concentration before cluster generation with a HiSeq 3000/4000 SR Cluster Kit (Illumina). Twelve samples were pooled per lane and sequenced in 150 bp single-end mode on a HiSeq 3000 platform (Illumina). On average, over 32 million reads per library were obtained (Supplementary Table S1).
Read mapping and annotation
Illumina reads were trimmed with Trimmomatic version 0.33 (Bolger et al., 2014), keeping only the reads longer than 36 bases. Subsequently, BLAT (Kent, 2002) in protein space (parameters: -t=dnax -q=dnax) was employed to map the trimmed reads against the reference transcriptome of Beta vulgaris, containing 28 721 transcripts in total (RefBeet-1.2.2; Dohm et al., 2014). The best BLAT hits (the lowest e-value and the highest bit score) were counted. On average, 70% mapping efficiency (i.e. T. triangulare reads matching a B. vulgaris target) was achieved, and 18 657 reference transcripts with at least one count were detected (Supplementary Table S1). Reference transcripts were annotated based on their homology to representative Arabidopsis peptides (Araport11, https://www.araport.org) via BlastX (BLAST 2.6.0+ suite, parameters: -evalue 1e-5 -max_target_seqs 1) (Camacho et al. 2009). Identified homologies were also used for assignment of MapMan functional categories (version X4_R1.0; Thimm et al., 2004) and TF families based on PlantTFDB (Jin et al., 2017). Putative protein localization was predicted with TargetP (Emanuelsson et al., 2000) based on B. vulgaris reference sequences. Selected transcripts were independently validated by quantitative real-time PCR (Supplementary Protocol S1).
Non-negative matrix factorization
Reads normalized to reads per kilobase million were used for non-negative matrix factorization (NMF), which was performed with altered variants of the Lee and Seung (euclidean distance), the Brunet, and the Kullback–Leibler (KL) algorithms (KL divergence) from the NMF package for R (Gaujoux and Seoighe, 2010). To facilitate interpretation, the coefficient matrix was scaled, so that the sum of all coefficients in a sample would sum to 1 in each iteration. Factorization was carried out in 50 iterations with random seeding and the factorization rank was determined using the elbow within the residual sum of squares and the average regression coefficient (R2). With all three algorithms, five factors were identified, with the KL and Brunet algorithms having a higher average R2 than the Lee and Seung method (Supplementary Fig. S4). The final factorization was performed 500 times and the result with the lowest residual sum of squares was used for further analysis. The factorized data were z-scored and clustered via the Hartigan and Wong algorithm for k-means clustering (Hartigan and Wong, 1979). The enrichment of biological functions was analysed using Fisher’s exact test on a set of reduced MapMan categories (Supplementary Table S2). All P-values were adjusted for multiple hypothesis testing by Benjamini–Hochberg correction (Benjamini and Hochberg, 1995).
Data analysis
Downstream analyses were performed in R version 3.5.1 (https://www.R-project.org/) and required packages. Raw counts were used for analysis of differential gene expression with the DESeq2 package version 1.22.2 (Love et al., 2014) in the default mode, with Benjamini–Hochberg P-value adjustment and a 0.01 significance threshold. Under- and over-represented biochemical pathways were identified using MapMan categorization and the Wilcoxon rank sum test with false discovery rate adjustment according to Benjamini and Hochberg (1995) and q-value≤0.05 as a significance threshold. Reads per million were used to identify genes with a significantly different temporal pattern between the mock and ABA treatments using the maSigPro package (Nueda et al., 2014) (see Supplementary Protocol S2 for more details).
Accession numbers
The read data as well as processed files are deposited at the National Center for Biotechnology Information Gene Expression Omnibus under accession number GSE116590 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE116590).
Results
Exogenous ABA induces rapid changes at the level of the transcriptome
To assess the immediate response of the facultative CAM species T. triangulare to foliar application of ABA, we performed an RNA-seq time-course experiment spanning 40–1280 min after the treatment (Fig. 1A). DESeq analysis of transcript abundances in ABA-treated compared with mock-treated leaves revealed that the number of differentially expressed genes (DEGs) generally increased with time. While the effect of ABA on the number of DEGs was negligible after 40 min, 25% of mapped genes were differentially expressed after 1280 min (Fig. 1B).
For a closer analysis of the observed changes and their biological effect, the following approaches were taken. First, analysis of steady-state transcript abundances was performed, focusing especially on CAM genes and components of the ABA signalling pathway. Overall patterns of transcript abundance of all mapped genes were analysed to identify genes whose temporal patterns changed in response to ABA. Combining both analyses, genes with significantly altered steady-state transcript abundances (q-value ≤0.01) and temporal pattern were identified. NMF combined with k-means clustering was applied to identify genes and biological processes with similar time-dependent co-expression. Finally, we aimed to select candidates relevant for CAM induction by comparing ABA- and drought-induced responses of T. triangulare with ABA treatment of the C3 model plant Arabidopsis.
Altered transcript levels of CAM genes
Out of 23 core CAM genes (Supplementary Table S2; Supplementary Dataset S1), a homologous gene for 20 of them was found in the B. vulgaris reference, of which five were up-regulated after 160 min and six after 320 min, (Supplementary Fig. S5A). Transcript levels of PHOSPHOENOLPYRUVATE CARBOXYLASE 3 (PPC3) accumulated 2.1-fold in ABA-treated leaves after 160 min and remained increased at the remaining time points (5.1-fold after 1280 min). Transcripts of the primary PPC isoform, PPC1, accumulated likewise (Supplementary Fig. S6), but the overall abundance pattern did not change significantly in response to ABA (maSigPro analysis). PYRUVATE ORTHOPHOSPHATE DIKINASE (PPDK) transcripts started to accumulate after 80 min (3.6-fold), reaching a 12.8-fold higher level after 640 min. In addition, transcript levels of genes encoding carbonic anhydrases (CAs) were altered after ABA treatment. While ALPHA CARBONIC ANHYDRASE 1 (ACA1) transcript diminished after 160 min (3.5-fold decrease) and remained reduced throughout the remaining sampling points (5.3-fold after 1280 min), transcripts of chloroplastic BETA CARBONIC ANHYDRASE 5 (BCA5) accumulated up to 2.9-fold after 320 min (Fig. 2). To avoid futile cycling, PPC activity is post-translationally regulated by PPCK. In T. triangulare, PPCK1 transcript levels were reduced 2.6-fold in ABA-treated leaves after the dark period (Fig. 2). Transcripts of decarboxylating MALIC ENZYMEs (MEs), chloroplastic NADP-ME4 and cytosolic NADP-ME1, accumulated in response to ABA. NADP-ME4 was the faster-responding isoform, while NADP-ME1 transcripts accumulated strongly after 320 min, reaching 2.8-fold up-regulation after 1280 min (Fig. 2).
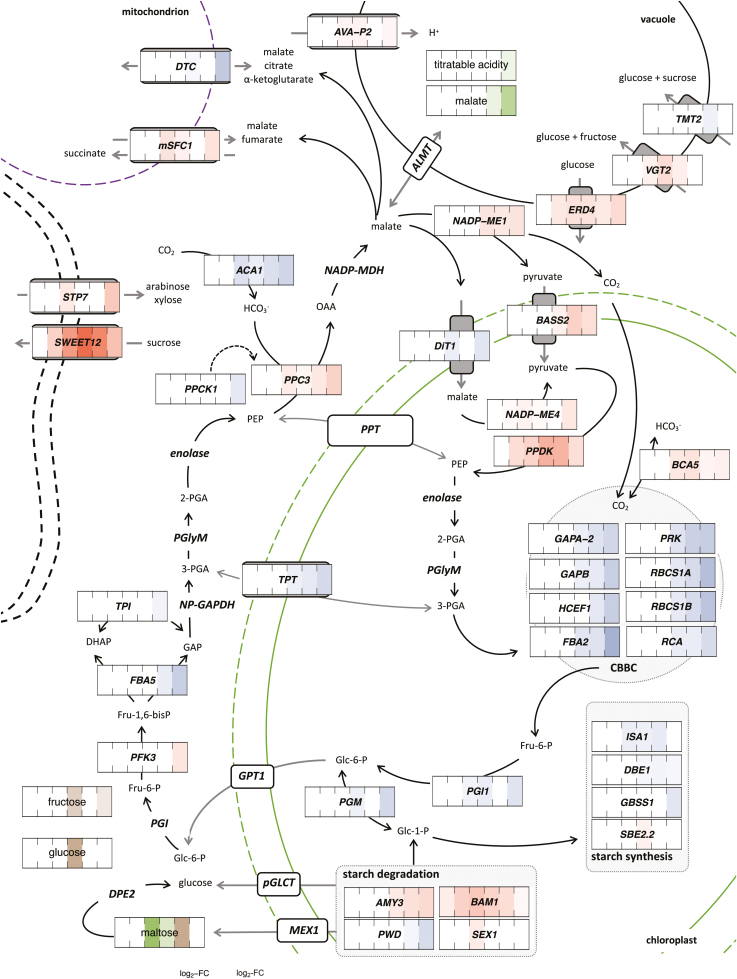
Fig. 2.
Exogenous ABA altered the transcript levels of core CAM genes and genes of related pathways, correlating with altered levels of selected metabolites in Talinum triangulare. Transcript abundances and metabolite amounts are expressed as log2-fold changes of ABA-treated compared with mock-treated leaves on blue–red and yellow–green scales, respectively (expression, n=2; metabolites, n=4). Only genes with significantly different transcript abundances (DESeq with Benjamini–Hochberg correction, q-value ≤0.01) and significantly altered temporal patterns (maSigPro with Benjamini–Hochberg correction, q-value ≤0.01 and R2>0.85) are shown. The significance level for metabolites was 0.05 after Benjamini–Hochberg correction. Substrate conversions are depicted with solid lines, post-translation regulations with dashed lines, and transport processes are shown in grey. Genes whose involvement in the depicted pathways is expected are also included. Protein subcellular localization is based on prediction from Beta vulgaris (TargetP). 1,3-BPG, 1,3-bisphosphoglycerate; 2-PGA, 2-phosphoglycerate; 3-PGA, 3-phosphoglycerate; CBBC, Calvin–Benson–Bassham cycle; DHAP, dihydroxyacetone phosphate; Fru-6-P, fructose-6-phosphate; Fru-1,6-bisP, fructose-1,6-bisphosphate; GAP, glyceraldehyde-3-phosphate; Glc-1-P, glucose-1-phosphate; Glc-6-P, glucose-6-phosphate; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PGlyM, phosphoglycerate mutase.
Flow of metabolites through the CAM pathway requires numerous known and putative transporters. The chloroplastic pyruvate transporter BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 2 (BASS2) was up-regulated, especially after 640 min (5.2-fold), while transcript levels of malate-transporting DICARBOXYLATE TRANSPORTER 1 (DiT1) declined at 320 and 1280 min (2.1-fold and 2.9-fold, respectively) (Fig. 2). Mitochondrial organic acid transporters were affected by ABA as well: DICARBOXYLATE/TRICARBOXYLATE TRANSPORTER (DTC) transcripts declined up to 8.1-fold at 1280 min, while MITOCHONDRIAL SUCCINATE-FUMARATE CARRIER 1 (mSFC1) transcripts accumulated at 160, 320, and 1280 min (2.8-, 1.8-, and 2.9-fold, respectively) (Fig. 2).
Carbohydrate metabolism and transport
ABA treatment also affected the levels of transcripts encoding enzymes of starch and sucrose metabolism. Compared with the CAM genes, these responded later (after 640 and 1280 min) and they were generally down-regulated (Supplementary Fig. S5B). Of the genes encoding starch-metabolism enzymes, transcripts of ISOAMYLASE 1 (ISA1) declined 2.2-fold after 160 min, followed by 1.6-fold depletion of DEBRANCHING ENZYME 1 (DBE1) transcripts after 320 min and finally by 2.8-fold depletion of GRANULE BOUND STARCH SYNTHASE 1 (GBSS1) transcripts, the depletion of which was restricted to the early hours of the light period (Fig. 2). In contrast, BETA-AMYLASE 1 (BAM1) transcripts accumulated 2-fold in ABA-treated leaves as soon as after 40 min and remained increased throughout the remaining sampling points. Additionally, ISOAMYLASE 3 (AMY3) transcripts accumulated 2.3-fold after 320 min (Fig. 2).
Multiple genes encoding glycolytic/CBBC enzymes were down-regulated, including at 1280 min (i.e. early in the dark period), among them chloroplastic PHOSPHOGLYCERATE/BISPHOSPHOGLYCERATE MUTASE (PGM) (4.7-fold), PHOSPHOGLUCOSE ISOMERASE 1 (PGI1) (3.2-fold), HIGH CYCLIC ELECTRON FLOW 1 (HCEF1) (6.7-fold), and FRUCTOSE-BISPHOSPHATE ALDOLASE 2 (FBA2) (3.9-fold) (Fig. 2). Additionally, TRIOSE-PHOSPHATE/PHOSPHATE TRANSLOCATOR (TPT) transcripts declined 4.5-fold after 320 min and remained at a reduced level also at the last sampling (Fig. 2).
In contrast to the pronounced down-regulation of enzyme-coding transcripts, there was up-regulation of sugar transporters localized in both the plasma membrane and the tonoplast. Transcript accumulation of SUGAR TRANSPORTER PROTEIN 7 (STP7) was found only at time points 160 and 1280 min (2.3- and 6.7-fold, respectively). Transcript levels of HOMOLOG OF MEDICAGO TRUNCATULA MTN3 (SWEET12) accumulated 5.4-fold after 80 min, reaching an 80-fold increase late in the light period (320 min after the treatment) (Fig. 2). Transcripts of EARLY-RESPONSIVE TO DEHYDRATION 4 (ERD4) and VACUOLAR GLUCOSE TRANSPORTER 2 (VGT2) accumulated in ABA-treated leaves 2.4-fold after 80 min and 1.8-fold after 160 min, respectively (Fig. 2).
Changes in metabolite levels in response to exogenous ABA
The above-described changes in transcript levels correlated with altered levels of several metabolites. End-of-night titratable acidity increased in ABA-treated leaves to 52.7±4.1 µmol H+ g–1 FW, representing a 1.6-fold increase compared with mock-treated leaves (Fig. 2). GC-MS analysis of leaf extracts revealed malate and citrate as the primary organic acids that accumulated during the dark period. At the last time point (1280 min), relative amounts of malate and citrate were 4- and 16-fold higher than levels in mock-treated leaves, respectively. Additionally, α-ketoglutarate, succinate, and fumarate accumulated 11.3-, 5.7-, and 3-fold, respectively (Supplementary Dataset S4; Supplementary Fig. S7). Besides organic acids, levels of several carbohydrates were affected. Maltose accumulated transiently at 320 min, followed by a decline at 640 min. At 640 min, glucose was reduced as well (Fig. 2). A reduction of fructose levels occurred specifically after the dark period. A trend towards a time-dependent decline in starch and sucrose levels was observed (Supplementary Fig. S8). Total protein levels also declined up to 1.3-fold in response to ABA (at 1280 min) (Supplementary Fig. S7).
As ABA plays a major role in stress signalling, the levels of stress-related compounds were assessed. γ-aminobutyric acid (GABA), myo-inositol, and raffinose levels declined transiently after 640 min, or after 320 min in the case of myo-inositol (Supplementary Dataset S4). In contrast, levels of amino acids were not affected by ABA treatment (Supplementary Dataset S4).
Exogenous ABA alters ABA signalling
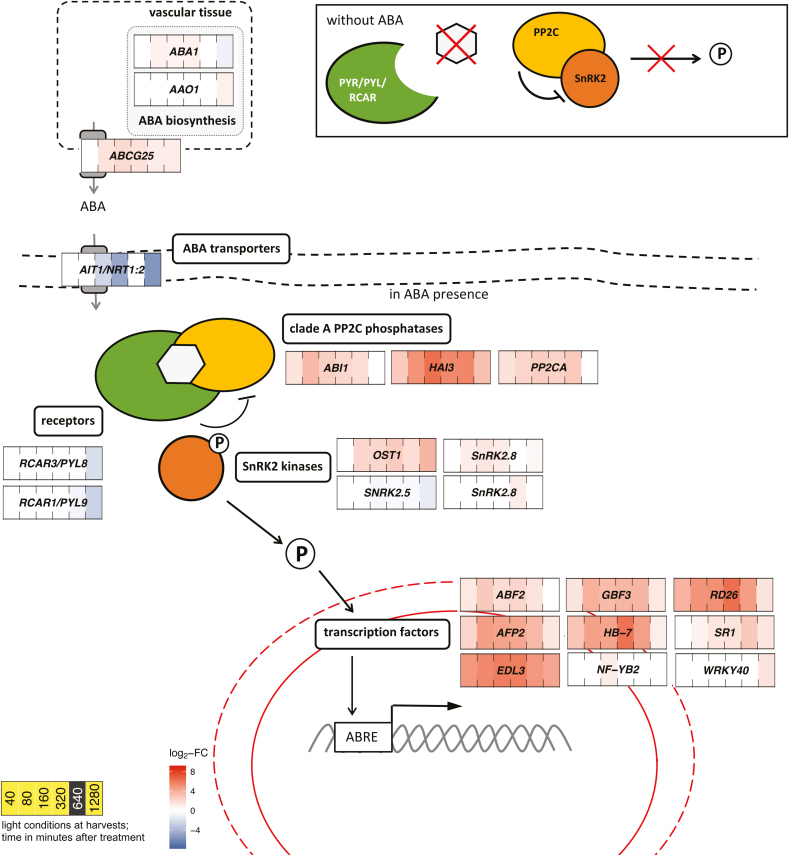
Transcript levels of REGULATORY COMPONENT OF ABA RECEPTOR 1 (RCAR1/PYL9) and RCAR3/PYL8 showed altered temporal patterns in response to ABA, and their abundance declined 5.9- and 4-fold, respectively, after 1280 min (Fig. 3). In contrast, transcripts encoding downstream components of the ABA signalling pathway were affected more rapidly. Transcripts of the phosphatases PROTEIN PHOSPHATASE 2CA (PP2CA), ABA INSENSITIVE 1 (ABI1), and HIGHLY ABA-INDUCED PP2C GEN3 (HAI3) accumulated 3.1- 2.9-, and 6.5-fold, respectively, starting at 40 min, and remained up-regulated as time progressed. Transcript levels of SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASEs 2 (SnRK2) were influenced by ABA with similar kinetics, but the temporal patterns of only SnRK2.5, SnRK2.6/OST1, and SnRK2.8 were altered. While SnRKs 2.6 and 2.8 were up-regulated 4.5- and 2.1-fold, respectively, in response to ABA, there was 1.5-fold down-regulation of SnRK2.5 after 160 min (Fig. 3).
Fig. 3.
Transcript levels of genes involved in ABA signalling and biosynthesis were influenced by exogenous ABA in Talinum triangulare. Transcript abundances are expressed as log2-fold changes of ABA-treated compared with mock-treated leaves (n=2). Only genes with significantly different transcript abundances (DESeq with Benjamini–Hochberg correction, q-value≤0.01) and significantly altered temporal patterns (maSigPro with Benjamini–Hochberg correction, q-value≤0.01 and R2>0.85) are shown. The core signalling pathway comprises PYR/PYL/RCAR receptors, clade A phosphatases PP2C, and SnRK2 kinases. In the absence of ABA, PP2C phosphatases bind SnRK2s and prevent them from phosphorylating downstream targets. Upon binding of ABA to the receptors, they capture PP2Cs, releasing phosphorylated SnRK2s. Targets of SnRK2s include transcription factors in particular, many of which regulate gene expression through binding to ABA-responsive element (ABRE) motifs in promoter sequences of target genes. OST1 is involved in the control of stomatal movement. P, phosphorylation.
The temporal patterns of numerous TFs acting downstream were influenced by ABA. Transcripts of ABSCISIC ACID RESPONSIVE ELEMENT-BINDING FACTOR 2 (ABF2), ABI FIVE BINDING PROTEIN 2 (AFP2), EID1-LIKE 3 (EDL3), HOMEOBOX PROTEIN 7 (HB7), and RESPONSIVE TO DESICCATION (RD26) accumulated 2.5-, 4.8-, 28.6-, 7.7-, and 12.7-fold, respectively, as soon as after 40 min, and remained increased at the remaining sampling times (Fig. 3). Levels of several transcripts encoding ABA synthesis and transport proteins were also affected by the ABA treatment. Transcripts of ABA DEFICIENT 1 (ABA1) increased 1.8-fold after 80 min, but transcripts of ABSCISIC ALDEHYDE OXIDASE 1 (AAO1) accumulated only after 1280 min (1.9-fold) (Fig. 3). Transcript levels of ATP-BINDING CASETTE G25 (ABCG25), an ABA exporter from the site of synthesis, accumulated 2.9-fold starting 80 min after the treatment (Fig. 3). In contrast, ABA-IMPORTING TRANSPORTER 1 (AIT1/NRT1:2) was 5.2-, 25.2-, and 45.9-fold down-regulated after 160, 320, and 1280 min.
Factorization and k-means clustering of temporal transcript patterns
NMF was performed to obtain insights into time-dependent abundance patterns. Based on the transcriptional contributions of the mapped genes, five factors were identified (Fig. 4A). In factor 1, most of the transcriptional contribution occurred at 320 and 640 min in the mock treatment. Similarly, factor 2 was most pronounced at 320 and 640 min, but was specific to the ABA treatment. Factor 3 was characterized by basal transcriptional contribution over time with major expression in the ABA treatment specifically after 1280 min. In factor 4, the contribution declined with time until 640 min, followed by a pronounced contribution only in the mock treatment after 1280 min. The transcriptional contribution in factor 5 changed over time but did not differ between treatments (Fig. 4A).
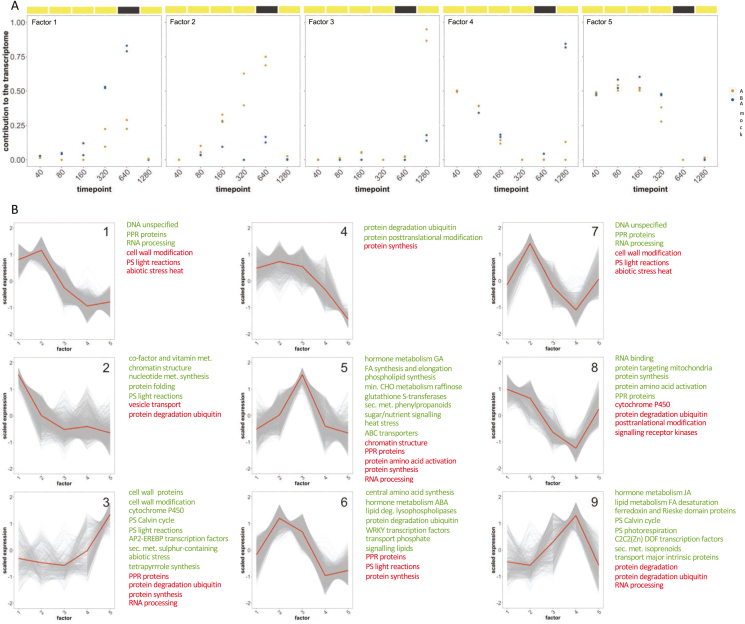
Fig. 4.
Non-negative matrix factorization and k-means clustering of the Talinum triangulare transcriptome. (A) Five factors were identified based on time-dependent expression of individual genes (for details, see Materials and Methods). (B) Based on the transcriptional contribution of each factor, mapped genes were assigned to nine clusters (for details, see Materials and Methods). Enrichment of MapMan categories in each cluster is shown in green (over-represented categories) and red (under-represented categories). deg, Degradation; FA, fatty acids; GA, gibberellic acid; JA, jasmonic acid; met, metabolism; min. CHO, minor carbohydrate; misc, miscellaneous; PPR, pentatricopeptide repeat; PS, photosynthesis; sec. met., secondary metabolism.
Based on these factors, subsequent k-means clustering identified nine clusters, three of them comprising significant contributions of either of the ABA-specific factors. The early ABA factor contributed to clusters 6 and 7, while the late ABA factor was present in cluster 5 (Fig. 4B). Enrichment analysis of MapMan categories identified ABA metabolism, WRKY TFs, phosphate transport, and ubiquitin-mediated protein degradation as over-represented in the early cluster 6. In the early cluster 7, vesicle transport, protein glycosylation, and reductive-oxidative thioredoxin reactions were over-represented. The late cluster 5 was enriched for sugar and nutrient signalling, ABC transporters, and glutathione S-transferases, but also heat stress response, gibberellin metabolism, and raffinose and phenylpropanoid metabolism pathways (Fig. 4B). Besides their specific enrichments, all three clusters shared under-representation of protein synthesis, and clusters 6 and 7 both had under-representation of photosynthesis.
Due to the large proportion of factor 5 in cluster 3, this cluster could explain time-dependent changes to the transcriptome in both treatments. This cluster was enriched for CBBC, light photosynthetic reactions, and AP2-EREBP TFs (Fig. 4B). Over-representation of photosynthetic processes was shared by clusters 1, 2, and 9, which comprised large contributions of mock-treatment-specific factors 1 and 4, as well as early ABA factor 2 in cluster 1 (Fig. 4B).
Candidate regulators of CAM induction
ABA-induced changes in both the transcriptome and the metabolome hinted at activation of the CAM pathway. This prompted the question of which gene products can translate the exogenous ABA signal into CAM induction. To answer this question, we performed a DESeq analysis, comparing a pool of all ABA-treated samples against a pool of mock-treated samples, thus reducing the changes in transcript abundance down to the direction and degree of the changes. In this way, 773 DEGs were obtained (Supplementary Dataset S3).
For further filtering, temporal transcript abundances were considered, limiting the search only to clusters 5, 6, and 7 (i.e. the clusters of major ABA responsiveness; Fig. 4B). Additionally, to be considered a CAM regulator, a gene had to show an altered temporal pattern in the maSigPro analysis (Supplementary Dataset S2). Finally, the transcript abundance of such a gene had to be altered with progressing time during drought induction of CAM and reversible to non-stress levels 2 days after rewatering in the same experiment (Brilhaus et al., 2016). As we searched for genes responsive to ABA specifically in the facultative CAM species T. triangulare, we compared the DEGs identified here to ABA-responsive genes in Arabidopsis (Song et al., 2016). Genes responsive in both species were not considered to be CAM regulators. When all these filtering criteria were applied, 32 candidate genes were identified; among them were nine transcription factors, six genes of amino acid metabolism, and five transporters (Table 1). Among the genes differentially expressed between ABA- and mock-treated leaves, there were 36 genes without a known Arabidopsis homolog (Supplementary Dataset S3) but only five of them also showed an altered temporal pattern of transcript abundance (Table 2).
Table 1.
Genes identified as possible CAM regulators in Talinum triangulare
| Transcript ID | AGI | Name | TF family | log2-FC pool (ABA/mock) | Cluster (NMF) | |
|---|---|---|---|---|---|---|
| Transcriptional activators | KMT03072 | AT5G19650 | OFP8 | –1.30 | 2 | |
| KMT14500 | AT3G20910 | NF-YA9 | NF-YA | 1.38 | 5 | |
| KMT02260 | AT4G14540 | NF-YB3 | NF-YB | 0.71 | 6 | |
| KMT18169 | AT3G11090 | LBD21 | LBD | –1.88 | 2 | |
| KMT07141 | AT3G01470 | HB-1 | HD-ZIP | 1.02 | 6 | |
| KMS96841 | AT2G26150 | HSFA2 | HSF | 2.58 | 5 | |
| KMS95225 | AT1G25440 | BBX15 | CO-like | –1.80 | 2 | |
| KMS99638 | AT1G19850 | MP | ARF | 1.23 | 6 | |
| KMT11833 | AT2G44730 | Trihelix | 0.41 | 6 | ||
| KMT10399 | AT4G00990 | 0.62 | 7 | |||
| KMT03067 | AT3G24520 | HSFC1 | HSF | 1.63 | 7 | |
| Amino acid metabolism | KMT00186 | AT1G29900 | CARB | 1.06 | 5 | |
| KMT12470 | AT1G64660 | MGL | 2.67 | 5 | ||
| KMT14300 | AT1G55510 | BCDH BETA1 | 1.50 | 6 | ||
| KMT10213 | AT1G03090 | MCCA | 1.57 | 6 | ||
| KMT12470 | AT1G64660 | MGL | 2.67 | 5 | ||
| KMS98028 | AT1G08630 | THA1 | 1.49 | 5 | ||
| Solute transport | KMT02985 | AT2G26900 | BASS2 | 1.05 | 6 | |
| KMT13799 | AT1G08960 | CAX11 | 1.02 | 6 | ||
| KMT03912 | AT1G30360 | ERD4 | 1.60 | 5 | ||
| KMT03911 | AT1G30360 | ERD4 | 1.50 | 5 | ||
| KMS95179 | AT1G19910 | AVA-P2 | 0.45 | 6 | ||
| Carbohydrate metabolism | KMT07128 | AT1G69830 | AMY3 | 1.08 | 6 | |
| Chromatin organisation | KMT10399 | AT4G00990 | 0.62 | 7 | ||
| Lipid metabolism | KMT19551 | AT1G43620 | UGT80B1 | 0.48 | 6 | |
| Nucleotide metabolism | KMT00186 | AT1G29900 | CARB | 1.06 | 5 | |
| Protein degradation | KMT12139 | AT3G61180 | 1.28 | 5 | ||
| Protein modification | KMT01924 | AT1G28480 | GRX480 | 2.88 | 5 | |
| Secondary metabolism | KMS96359 | AT2G20340 | AAS | 4.15 | 5 | |
| KMT17647 | AT2G07050 | CAS1 | 1.03 | 6 | ||
| Vesicle trafficking | KMT15610 | AT2G44140 | 0.46 | 6 |
These genes were identified based on their differential expression between mock treatment and ABA treatment, time-dependent pattern of transcript abundance (NMF analysis), altered temporal pattern of transcript abundance (maSigPro analysis), differential transcript accumulation in drought-induced CAM in T. triangulare (Brilhaus et al., 2016), and lack of ABA responsiveness in C3 Arabidopsis (Song et al., 2016). Log2-FC pool (ABA/mock) is based on DESeq analysis of all ABA-treated samples against all mock-treated samples, i.e. across all time points. All shown log2-FC are significant at the 0.01 level. AGI, Arabidopsis Genome Initiative; TF, transcription factor.
Table 2.
ABA-responsive genes in Talinum triangulare without a known Arabidopsis homolog
| Transcript ID | log2-FC pool (ABA/mock) | q-value | NMF cluster | Domain | Description |
|---|---|---|---|---|---|
| KMT11995 | –1.36 | 4.30E-04 | 2 | IPR005079 | Peptidase C45 |
| KMT13506 | 1.32 | 3.24E-03 | 6 | n.d. | n.d. |
| KMT00528 | 1.51 | 2.85E-04 | 6 | n.d. | n.d. |
| KMT09356 | 2.59 | 5.72E-03 | 5 | n.d. | n.d. |
| KMS99845 | 3.31 | 2.46E-05 | 6 | n.d. | n.d. |
Analysis of differential expression between mock and ABA treatments, considering only genes with altered temporal patterns of transcript abundance (maSigPro analysis). Log2-FC pool (ABA/mock) is based on DESeq analysis of all ABA-treated samples against all mock-treated samples, i.e. across all time points. Protein domains were determined using InterProScan. n.d., Not determined.
Discussion
In this study, we applied ABA to induce CAM in a controlled manner in our facultative CAM model T. triangulare. To the best of our knowledge, this is the first study to describe the earliest changes in both the transcriptome and the metabolome during the induction phase. By following transcript abundances in ABA-treated and mock-treated leaves in parallel, we aimed to reduce the effect of circadian regulation when identifying DEGs. However, it should be noted that most pronounced changes (Fig. 1A) were observed after the dark period, suggesting a combined effect of ABA and darkness on transcript abundances. At the same time, it seems unlikely that these changes were caused by alterations to circadian clock genes. Out of 34 genes involved in circadian regulation (according to MapMan), nine showed a significantly altered temporal pattern of transcript abundances, but only LATE ELONGATED HYPOCOTYL (LHY) and ELF4-LIKE 4 (ELF4-L4) were assigned to ABA-specific clusters 5 and 7, respectively (Supplementary Table S3), suggesting that (i) the central circadian clock was not affected by the ABA treatment and (ii) the detected and further discussed changes in transcript abundances are a response to ABA or ABA and darkness, but not the diel cycle alone.
ABA induced changes in transcript abundance of CAM core and related genes as well as nocturnal acidification
Amounts of transcripts encoding central CAM enzymes were influenced by exogenous ABA. These transcripts included PPC1 (Supplementary Fig. S6) and PPC3 (Fig. 2), in agreement with ABA-induced transcript accumulation (Taybi and Cushman, 1999) and increased measurable PPC activity in M. crystallinum (Dai et al., 1994). While PPC1 was the most abundant isoform (Supplementary Dataset S1), only the temporal pattern of PPC3 was significantly altered by ABA (Supplemental Dataset S2). In contrast, exogenous ABA induced the depletion of PPC3 transcripts in Arabidopsis (Song et al. 2016), indicating different transcriptional responses to ABA between a C3 species and a facultative CAM species. It might therefore be of interest to further investigate the contribution of different PPC isoforms to CAM induction. Transcript abundance of PPCK1, which post-translationally regulates PPC activity, has been reported to be under circadian control (Nimmo et al., 1999; Taybi et al., 2000; Boxall et al., 2017); here, the amount of PPCK1 transcript in ABA-treated leaves declined after 1280 min (i.e. shortly after the onset of light).
Assuming a requirement for active CA together with PPC, co-expression of their genes could be expected. This was observed for chloroplastic BCA5 only, transcripts of which started to accumulate after 160 min (Fig. 2). In a comparison of four CAM species, CA activity was detected in chloroplasts of ME species, while activity in the cytosol was detected in phosphoenolpyruvate carboxykinase species (Tsuzuki et al., 1982). As T. triangulare relies on ME (Brilhaus et al., 2016), we hypothesize that BCA5 is the CAM-specific isoform. In this context, it will be of interest to investigate the role of CAs in the CAM pathway, their localization, and their regulatory mechanism(s). Transcript levels of both cytosolic and chloroplastic isoforms of NADP-ME were responsive to exogenous ABA. Transcript accumulation of chloroplastic NADP-ME4 was restricted to 160 and 1280 min, while transcript levels of cytosolic NADP-ME1 were increased from 320 to 1280 min (Fig. 2), further indicating adjustment of the CAM enzymatic toolkit.
Increased transcript levels of carboxylating enzymes correlated with the nocturnal increase in titratable acidity and organic acid content (Fig. 2). Nocturnal titratable acidity of T. triangulare was previously reported to reach 10–30 µmol H+ g–1 FW with progressing drought (Brilhaus et al., 2016) and 40–60 µmol H+ g–1 FW in salt-induced recycling CAM (Montero et al., 2018). Here, during the first dark period after the ABA treatment, end-of-night titratable acidity reached 52.7±4.1 µmol H+ g–1 FW, and the difference in titratable acidity between the early hours of the dark period and the subsequent morning was comparable to that observed by Herrera et al. (1991) in sun-exposed leaves after 4 days of water withdrawal. It has been shown in both obligate and facultative CAM species that the major storage acid is malate (Chen and Nose, 2004; Abraham et al., 2016; Rainha et al., 2016). However, diel citrate fluctuations during early phases of CAM (e.g. in M. crystallinum and Clusia minor) (Herppich et al., 1995; Borland et al., 1998) and a contribution of succinate to nocturnal acidification [e.g. in Ananas comosus (pineapple) and Agave americana] have also been reported (Medina et al., 1993; Abraham et al., 2016; Rainha et al., 2016). In ABA-induced CAM in T. triangulare, the increase in succinate levels was also accompanied by α-ketoglutarate accumulation (Fig. 2).
Transcript levels of several transport proteins were affected by exogenous ABA, as observed in CAM-performing M. crystallinum (Cushman et al., 2008). The reduced transcript levels of the chloroplast malate transporter DiT1 in ABA-treated leaves after 320 and 1280 min (Fig. 2) could indicate reduced flow of malate into the chloroplast towards the end of the light period and in the dark period, suggesting increased flow into the vacuole. However, transcripts of ALUMINIUM-ACTIVATED MALATE TRANSPORTERs did not accumulate significantly upon ABA treatment and were of low abundance (Supplementary Dataset S1), an observation also made in drought-induced CAM (Brilhaus et al., 2016). In contrast, BASS2 transcripts accumulated promptly (starting at 320 min, Fig. 2; cluster 6, Supplementary Dataset S1), suggesting adjustment for increased transport of pyruvate originating from malate decarboxylation to the chloroplast.
Storage of malate in the vacuole requires a proton gradient to fuel its transport (Lüttge, 1987). Indeed, VACUOLAR-TYPE H+ATPASE C2 (AVA-P2) (cluster 6) was slightly but significantly up-regulated in response to ABA. In addition, transcripts of a putative ATPase activator AT5G58110 (cluster 5) accumulated (Supplementary Dataset S1). Although it is not clear whether V-ATPase activity is controlled at the level of transcript abundance, ABA-induced activity of V-ATPase was observed in M. crystallinum (Barkla et al., 1999).
Utilization of carbohydrate reserves
Reprogramming of carbohydrate metabolism ensures the flow of carbon skeletons between storage carbohydrates and the PEP pool. Genes involved in starch metabolism were down-regulated in ABA-treated leaves, especially at the final two time points (Supplementary Fig. S5B). Transcript levels of ISA1 (after 160 min), DBE1 (320 min), and GBBS1 (1280 min) were depleted (Fig. 2). Glycolytic genes were also down-regulated (Supplementary Fig. S5B), including early-responsive PGI1 (after 160 min) and FBA2, which showed the strongest transcript depletion after 1280 min (Fig. 2). While the time-course experiment presented here covered a period of time too short to follow full diel cycling of transcript abundances, these early changes suggest ABA- and time-dependent adjustment of glycolysis, as observed in M. crystallinum for FRUCTOSE-BISPHOSPHATE ALDOLASE 1 and GLUCOSE-6-PHOSPHATE ISOMERASE 1 (Cushman et al., 2008) and by fluctuating mid-day and mid-night transcript levels of glycolytic genes in drought-induced CAM in T. triangulare (Brilhaus et al., 2016).
Starch breakdown products enter the cytosolic glycolytic pathway and, while GLUCOSE-6-PHOSPHATE TRANSPORTER (GPT2) transcripts accumulated (after 640 and 1280 min; Supplementary Dataset S1), the overall pattern of abundance did not differ significantly between the treatments. The nocturnal accumulation of GPT2 transcripts agrees with observations in pineapple (Borland et al., 2016) and M. crystallinum (Neuhaus and Schulte, 1996; Häusler et al., 2000). PPDK transcripts accumulated rapidly (after 320 and 640 min; Fig. 2), suggesting production of the PEP pool, which correlates with the trend towards declining amounts of starch, also starting after 320 min (Supplementary Fig. S8), and accumulating transcripts of starch-degrading enzymes such as BAM1 and AMY3 (Fig. 2). Besides starch, sucrose levels also showed a declining trend with progressing time (Supplementary Fig. S8), suggesting the utilization of sucrose as an additional source of carbon backbones. Alternatively, carbohydrate reserves could be used in respiration to provide energy for synthesis and necessary transcriptional and translational adjustment during the induction phase of CAM.
Transcripts of the vacuolar hexose transporters VGT2 and ERD4 accumulated in ABA-treated leaves (Fig. 2), indicating increased sugar flow between the cytosol and vacuole. Depending on current demands, glucose stored in the vacuole either supports PEP regeneration or provides energy for maintenance processes and growth (Borland et al., 2016). In addition, exogenous ABA induced accumulation of transcripts of plasma membrane-localized STP7 and SWEET12 (Fig. 2). While diel cycling between synthesis and degradation of storage carbohydrates is well described (Borland and Taybi, 2004; Taybi et al., 2017), transcriptional responsiveness of numerous transporters suggests the requirement for proper partitioning of sugar resources between compartments and possibly cells or tissues. In addition, sugars could also play a signalling role during CAM induction, as supported by the enrichment of sugar signalling in cluster 5 (Fig. 4B).
Transcript accumulation of components of ABA biosynthetic and signalling pathways
Exogenous ABA affected the abundance patterns of several transcripts encoding components of ABA signalling, ABA-synthesizing enzymes, and ABA transporters. Transcript levels of the ABA receptors RCAR1/PYL9 and RCAR3/PYL8 declined in response to ABA after 640 and 1280 min, respectively. PYL4, PYR1, and PYR4 were also down-regulated at least at one time point (Supplementary Dataset S1), but the overall expression pattern did not differ significantly between the treatments. While depletion of ABA receptor transcripts is a common response to both ABA and abiotic stress, occurring in a wide range of species such as Arabidopsis (Chan, 2012; Gonzalez-Guzman et al., 2012; Song et al., 2016), cotton (Zhang et al., 2017), and maize (Fan et al., 2016; Li et al., 2017), diverse responses have been observed. For example, reversed expression patterns between maize roots and shoots (Fan et al., 2016) and time-dependent expression patterns in cotton (Zhang et al., 2017) were described. Given the spatiotemporal expression of ABA receptors observed in other species, it is intriguing to hypothesize that differentiation between the stress response and CAM induction could occur at the level of ABA interaction partners.
In contrast to the transcript levels of genes encoding ABA receptors, transcript levels of downstream components changed more promptly and were generally up-regulated; among them were the PP2CA phosphatases ABI1, HAI3, and PP2CA, as well as their target kinases SnRK2.6/OST1 SnRK2.8. In contrast, SnRK2.5 transcripts diminished upon ABA treatment (Fig. 3). This finding is not necessarily contradictory, as only selected SnRK2s were shown to act in an ABA-dependent manner under hyperosmotic conditions in Arabidopsis (Boudsocq et al., 2004) and differentiation in SnRK2 transcriptional responsiveness depending on the type of stress was observed in Zea mays (Huai et al., 2008).
Transcripts of several genes encoding ABA-synthesizing enzymes accumulated in response to ABA (Supplementary Dataset S1), but only ABA1 and AAO1 showed altered temporal patterns (Fig. 3). These changes were accompanied by transcript accumulation of ABCG25, which encodes an ABA exporter from the site of synthesis (Kuromori et al., 2010). Induction of AAO3 and NCED3 expression by exogenous ABA has been observed in Arabidopsis, even though the latter was detected only in the Landsberg erecta background (Xiong and Zhu, 2003). Exogenous ABA led to increased endogenous ABA levels in M. crystallinum (Taybi and Cushman, 2002), suggesting that ABA-induced ABA biosynthesis is conserved between species.
Protein degradation during CAM induction
Previous studies have revealed the importance of de novo protein synthesis in the adjustment of enzymatic machinery and in signalling during CAM induction in M. crystallinum (Höfner et al., 1987; Thomas et al., 1992; Taybi and Cushman, 2002). In the present study, the total protein amount declined after 320 min in response to ABA treatment, which correlated with the observed transcriptional changes. Transcripts encoding protein-degrading enzymes accumulated in ABA-treated leaves, starting at 40 min and peaking after 1280 min (Supplementary Fig. S7). Besides up-regulation of protein degradation, the enrichment analysis revealed down-regulation of protein-synthesis enzymes (Supplementary Fig. S5). These findings are consistent with NMF, which identified under-representation of protein biosynthesis in all three clusters composed of ABA-specific factors (clusters 5–7; Fig. 4B).
Protein degradation could serve multiple purposes. First, protein breakdown would release amino acids for the synthesis of new proteins, including CAM-specific enzymes. Rapid utilization of released amino acids is supported by the lack of accumulation of free amino acids (Supplementary Dataset S3). Second, protein degradation may contribute to adjustment of the enzymatic machinery by degrading competing C3 enzymes or proteins with diurnally fluctuating amounts, such as PPCK (Nimmo, 2003). For completeness, turnover of components of the ABA signalling pathway should be mentioned (Wu et al., 2016; Li et al., 2017). Finally, degraded proteins might provide energy to fuel synthesis and transport processes. Nocturnal accumulation of TCA intermediates in ABA-treated leaves suggest increased flow through this pathway, as proposed by flux-balance analysis by Cheung et al. (2014). Under conditions of rapid CAM induction, this might enable preferential use of carbohydrate reserves for PEP synthesis.
Short-term ABA treatment did not induce a general stress response
Given the altered transcript levels of ABA signalling components, the next question was whether the metabolome of ABA-treated leaves showed a general stress response, manifested through the accumulation of sugars, sugar alcohols, or amino acids (Papageorgiou and Murata, 1995; Bohnert and Jensen, 1996; Hare et al., 1998). In Arabidopsis, increased salinity as well as exogenous ABA reduced sucrose and starch content, while maltose content increased (Kempa et al., 2008). This result agrees with the response observed in ABA-treated leaves of T. triangulare. However, given the nocturnal increase in titratable acidity (Fig. 2), it is likely that starch degradation primarily served PEP generation. Raffinose levels remained largely unchanged in ABA-treated leaves of T. triangulare (Supplementary Dataset S4B), indicating that raffinose accumulation after water withdrawal reported in the same species (Brilhaus et al., 2016) was (i) independent of ABA and (ii) unrelated to CAM induction. The former agrees with observations made in Arabidopsis, where raffinose accumulation is also independent of ABA (Kempa et al., 2008; Urano et al., 2009). Amounts of the sugar alcohols glycerol, mannitol, myo-inositol, and sorbitol did not increase after ABA treatment (Supplementary Dataset S4B). This is not surprising at least in the case of myo-inositol, as its biosynthesis was shown to be independent of ABA in Arabidopsis (Urano et al., 2009). GABA accumulates in response to various stresses, possibly having a signalling role (Bouché and Fromm, 2004), but no increase in GABA levels was observed in ABA-treated T. triangulare leaves. In fact, GABA levels even decreased transiently, an observation also made in Arabidopsis (Kempa et al., 2008; Urano et al., 2010). In Arabidopsis, stress-induced accumulation of amino acids was observed within 15–18 hours (Nambara et al., 1998; Urano et al., 2009). While the accumulation of branched-chain amino acids, tyrosine, and histidine depends on ABA, dehydration-induced accumulation of proline can also occur independently of ABA (Kempa et al., 2008; Urano et al., 2009, 2010). In T. triangulare, no significant accumulation of amino acids was observed either under drought stress (Brilhaus et al., 2016) or in response to exogenous ABA (Supplementary Dataset S4B).
In summary, among the stress-related metabolites, those most affected by ABA application were starch and sucrose, but we attribute their reduced levels to the PEP regeneration typical of CAM and energy provision to fuel the adjustment of metabolism. The lack of stress response may be due to the use of exogenous ABA as the only stimulus. It has been proposed that stress (e.g. drought) is sensed by multiple sensors, which induce a variety of secondary signals (Xiong et al., 2002). Besides ABA, stress signalling also relies on Ca2+ and reactive oxygen species, and it is possible that the sole application of ABA was not enough to induce a stress response as rapidly as it induced the CAM pathway. Measurements of photosynthetic yield, which is sensitive to environmental and other stresses (Maxwell and Johnson, 2000), did not reveal any damage to photosystem II (Supplementary Protocol S3), in agreement with the metabolome analysis.
Candidate regulators of CAM induction
The currently available transcriptome data with ~25% ABA-responsive genes (this study) and ~39% drought-responsive genes (Brilhaus et al., 2016) in T. triangulare represent a rich source for the identification of regulators downstream of the two, partially overlapping, signalling networks. Based on the DESeq analysis and additional filtering criteria, seven candidate transcriptional regulators were identified (Table 1). HEAT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2), the dominant HSF involved in thermotolerance of both Arabidopsis and tomato (Charng et al., 2007; Chan-Schaminet et al., 2009), also contributes to salt and osmotic stress tolerance (Ogawa et al., 2007). Since CAM can be induced by both drought and salinity, a TF responsive to various environmental stresses in an ABA-dependent manner would be a meaningful CAM inducer. HSFC1 was also up-regulated. In Arabidopsis, HSFC1 has been shown to be heat- and drought-responsive (Rizhsky et al., 2004).
NUCLEAR FACTOR Y (NF-Y) SUBUNITS A9 and B3 were up-regulated in ABA-treated leaves (Table 1). NF-Ys act as heterodimers and heterotrimers of NF-YA, NF-YB, and NF-YC subunits and form complexes with other proteins, including bZIP-family TFs and components of phytochrome, gibberellic acid, and ABA signalling. NF-Y TFs regulate plant growth, development, and stress responses (Zhao et al., 2017). This is also true for NF-YB3, which is part of the complex binding to the DREB2A promoter motif, thus regulating the heat stress response (Sato et al., 2014). Over-expression of poplar NF-YA9 in Arabidopsis revealed its role in promoting stomatal closure in an ABA-dependent manner (Lian et al., 2018). While NF-YB3 up-regulation might suggest a developing stress response, it could also connect CAM induction with the necessary adjustment of stomatal behaviour.
JmjC DOMAIN-CONTAINING PROTEIN 27 (JMJ27) acts as a demethylase targeting histone H3 at lysine 9 (H3K9) and plays a role in both pathogen response and flowering time regulation in Arabidopsis. In the defence response, JMJ27 acts as a suppressor of WRKY TF expression (Dutta et al., 2017). Transcript abundances of several WRKYs were affected in ABA-treated leaves, but these were primarily up-regulated, including WRKY33 (Supplementary Dataset S1). MONOPTEROS/AUXIN RESPONSE FACTOR 5 (MP/ARF5) regulates development in an auxin-dependent manner (Hardtke et al., 1998; Krogan et al., 2012). Since phytohormone signalling pathways frequently overlap (Jaillais and Chory, 2010), the involvement of MP in the ABA response, at least under some circumstances, cannot be excluded. TFs identified as putative CAM regulators share their involvement in responses to stressors, including heat, a biological function that was over-represented in the late ABA cluster 5 (Fig. 4B). In addition, numerous identified TFs are related to development and response to other phytohormones.
The abundances of transcripts encoding transport proteins and enzymes of amino acid metabolism were affected by exogenous ABA as well, but it can only be speculated whether transcription of any of these genes is regulated by either of the abovementioned TFs. The altered flow of metabolites could directly contribute to their flow through the CAM pathway, as in the case of BASS2 (Table 1), transcripts of which are highly abundant in the C4 species Flaveria, which employs a type of photosynthesis that shares many similarities with CAM (Furumoto et al., 2011). Alternatively, transporters may contribute to signalling, which might be the role of CATION EXCHANGER 11 (CAX11), even though its role in Ca2+ signalling under stress conditions has been described only in Arabidopsis roots so far (Wang et al., 2016).
The number of ABA-responsive transcripts encoding enzymes of amino acid metabolism could contribute to protein synthesis. For example, METHIONINE GAMMA-LYASE (MGL) has been shown to catalyse conversion of methionine to cysteine (Goyer et al., 2007) and isoleucine synthesis (Joshi and Jander, 2009). Moreover, amino acid metabolism could play a more global role in the adjustment of metabolism. Mitochondrial MCCA, encoding methylcrotonyl-CoA carboxylase alpha chain, is involved in the catabolism of leucine and branched-chain amino acids, thus providing a respiratory substrate to satisfy energy demands. Besides MCCA, THA1 was up-regulated in response to both ABA and drought (Table 1). In Arabidopsis, both MCCA and THA1 are targets of bZIP TFs (Doidy et al., 2016), leading to the hypothesis that both genes could be co-regulated in T. triangulare as well.
Based on Arabidopsis coexpression network analysis, a hierarchical structure of catabolism genes was suggested in which smaller pathways, such as single amino acid degradation, form a part of larger biochemical modules, meaning that specialized genes also possesses a broader metabolic function (Mentzen et al., 2008). However, whether such regulation could also contribute to CAM induction (e.g. via amino acid catabolism genes) has not been investigated yet.
Conclusions
Based on the observed transcriptional changes and increased nocturnal acidification, foliar application of 200 µM ABA was sufficient to induce CAM in the facultative CAM species T. triangulare within 24 hours. CAM induction by exogenous ABA has been reported previously (Ting, 1981; Chu et al., 1990; Dai et al., 1994; Taybi et al., 1995; Minardi et al., 2014) but we are not aware of any other work showing the rapid pace of adjustment of the carbon assimilation strategy. Establishing ABA-inducible CAM models could boost research in the field and expand our knowledge about this pathway, including but not limited to functional confirmation of proposed ‘CAM switches’, gas exchange studies and labelling experiments to disentangle carbon flow during the induction phase of CAM.
Supplementary data
Supplementary data are available at JXB online.
Dataset S1. Quantitative information and annotation for all Talinum triangulare reads mapped on to the reference transcriptome of Beta vulgaris.
Dataset S2. Genes with significantly altered temporal pattern of transcript abundances upon ABA treatment of Talinum triangulare; analysis with maSigPro package for R.
Dataset S3. A complete list of significantly differentially expressed genes based on comparison of the pool of ABA-treated leaves of Talinum triangulare with the pool of mock-treated leaves.
Dataset S4. Relative metabolite amounts in Talinum triangulare leaves as determined by GC-MS, and amino acid amounts in Talinum triangulare leaves as determined by LC-MS.
Fig. S1. Titratable acidity in mature Talinum triangulare leaves at the end of the light and dark phases upon daily treatments with 10–600 µM ABA.
Fig. S2. Principal component analysis of Talinum triangulare RNA-seq data.
Fig. S3. Principal component analysis of metabolite amounts detected in leaves of Talinum triangulare harvested 40, 80, 160, 320, 640, and 1280 min after the first ABA treatment.
Fig. S4. Comparison of performance of Lee and Seung, Brunet, and KL algorithms on the transcriptome dataset of Talinum triangulare.
Fig. S5. Over-representation of differentially expressed genes for biological functions in Talinum triangulare at individual sampling time points.
Fig. S6. Transcript abundance of three detected isoforms of PPC in Talinum triangulare.
Fig. S7. ABA-induced changes in abundances of transcripts encoding components of protein degradation/synthesis and amino acid degradation/synthesis pathways in Talinum triangulare.
Fig. S8. Diurnal pattern of sucrose and starch abundance in ABA- and mock-treated leaves of Talinum triangulare.
Protocol S1. Validation of transcript levels determined by RNA-seq.
Protocol S2. Time series RNA-seq analysis.
Protocol S3. Pulse-amplitude modulated (PAM) fluorescence measurements.
Table S1. Sequencing statistics of the RNA-seq experiment in Talinum triangulare.
Table S2. Gene assignment to MapMan and to homemade reduced categories with expanded Photosynthesis.CAM/C4 photosynthesis category.
Table S3. Genes involved in clock regulation and analysis of time-dependent abundances of their transcript levels in Talinum triangulare.
Acknowledgements
We thank M. Graf, E. Klemp, and K. Weber for their expert technical assistance with the GC-MS and LC-MS measurements. This work was funded by grants (to APMW) of the Deutsche Forschungsgemeinschaft (IRTG 1525, and under Germany’s Excellence Strategy – EXC-2048/1 – Project ID: 390686111), and a graduate fellowship by the German Academic Exchange Service (DAAD) to EM.
Glossary
Abbreviations
- ABA
abscisic acid
- CA
carbonic anhydrase
- CAM
crassulacean acid metabolism
- CBBC
Calvin–Benson–Bassham cycle
- DEG
differentially expressed gene
- ME
malic enzyme
- NMF
non-negative matrix factorization
- PEP
phosphoenolpyruvate
- TCA
tricarboxylic acid
- TF
transcription factor.
References
- Abraham PE, Yin H, Borland AM, et al. 2016. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nature Plants 2, 16178. [DOI] [PubMed] [Google Scholar]
- Antoni R, Rodriguez L, Gonzalez-Guzman M, Pizzio GA, Rodriguez PL. 2011. News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Current Opinion in Plant Biology 14, 547–553. [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Maldonado-Gama M, Pantoja O. 1999. Abscisic acid induction of vacuolar H+-ATPase activity in Mesembryanthemum crystallinum is developmentally regulated. Plant Physiology 120, 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Bohnert HJ, Jensen RG. 1996. Strategies for engineering water-stress tolerance in plants. Trends in Biotechnology 14, 89–97. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland AM, Guo HB, Yang X, Cushman JC. 2016. Orchestration of carbohydrate processing for crassulacean acid metabolism. Current Opinion in Plant Biology 31, 118–124. [DOI] [PubMed] [Google Scholar]
- Borland AM, Hartwell J, Weston DJ, Schlauch KA, Tschaplinski TJ, Tuskan GA, Yang X, Cushman JC. 2014. Engineering crassulacean acid metabolism to improve water-use efficiency. Trends in Plant Science 19, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland AM, Taybi T. 2004. Synchronization of metabolic processes in plants with Crassulacean acid metabolism. Journal of Experimental Botany 55, 1255–1265. [DOI] [PubMed] [Google Scholar]
- Borland AM, Técsi LI, Leegood RC, Walker RP. 1998. Inducibility of crassulacean acid metabolism (CAM) in Clusia species; physiological/biochemical characterisation and intercellular localization of carboxylation and decarboxylation processes in three species which exhibit different degrees of CAM. Planta 205, 342–351. [Google Scholar]
- Bouché N, Fromm H. 2004. GABA in plants: just a metabolite? Trends in Plant Science 9, 110–115. [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Barbier-Brygoo H, Laurière C. 2004. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. The Journal of Biological Chemistry 279, 41758–41766. [DOI] [PubMed] [Google Scholar]
- Boxall SF, Dever LV, Kneřová J, Gould PD, Hartwell J. 2017. Phosphorylation of phosphoenolpyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate crassulacean acid metabolism species Kalanchoë fedtschenkoi. The Plant Cell 29, 2519–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhaus D, Bräutigam A, Mettler-Altmann T, Winter K, Weber AP. 2016. Reversible burst of transcriptional changes during induction of crassulacean acid metabolism in Talinum triangulare. Plant Physiology 170, 102–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Z. 2012. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 100, 110–115. [DOI] [PubMed] [Google Scholar]
- Chan-Schaminet KY, Baniwal SK, Bublak D, Nover L, Scharf KD. 2009. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. The Journal of Biological Chemistry 284, 20848–20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. 2007. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiology 143, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Lin Q, Nose A. 2002. A comparative study on diurnal changes in metabolite levels in the leaves of three crassulacean acid metabolism (CAM) species, Ananas comosus, Kalanchoë daigremontiana and K. pinnata. Journal of Experimental Botany 53, 341–350. [DOI] [PubMed] [Google Scholar]
- Chen LS, Nose A. 2004. Day-night changes of energy-rich compounds in crassulacean acid metabolism (CAM) species utilizing hexose and starch. Annals of Botany 94, 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Poolman MG, Fell DA, Ratcliffe RG, Sweetlove LJ. 2014. A diel flux balance model captures interactions between light and dark metabolism during day-night cycles in C3 and crassulacean acid metabolism leaves. Plant Physiology 165, 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Dai Z, Ku MS, Edwards GE. 1990. Induction of crassulacean acid metabolism in the facultative halophyte Mesembryanthemum crystallinum by abscisic acid. Plant Physiology 93, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman JC, Tillett RL, Wood JA, Branco JM, Schlauch KA. 2008. Large-scale mRNA expression profiling in the common ice plant, Mesembryanthemum crystallinum, performing C3 photosynthesis and Crassulacean acid metabolism (CAM). Journal of Experimental Botany 59, 1875–1894. [DOI] [PubMed] [Google Scholar]
- Dai Z, Ku MSB, Zhang D, Edwards GE. 1994. Effects of growth regulators on the induction of Crassulacean acid metabolism in the facultative halophyte Mesembryanthemum crystallinum L. Planta 192, 287–294. [Google Scholar]
- Di Martino C, Delfine S, Pizzuto R, Loreto F, Fuggi A. 2003. Free amino acids and glycine betaine in leaf osmoregulation of spinach responding to increasing salt stress. New Phytologist 158, 455–463. [DOI] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgrwe D, et al. 2014. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549. [DOI] [PubMed] [Google Scholar]
- Doidy J, Li Y, Neymotin B, Edwards MB, Varala K, Gresham D, Coruzzi GM. 2016. “Hit-and-Run” transcription: de novo transcription initiated by a transient bZIP1 “hit” persists after the “run”. BMC Genomics 17, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Choudhary P, Caruana J, Raina R. 2017. JMJ27, an Arabidopsis H3K9 histone demethylase, modulates defense against Pseudomonas syringae and flowering time. The Plant Journal 91, 1015–1028. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Fan W, Zhao M, Li S, Bai X, Li J, Meng H, Mu Z. 2016. Contrasting transcriptional responses of PYR1/PYL/RCAR ABA receptors to ABA or dehydration stress between maize seedling leaves and roots. BMC Plant Biology 16, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. 2000. Metabolite profiling for plant functional genomics. Nature Biotechnology 18, 1157–1161. [DOI] [PubMed] [Google Scholar]
- Furumoto T, Yamaguchi T, Ohshima-Ichie Y, et al. 2011. A plastidial sodium-dependent pyruvate transporter. Nature 476, 472–475. [DOI] [PubMed] [Google Scholar]
- Gaujoux R, Seoighe C. 2010. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics 11, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, et al. 2012. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. The Plant Cell 24, 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyer A, Collakova E, Shachar-Hill Y, Hanson AD. 2007. Functional characterization of a methionine γ-lyase in Arabidopsis and its implication in an alternative to the reverse trans-sulfuration pathway. Plant & Cell Physiology 48, 232–242. [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. 1998. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. The EMBO Journal 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare PD, Cress WA, van Staden J. 1998. Dissecting the roles of osmolyte accumulation during stress. Plant, Cell and Environment 21, 535–553. [Google Scholar]
- Hartigan JA, Wong MA. 1979. Algorithm AS 136: A K-means clustering algorithm. Journal of the Royal Statistical Society. Series C (Applied Statistics) 28, 100–108. [Google Scholar]
- Hartwell J, Dever LV, Boxall SF. 2016. Emerging model systems for functional genomics analysis of Crassulacean acid metabolism. Current Opinion in Plant Biology 31, 100–108. [DOI] [PubMed] [Google Scholar]
- Häusler RE, Baur B, Scharte J, Teichmann T, Eicks M, Fischer KL, Flügge UI, Schubert S, Weber A, Fischer K. 2000. Plastidic metabolite transporters and their physiological functions in the inducible crassulacean acid metabolism plant Mesembryanthemum crystallinum. The Plant Journal 24, 285–296. [DOI] [PubMed] [Google Scholar]
- Herppich M, von Willert DJ, Herppich WB. 1995. Diurnal rhythm in citric acid content preceded the onset of nighttime malic acid accumulation during metabolic changes from C3 to CAM in salt-stressed plants of Mesembryanthemum crystallinum. Journal of Plant Physiology 147, 38–42. [Google Scholar]
- Herrera A, Delgado J, Paraguatey I. 1991. Occurrence of inducible crassulacean acid metabolism in leaves of Talinum triangulare (Portulacaceae). Journal of Experimental Botany 42, 493–499. [Google Scholar]
- Höfner R, Vazquez-Moreno L, Winter K, Bohnert HJ, Schmitt JM. 1987. Induction of crassulacean acid metabolism in Mesembryanthemum crystallinum by high salinity: mass increase and de novo synthesis of PEP-carboxylase. Plant Physiology 83, 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtum JAM, Smith JAC, Neuhaus E. 2005. Intracellular transport and pathways of carbon flow in plants with crassulacean acid metabolism. Functional Plant Biology 32, 429–449. [DOI] [PubMed] [Google Scholar]
- Holtum JA, Winter K. 1982. Activity of enzymes of carbon metabolism during the induction of Crassulacean acid metabolism in Mesembryanthemum crystallinum L. Planta 155, 8–16. [DOI] [PubMed] [Google Scholar]
- Huai J, Wang M, He J, Zheng J, Dong Z, Lv H, Zhao J, Wang G. 2008. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Reports 27, 1861–1868. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Chory J. 2010. Unraveling the paradoxes of plant hormone signaling integration. Nature Structural & Molecular Biology 17, 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. 2017. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research 45, D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Jander G. 2009. Arabidopsis methionine γ-lyase is regulated according to isoleucine biosynthesis needs but plays a subordinate role to threonine deaminase. Plant Physiology 151, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C. 2008. A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS ONE 3, e3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Research 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Ckurshumova W, Marcos D, Caragea AE, Berleth T. 2012. Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytologist 194, 391–401. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K. 2010. ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proceedings of the National Academy of Sciences, USA 107, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Cao W, Fang H, et al. 2017. Transcriptomic profiling of the maize (Zea mays L.) leaf response to abiotic stresses at the seedling stage. Frontiers in Plant Science 8, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C, Li Q, Yao K, Zhang Y, Meng S, Yin W, Xia X. 2018. Populus trichocarpa PtNF-YA9, a multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis. Frontiers in Plant Science 9, 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttge U. 1987. Carbon dioxide and water demand: crassulacean acid metabolism (CAM), a versatile ecological adaptation exemplifying the need for integration in ecophysiological work. New Phytologist 106, 593–629. [DOI] [PubMed] [Google Scholar]
- Lüttge U. 1988. Day-night changes of citric-acid levels in crassulacean acid metabolism: phenomenon and ecophysiological significance. Plant, Cell and Environment 11, 445–451. [Google Scholar]
- Lüttge U. 2004. Ecophysiology of Crassulacean acid metabolism (CAM). Annals of Botany 93, 629–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. 2000. Chlorophyll fluorescence—a practical guide. Journal of Experimental Botany 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Medina E, Popp M, Olivares E, Janett H-P, Lüttge U. 1993. Daily fluctuations of titratable acidity, content of organic acids (malate and citrate) and soluble sugars of varieties and wild relatives of Ananas comosus L. growing under natural tropical conditions. Plant, Cell and Environment 16, 55–63. [Google Scholar]
- Mentzen WI, Peng J, Ransom N, Nikolau BJ, Wurtele ES. 2008. Articulation of three core metabolic processes in Arabidopsis: fatty acid biosynthesis, leucine catabolism and starch metabolism. BMC Plant Biology 8, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minardi BD, Voytena APL, Santos M, Randi ÁM. 2014. Water stress and abscisic acid treatments induce the CAM pathway in the epiphytic fern Vittaria lineata (L.) Smith. Photosynthetica 52, 404–412. [Google Scholar]
- Montero E, Francisco AM, Montes E, Herrera A. 2018. Salinity induction of recycling Crassulacean acid metabolism and salt tolerance in plants of Talinum triangulare. Annals of Botany 121, 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S. 1998. Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant & Cell Physiology 39, 853–858. [DOI] [PubMed] [Google Scholar]
- Neuhaus HE, Schulte N. 1996. Starch degradation in chloroplasts isolated from C3 or CAM (crassulacean acid metabolism)-induced Mesembryanthemum crystallinum L. Biochemical Journal 318, 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo HG. 2003. How to tell the time: the regulation of phosphoenolpyruvate carboxylase in Crassulacean acid metabolism (CAM) plants. Biochemical Society Transactions 31, 728–730. [DOI] [PubMed] [Google Scholar]
- Nimmo HG, Fontaine V, Hartwell J, Jenkins GI, Nimmo GA, Wilkins MB. 1999. PEP carboxylase kinase is a novel protein kinase controlled at the level of expression. New Phytologist 151, 91–97. [DOI] [PubMed] [Google Scholar]
- Nueda MJ, Tarazona S, Conesa A. 2014. Next maSigPro: updating maSigPro bioconductor package for RNA-seq time series. Bioinformatics 30, 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T. 2007. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. Journal of Experimental Botany 58, 3373–3383. [DOI] [PubMed] [Google Scholar]
- Papageorgiou GC, Murata N. 1995. The unusually strong stabilizing effects of glycine betaine on the structure and function of the oxygen-evolving Photosystem II complex. Photosynthesis Research 44, 243–252. [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainha N, Medeiros VP, Ferreira C, Raposo A, Leite JP, Cruz C, Pacheco CA, Ponte D, Silva AB. 2016. Leaf malate and succinate accumulation are out of phase throughout the development of the CAM plant Ananas comosus. Plant Physiology and Biochemistry 100, 47–51. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R. 2004. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiology 134, 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Mizoi J, Tanaka H, et al. 2014. Arabidopsis DPB3-1, a DREB2A interactor, specifically enhances heat stress-induced gene expression by forming a heat stress-specific transcriptional complex with NF-Y subunits. The Plant Cell 26, 4954–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera K, Neubig KM, Whitten WM, Williams NH, Winter K, Cushman JC. 2010. Evolution along the crassulacean acid metabolism continuum. Functional Plant Biology 37, 995–1010. [Google Scholar]
- Song L, Huang SC, Wise A, Castanon R, Nery JR, Chen H, Watanabe M, Thomas J, Bar-Joseph Z, Ecker JR. 2016. A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Cushman JC. 1999. Signaling events leading to crassulacean acid metabolism induction in the common ice plant. Plant Physiology 121, 545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Cushman JC. 2002. Abscisic acid signaling and protein synthesis requirements for phosphoenolpyruvate carboxylase transcript induction in the common ice plant. Journal of Plant Physiology 159, 1235–1243. [Google Scholar]
- Taybi T, Cushman JC, Borland AM. 2017. Leaf carbohydrates influence transcriptional and post-transcriptional regulation of nocturnal carboxylation and starch degradation in the facultative CAM plant, Mesembryanthemum crystallinum. Journal of Plant Physiology 218, 144–154. [DOI] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC. 2000. A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiology 123, 1471–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Sotta B, Gehrig H, Güçlü S, Kluge M, Brulfert J. 1995. Differential effects of abscisic acid on phosphoenolpyruvate carboxylase and CAM operation in Kalanchoë blossfeldiana. Botanica Acta 108, 240–246. [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Thomas JC, McElwain EF, Bohnert HJ. 1992. Convergent induction of osmotic stress-responses: abscisic acid, cytokinin, and the effects of NaCl. Plant Physiology 100, 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting IP. 1981. Effects of abscisic acid on CAM in Portulacaria afra. Photosynthesis Research 2, 39–48. [DOI] [PubMed] [Google Scholar]
- Tsuzuki M, Miyachi S, Winter K, Edwards GE. 1982. Localization of carbonic anhydrase in Crassulacean acid metabolism plants. Plant Science Letters 24, 211–218. [Google Scholar]
- Urano K, Kurihara Y, Seki M, Shinozaki K. 2010. ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Current Opinion in Plant Biology 13, 132–138. [DOI] [PubMed] [Google Scholar]
- Urano K, Maruyama K, Ogata Y, et al. 2009. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. The Plant Journal 57, 1065–1078. [DOI] [PubMed] [Google Scholar]
- Wang F, Chen ZH, Liu X, Colmer TD, Zhou M, Shabala S. 2016. Tissue-specific root ion profiling reveals essential roles of the CAX and ACA calcium transport systems in response to hypoxia in Arabidopsis. Journal of Experimental Botany 67, 3747–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K, Garcia M, Holtum JA. 2008. On the nature of facultative and constitutive CAM: environmental and developmental control of CAM expression during early growth of Clusia, Kalanchöe, and Opuntia. Journal of Experimental Botany 59, 1829–1840. [DOI] [PubMed] [Google Scholar]
- Winter K, Holtum JA. 2014. Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis. Journal of Experimental Botany 65, 3425–3441. [DOI] [PubMed] [Google Scholar]
- Wu Q, Zhang X, Peirats-Llobet M, et al. 2016. Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. The Plant Cell 28, 2178–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK. 2002. Cell signaling during cold, drought, and salt stress. The Plant Cell 14Suppl 1, S165– S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. 2003. Regulation of abscisic acid biosynthesis. Plant Physiology 133, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2010. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant Journal 61, 672–685. [DOI] [PubMed] [Google Scholar]
- Zhang G, Lu T, Miao W, Sun L, Tian M, Wang J, Hao F. 2017. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ 5, e4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Wu D, Kong F, Lin K, Zhang H, Li G. 2017. The Arabidopsis thaliana nuclear factor Y transcription factors. Frontiers in Plant Science 7, 2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.