Abstract
Anaerobic fungi (Neocallimastigomycota) are emerging non-model hosts for biotechnology due to their wealth of biomass-degrading enzymes, yet tools to engineer these fungi have not yet been established. Here, we show that the anaerobic gut fungi have the most GC depleted genomes among 443 sequenced organisms in the fungal kingdom, which has ramifications for heterologous expression of genes as well as for emerging CRISPR-based genome engineering approaches. Comparative genomic analyses suggest that anaerobic fungi may contain cellular machinery to aid in sexual reproduction, yet a complete mating pathway was not identified. Predicted proteomes of the anaerobic fungi also contain an unusually large fraction of proteins with homopolymeric amino acid runs consisting of five or more identical consecutive amino acids. In particular, threonine runs are especially enriched in anaerobic fungal carbohydrate active enzymes (CAZymes) and this, together with a high abundance of predicted N-glycosylation motifs, suggests that gut fungal CAZymes are heavily glycosylated, which may impact heterologous production of these biotechnologically useful enzymes. Finally, we present a codon optimization strategy to aid in the development of genetic engineering tools tailored to these early-branching anaerobic fungi.
Keywords: Codon optimization, Amino acid distribution, Anaerobe, Genome sequencing, Fungi, Neocallimastigomycota
Highlights
-
•
Anaerobic fungi are emerging non-model hosts to produce biomass-degrading enzymes.
-
•
Comparative genomics reveals that anaerobic fungi are the most AT-rich organisms in the fungal kingdom.
-
•
Predicted gut fungal proteomes contain a large fraction of proteins with homopolymeric amino acid runs.
-
•
AT-richness of anaerobic fungi dictates non-conventional codon optimization and genetic engineering strategies.
1. Introduction
Metabolic engineering strives to streamline and sculpt microorganisms for the optimal production of valuable fuels and chemicals. To date, most metabolic engineering efforts have targeted well-characterized microorganisms such as E. coli and S. cerevisiae, but it is well recognized that non-model microorganisms hold tremendous biotechnological potential (Bonugli-Santos et al., 2015; Coker, 2016; Podolsky et al., 2019; Seppälä et al., 2017). In this regard, the anaerobic fungi in the clade Neocallimastigomycota possess an unparalleled collection of carbohydrate active enzymes (CAZymes) that can be leveraged to convert plant biomass into value-added commodity and fine chemicals (Haitjema et al., 2017; Morrison et al., 2016; Solomon et al., 2016; Youssef et al., 2013). The Neocallimastigomycota fungi are primarily found in the digestive tracts of herbivorous animals where they break down ingested lignocellulosic plant biomass (Liggenstoffer et al., 2010; Orpin, 1975; Theodorou et al., 1996) and although their importance for animal welfare is well established, anaerobic gut fungi have not yet been adapted for metabolic engineering or bioprocessing applications.
To fully exploit the biotechnological potential of anaerobic fungi, it is first necessary to understand the functional properties of their proteins, especially their diverse set of biotechnologically important CAZymes (Podolsky et al., 2019; Seppälä et al., 2017). To achieve this goal, there is a critical need to (1) develop strategies to transfer gut fungal genes to heterologous hosts, and (2) develop molecular tools to modify the genomic content of the gut fungi. The recently acquired high-resolution transcriptomes and genomes of several gut fungal strains aid in this regard as they not only reveal the enzymatic and proteomic potential of these fungi, but also the genomic guanine-cytosine (GC)/adenine-thymine (AT) nucleotide content, apparent codon-usage patterns, and the amino acid composition of encoded proteins (Haitjema et al., 2017; Henske et al., 2018; Solomon et al., 2016; Youssef et al., 2013). In particular, the GC content of any genome often dictates genetic engineering strategies, whether the aim is to transfer genes to a more easily manipulated organism or to engineer the genome of the non-model organism directly.
While a handful of gut fungal proteins have already been produced in model microorganisms (reviewed in (Seppälä et al., 2017)) (Dollhofer et al., 2019; O’Malley et al., 2012), the vast majority remains uncharacterized, and recent reports suggest that at least some gut fungal genes must be codon optimized for the successful expression in heterologous hosts like yeast (Seppälä et al., 2019; Solomon et al., 2016). Likewise, production of non-native proteins (e.g. reporter proteins) and exogenous metabolic pathways in the anaerobic fungi is likely aided if the codon composition of the exogeneous gene is matched to the apparent codon preference of the host, as has been demonstrated in other fungi (Wang et al., 2019). Moreover, the genomic nucleotide composition may affect how efficiently a genome can be engineered using endonucleases that recognize specific, often G-rich, nucleic acid motifs, such as the increasingly popular Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-CRISPR Associated protein (Cas) system.
Here, we have analyzed the genomes and proteomes of anaerobic fungi to establish a framework for metabolic engineering in these non-model organisms. The nucleotide composition of all published anaerobic fungal genomes was used to determine gut fungal codon preferences, amino acid distributions, and identify low-complexity regions in the predicted proteomes with emphasis on their rich repertoire of biotechnologically interesting CAZymes. Currently, five species of anaerobic fungi have published nuclear genomes: Orpinomyces sp. C1A (later re-classified as Pecoramyces ruminantium) (Hanafy et al., 2017; Youssef et al., 2013), Piromyces sp. E2, and the high-quality genomes of Neocallimastix californiae, Anaeromyces robustus and Piromyces finnis (Haitjema et al., 2017). Using the Joint Genome Institute’s (JGI) Mycocosm fungal genomes repository, we compared these anaerobic fungal genomes to 438 other sequenced fungi, spanning the fungal tree of life (I. V. Grigoriev et al., 2014). Overall, we find that the coding genomes of anaerobic fungi are exceptionally GC depleted, which significantly impacts codon and amino acid usage in anaerobic gut fungi and limits the application of certain CRISPR variants. Based on these native biases, we introduce a codon optimization table for use in expressing non-native genes in the gut fungi. Analysis of the genomes also reveals genetic machinery implicated in sexual reproduction, and shows that gut fungal CAZymes are highly enriched in repetitive sequences that are linked to glycosylation motifs. Overall, this comparative analysis will aid in the development of metabolic engineering strategies by identifying common pitfalls and suggesting possible solutions to genetically manipulate Neocallimastigomycota fungi.
2. Results and discussion
2.1. Anaerobic gut fungi have the most GC depleted genomes in the fungal kingdom
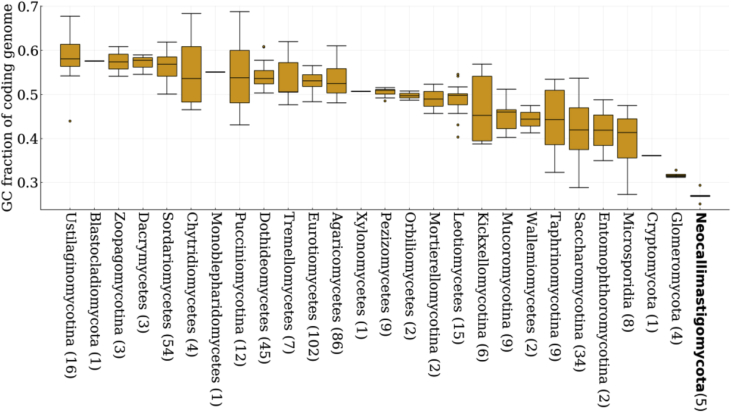
Biased genomic GC content has significant implications for modern genome sequencing and engineering techniques. For example, it has been shown that regions with extreme nucleotide content hamper next-generation sequencing techniques owing to poor read coverage and difficulties in assembly (Oyola et al., 2012). Moreover, the apparent preferred codon usage of an organism may affect how efficiently genes can be transferred between organisms, in particular those that exhibit extreme codon biases (Seppälä et al., 2019). An analysis of 443 published fungal genomes, sourced from the JGI Mycocosm database and covering 278 genera from across the fungal kingdom, reveals a large variation in the GC content of fungal protein coding genomes, ranging from ~25% GC to ~69% GC (Fig. 1, Table S1). Among these, the obligate anaerobic Neocallimastigomycota consistently have the most GC depleted coding genomes of all sequenced fungi, ranging from ~25% GC in A. robustus to ~29% GC in Piromyces sp. E2 (Haitjema et al., 2017; Youssef et al., 2013). The GC content of the intergenic, non-coding regions in the anaerobic gut fungi is even lower (~16% on average): causing the whole-genome GC content of Neocallimastigomycota to range from ~16% to ~22%. This peculiar nucleotide composition of anaerobic fungi was suggested by thermal denaturation studies more than two decades ago, and is a contributing factor to why the first high-resolution genomes were only recently acquired via long-read sequencing technologies (Brownlee, 1989; Nicholson et al., 2005; Oyola et al., 2012).
Fig. 1.
Neocallimastigomycota are characterized by extremely GC-depleted genomes and proteomes. GC content within the predicted proteome of 443 fungal genomes is plotted as a function of fungal clade, and varies significantly across the fungal kingdom. The number of species analyzed per clade is indicated in parentheses on the x-axis. The box-and-whisker plots show outliers as points, minima and maxima as whiskers, and the inter-quartile ranges inside the boxes.
Figure 1 also illustrates that the GC content of the fungal protein coding genomes is not readily explained by phylogenetic relationships, as has also been noted for other kingdoms (Knight et al., 2001; Reichenberger et al., 2015; Wu et al., 2012). For example, while Neocallimastigomycota appears to be the most GC depleted fungal phylum, the phylogenetically-related Chytridiomycetes is rather GC rich at ~56% based on 4 genomes from 4 genera. Possibly confounding this analysis is the number of sequenced genomes analyzed in each clade, as some clades are extremely under-sampled to date (Fig. 1).
2.2. The extreme AT-richness of anaerobic fungal genomes limits CRISPR and other genetic engineering strategies
The GC content of a genome can be affected by mutation rates, recombination and selection (Birdsell, 2002; Duret and Galtier, 2009; Hershberg and Petrov, 2010; Hildebrand et al., 2010). While it remains unclear how the anaerobic gut fungal genomes became GC depleted, their AT-richness has several implications for genetic engineering approaches. For example, plasmid-based expression systems are hampered by difficulties associated with identifying promotors and regulatory elements in these genomes, and AT-rich sequences are associated with non-specific binding affecting both primer design and homologous recombination approaches.
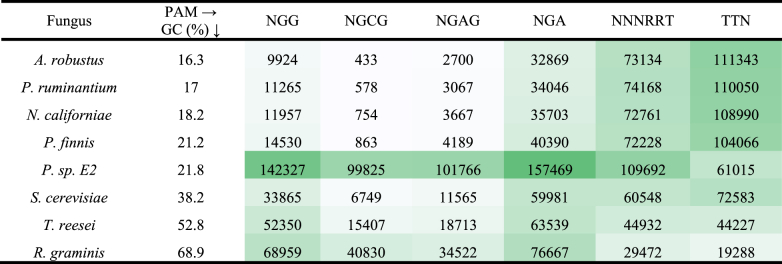
Moreover, most genome editing approaches use specific DNA sequence motifs to guide nucleases to the genome. For example, technologies using transcription-factor-like-endonucleases (TALENs) (Arazoe et al., 2015) and zinc finger nucleases (ZFN) (Boch et al., 2009) depend on DNA-binding protein domains, and the CRISPR-Cas9 system depends on a guide-RNA that brings the nuclease to the desired site (Gasiunas et al., 2012). Genomes that contain regions with extreme nucleotide content may cause poor or nonspecific targeting. For example, the canonical CRISPR-Cas9 system makes use of a G-rich (NGG) protospacer-adjacent motif (PAMs) to target genes for editing (Jiang et al., 2013), however recent engineering efforts have broadened the diversity of PAM sites that can be targeted to include: TTN (Zetsche et al., 2015); NGG, NGA, NGAG, and NGCG (Kleinstiver et al., 2015b); as well as NNNRRT (Kleinstiver et al., 2015a) among others. Table 1 shows that the frequency of observing GC-rich PAMs increases accordingly with genomic GC content. Conversely, PAMs that are richer in AT bases are much more abundant in genomes with lower GC content. The relative paucity of GC-rich PAM sites in anaerobic fungal genomes is likely to limit the ability of certain endonucleases to target specific positions of interest, and suggests that AT-rich PAM targeting Cas enzymes may be the most appropriate choice for CRISPR engineering efforts.
Table 1.
Increasing GC content of fungal genomes increases the number of PAM sequences with higher GC content. The number of PAMs (PAM sequences ordered in decreasing GC content from left to right) found per mega base pair in the genomes (coding and non-coding regions) of fungi in Neocallimastigomycota, as well as Saccharomyces cerevisiae, Trichoderma reesei and Rhodotorula graminis. The fungi are ordered in increasing GC content. The color scale shows the abundance of PAM sequences found in the genome of each fungus, with darker cells corresponding to more PAM sequences identified.
On the other hand, one of the most GC-depleted non-fungal eukaryotic microbes are the Apicomplexan Plasmodium spp., which include avian and human malaria parasites P. gallinaceum (~21% GC) and P. falciparum (~24% GC) (Videvall, 2018). Recently, it was suggested that the genome of P. falciparum undergoes an extremely high rate of mutations, associated with sequences that have an extreme GC or AT bias, and that this phenomenon may contribute to adaptive evolution (Hamilton et al., 2017). Although the mutation rate of Neocallimastigomycota is unknown, it is tempting to speculate that similar mechanisms are in place for anaerobic fungi, possibly facilitating horizontal gene transfer of enzymes from ruminal bacteria to the anaerobic fungi (Duarte and Huynen, 2019; Haitjema et al., 2017; Murphy et al., 2019) that could be harnessed for genome engineering. Nevertheless, the possibility of high mutation rates could negatively impact the efficacy of the highly specific edits made by CRISPR-Cas like systems.
2.3. Anaerobic fungal genomes contain genes used in sexual reproduction
Sexual reproduction is often leveraged for engineering industrial fungal strains, thus identification of a putative mating pathway in anaerobic gut fungi could inform future approaches to generate genetic variants (Mertens et al., 2015; Solieri et al., 2015; Steensels et al., 2014b, 2014a). Inducing breeding events in yeast strains, and other biotechnologically relevant fungi, rapidly increases the diversity of mutant libraries through naturally occurring homologous recombination. This mode of diversity generation has advantages over direct genetic engineering; it is straightforward, rapid, and generates genetic variants that are not considered as genetically modified organisms by regulatory frameworks (Steensels et al., 2014b).
Sexual reproduction is associated with several genomic signatures, including the presence of genes required for mating events and GC content enrichment in genomes and genomic regions that are prone to homologous recombination (Galtier, 2001; Glémin, 2015; Hull et al., 2000; Kiktev et al., 2018; Liu et al., 2018; Magee, 2002; Meunier and Duret, 2004; Ropars et al., 2016). These genomic signatures have successfully been leveraged to interrogate the existence of sexual reproduction in other fungi (Hull et al., 2000). Further, the positive relationship between GC content and the different rates of outcrossing among fungi could help rationalize why GC content within the fungal kingdom is not readily explained by phylogeny (Hartfield, 2016; Nieuwenhuis and James, 2016) (Fig. 1). While many variables likely influence GC content in fungal genomes, the gut fungi standout as being particularly GC depleted, possibly suggesting very infrequent outcrossing (Fig. S1).
However, some organisms with GC content near that of Neocallimastigomycota were until recently erroneously thought to be asexual, bolstering the idea that the anerobic gut fungi might be able to outcross. For example, sexual reproduction was demonstrated in the opportunistic human pathogen Candida albicans (35% GC) (Hull et al., 2000; Magee, 2002). Likewise, fungi in the phylum Glomeromycota (~32% GC) were also thought to be asexual, yet recent sequencing of several genomes revealed genes encoding the molecular machinery required for sexual reproduction (Ropars et al., 2016). These findings seem to support the hypothesis that sexual reproduction is an ability shared by all fungi, even those that infrequently outcross.
While experimental evidence has thus far failed to confirm a sexual cycle in anaerobic gut fungi, we find genes with high homology to sex-implicated proteins in every high quality anaerobic fungal genome sequenced to date. Using well characterized proteins from Saccharomyces cerevisiae, we are able to identify homologs to kinases and accessory proteins heavily implicated in sexual reproduction (STE20, STE6, GPA1), as well as several meiosis specific genes such as the meiotic recombinase DMC1 (Table S2). Notably absent in the Neocallimastigomycota genomes are genes homologous to those coding for peptide mating factors deployed by S. cerevisiae, but regions of homology to the N. crasssa mating type “a” pheromone are detected in each genome (Table S2).
The presence of genes implicated in sexual reproduction indicate that Neocallimastigomycota can, or at one point in evolutionary time were able to, sexually reproduce. However, low GC content in anaerobic fungal genomes could imply that these organisms outcross with extreme discretion. Further experimentation is needed to determine whether sexual reproduction will be a useful tool to generate anaerobic fungal variants. Specifically, elucidation of viable signaling pathways that lead to induction of mating type cells should be tested to determine how such a mating event could be induced and used for metabolic engineering applications (Magee, 2002).
2.4. Codon usage preferences of Neocallimastigomycota are a recipe for genetic engineering and expression optimization
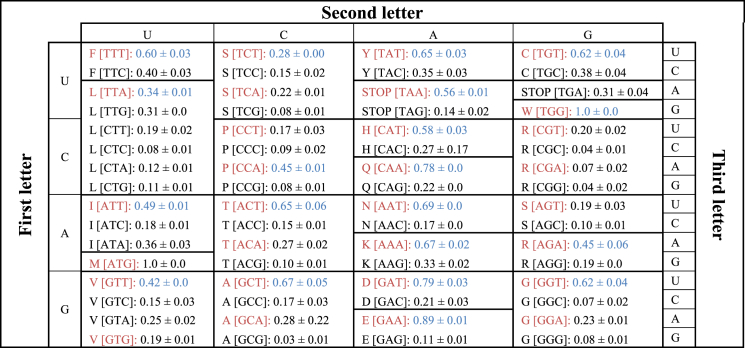
Although the fungi in Neocallimastigomycota are not yet genetically tractable, efforts are being made to develop genetic transformation methodologies (Calkins et al., 2018; Durand et al., 1997). The introduction of novel genes encoding reporter proteins and selection markers into anaerobic gut fungi will likely require codon optimization such that the gut fungal machinery properly maintains and decodes the material. Codon optimization will likely be an important tool to aid in this regard, as has been shown for other fungal clades (Camiolo et al., 2019; Wang et al., 2019). Analysis of the preferred codon usage in highly expressed genes in Neocallimastigomycota suggests that the anaerobic fungi have a strong preference for AT-rich codons (Table 2), consistent with their GC-depleted genomes (Fig. 1). Table S3 shows the individual codon usage for highly expressed transcripts, as well as the predicted tRNA counts, for both the fungi with transcriptomic expression level data available (Henske et al., 2018).
Table 2.
Codon optimization table for Neocallimastigomycota. Fraction of the proteome encoded for by each codon in highly expressed transcripts of N. californiae and A. robustus averaged, with standard deviation noted. The most AT rich codon for each amino acid is shown in red font, while the most abundant codon within the transcriptome is shown in blue font. AT-rich codons are invariably preferred in anaerobic fungi, in line with the predicted low GC content of the clade.
While codon optimization may be necessary to express exogenous genes in the gut fungi, their very strong codon bias has implications for heterologous expression of gut fungal genes in model microorganisms. For example, codon optimization to increase the GC content of genes was shown to be a prerequisite for the expression of gut fungal genes in some hosts (Li et al., 2007; Seppälä et al., 2019). However, this does not seem to be a universal constraint as other genes from gut fungi have been expressed without codon optimization (Kuyper et al., 2003; Wang et al., 2011). Nevertheless, codon optimization may prove to be an important consideration for genetic exploitation of gut fungi because their genomes, and consequently their genes, are so extremely GC depleted. Interestingly, Table S3 also shows that the most abundant codon does not always correspond to the most abundant associated tRNA. For example, across both anaerobic fungi analyzed, AAT is the most common asparagine codon, however only AAC (a synonymous asparagine codon) matching tRNAs (TTG anticodons) were identified on the genome. It is likely that tRNA wobbling in the third base position explains this phenomena, as had been noted in other filamentous fungi (Chen et al., 2012).
2.5. Amino acids coded by AT rich codons are favored by Neocallimastigomycota
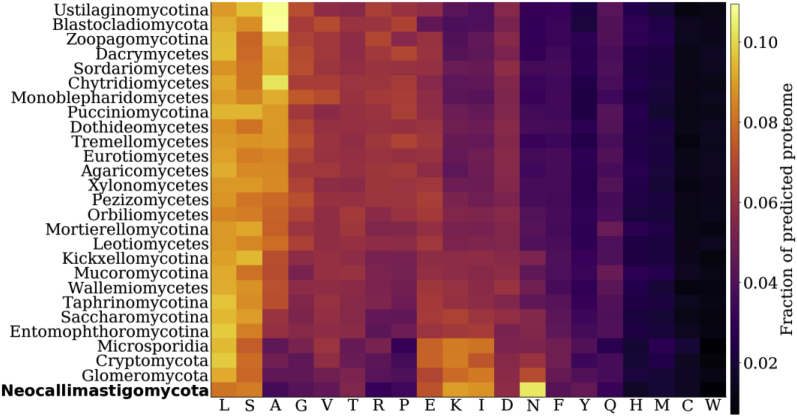
Amino acid composition is important for protein production, stability and post-translational modifications, which has implications for heterologous production. As shown in Fig. 2, there is a clear correlation between GC content and predicted amino acid distribution in the fungi. The GC depleted fungi, including the Neocallimastigomycota, appear to be enriched in amino acids that are encoded for by AT rich codons (lysine, isoleucine and asparagine) and depleted in amino acids that are encoded for by GC rich codons (alanine, glycine, arginine). Conversely, fungi with GC rich coding genomes have a higher proportion of amino acids that are encoded for by GC rich codons. This is consistent with previous cross-kingdom analyses suggesting that the relative abundances of amino acids in a proteome is largely determined by the GC content of the genome (Knight et al., 2001).
Fig. 2.
GC-depleted fungal proteomes are enriched in lysine, isoleucine and asparagine. Average predicted amino acid abundance per clade, ordered in decreasing GC content, is shown across the fungal kingdom. GC-rich fungal phyla are enriched in alanine, glycine, arginine, proline and valine. Asparagine is particularly enriched in Neocallimastigomycota, similar to P. falciparum, another extremely GC depleted organism.
Unique glycosylation patterns, influenced by the amino acid composition of a protein, are often difficult to mimic in heterologous hosts and may affect function (Gerngross, 2004). The high abundance of serine, threonine and asparagine in the gut fungal proteomes suggest that glycosylation could be an important component in protein production, and perhaps activity and stability. Additionally, amino acid composition of enzymes has been shown to correlate with, amongst other properties, thermal stability. The gut fungi grow optimally at 39 °C, suggesting that their enzymes, and specifically their biotechnologically relevant CAZymes, are tailored for this temperature (Haitjema et al., 2014). In contrast, T. reesei grows optimally at 28 °C, but significant protein engineering efforts have improved the thermal stability of its cellulases to function in the range of 50–70 °C (Chokhawala et al., 2015). It is tempting to speculate that gut fungal enzymes may also be amenable to such engineering efforts and, combined with their natively very diverse cellulolytic enzyme repertoire, might increase the efficiency of high temperature biomass conversion processes even further. Finally, the nitrogen content of media has been shown to influence the growth rates of gut fungi, possibly due to amino acid biosynthesis (in particular lysine) bottlenecks (Atasoglu and Wallace, 2002). Since lysine is one of the most abundantly used amino acids in the gut fungal proteome, it suggests that media supplementation strategies could be beneficial for protein production in the gut fungi.
2.6. Anaerobic fungal CAZymes are enriched in homopolymeric amino acid runs
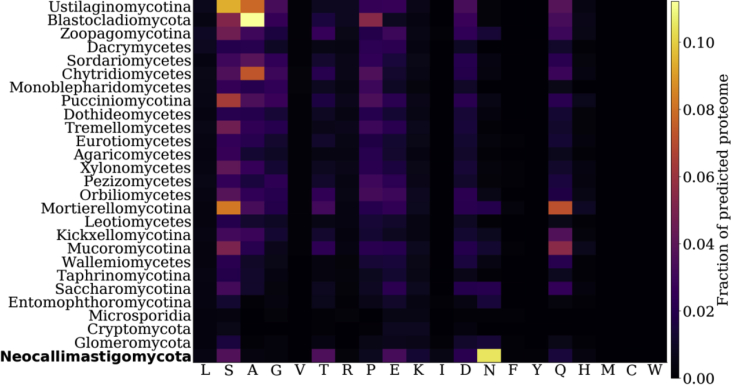
Homopolymeric runs of five or more consecutive identical amino acids are common in eukaryotic proteins (Albà et al., 2007). While their evolutionary origin is debated, it has been suggested that these low-complexity regions provide eukaryotes with a major source of phenotypic variation (Fondon III and Garner, 2004) and are associated with functionally important intrinsically disordered regions (Wright and Dyson, 2015). All fungal clades we investigated here have proteins with runs, where the average fraction of the proteome with runs ranges from 3% in Microsporidia (based on 8 genomes, 5 genera), to 30% in Neocallimastigomycota (based on 5 genomes, 4 genera), and finally to 37% in Ustilaginomycotina (based on 16 genomes, 14 genera) for each clade (Table S4). However, these runs are not evenly distributed across all the amino acids. Fig. 3 shows that runs with leucine, valine, isoleucine, arginine, phenylalanine, tyrosine, methionine, cysteine and tryptophan are largely absent. The absence of proteins with bulky aromatic or hydrophobic amino acids implies that there is likely a cost associated with having long stretches of these residues. In the case of hydrophobic amino acids (valine, leucine, methionine and isoleucine) protein aggregation likely plays a role in preventing such runs from occurring. Smaller amino acids, like glycine, serine and alanine are more frequently found in runs, along with most of the polar amino acids. Cysteine is an exception to this, likely due to its reactive sulphur side chain.
Fig. 3.
Proteins with asparagine runs constitute an unusually large fraction of the Neocallimastigomycota proteome. Average amino acid run (five or more of the same amino acid consecutively in a protein) fraction per clade, ordered in decreasing GC content, in the fungal kingdom. Hydrophobic (valine, leucine, methionine and isoleucine) and bulky (phenylalanine, tyrosine and tryptophan) amino acids are noticeably absent in runs, while smaller (alanine) uncharged, polar (serine, threonine, proline, glutamine) amino acids are frequently found in runs.
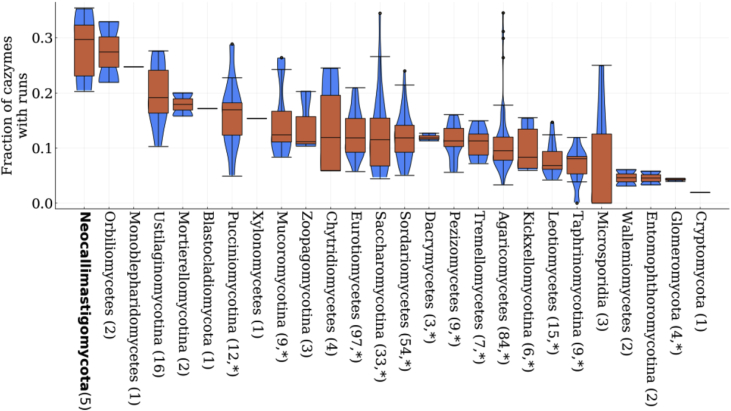
Despite the likely complex evolutionary origin of these runs in proteins, analysis of all the CAZymes found in Neocallimastigomycota revealed that more than a quarter of all CAZymes contained a run motif. Interestingly, a large variation in the fraction of CAZymes with runs were found throughout the fungal kingdom (Fig. 4). Neocallimastigomycota (28% with 5 genomes), Orbiliomycetes (27% with 2 genomes), and Monoblepharidomycetes (25% for a single genome) had the highest fractions of CAZymes with runs; all the other clades had less than 20% on average. Given that only the simplest repetitive structure was searched for, this is likely an underestimate of the CAZymes that contain such low-complexity regions. Furthermore, using transcriptomic data for N. californiae and A. robustus (Henske et al., 2018; Solomon et al., 2016) it was found that there are no significant differences in expression levels between CAZymes with and without run motifs (using the two sample Kolmogorov-Smirnov test). However, there is a significant difference (using the two-sample unequal variance t-test, P < 0.01) in the ratio of CAZymes with runs versus total number of CAZymes between the fungi in Neocallimastigomycota and the genera Trichoderma and Aspergillus, which contain biotechnologically relevant organisms, (mean ratio of 0.28, 0.11, and 0.14 CAZymes with runs versus the total number CAZymes for each group respectively). Given the wide spread usage of Trichoderma reesei and Aspergillus niger in cellulase production (Sukumaran et al., 2009), it raises the question of the function of these runs, and if they impart some benefit to enzyme effectiveness.
Fig. 4.
Neocallimastigomycota have significantly more CAZymes with amino acid repeatrunsthan other fungal clades. Here the distribution of the fraction of CAZymes with runs relative to all the CAZymes in each fungus, grouped by clade, is plotted. The number in parentheses is the number of fungi included in each clade. Statistically significant differences in the distributions between Neocallimastigomycota and all the other clades are indicated by * using the two sample Kolmogorov-Smirnoff test (P < 0.05). The distribution of the fraction of CAZymes with runs in each clade is shown in the blue violin plots overlaid by orange box-and-whisker plots where outliers are shown as points, minima and maxima as whiskers, and the inter-quartile ranges inside the boxes. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.7. Homopolymeric amino acid runs in CAZymes are enriched in threonine and serine, suggesting these enzymes are heavily glycosylated
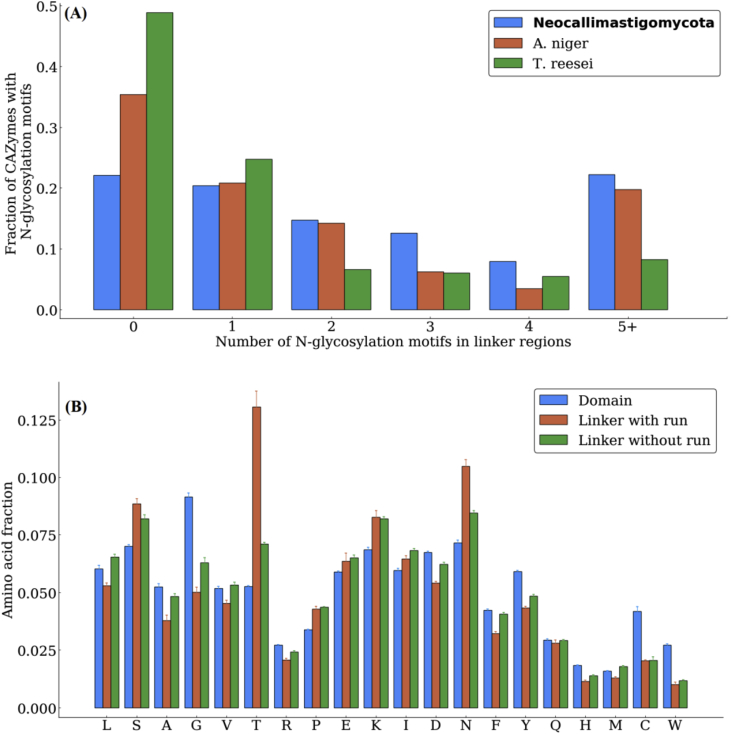
While the organisms belonging to Neocallimastigomycota are still genetically intractable, heterologous expression of their CAZymes is likely the most expedient route to unlocking their biotechnological promise. However, expressing CAZymes heterologously is not straightforward, in part due to glycosylation patterns that are difficult to mimic outside of the native host (Greene et al., 2015). Moreover, recent work highlighting the role of processive enzymes attached to the cellulosome produced by members of Neocallimastigomycota showed that its CAZymes are heavily glycosylated (Haitjema et al., 2017). Indeed, genomic data indicate (Table S5) that the machinery for both N- and O-linked glycosylation is present in each sequenced genome of Neocallimastigomycota. Furthermore, by scanning the linker regions of all the CAZymes found in Neocallimastigomycota, T. reesei, and A. niger (Fig. 5A) for the canonical N-X-(S or T) (where X is any amino acid except proline) N-glycosylation motif, it becomes apparent that the motif is more abundant in the anaerobic gut fungi than in the latter two organisms. Only ~22% of the CAZymes found in the high-quality genomes of N. californiae, A. robustus, and P. finnis lack N-glycosylation motifs, compared to ~49% and ~35% for T. reesei and A. niger, respectively.
Fig. 5.
(A) CAZymes in Neocallimastigomycota have more N-glycosylation motifs relative to other industrially important cellulolytic fungi. The fraction of CAZymes with a specified number of N-glycosylation motifs (N-X-S/T where X is not proline) on the x-axis in N. californiae, A. robustus and P. finnis (grouped as Neocallimastigomycota here, the other members are not shown due to their lower quality genomes), T. reesei, and A. niger. Linker regions are defined as the inter-domain regions of proteins. Neocallimastigomycota has a higher proportion of CAZymes with 2 or more N-glycosyaltion motifs than either T. reesei or A. niger. (B) Threonine is disproportionately abundant in the linker region of CAZymes in Neocallimastigomycota, suggesting O-glycosylation sites may be abundant. Amino acid fraction in all proteins with at least one CAZyme domain divided into three groups: domains, linker regions of proteins with runs (five or more of the same amino acid consecutively in a protein), and linker regions of proteins without runs. Linker regions are defined as the inter-domain regions. Serines, and especially threonines, are highly enriched in the inter-domain regions of CAZymes with runs and without runs.
While N-linked glyosylation sites are straightforward to predict, no such recognition site has yet been identified for O-linked glycosylation. However, threonine and serine rich regions in the linker region of cellulase proteins are likely candidates for O-glycosylation (Beckham et al., 2010; Sammond et al., 2012). Figure 5B shows the amino acid abundance in CAZymes split into domains and the inter-domain (linker) regions, which are further separated into linker regions of proteins with and without runs. It is clear that asparagine, serine, and especially threonine, are significantly enriched in the linker regions of Neocallimastigomycota. The threonine enrichment is even more pronounced in the linker regions of proteins that have runs, reflecting the disproportionate abundance of threonine runs in CAZymes.
Given that glycosylation is a mechanism used by cells to protect CAZymes from proteolytic cleavage, and that the rumen of herbivores is heavily populated with proteases (Bach et al., 2005), is reasonable to hypothesize that these regions are indeed glycosylated in vivo. Prior work has shown that CAZymes within the cellulosomes of Neocallimastigomycota are indeed heavily glycosylated (Haitjema et al., 2017). Additionally, marked increases in CAZyme activity have been observed when the expression host is changed to an organism capable of glycosylating its enzymes (Cheng et al., 2015, 2014; Ximenes et al., 2005). Finally, the importance of linker regions in cellulase function (Sonan et al., 2007) reinforces the idea that metabolic engineering strategies should take these features into account to optimally leverage the CAZyme machinery of Neocallimastigomycota.
3. Conclusions
While the underlying reasons for GC depletion in Neocallimastigomycota remain unclear, the consequences of this AT-richness for metabolic engineering are numerous. The possibility that the anaerobic gut fungi have high mutational rates due to their GC depletion has interesting implications for strain evolution, engineering, and stability. Understanding how, and at what rate, their genomes evolve will provide an improved roadmap to engineer these organisms (Nørholm, 2019; Sekowska et al., 2016). While functional genetic tools to modify the anaerobic fungi are in development, the codon optimization strategy presented here may attenuate the current difficulties associated with expressing non-native genes in these hosts. The GC depleted genomes likely also limit the use of G-rich PAM targeting Cas enzymes in the CRISPR system, suggesting that Cas enzymes engineered to target T-rich PAM sites should be prioritized for engineering anaerobic fungi. Comparative genomic analyses have shown that homopolymeric runs of amino acids are unusually common in anaerobic fungi, especially in their CAZyme machinery. These motifs likely serve important functions, e.g. glycosylation sites that prevent proteolytic cleavage, suggesting the importance of understanding their role if gut fungal CAZymes are heterologously produced in a model organism.
4. Materials and methods
4.1. Collection and processing of genomic data
The Mycocosm database, curated by the Joint Genome Institute (JGI), was used to download 443 sequenced fungal genomes (listed in Tables S1 and (I. V Grigoriev et al., 2014)), as well as their predicted protein coding genes, predicted proteomes and associated PFAM annotations. The whole genomes (protein coding and non-coding regions) were also downloaded for all the sequenced fungi in Neocallimastigomycota, as well as Saccharomyces cerevisiae, Trichoderma reesei and Rhodotorula graminis (accessed November 2018) (I. V. Grigoriev et al., 2014). The de novo assembled transcriptomes of A. robustus and N. californiae (Solomon et al., 2016) and the associated differential transcriptomic datasets for each these fungi grown in isolation on reed canary grass (Henske et al., 2018) were also used as described below. Scripts using the Julia programming language, and the associated BioSequences and Hypothesis Tests packages, were used to process and analyze the data (Bezanson et al., 2017). Code is available on Mendeley Data, https://doi.org/10.17632/26vywtfkrz.1.
4.2. Nucleotide content, genome analysis and construction of codon tables
Using genomic data, the nucleotide content of the protein coding genes for all 443 fungi, as well as the unmasked whole fungal genomes for the fungi in Neocallimastigomycota, were calculated by counting each nucleotide base (G, C, T, A) and ignoring gaps and indeterminate bases (N). The GC fraction was then calculated by dividing the total number of G and C bases by the total number of G, C, T, and A bases ((G + C)/(G + C + A + T)). Similarly, the amino acid abundances were calculated by counting the number of each amino acid found in the predicted proteome, which is the translated protein coding gene, relative to the total number of amino acids in the same predicted proteome for each organism. Phylogenetic classifications were based on the taxonomic assignments as defined by the JGI. The number of protospacer adjacent motif (PAM) sites per genome was counted by parsing through each scaffold on the whole genomes of all the sequenced fungi in Neocallimastigomycota, as well as Saccharomyces cerevisiae, Trichoderma reesei and Rhodotorula graminis, and counting the number of matches to a particular motif, e.g. TTN would match to TTA, TTG, TTT and TTC, using the Julia BioSequences package. The number of hits was then divided by the total number of bases in each fungal genome.
Codon optimization tables for anaerobic fungi were calculated by first identifying the top 1000 most expressed genes in the fungal isolates N. californiae and A. robustus using the differential transcriptomic data for these fungi grown on reed canary grass from Henske et al. (2018) (Henske et al., 2018). BLASTn was then used to align these transcripts to their predicted genes (Camacho et al., 2009). Only genes with coverage greater than 90% and e-values less than 10−60 were decomposed into codons. The frequency of each codon was then calculated by counting the number of times it appears relative to all the other synonymous codons. The tRNA gene counts in the genomes of A. robustus and N. californiae were found by tRNAscan-SE using the eukaryotic specific parameters (Chan and Lowe, 2019).
4.3. Identification of homopolymeric amino acid runs and glycosylation motifs in fungi
Using the Julia BioSequences package, the predicted proteomes from downloaded Mycocosm fungal genomes were searched for homopolymeric runs of 5 or more consecutive amino acids of the same type (Karlin et al., 2002) through the regular expression “X{5,}” where X is the amino acid query. This bioinformatic search returns the longest uninterrupted hit of 5 of more amino acids (X) in succession within the proteome. For example, the hypothetical protein, “MGKTTTTTLTTTTTTTF”, has two threonine runs of length five and six. The canonical N-glycosylation motif, N-X-(S or T) (where N is asparagine, X is any amino acid except proline, S is serine and T is threonine) (Deshpande et al., 2008), was found by searching each protein using the regular expression “N[ˆP](S|T)”.
4.4. CAZyme identification and transcriptomic expression analysis
CAZymes were identified by matching the predicted protein family annotations from the PFAM annotation files in the 443 sequenced fungal genomic datasets to a list of CAZyme family domains. See Table S6 for the PFAM to CAZyme family domain association table (Carlson et al., 2019). A protein was designated as a CAZyme if at least one annotated PFAM domain was found in the CAZyme family domain list. For each fungus, this filtered list of CAZymes was used to search for amino acid runs as described above, to determine the amino acid composition of the CAZymes and to find predicted N-glycosylation motifs. Furthermore, only predicted CAZymes that had a coverage greater than 90% and an e-value less than 10−40 (using BLASTn (Camacho et al., 2009) to match the associated gene against the transcriptomes in (Solomon et al., 2016)) were included in the CAZyme expression analysis using the reed canary grass condition data of (Henske et al., 2018).
4.5. Annotation of sexual reproduction and glycosylation genomic machinery in fungal genomes
To evaluate the potential for sexual reproduction by clade Neocallimastigomycota, we searched member genomes for a subset of proteins required by other fungi for sexual reproduction (Hull et al., 2000). Using S. cerevisiae peptide sequences obtained from “The Saccharomyces Genome Database (SGD)” (www.yeastgenome.org) as queries we searched for mating factors, proteins involved in sexual reproduction, and proteins involved in meiosis. Specific Saccharomyces genes queried included sex-implicated kinases (STE20) (Leberer et al., 1996), sex-signal transduction proteins (STE6, GPA1) (Raymond et al., 1998; Sadhu et al., 1992), meiosis specific recombinases (DMC1) (Diener and Fink, 1996), and mating factors (MATa/MATα) (Hull et al., 2000). Additionally, peptide mating factors of N. crassa (MATA/MATa), and pheromone receptor domain containing proteins from cryptically sexual fungi were queried against anaerobic fungal genomes (Glass et al., 1990; Ropars et al., 2016; Staben and Yanofsky, 1990). The tBLASTn algorithm with a BLOSUM62 substitution matrix was used to score peptide alignments against genomes using an expected e-value of 10−25 (Camacho et al., 2009).
The glycosylation machinery in fungi is highly conserved, as such S. cerevisiae’s canonical genes were used as benchmarks for the identification of putative glycosylation pathways (Deshpande et al., 2008). The predicted proteins of the gut fungi were compared to benchmark proteins found in S. cerevisiae (downloaded from Uniprot (“UniProt: a worldwide hub of protein knowledge,” 2019)) using BLASTp (Camacho et al., 2009). A gene was deemed present if the coverage was greater than 50% and the e-value less than 10−20. O-glycosylation was deemed possible if all PMT1-4 genes were found (Gentzsch and Tanner, 1996) and N-glycosylation was deemed possible if at least three of DPM1, ALG3, ALG9, ALG12, OST1, OST3 and STT3 were found (Deshpande et al., 2008; Knauer and Lehle, 1999), see Table S5 for a collation of the blast results.
Acknowledgements
The authors acknowledge funding support from the National Science Foundation (NSF) (MCB-1553721), the Office of Science (BER) the US Department of Energy (DOE) (DE-SC0010352), the Institute for Collaborative Biotechnologies through grants W911NF-19–D-0001 and W911NF-19-2-0026 from the US Army Research Office, and the Camille Dreyfus Teacher-Scholar Awards Program. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the Office of Biological and Environmental Research of the DOE Office of Science through contract DE-AC02–05CH11231 between Lawrence Berkeley National Laboratory and the DOE.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00107.
Contributor Information
St. Elmo Wilken, Email: stelmo@ucsb.edu.
Susanna Seppälä, Email: sseppala@ucsb.edu.
Thomas S. Lankiewicz, Email: tom.lankiewicz@ucsb.edu.
Mohan Saxena, Email: mohan_saxena@ucsb.edu.
John K. Henske, Email: john.henske@gmail.com.
Asaf A. Salamov, Email: aasalamov@lbl.gov.
Igor V. Grigoriev, Email: ivgrigoriev@lbl.gov.
Michelle A. O’Malley, Email: momalley@ucsb.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Albà M.M., Tompa P., Veitia R.A. Gene and Protein Evolution. KARGER; Basel: 2007. Amino acid repeats and the structure and evolution of proteins; pp. 119–130. [DOI] [PubMed] [Google Scholar]
- Arazoe T., Ogawa T., Miyoshi K., Yamato T., Ohsato S., Sakuma T., Yamamoto T., Arie T., Kuwata S. Tailor-made TALEN system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol. Bioeng. 2015;112:1335–1342. doi: 10.1002/bit.25559. [DOI] [PubMed] [Google Scholar]
- Atasoglu C., Wallace R.J. De novo synthesis of amino acids by the ruminal anaerobic fungi, Piromyces communis and Neocallimastix frontalis. FEMS Microbiol. Lett. 2002;212:243–247. doi: 10.1111/j.1574-6968.2002.tb11273.x. [DOI] [PubMed] [Google Scholar]
- Bach A., Calsamiglia S., Stern M.D. Nitrogen metabolism in the Rumen. J. Dairy Sci. 2005;88:E9–E21. doi: 10.3168/jds.S0022-0302(05)73133-7. [DOI] [PubMed] [Google Scholar]
- Beckham G.T., Bomble Y.J., Matthews J.F., Taylor C.B., Resch M.G., Yarbrough J.M., Decker S.R., Bu L., Zhao X., McCabe C., Wohlert J., Bergenstråhle M., Brady J.W., Adney W.S., Himmel M.E., Crowley M.F. The O-glycosylated linker from the Trichoderma reesei family 7 cellulase is a flexible, disordered protein. Biophys. J. 2010;99:3773–3781. doi: 10.1016/j.bpj.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanson J., Edelman A., Karpinski S., Shah V.B. Julia: a fresh approach to numerical computing *. Soc. Ind. Appl. Math. 2017;59 [Google Scholar]
- Birdsell J. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 2002;19:1181–1197. doi: 10.1093/oxfordjournals.molbev.a004176. [DOI] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bonugli-Santos R.C., dos Santos Vasconcelos M.R., Passarini M.R.Z., Vieira G.A.L., Lopes V.C.P., Mainardi P.H., dos Santos J.A., de Azevedo Duarte L., Otero I.V.R., da Silva Yoshida A.M., Feitosa V.A., Pessoa A., Sette L.D. Marine-derived fungi: diversity of enzymes and biotechnological applications. Front. Microbiol. 2015;6:269. doi: 10.3389/fmicb.2015.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee A.G. Remarkably AT-rich genomic DNA from the anaerobic fungus Neocallimastix. Nucleic Acids Res. 1989;17:1327–1335. doi: 10.1093/nar/17.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins S.S., Elledge N.C., Mueller K.E., Marek S.M., Couger M., Elshahed M.S., Youssef N.H. Development of an RNA interference (RNAi) gene knockdown protocol in the anaerobic gut fungus Pecoramyces ruminantium strain C1A. PeerJ. 2018;6:e4276. doi: 10.7717/peerj.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camiolo S., Toome-Heller M., Aime M.C., Haridas S., Grigoriev I.V., Porceddu A., Mannazzu I. An analysis of codon bias in six red yeast species. Yeast. 2019;36:53–64. doi: 10.1002/yea.3359. [DOI] [PubMed] [Google Scholar]
- Carlson M., Liu T., Lin C., Falcon S., Zhang J., MacDonald J. 2019. PFAM.db: a set of protein ID mappings for PFAM. R Packag. version 3.8.2. [Google Scholar]
- Chan P.P., Lowe T.M. Methods in Molecular Biology (Clifton, N.J.) 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences; pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Xie T., Shao Y., Chen F. Genomic characteristics comparisons of 12 food-related filamentous fungi in tRNA gene set, codon usage and amino acid composition. Gene. 2012;497:116–124. doi: 10.1016/j.gene.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Cheng Y.S., Chen C.C., Huang C.H., Ko T.P., Luo W., Huang J.W., Liu J.R., Guo R.T. Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme. J. Biol. Chem. 2014;289:11020–11028. doi: 10.1074/jbc.M114.550905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.S., Chen C.C., Huang J.W., Ko T.P., Huang Z., Guo R.T. Improving the catalytic performance of a GH11 xylanase by rational protein engineering. Appl. Microbiol. Biotechnol. 2015;99:9503–9510. doi: 10.1007/s00253-015-6712-0. [DOI] [PubMed] [Google Scholar]
- Chokhawala H.A., Roche C.M., Kim T.-W., Atreya M.E., Vegesna N., Dana C.M., Blanch H.W., Clark D.S. Mutagenesis of Trichoderma reesei endoglucanase I: impact of expression host on activity and stability at elevated temperatures. BMC Biotechnol. 2015;15:11. doi: 10.1186/s12896-015-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coker J.A. Extremophiles and biotechnology: current uses and prospects. F1000Res. 2016;5 doi: 10.12688/f1000research.7432.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande N., Wilkins M.R., Packer N., Nevalainen H. Protein glycosylation pathways in filamentous fungi. Glycobiology. 2008;18:626–637. doi: 10.1093/glycob/cwn044. [DOI] [PubMed] [Google Scholar]
- Diener A.C., Fink G.R. DLH1 is a functional Candida albicans homologue of the meiosis-specific gene DMC1. Genetics. 1996;143:769–776. doi: 10.1093/genetics/143.2.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollhofer V., Young D., Seppälä S., Hooker C., Youssef N., Podmirseg S.M., Nagler M., Reilly M., Li Y., Fliegerová K., Cheng Y., Griffith G.W., Elshahed M., Solomon K.V., O’Malley M.A., Theodorou M.K. The Mycota. 2019. The biotechnological potential of the anaerobic gut fungi. [Google Scholar]
- Duarte I., Huynen M.A. 2019. Contribution of Lateral Gene Transfer to the Evolution of the Eukaryotic Fungus Piromyces sp. E2: Massive Bacterial Transfer of Genes Involved in Carbohydrate Metabolism. bioRxiv 514042. [Google Scholar]
- Durand R., Rascle C., Fischer M., Fèvre M. Transient expression of the beta-glucuronidase gene after biolistic transformation of the anaerobic fungus Neocallimastix frontalis. Curr. Genet. 1997;31:158–161. doi: 10.1007/s002940050190. [DOI] [PubMed] [Google Scholar]
- Duret L., Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genom. Hum. Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Fondon J.W., III, Garner H.R. Harvard Medical School.; 2004. Molecular Origins of Rapid and Continuous Morphological Evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzsch M., Tanner W. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996;15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- Gerngross T.U. Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat. Biotechnol. 2004;22:1409–1414. doi: 10.1038/nbt1028. [DOI] [PubMed] [Google Scholar]
- Glass N.L., Grotelueschen J., Metzenberg R.L. Neurospora crassa A mating-type region. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4912–4916. doi: 10.1073/pnas.87.13.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S. Quantification of GC-biased gene conversion in the human genome. Genome Res. 2015:1215–1228. doi: 10.1101/gr.185488.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene E.R., Himmel M.E., Beckham G.T., Tan Z. Glycosylation of cellulases: engineering better enzymes for biofuels. Adv. Carbohydr. Chem. Biochem. 2015;72:63–112. doi: 10.1016/bs.accb.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., Smirnova T., Nordberg H., Dubchak I., Shabalov I. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haitjema C.H., Solomon K.V., Henske J.K., Theodorou M.K., O’Malley M.A. Anaerobic gut fungi: advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol. Bioeng. 2014;111:1471–1482. doi: 10.1002/bit.25264. [DOI] [PubMed] [Google Scholar]
- Haitjema C.H., Gilmore S.P., Henske J.K., Solomon K.V., De Groot R., Kuo A., Mondo S.J., Salamov A.A., LaButti K., Zhao Z., Chiniquy J., Barry K., Brewer H.M., Purvine S.O., Wright A.T., Hainaut M., Boxma B., Van Alen T., Hackstein J.H.P., Henrissat B., Baker S.E., Grigoriev I.V., O’Malley M.A. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2017;2:1–8. doi: 10.1038/nmicrobiol.2017.87. [DOI] [PubMed] [Google Scholar]
- Hamilton W.L., Claessens A., Otto T.D., Kekre M., Fairhurst R.M., Rayner J.C., Kwiatkowski D. Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res. 2017;45:1889–1901. doi: 10.1093/nar/gkw1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafy R.A., Elshahed M.S., Liggenstoffer A.S., Griffith G.W., Youssef N.H. Pecoramyces ruminantium, gen. nov., sp. nov., an anaerobic gut fungus from the feces of cattle and sheep. Mycologia. 2017;109:231–243. doi: 10.1080/00275514.2017.1317190. [DOI] [PubMed] [Google Scholar]
- Hartfield M. Evolutionary genetic consequences of facultative sex and outcrossing. J. Evol. Biol. 2016;29:5–22. doi: 10.1111/jeb.12770. [DOI] [PubMed] [Google Scholar]
- Henske J.K., Wilken S.E., Solomon K.V., Smallwood C.R., Shutthanandan V., Evans J.E., Theodorou M.K., O’Malley M.A. Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose. Biotechnol. Bioeng. 2018;115:874–884. doi: 10.1002/bit.26515. [DOI] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001115. e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F., Meyer A., Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6:e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C.M., Raisner R.M., Johnson A.D. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science (80-.) 2000;289:307–310. doi: 10.1126/science.289.5477.307. [DOI] [PubMed] [Google Scholar]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Brocchieri L., Bergman A., Mrazek J., Gentles A.J. Amino acid runs in eukaryotic proteomes and disease associations. Proc. Natl. Acad. Sci. U. S. A. 2002;99:333–338. doi: 10.1073/pnas.012608599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiktev D.A., Sheng Z., Lobachev K.S., Petes T.D. GC content elevates mutation and recombination rates in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 2018;115:E7109–E7118. doi: 10.1073/pnas.1807334115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver Benjamin P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver Benjamin P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J., Aryee M.J., Joung J.K. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer R., Lehle L. The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta - Gen. Subj. 1999;1426:259–273. doi: 10.1016/s0304-4165(98)00128-7. [DOI] [PubMed] [Google Scholar]
- Knight R.D., Freeland S.J., Landweber L.F. A simple model based on mutation and selection explains trends in codon and amino-acid usage and GC composition within and across genomes. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-4-research0010. research0010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyper M., Harhangi H.R., Stave A.K., Winkler A.A., Jetten M.S.M., De Laat W.T.A.M., Den Ridder J.J.J., Op Den Camp H.J.M., Van Dijken J.P., Pronk J.T. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 2003;4:69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Leberer E., Harcus D., Broadbent I.D., Clark K.L., Dignard D., Ziegelbauer K., Schmidt A., Gow N.A., Brown A.J., Thomas D.Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.L., Skory C.D., Ximenes E.A., Jordan D.B., Dien B.S., Hughes S.R., Cotta M.A. Expression of an AT-rich xylanase gene from the anaerobic fungus Orpinomyces sp. strain PC-2 in and secretion of the heterologous enzyme by Hypocrea jecorina. Appl. Microbiol. Biotechnol. 2007;74:1264–1275. doi: 10.1007/s00253-006-0787-6. [DOI] [PubMed] [Google Scholar]
- Liggenstoffer A.S., Youssef N.H., Couger M.B., Elshahed M.S. Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. ISME J. 2010;4:1225–1235. doi: 10.1038/ismej.2010.49. [DOI] [PubMed] [Google Scholar]
- Liu H., Huang J., Sun X., Li J., Hu Y., Yu L., Liti G., Tian D., Hurst L.D., Yang S. Tetrad analysis in plants and fungi finds large differences in gene conversion rates but no GC bias. Nat. Ecol. Evol. 2018;2:164–173. doi: 10.1038/s41559-017-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee B.B. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science (80-.) 2002;289:310–313. doi: 10.1126/science.289.5477.310. [DOI] [PubMed] [Google Scholar]
- Mertens S., Steensels J., Saels V., De Rouck G., Aerts G., Verstrepen K.J. A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl. Environ. Microbiol. 2015;81:8202–8214. doi: 10.1128/AEM.02464-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier J., Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Morrison J.M., Elshahed M.S., Youssef N.H. Defined enzyme cocktail from the anaerobic fungus Orpinomyces sp. strain C1A effectively releases sugars from pretreated corn stover and switchgrass. Sci. Rep. 2016;6:29217. doi: 10.1038/srep29217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C.L., Youssef N.H., Hanafy R.A., Couger M.B., Stajich J.E., Wang Y., Baker K., Dagar S.S., Griffith G.W., Farag I.F., Callaghan T.M., Elshahed M.S. Horizontal gene transfer as an indispensable driver for evolution of Neocallimastigomycota into a distinct gut-dwelling fungal lineage. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.00988-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson M.J., Theodorou M.K., Brookman J.L. Molecular analysis of the anaerobic rumen fungus Orpinomyces - insights into an AT-rich genome. Microbiology. 2005;151:121–133. doi: 10.1099/mic.0.27353-0. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis B.P.S., James T.Y. The frequency of sex in fungi. Philos. Trans. R. Soc. B Biol. Sci. 2016 doi: 10.1098/rstb.2015.0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørholm M.H.H. Meta synthetic biology: controlling the evolution of engineered living systems. Microbiol Biotechnol. 2019;12:35–37. doi: 10.1111/1751-7915.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orpin C.G. Studies on the rumen flagellate Neocallimastix frontalis. J. Gen. Microbiol. 1975;91:249–262. doi: 10.1099/00221287-91-2-249. [DOI] [PubMed] [Google Scholar]
- Oyola S.O., Otto T.D., Gu Y., Maslen G., Manske M., Campino S., Turner D.J., MacInnis B., Kwiatkowski D.P., Swerdlow H.P., Quail M.A. Optimizing illumina next-generation sequencing library preparation for extremely at-biased genomes. BMC Genomics. 2012;13:1. doi: 10.1186/1471-2164-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley M.A., Theodorou M.K., Kaiser C.A. Evaluating expression and catalytic activity of anaerobic fungal fibrolytic enzymes native topiromyces sp E2 in Saccharomyces cerevisiae. Environ. Prog. Sustain. Energy. 2012;31:37–46. [Google Scholar]
- Podolsky I., Seppälä S., Lankiewicz T., Brown J., Swift C., O’Malley M. Harnessing nature’s anaerobes for biotechnology and bioprocessing. Annu. Rev. Chem. Biomol. Eng. 2019 doi: 10.1146/annurev-chembioeng-060718-030340. [DOI] [PubMed] [Google Scholar]
- Raymond M., Dignard D., Alarco A.M., Mainville N., Magee B.B., Thomas D.Y. A Ste6p/P-glycoprotein homologue from the asexual yeast Candida albicans transports the a-factor mating pheromone in Saccharomyces cerevisiae. Mol. Microbiol. 1998;27:587–598. doi: 10.1046/j.1365-2958.1998.00704.x. [DOI] [PubMed] [Google Scholar]
- Reichenberger E.R., Rosen G., Hershberg U., Hershberg R. Prokaryotic nucleotide composition is shaped by both phylogeny and the environment. Genome Biol. Evol. 2015;7:1380–1389. doi: 10.1093/gbe/evv063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropars J., Toro K.S., Noel J., Pelin A., Charron P., Farinelli L., Marton T., Krüger M., Fuchs J., Brachmann A., Corradi N. Evidence for the sexual origin of heterokaryosis in arbuscular mycorrhizal fungi. Nat. Microbiol. 2016;1:1–9. doi: 10.1038/nmicrobiol.2016.33. [DOI] [PubMed] [Google Scholar]
- Sadhu C., Hoekstra D., McEachern M.J., Reed S.I., Hicks J.B. A G-protein alpha subunit from asexual Candida albicans functions in the mating signal transduction pathway of Saccharomyces cerevisiae and is regulated by the a1-alpha 2 repressor. Mol. Cell. Biol. 1992;12:1977–1985. doi: 10.1128/mcb.12.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammond D.W., Payne C.M., Brunecky R., Himmel M.E., Crowley M.F., Beckham G.T. Cellulase linkers are optimized based on domain type and function: insights from sequence analysis, biophysical measurements, and molecular simulation. PLoS One. 2012;7:e48615. doi: 10.1371/journal.pone.0048615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekowska A., Wendel S., Fischer E.C., Nørholm M.H.H., Danchin A. Generation of mutation hotspots in ageing bacterial colonies. Sci. Rep. 2016;6:2. doi: 10.1038/s41598-016-0005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä S., Wilken S.E., Knop D., Solomon K.V., O’Malley M.A. The importance of sourcing enzymes from non-conventional fungi for metabolic engineering and biomass breakdown. Metab. Eng. 2017;44:45–49. doi: 10.1016/j.ymben.2017.09.008. [DOI] [PubMed] [Google Scholar]
- Seppälä S., Yoo J.I., Yur D., O’Malley M.A. Heterologous transporters from anaerobic fungi bolster fluoride tolerance in Saccharomyces cerevisiae. Metab. Eng. Commun. 2019;9 doi: 10.1016/j.mec.2019.e00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solieri L., Verspohl A., Bonciani T., Caggia C., Giudici P. Fast method for identifying inter- and intra-species Saccharomyces hybrids in extensive genetic improvement programs based on yeast breeding. J. Appl. Microbiol. 2015;119:149–161. doi: 10.1111/jam.12827. [DOI] [PubMed] [Google Scholar]
- Solomon K.V., Haitjema C.H., Henske J.K., Gilmore S.P., Borges-Rivera D., Lipzen A., Brewer H.M., Purvine S.O., Wright A.T., Theodorou M.K., Grigoriev I.V., Regev A., Thompson D.A., O’Malley M.A. Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science (80-.) 2016;351:1192–1196. doi: 10.1126/science.aad1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonan G.K., Receveur-Brechot V., Duez C., Aghajari N., Czjzek M., Haser R., Gerday C. The linker region plays a key role in the adaptation to cold of the cellulase from an Antarctic bacterium. Biochem. J. 2007;407:293–302. doi: 10.1042/BJ20070640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staben C., Yanofsky C. Neurospora crassa a mating-type region (sexual reproduction/vegetative incompatibility/perithecium formation/filamentous fungus) Genetics. 1990;87:4917–4921. doi: 10.1073/pnas.87.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels J., Meersman E., Snoek T., Saels V., Verstrepen K.J. Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 2014;80:6965–6975. doi: 10.1128/AEM.02235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels J., Snoek T., Meersman E., Picca Nicolino M., Voordeckers K., Verstrepen K.J. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014;38:947–995. doi: 10.1111/1574-6976.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran R.K., Singhania R.R., Mathew G.M., Pandey A. Cellulase production using biomass feed stock and its application in lignocellulose saccharification for bio-ethanol production. Renew. Energy. 2009;34:421–424. [Google Scholar]
- Theodorou M.K., Mennim G., Davies D.R., Zhu W.-Y.Y., Trinci A.P.J., Brookman J.L. Anaerobic fungi in the digestive tract of mammalian herbivores and their potential for exploitation. Proc. Nutr. Soc. 1996;55:913–926. doi: 10.1079/pns19960088. [DOI] [PubMed] [Google Scholar]
- UniProt: a worldwide hub of protein knowledge Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videvall E. Plasmodium parasites of birds have the most AT-rich genes of eukaryotes. Microb. Genomics. 2018;4 doi: 10.1099/mgen.0.000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T.Y., Chen H.L., Lu M.Y.J., Chen Y.C., Sung H.M., Mao C.T., Cho H.Y., Ke H.M., Hwa T.Y., Ruan S.K., Hung K.Y., Chen C.K., Li J.Y., Wu Y.C., Chen Y.H., Chou S.P., Tsai Y.W., Chu T.C., Shih C.C.A., Li W.H., Shih M.C. Functional characterization of cellulases identified from the cow rumen fungus Neocallimastix patriciarum W5 by transcriptomic and secretomic analyses. Biotechnol. Biofuels. 2011;4 doi: 10.1186/1754-6834-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Su X., Sun M., Zhang M., Wu J., Xing J., Wang Y., Xue J., Liu X., Sun W., Chen S. Efficient production of glycyrrhetinic acid in metabolically engineered Saccharomyces cerevisiae via an integrated strategy. Microb. Cell Factories. 2019;18:95. doi: 10.1186/s12934-019-1138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhang Z., Hu S., Yu J. On the molecular mechanism of GC content variation among eubacterial genomes. Biol. Direct. 2012;7:2. doi: 10.1186/1745-6150-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ximenes E.A., Chen H., Kataeva I.A., Cotta M.A., Felix C.R., Ljungdahl L.G., Li X.L. A mannanase, ManA, of the polycentric anaerobic fungus Orpinomyces sp. strain PC-2 has carbohydrate binding and docking modules. Can. J. Microbiol. 2005;51:559–568. doi: 10.1139/w05-033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef N.H., Couger M.B., Struchtemeyer C.G., Liggenstoffer A.S., Prade R.A., Najar F.Z., Atiyeh H.K., Wilkins M.R., Elshahed M.S. The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 2013;79:4620–4634. doi: 10.1128/AEM.00821-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., Koonin E.V., Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.