Abstract
It is well known that cells rely on mitochondrial respiration for survival. However, the effect of microRNAs (miRNAs) on mitochondria of cells has not been extensively explored. Our results indicated that the overexpression of a miRNA (miR-1) could destroy mitochondria of cancer stem cells. miR-1 was downregulated in melanoma stem cells (MSCs) and breast cancer stem cells (BCSCs) compared with cancer non-stem cells. However, the upregulation of miR-1 in cancer non-stem cells did not induce mitochondrial damage. miR-1 overexpression caused mitochondrial damage of cancer stem cells by directly targeting the 3′ UTRs of MINOS1 (mitochondrial inner membrane organizing system 1) and GPD2 (glycerol-3-phosphate dehydrogenase 2) genes and interacting with LRPPRC (leucine-rich pentatricopeptide-repeat containing) protein, a protein localized in mitochondria. MINOS1, GPD2, and LRPPRC in mitochondria were required for mitochondrial inner membrane. The results of in vitro and in vivo assays demonstrated that miR-1 overexpression induced mitophagy of cancer stem cells. Therefore, our study contributed novel insights into the mechanism of miRNA-mediated regulation of mitochondria morphology of cancer stem cells.
Keywords: miR-1, mitochondria, target gene, target protein, mitophagy
Introduction
Cancer is the leading cause of human death in the world. Despite continuous efforts to improve the therapy of cancer, the overall survival rate for different malignant cancers is still very low, due at least in part to the existence of cancer stem cells. As reported, tumors are composed of a heterogeneous group of cells, which form a cellular hierarchy with different biological and phenotypic characteristics during tumor cell differentiation.1,2 At the apex of this hierarchy, there exist cancer stem cells, serving as the source of newly formed tumor cells. The generation of cancer stem cells is usually related to the aberrant expression of a wide range of genes. As is well known, microRNAs (miRNAs) play crucial roles in regulating gene expressions. miRNAs, 21- to 23-nt noncoding RNAs, are transcribed by RNA polymerase II and then processed by RNase III-like enzymes Drosha and Dicer.3 Mature miRNAs guide the RNA-induced silencing complex (RISC) to suppress the translation of target mRNA or mediate target mRNA’s degradation.4 Increasing evidence has indicated the involvement of miRNAs in cancer stem cells.5 Tumor suppressor miR-34 can alter the balance between self-renewal and differentiation in colon cancer stem cells.6 The miRNA profile of Ewing sarcoma family tumor stem cells is shared by embryonic stem cells and cancer stem cells from divergent tumor types.5 It is found that miRNA-146a directs the symmetric division of colorectal cancer stem cells through targeting the epithelial-mesenchymal transition (EMT) inducer Snail.7 Although miRNAs can regulate the stemness of cancer stem cells during tumor progression, the miRNA-mediated mechanism is still poorly understood.
In general, miRNAs are loaded onto RISC and then one of the two miRNA strands is bound to Argonaute (Ago) protein to repress target genes’ expressions.8 A single miRNA can target multiple mRNAs and, in turn, an mRNA can be targeted by multiple miRNAs.9,10 Recently, it was found that except for Ago2 protein, miRNAs can interact with other RNA-binding proteins, such as TAR DNA-binding protein 43, heterogeneous nuclear ribonucleoprotein E2, Toll-like receptor 7/8, and the HIV Gag protein.5 TAR DNA-binding protein 43 can negatively regulate the activity of the miR-1 family by directly binding to miR-1, limiting their bioavailability for RISC loading.11 The restoration of miR-328 expression rescues differentiation and impairs survival of leukemic blasts by interacting with the translational regulator heterogeneous ribonucleoprotein (hnRNP) E2.12 Toll-like receptor 7/8 and the HIV Gag protein are participated in interacting with miRNAs (miR-29 and miR-146, respectively13,14). At present, however, the role of miRNA-protein interactions in cancer stem cells has not been explored.
Our previous investigation demonstrated that miR-1 inhibited the growth of breast and gastric cancer cells through simultaneously targeting six target genes (CDK4, TWF1, CNN3, CORO1C, WASF2, and TMSB4X).15 As reported, the expression of miR-1 is downregulated in various types of human cancers, such as breast cancer, gastric cancer, and prostate cancer.16 miR-1 can regulate EMT and function in tumor-suppressive programs by directly targeting EGFR and Slug.17,18 In this context, to further explore the influence of miR-1 on cancer stem cells, the underlying molecular mechanism of miR-1 in breast cancer and melanoma stem cells was characterized in this study. The results showed that miR-1 destroyed mitochondria of cancer stem cells by binding LRPPRC (leucine-rich pentatricopeptide-repeat containing) protein and targeting MINOS1 (mitochondrial inner membrane organizing system 1) and GPD2 (glycerol-3-phosphate dehydrogenase 2) genes, which were required for mitochondrial organization. Therefore, our study shed novel light on the mechanism of miRNA-mediated regulation of mitochondrial morphology in cancer stem cells.
Results
The Influence of miR-1 on Mitochondrial Cristae Organization of Cancer Stem Cells
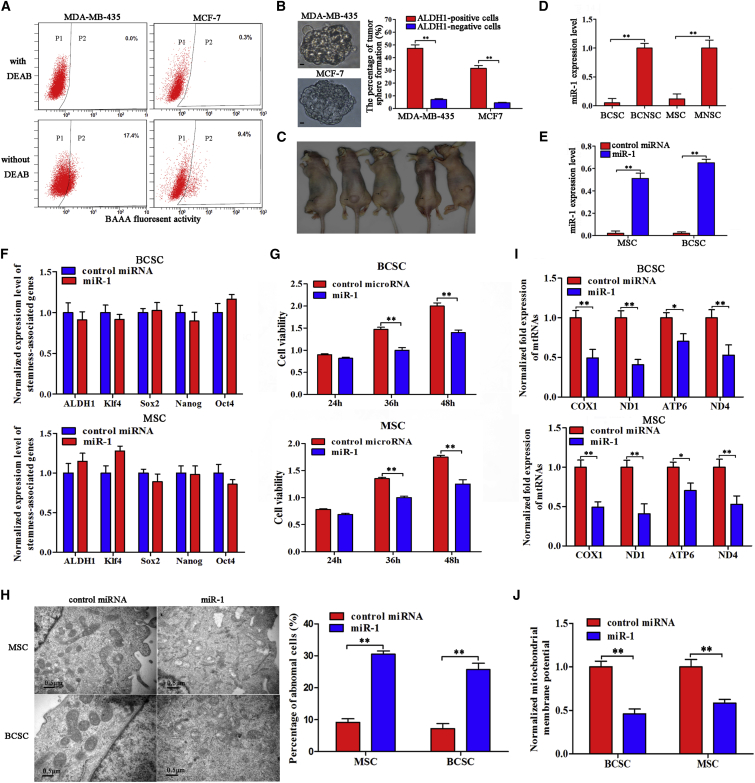
To explore the roles of miRNAs in tumorigenesis of melanoma stem cells (MSCs) and breast cancer stem cells (BCSCs), aldehyde dehydrogenase 1 (ALDH1)-positive cancer stem cells and ALDH1-negative cancer non-stem cells were sorted from the MDA-MB-435 melanoma cell line and MCF7 breast cancer cell line, respectively (Figure 1A). Then, the self-renewal capability of ALDH1-positive and ALDH1-negative cells was determined using sphere-forming assays. The results indicated that the ALDH1-positive cells but not the ALDH1-negative cells were capable of generating tumorspheres with a much higher frequency in three consecutive passages (Figure 1B). The data of tumorigenicity of ALDH1-positive and ALDH1-negative MDA-MB-435 cells in vivo revealed that tumors formed in all five mice injected with ALDH1-positive cells (Figure 1C), while no tumor was observed for ALDH1-negative cells. These data indicated that the ALDH1-positive cells were melanoma or breast cancer stem cells.
Figure 1.
The Influence of miR-1 on Mitochondrial Cristae Organization of Cancer Stem Cells
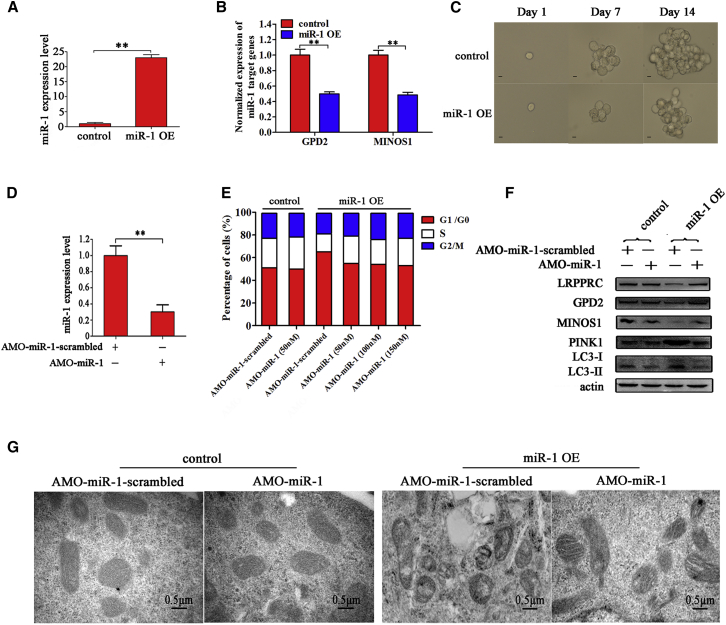
(A) The sorting of ALDH1-positive cells. The baseline fluorescence was established by cells (P1 region) incubated with ALDEFLUOR substrate (BAAA) and ALDH1 inhibitor (DEAB). DEAB was used to block the background signal by inhibiting ALDH1 enzyme activity. Incubation of cells with ALDEFLUOR substrate in the absence of DEAB defined the ALDH1-positive population (P2 region). (B) Representative photographs of ALDH1-positive tumorspheres (top) and the percentages of tumor sphere formation of ALDH1-positive and ALDH1-negative cells (bottom). Scale bars, 10 μm. (C) Tumorigenicity of cancer stem cells (MDA-MB-435) in nude mice. Five mice were subcutaneously injected with the cells isolated from the spheres of tumorsphere formation assays (the ALDH1-positive cells). As controls, the ALDH1-negative cells were subcutaneously injected into five mice. Forty days later, the tumors were examined. The arrows indicate the tumors. (D) Differential expression of miR-1 in cancer stem cells and cancer non-stem cells. Quantitative real-time PCR was conducted to detect the expression level of miR-1 in melanoma stem cells (MSCs), melanoma non-stem cells (MNSCs), breast cancer stem cells (BCSCs), and breast cancer non-stem cells (BCNSCs) (**p < 0.01). U6 was used as an internal reference. (E) Overexpression of miR-1 in cancer stem cells. Cancer stem cells were transfected with miR-1 or control miRNA, followed by detection of miR-1 with quantitative real-time PCR (**p < 0.01). U6 was used as an internal reference. (F) Detection of stemness-associated genes in miR-1-overexpressing cancer stem cells. In the miR-1-transfected melanoma or breast cancer stem cells, the levels of stemness-associated genes’ expressions were examined by quantitative real-time PCR. (G) Influence of miR-1 overexpression on the viability of cancer stem cells. Cancer stem cells were transfected with miR-1. At different times after transfection, the viability of cancer stem cells was examined (**p < 0.01). (H) Effects of miR-1 overexpression on morphology of mitochondrial cristae of cancer stem cells. The mitochondrial cristae of miR-1-overexpressing cancer stem cells were examined under transmission electron microscopy (TEM) (left). The statistical data are indicated on the right (**p < 0.01). Scale bars, 0.5 μm. (I) Influence of miR-1 overexpression on mitochondrial transcripts of cancer stem cells. Mitochondrial transcripts of miR-1-overexpressing cancer stem cells were determined using quantitative real-time PCR (*p < 0.05; **p < 0.01). (J) Mitochondrial membrane potential analysis. At 36 h after miR-1 transfection, cancer stem cells were subjected to flow cytometry analysis and the value of mitochondrial membrane potential was calculated (**p < 0.01). Cancer stem cells transfected with control miRNA were used as controls. All assays were biologically repeated three times.
The quantitative real-time PCR data indicated that miR-1 was significantly downregulated in MSCs and BCSCs compared with cancer non-stem cells (Figure 1D), suggesting that miR-1 was involved in tumorigenesis of melanoma and breast cancer.
To reveal the role of miR-1 in cancer stem cells, miR-1 was overexpressed in MSCs and BSCSs (Figure 1E). The results showed that the expression levels of stemness-associated genes (ALDH1, Sox2, Oct4, Nanog, and Klf4) in miR-1-overexpressing melanoma and breast cancer stem cells were not significantly changed compared with those of the controls (Figure 1F), indicating that miR-1 did not participate in the maintenance of stemness of cancer stem cells. The data from cell viability assays revealed that miR-1 overexpression significantly inhibited the viability of MSCs and BCSCs compared with the controls (Figure 1G), suggesting that miR-1 had important roles in cancer stem cells. To explore the influence of miR-1 on cancer stem cell proliferation, the morphology of cancer stem cells was examined. The transmission electron microscopy data indicated that the mitochondrial cristae of miR-1-overexpressing BCSCs and MSCs were disordered and circular compared with the controls (Figure 1H). In more than 20% of miR-1-overexpressing cancer stem cells, the mitochondria became abnormal (Figure 1H). These results showed that miR-1 overexpression could destroy the mitochondria of cancer stem cells. To evaluate the influence of the miR-1-mediated destruction of mitochondria on the mitochondrial transcripts, the expressions of genes COX1 (cytochrome C oxidase subunit 1), ND1 (NADH dehydrogenase subunit 1), ATP6 (ATP synthase F0 subunit 6), and ND4 (NADH dehydrogenase 4), which are required for the organization and morphology of mitochondrial cristae,19 were detected. Quantitative real-time PCR data showed that the transcript levels of COX1, ND1, ATP6, and ND4 in cancer stem cells transfected with miR-1 were significantly decreased compared with the controls (Figure 1I), indicating that the miR-1-mediated destruction of mitochondria affected the function of mitochondria. At the same time, the mitochondrial membrane potential analysis showed an obvious reduction after miR-1 overexpression in cancer stem cells compared with the controls (cancer stem cells transfected with control miRNA) (Figure 1J).
Taking the above data together, the findings revealed that miR-1 overexpression could destroy mitochondria of cancer stem cells.
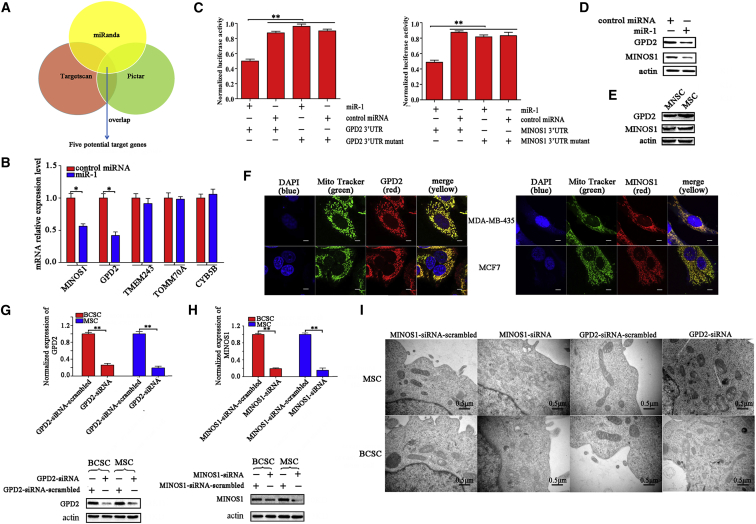
The Induction of miR-1-Mediated Mitochondrial Damage by Targeting GPD2 and MINOS1 Genes
In an attempt to explore the mechanism of miR-1-mediated mitochondrial morphologic alteration, the target genes of miR-1 were predicted. The target prediction analysis showed that 85 genes might be the targets of miR-1. As is well known, miRNAs mediate translational repression and mRNA degradation of target genes in cytoplasm. The potential mitochondrial target genes of miR-1 were excluded in this study. Our results indicated that miR-1 could affect mitochondrial organization. In this context, five genes might be the targets of miR-1 (Figure 2A). In the miR-1-overexpressing cells, the expressions of MINOS1 and GPD2, two genes of the five potential targets of miR-1, were significantly decreased (Figure 2B), indicating that MINOS1 and GPD2 were possible target genes of miR-1. To evaluate the direct interaction between miR-1 and its two target genes, miR-1 and the 3′ UTR of MINOS1 or GPD2 mRNA were co-transfected into MDA-MB-435 cells. The results showed that the luciferase activity of the cells co-transfected with miR-1 and GPD2 or MINOS1 3′ UTR was significantly decreased compared with the controls (Figure 2C), indicating that miR-1 directly interacted with GPD2 and MINOS1. When miR-1 was overexpressed in melanoma stem cells, the protein level of GPD2 or MINOS1 was significantly decreased compared with the control (Figure 2D), showing the interaction between miR-1 and GPD2 or MINOS1 in MSCs. The interaction between miR-1 and GPD2 in MSCs revealed in this study was consistent with that in lung cancer cells as reported previously.20 Compared with MSCs, melanoma non-stem cells also showed reduced protein levels of GPD2 and MINOS1 (Figure 2E). The above data revealed that GPD2 and MINOS1 were the target genes of miR-1.
Figure 2.
The Induction of miR-1-Mediated Mitochondrial Damage by Targeting GPD2 and MINOS1 Genes
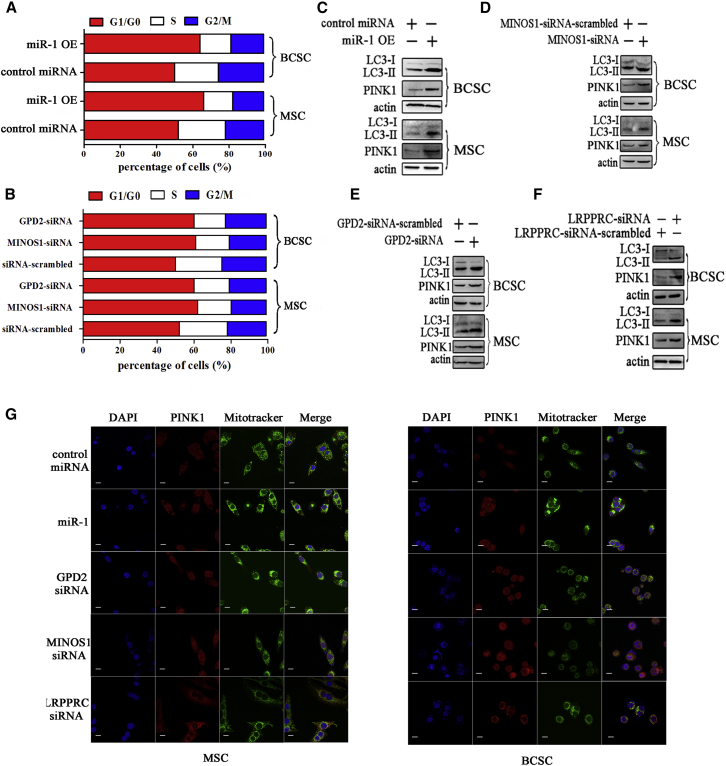
(A) Prediction of miR-1 target genes. As predicted, five genes (MINOS1, GPD2, TMEM243, TOMM70A, and CYB5B) associated with mitochondria were the potential targets of miR-1. (B) The influence of miR-1 on the expressions of potential target genes. The melanoma stem cells (MSCs) were transfected with miR-1. At 48 h after transfection, the expression levels of potential target genes were examined using quantitative real-time PCR (**p < 0.01). (C) The direct interactions of miR-1 with its target genes. MDA-MB-435 cells were co-transfected with miR-1 and a luciferase reporter fused with MINOS1 or GPD2 3′ UTR. At 36 h after co-transfection, the firefly and renilla luciferase activities were analyzed. As controls, control miRNA, MINOS1 3′ UTR mutant, and GPD2 3′ UTR mutant were included in the co-transfections (**p < 0.01). (D) Western blot analysis of MINOS1 and GPD2 in miR-1-overexpressed melanoma stem cells (MSCs). At 36 h after miR-1 or control miRNA transfection, the protein level of MINOS1 or GPD2 was determined by western blot. Actin was used as a control. (E) Western blot analysis of MINOS1 and GPD2 in MSCs and melanoma non-stem cells (MNSCs). (F) Co-localization of MINOS1 and GPD2 with mitochondria of cancer stem cells. MSCs (MDA-MB-435) and breast cancer stem cells (MCF7) were respectively incubated with mitochondria tracker (MitoTracker). Then, the cells were fixed and stained with GPD2-specific or MINOS1-specific antibody. The nucleus was labeled with DAPI. The images were obtained using confocal microscopy. Scale bars, 10 μm. (G) The knockdown of GPD2 in cancer stem cells. The cancer stem cells were transfected with GPD2-siRNA. As a control, GPD2-siRNA-scrambled was included in the assays. The knockdown efficiency of GPD2 was investigated using quantitative real-time PCR (top) and western blot (bottom) (**p < 0.01). Actin was used as a negative control. (H) The silencing of MINOS1 in cancer stem cells. Quantitative real-time PCR (top) and western blot (bottom) were used to detect the expression level of MINOS1 in cancer stem cells (**p < 0.01). (I) The effects of GPD2 and MINOS1 silencing on the mitochondrial ultrastructure of cancer stem cells. Melanoma or breast cancer stem cells (MSCs and BCSCs) treated with sequence-specific siRNA were imaged under transmission electron microscopy. Scale bars, 0.5 μm.
To characterize the roles of GPD2 and MINOS1 in mitochondria of cancer stem cells, the co-localization of GPD2 and MINOS1 proteins with mitochondria was conducted. Confocal microscopy assays indicated that the GPD2 and MINOS1 proteins were co-localized with mitochondria in MSCs and BCSCs (Figure 2F), which was consistent with the results of tumor cells as previously described,21,22 suggesting that the functions of GPD2 and MINOS1 proteins were associated with mitochondria. Then, the expression of GPD2 and MINOS1 was knocked down using sequence-specific siRNA in MSCs and BCSCs, respectively. Quantitative real-time PCR and western blot data showed that the expression levels of GPD2 and MINOS1 were significantly reduced in siRNA-transfected cancer stem cells compared with the control (Figures 2G and 2H), indicating that the expressions of GPD2 and MINOS1 were silenced in cancer stem cells. The results of transmission electron microscopy revealed that the silencing of GPD2 or MINOS1 led to the destruction of mitochondrial cristae of 28.6% or 32.8% of cancer stem cells, while the cells treated with GPD2-siRNA-scrambled or MINOS1-siRNA-scrambled displayed typical sheet-like mitochondrial cristae organization (Figure 2I).
The above findings indicated that miR-1 could target mitochondrial MINOS1 and GPD2 genes, leading to mitochondria damage of MSCs and BCSCs.
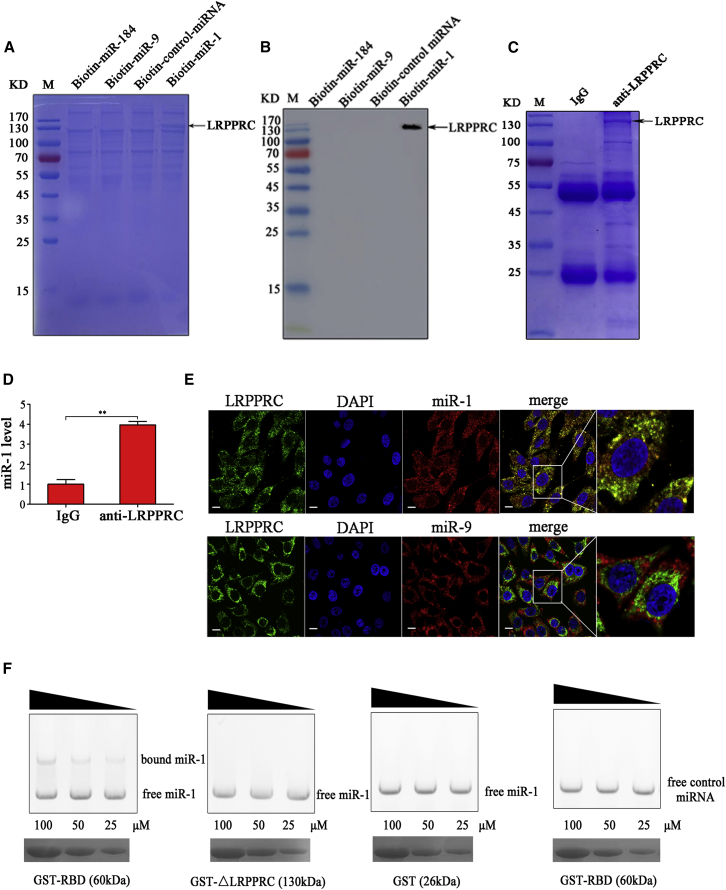
The Interaction between miR-1 and LRPPRC Protein
To explore whether miR-1 could interact with some proteins to influence mitochondrial morphology, an RNA pulldown assay of biotin-labeled miR-1 was conducted using the lysate of MSCs. A specific protein bound to miR-1 was observed compared with the controls (Figure 3A). Mass spectrometry identification revealed that this protein was LRPPRC. As reported, LRPPRC is associated with mitochondrial cristae morphology.23 Therefore, LRPPRC was further characterized.
Figure 3.
The Interaction between miR-1 and LRPPRC Protein
(A) The proteins bound to miR-1. The biotinylated miR-1 (biotin-miR-1) was incubated with the lysate of cancer stem cells (MDA-MB-435). The biotinylated control miRNA (biotin-control miRNA), miR-9 (biotin-miR-9), and miR-184 (biotin-miR-184) were used as controls. The proteins bound to miR-1 were separated using 12% SDS-PAGE and then identified by mass spectrometry. The arrow indicates the protein bound to miR-1. M, protein marker. (B) Detection of the protein bound to miR-1. The products of RNA pulldown were analyzed using western blot with anti-LRPPRC IgG. (C) The interaction between miR-1 and LRPPRC protein in melanoma stem cells. RNA crosslinking and immunoprecipitation (CLIP) assay using LRPPRC-specific antibody or goat anti-mouse IgG was conducted in melanoma stem cells. The immunoprecipitated complex was examined by SDS-PAGE with Coomassie staining. (D) miR-1 in the immunoprecipitated complex was determined using quantitative real-time PCR (**p < 0.01). (E) The co-localization of LRPPRC protein (green) with miR-1 (red). Melanoma stem cells were fixed, followed by labeling with LRPPRC-specific antibody and miR-1 probe. Nuclei were stained with DAPI. The Pearson coefficient of the signals for co-localization of miR-1 and LRPPRC protein was 0.4125. As a control, miR-9 (red) was included in the assays. The yellow shows the interaction of miR-1 and LRPPRC protein. Scale bars, 10 μm. (F) The direct interaction between miR-1 and RNA-binding domain (RBD) of LRPPRC. miR-1 or control miRNA was incubated with GST-RBD at different concentrations. The mutant of LRPPRC protein (ΔLRPPRC) lacking RBD was included in the assays. GST alone was used as a control. Subsequently, the mixture was separated by agarose gel, followed by staining with ethidium bromide (top). At the same time, the mixture was separated by SDS-PAGE and stained with Coomassie blue (bottom). The wedges show the concentration gradients of recombinant proteins used. Data shown are representative of three experiments performed.
Western blot data showed that the LRPPRC protein was detected in the product of RNA pulldown using miR-1, but not in that using control miRNAs (Figure 3B), indicating that the LRPPRC protein was specifically bound to miR-1. To confirm the binding of miR-1 to the LRPPRC protein in cells, RNA crosslinking and immunoprecipitation (CLIP) assays using the LRPPRC-specific antibody were performed in melanoma stem cells. SDS-PAGE results showed that the LRPPRC protein was immunoprecipitated (Figure 3C). miR-1 was detected in the immunoprecipitated complex of LRPPRC, but not in that of the control (immunoglobulin G [IgG]) (Figure 3D). At the same time, the confocal microscopy data demonstrated that the LRPPRC protein was co-localized with miR-1 but not the control miR-9 in the cells (Figure 3E). The Pearson coefficient of the signals for co-localization was 0.4125, showing the direct interaction of miR-1 with LRPPRC in vivo (Figure 3E). These findings indicated that miR-1 interacted with the LRPPRC protein in melanoma stem cells.
As reported, the RNA-binding domain (RBD) of LRPPRC is essential for conferring RNA binding activity.24 Therefore, the binding capacity of LRPPRC RBD to miR-1 was explored. Electrophoretic mobility shift assay (EMSA) data showed that miR-1 could bind to the recombinant glutathione S-transferase (GST)-RBD of LRPPRC in a dose-dependent manner, but not with the LRPPRC mutant (ΔLRPPRC) without RBD and GST alone (Figure 3F). At the same time, the control miRNA was not bound to GST-RBD (Figure 3F). The apparent dissociation constant for the binding of miR-1 with the LRPPRC RBD was estimated to be 58 ± 5 μM. These results demonstrated that miR-1 specifically interacted with the LRPPRC protein by binding to its RBD in MSCs.
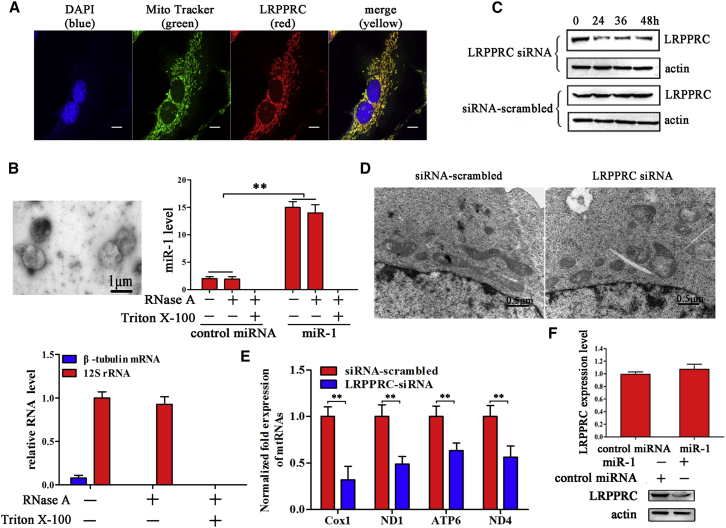
The Effects of miR-1-Protein Interactions on Mitochondrial Damage
To reveal the role of LRPPRC in mitochondria, the localization of LRPPRC protein in MSCs was evaluated. Confocal microscopy data showed that the LRPPRC protein was localized in mitochondria of cancer stem cells (Figure 4A), which was consistent with the previous report in tumor cells,23 suggesting that LRPPRC has a role in mitochondria. To better understand whether LRPPRC could mediate the localization of miR-1 into mitochondria, we isolated mitochondria from cancer stem cells after miR-1 transfection. The transmission electron microscope analysis showed the good integrity of isolated mitochondria (Figure 4B). After RNase or/and Triton X-100 treatment, the mitochondria were subjected to RNA extraction. Quantitative real-time PCR results showed that miR-1 was significantly increased after miR-1 transfection in the isolated mitochondria (Figure 4B). However, miR-1 was not detected after the isolated mitochondria were treated with RNase and Triton X-100 (Figure 4B). As a control, the cytosolic β-tubulin mRNA was not detected in the isolated mitochondria treated with RNase and/or Triton X-100 (Figure 4B). These data demonstrated that the interaction of miR-1 with LRPPRC could occur in mitochondria, resulting in mitochondrial damage.
Figure 4.
The Effects of miR-1-Protein Interactions on Mitochondrial Damage
(A) The co-localization of LRPPRC protein (red) with MitoTracker (green). Melanoma stem cells were incubated with MitoTracker and fixed, followed by labeling with LRPPRC-specific antibody. Nuclei were stained with DAPI. Scale bar, 10 μm. (B) The detection of miR-1 existed in mitochondria. Mitochondria were isolated from cancer stem cells using ultracentrifugation (left). After RNase or/and Triton X-100 treatment, the level of miR-1 in the isolated mitochondria was investigated using quantitative real-time PCR (right) (**p < 0.01). As controls, the cytosolic β-tubulin mRNA and the mitochondrial 12S rRNA were also detected. (C) The silencing of LRPPRC in melanoma stem cells. Melanoma stem cells were transfected with LRPPRC-specific siRNA. The siRNA-scrambled was included as a control. At different times after transfection, the expression level of LRPPRC was examined with western blot using LRPPRC-specific antibody. Actin was used as a control. (D) The influence of LRPPRC silencing on mitochondrial damage. The siRNA-treated melanoma stem cells were observed under an electron microscope. Scale bars, 0.5 μm. (E) Influence of LRPPRC silencing on mitochondrial transcripts of melanoma stem cells (**p < 0.01). (F) The effects of LRPPRC silencing on the miR-1 level in mitochondria. In the presence of LRPPRC-siRNA, miR-1 was transfected into melanoma stem cells. The level of mitochondrial miR-1 was detected using quantitative real-time PCR (**p < 0.01). (G) The impact of miR-1 overexpression (miR-1 OE) on the stability of LRPPRC protein. Melanoma stem cells were transfected with miR-1. At 36 h after transfection, the mRNA level (left) and protein level (right) of LRPPRC were determined using quantitative real-time PCR and western blot, respectively.
When the expression of LRPPRC was knocked down by LRPPRC-specific siRNA in MSCs (Figure 4C), mitochondrial damage was obviously observed in 30.5% of cancer stem cells (Figure 4D), and the mitochondrial transcripts were also downregulated (Figure 4E), showing that LRPPRC was required for mitochondrial function. In the presence of siRNA-LRPPRC, the mitochondrial miR-1 level of the miR-1-overexpressing melanoma stem cells did not change compared with the controls (Figure 4F), indicating that LRPPRC could not affect the miR-1 content in mitochondria.
To explore the influence of miR-1 on the stability of LRPPRC protein, the mRNA and protein levels of LRPPRC in miR-1-overexpressing cancer stem cells were examined. The results showed that miR-1 overexpression resulted in the decrease of LRPPRC protein level compared with the control, but it did not affect the LRPPRC mRNA level (Figure 4G). The sequence analysis revealed that there was no sequence of miR-1 complementary to the LRPPRC mRNA sequence. These data indicated that miR-1 overexpression might destabilize the LRPPRC protein in cancer stem cells, leading to the degradation of LRPPRC protein.
Taking the above data together, these findings revealed that miR-1 interacts with LRPPRC protein in mitochondria, and this interaction destabilizes LRPPRC protein, leading to mitochondrial damage in MSCs.
Mitophagy of Cancer Stem Cells Mediated by miR-1
To elucidate the mechanism underlying the miR-1-mediated inhibition of cancer stem cell growth, the cell cycle of cancer stem cells transfected with miR-1 and control miRNA was analyzed. Flow cytometry data showed that the number of cells at the G0/G1 phase in the miR-1-overexpressing cells was significantly increased compared with the control (Figure 5A). In order to explore the influence of miR-1-mediated downregulation of GPD1 and MINOS1 on cell cycle, the cell cycle of MINOS1-silenced or GPD2-silenced cells was examined. The results indicated that MINOS1 silencing or GPD2 silencing led to cell cycle arrest in the G0/G1 phase (Figure 5B), showing that the knockdown of MINOS1 and GPD2 contributed to miR-1-mediated cycle arrest in cancer stem cells.
Figure 5.
Mitophagy of Cancer Stem Cells Mediated by miR-1
(A) Influence of miR-1 on cell cycle of cancer stem cells. At 36 h after miR-1 overexpression, the cell cycle of melanoma stem cells (MSCs) or breast cancer stem cells (BCSCs) was evaluated with flow cytometry. (B) Impact of miR-1 target gene silencing on cell cycle. At 36 h after siRNA transfection, the cell cycle of cancer stem cells was evaluated with flow cytometry. (C) Detection of miR-1-mediated autophagy in cancer stem cells. Melanoma or breast cancer stem cells were transfected with miR-1 or control miRNA. Thirty-six hours later, the LC3 and PINK1 protein levels of cancer stem cells were examined by western blot. Actin was used as a control. (D) Effects of MINOS1 knockdown on mitophagy of cancer stem cells. MSCs or BCSCs were transfected with MINOS1-siRNA. As a control, MINOS1-siRNA-scrambled was included in the transfection. At 36 h after siRNA transfection, the LC3 and PINK1 expression levels of cancer stem cells were determined by western blot. Actin was used as a loading control. (E) Impact of GPD2 silencing on mitophagy of cancer stem cells. Cancer stem cells were treated with GPD2-siRNA or GPD2-siRNA-scrambled. Thirty-six hours later, western blot was conducted to detect LC3 and PINK1 proteins of cancer stem cells. (F) Role of LRPPRC in mitophagy of cancer stem cells. MSCs or BCSCs were transfected with LRPPRC-siRNA or LRPPRC-siRNA-scrambled. Thirty-six hours later, the LC3 and PINK1 expression levels of cancer stem cells were detected by western blot. Actin was used as a loading control. (G) The fluorescent colocalization of PINK1 with mitochondria. After miR-1 or different siRNAs treatment, the level of PINK1 was detected using a confocal microscope. Scale bars, 10 μm.
Our previous study demonstrated that miR-1 has no effect on apoptosis of cancer cells. The mitochondrial damage mediated by miR-1 destabilization of mitochondrial LRPPRC protein expression might induce mitophagy, a kind of autophagy. Therefore, the levels of LC3 (light chain 3), a homolog of the yeast autophagy marker ATG8, and PINK1 (PTEN-induced kinase 1), a protein marker of mitophagy, were examined in miR-1-overexpressing cancer stem cells. Western blots showed that miR-1 overexpression obviously elevated the levels of LC3-II and PINK1 proteins in breast or melanoma stem cells compared with the control (Figure 5C), indicating that miR-1 overexpression could induce mitophagy of cancer stem cells. As predicted, no autophagy-related gene was the target gene of miR-1, indicating that miR-1 could not directly regulate the expression of autophagy-related genes.
To explore the roles of miR-1’s targets in mitophagy, the autophagy level of MINOS1-silenced or GPD2-silenced cancer stem cells was evaluated. The results showed that the levels of LC3-II and PINK1 proteins of BCSCs and MSCs were significantly increased by MINOS1-siRNA or GPD2-siRNA compared with the controls (Figures 5D and 5E). The data demonstrated that MINOS1 silencing and GPD2 silencing could induce autophagy of cancer stem cells.
To investigate the influence of LRPPRC, the interactor of miR-1, on mitophagy, the autophagy level of cancer stem cells treated with LRPPRC-siRNA was examined. Western blot results indicated that LRPPRC silencing significantly increased the level of autophagy of BCSCs and MSCs (Figure 5F). Confocal microscopy data also showed an elevated signal of PINK1, indicating increased mitophagy in MSCs and BCSCs after miR-1 overexpression, MINOS1-siRNA, GPD2-siRNA, or LRPPRC-siRNA treatment.
The Rescue Effects Mediated by miR-1 Inhibition on Mitochondria
To evaluate the impact of miR-1 on tumorigenesis of MSCs in vivo, a stable clone of MSCs with miR-1 overexpression was generated using lentiviral transduction. Quantitative real-time PCR results showed that the expression level of miR-1 was significantly increased in stable miR-1-overexpressing MSCs (miR-1 OE) compared with the control (Figure 6A). The miR-1 overexpression significantly downregulated the expressions of miR-1 target genes (MINOS1 and GPD2) (Figure 6B). Furthermore, tumorsphere formation assays revealed that the tumorsphere-forming capacity of MSCs was significantly suppressed when miR-1 was overexpressed (Figure 6C).
Figure 6.
The Rescue Effects Mediated by miR-1 Inhibition on Mitochondria
(A) Expression level of miR-1 in miR-1-overexpressing melanoma stem cells. Melanoma stem cells were transfected with lentiviral particles containing miR-1. Lentiviral vector alone was used as a control. At 48 h after transfection, the expression level of miR-1 in cells was examined with quantitative real-time PCR (**p < 0.01). (B) Influence of miR-1 overexpression on the expression of target genes in melanoma stem cells. The expression levels of miR-1 target genes in miR-1-overexpressing melanoma stem cells were determined by qRT- PCR (**p < 0.01). (C) Impact of miR-1 overexpression on the tumorsphere formation capacity of melanoma stem cells. The tumorsphere formation capacity of miR-1-overexpressing melanoma stem cells was examined at different times. Scale bars, 10 μm. (D) Inhibition of miR-1 in stable miR-1-overexpressing melanoma stem cells. The miR-1 expression level was investigated using qRT-PCR after anti-miR-1 oligonuoleotide (AMO-miR-1) transfection (**p < 0.01). (E) Influence of miR-1 depletion on cell cycle of cancer stem cells. After AMO-miR-1 transfection, the cell cycle of stable miR-1-overexpressing cells (miR-1 OE) or control cells was evaluated with flow cytometry. (F) The rescued effects on mitophagy and target proteins. After AMO-miR-1 transfection, the protein levels of GPD2, MINOS1, LRPPRC, and mitophagy markers were investigated by western blot. Actin was used as a control. (G) The effects of miR-1 inhibition on the mitochondrial ultrastructure of melanoma stem cells. The stable miR-1 OE or control cells treated with specific AMO were imaged under transmission electron microscopy. Scale bars, 0.5 μm.
To further explore the miR-1-mediated tumor suppression, the expression of miR-1 was significantly inhibited in stable miR-1 overexpressing cells using anti-miR-1 oligonucleotide (AMO-miR-1) (Figure 6D). Cell cycle analysis showed that the number of cells in the G0/G1 phase in miR-1-overexpressing cells, in which miR-1 was stably expressed, was moderately decreased after AMO-miR-1 transfection at different concentrations, but could not return to the level of control group, indicating that the cell cycle arrest induced by miR-1 was not a reversible process (Figure 6E). Western blot results showed that AMO-miR-1 treatment recovered the target gene expression in stable miR-1-overexpressing cells (Figure 6F). At the same time, the level of mitophagy was also rescued, indicating that the mitophagy that occurred in MSCs was really induced by miR-1 (Figures 6F and 6G).
The Role of miR-1 in Tumorigenesis of Melanoma Stem Cells In Vivo
When miR-1-overexpressing MSCs were subcutaneously injected into nude mice, the growth rate of tumors in mice overexpressing miR-1 was significantly reduced compared with the control (Figure 7A). The tumor sizes of miR-1-overexpressing mice were much smaller than those of control mice (Figure 7B). These results indicated that miR-1 could negatively regulate tumorigenesis of cancer stem cells.
Figure 7.
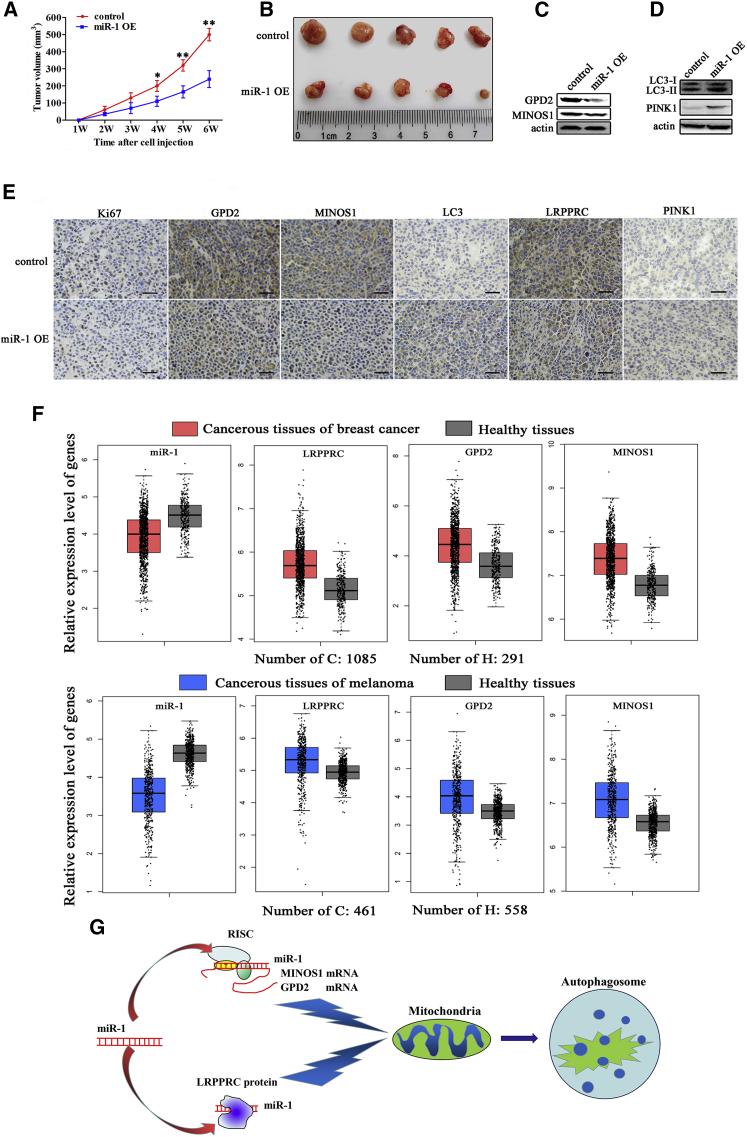
The Role of miR-1 in Tumorigenesis of Melanoma Stem Cells In Vivo
(A) Effects of miR-1 on solid tumor growth in mice. Melanoma stem cells overexpressing miR-1 or control cells were injected into nude mice. Two weeks later, the tumor volume in mice was measured every week. Each point represents the mean of five mice (*p < 0.1; **p < 0.01). (B) Evaluation of tumor size in miR-1-overexpressing mice. At 6 weeks after cell injection, the mice were sacrificed and the tumor volume was examined. (C) Western blot analysis of miR-1 targets in solid tumors of miR-1-overexpressing or control mice. Actin was used as a control. (D) Western blot analysis of LC3 and PINK1 proteins in solid tumors of miR-1-overexpressing or control mice. Actin was used as a control. (E) Immunohistochemical analysis of miR-1 targets mitophagy markers in solid tumors of miR-1-overexpressing or control mice. Brown represented Ki67, MINOS1, GPD2, LRPPRC, PINK1, or LC3 protein. Nuclei were stained with hematoxylin (blue). Scale bars, 50 μm. (F) The expression levels of miR-1 and its targets in melanoma and breast cancer tissues. Based on the GEO database, the expressions of miR-1 and its targets in the healthy and cancerous tissues of patients were evaluated. (G) Model for the miR-1-mediated mitophagy of cancer stem cells.
Western blot data of solid tumors showed that the expression levels of MINOS1, GPD2, and LRPPRC were significantly decreased in miR-1-overexpressing mice compared with the control (Figure 7C). Downregulations of GPD2, MINOS1, and LRPPRC were also observed in miR-1-overexpressing mice using immunohistochemical staining (Figure 7E), indicating that miR-1 suppressed the expression of its target genes in solid tumors. Based on the GEO database, it was found that miR-1 was significantly downregulated in the cancerous tissues of patients with breast cancer or melanoma compared with the corresponding healthy tissues, while the targets of miR-1 were significantly upregulated in the cancerous tissues (Figure 7F).
To explore the influence of miR-1-mediated mitochondrial damage on autophagy in vivo, the expression levels of LC3 and PINK1 proteins were determined using solid tumors. The results showed that the expression levels of LC3 and PINK1 were significantly increased in solid tumors of miR-1-overexpressing mice compared with the control (Figures 7D and 7E).
Taken together, the findings revealed that miR-1 overexpression could induce mitophagy of cancer stem cells through targeting MINOS1 and GPD2 genes and binding to LRPPRC protein to suppress cancer stem cell proliferation in vitro and in vivo (Figure 7G).
Discussion
Most cancer-related deaths occur after therapy resistance has emerged and/or after tumor cells have spread from the primary site. Characterized by high tumorigenic and metastatic capability, cancer stem cells have been advocated to constitute the sustaining force of tumorigenesis and tumor evolution.25 Thus the cancer stem cell-directed therapeutic approach may provide an attractive alternative in malignancies that are resistant to conventional chemotherapy.26 It has been found that cancer stem cells, having usually an altered metabolism relative to normal cells, have an enhanced reliance on mitochondrial metabolism.27 As reported, a subpopulation of dormant tumor cells, which has features of cancer stem cells, relies on mitochondrial respiration for survival.27,28 This renders these cancer stem cells susceptible to respiration inhibitors, providing a therapeutic strategy for delaying several cancer recurrences, i.e., pancreatic cancer. In invasive melanoma cells, oxidative phosphorylation, mitochondrial biogenesis, and the oxygen consumption rate are enhanced by upregulation of PGC-1α.29 MAPK inhibitor synergizes with a mitochondrion-targeted Hsp90 inhibitor and significantly decreases chemotherapy resistance and tumor recurrence in patients with melanoma tumors. Tigecycline, a mitochondrial translation inhibitor, selectively kills leukemia stem cells and progenitor cells compared to their normal counterparts.30 This may be attributable to the higher rate of mitochondrial biogenesis found in leukemic cells.31 In autochthonous models of breast and lung cancers, mitochondrial inhibition can suppress the proliferation of chemotherapy-resistant breast and lung tumor stem cells.32 These findings reveal the importance of mitochondria in cancer stem cells. In the present study, the results showed that the miR-1 overexpression could induce mitochondrial damage of BCSCs and MSCs by targeting MINOS1 and GPD2 genes and by binding to LRPPRC protein, leading to the suppression of cancer stem cell proliferation. Our data indicated that the mechanisms of miR-1-mediated destruction of mitochondria were consistent in two types of cancer stem cells (melanoma and breast cancer stem cells). However, the influence of miR-1 overexpression on mitochondria of other types of cancer cells remains unclear. This issue could be addressed in future investigations. The results of our study demonstrated that miR-1 was upregulated in cancer non-stem cells. However, the quantity of miR-1 in cancer non-stem cells was not enough to induce mitochondrial damage. The miR-1 overexpression could not induce the differentiation of cancer stem cells. In this context, our findings contribute to describe a novel role for miRNAs in cancer stem cell survival, and miR-1 could be proposed as a new mitochondrial inhibitor for restraining cancer progression. Growing evidence has indicated that cancer stem cells have more active mitochondria compared with tumor non-stem cells.28 The mitochondrial activity positively correlates with tumorigenic potential in cancer stem cells. Thus, the miR-1-induced mitochondrial damage of cancer stem cells revealed in this study would change the metabolism and suppress the tumorigenicity of cancer stem cells.
The underlying mechanisms of miRNAs have been well characterized in cytoplasm, but not in mitochondria.25 In cytoplasm, miRNAs can function through targeting complementary nucleotides in mRNA transcripts, usually in the 3′ UTR of an mRNA, resulting in mRNA degradation or destabilization.25 Therefore, the potential mitochondrial target genes of miR-1 were not explored in this study. In the present investigation, the findings revealed that miR-1 could directly bind the LRPPRC protein and target MINOS1 and GPD2 genes. Therefore, our study presented a novel mechanism of miRNA-mediated gene expression regulation that miR-1 could target the protein and genes to destroy mitochondria and induce mitophagy. MINOS1, a subunit of the mitochondrial inner membrane-organizing system complex, is preferentially localized at cristae junctions that connect the inner boundary membrane to lamellar cristae.33 The mitochondria of yeast with MINOS1 ablation display loss of inner membrane organization.21 As reported, GPD2 protein can catalyze the conversion of glycerol-3-phosphate to dihydroxyacetone phosphate, thus participating in the process of metabolism reprogramming in transformed cells.34 It has been found that the LRPPRC protein plays an important role in maintaining mitochondrial mRNA stability and mitochondrial translation.35 Interestingly, the documented data indicate that only Ago2 is imported into mitochondria, showing that miRNAs can function in mitochondria.36 It has been reported that miR-1 can be imported into mitochondria of myoblasts to promote the translation of some mitochondrial proteins.36 In our study, however, the miR-1 overexpression in cancer stem cells could induce mitochondrial damage by targeting MINOS1 and GPD2 genes and binding to LRPPRC protein. Our results indicated that the MINOS1, GPD2, and LRPPRC proteins were localized in mitochondria. The discrepancy of our results and previous data36 might result from the difference between myoblasts and cancer stem cells. In this context, our findings that miR-1 triggered mitophagy of cancer stem cells by binding to the LRPPRC protein and targeting MINOS1 and GPD2 mRNAs provided novel insights into the miRNA-mediated regulatory mechanism.
Materials and Methods
Sorting of Cancer Stem Cells and Non-Cancer Stem Cells
An ALDEFLUOR kit (STEMCELL Technologies, Durham, NC, USA) was used to isolate different cell populations according to the manufacturer’s instructions. Briefly, melanoma cells (MDA-MB-435) and breast cancer cells (MCF7), purchased from American Type Culture Collection (ATCC), were suspended in ALDEFLUOR assay buffer containing ALDH1 fluorescent substrate BODIPY-aminoacetate (BAAA, 1 μM) and then incubated for 40 min at 37°C. As a negative control, an aliquot of cells was treated with 50 mM ALDH1 inhibitor N,N-diethylaminobenzaldehyde (DEAB). After centrifugation at 250 × g for 5 min, the cells were resuspended in 0.5 mL of ALDEFLUOR assay buffer and stored on ice for fluorescence-activated cell sorting (FACS).
Cell Culture
Melanoma non-stem cells (MNSCs) were cultured in Leibovitz’s L-15 medium (Sigma, USA) supplemented with 10% fetal bovine serum (FBS) at 37°C with 100% humidified atmosphere. Breast cancer non-stem cells (BCNSCs) were cultured in DMEM basic medium (Sigma, USA) supplemented with 10% FBS at 37°C with 5% CO2 humidified atmosphere. Cancer stem cells were cultured in DMEM/F-12 medium (Invitrogen, USA) supplemented with 20 ng/mL epidermal growth factor (Beyotime Biotechnology, Jiangsu, China), 10 ng/mL basic fibroblast growth factor (Beyotime, China), 5 μg/mL insulin (Beyotime, China), and 2% B-27 (Sigma, USA) at 37°C in a humidified atmosphere with 5% CO2.
Tumorsphere Formation Assay
A tumorsphere formation assay was conducted under non-adherent and serum-free conditions. A single cell was plated into an ultra-low-adherent 96-well plate and cultured for 2 weeks in DMEM/F-12 medium (Invitrogen, USA) supplemented with 20 ng/mL epidermal growth factor (Beyotime Biotechnology), 10 ng/mL basic fibroblast growth factor (Beyotime, China), 5 μg/mL insulin (Beyotime, China), and 2% B-27 (Sigma, USA) at 37°C. The cells were examined under a light microscope. Subsequently, a tumorsphere was scattered and a scattered single cell was subjected to a tumorsphere formation assay. The assay was repeated three times.
Xenotransplantation Assay
The ALDH1-positive or ALDH1-negative cells were collected at 6 × 103 cells/mL in physiological saline (PBS). Matrigel (Becton Dickinson, USA) was added to the cell suspension at a ratio of 1:2. Subsequently, 250 μL of cell suspension was subcutaneously injected into non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice to induce tumor growth as described before.15 Forty days later, the mice were sacrificed to examine tumors. Animal experiments were approved by the Animal Experiment Center of Zhejiang University, China. All methods were carried out in accordance with the approved guidelines.
Quantitative Real-Time PCR of miRNA
Total RNAs were extracted from cells or tissues with a mirVana miRNA isolation kit (Ambion, USA). After treatment with DNase I, cDNA was reverse transcribed using a TaqMan miRNA reverse transcription kit (Applied Biosystems, USA). To quantify miR-1, quantitative real-time PCR was conducted according to the manufacturer’s instructions. U6 was used as an internal standard for normalization.
Northern Blot
RNAs, separated on a denaturing 15% polyacrylamide gel, were transferred to a Hybond-N membrane (Amersham Biosciences, Buckinghamshire, UK). The membrane was prehybridized in DIG Easy Hyb granule buffer (Roche, Basel, Switzerland) for 0.5 h after cross-linking with UV irradiation. Subsequently, the membrane was hybridized with a DIG-labeled probe (miR-1, 5′-DIG-ATACATACT TCTTTACATTCCA-3′; U6, 5′-DIG-AGTATATGTGCTGCCG AAGCGAGCAC-3′) for 12 h at 42°C. The signal detection was performed by following the DIG High Prime DNA Labeling and Detection Starter Kit II manual (Roche).
Overexpression and Silencing of miR-1 in Cells
Cells at 10%–30% confluence were transfected with the synthesized miR-1 using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The control miRNA and miR-1 were synthesized by GenePharma (Shanghai, China). At different times after transfection, the cells were collected for later use.
To silence miR-1 in miR-1-overexpressing cells, AMO-miR-1 was transfected into cells with Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) at a concentration of 100 nM. Prior to transfection, AMO (AMO-miR-1 or AMO-miR-1-scrambled) (100 nM) and Lipofectamine 2000 were mixed in Opti-MEM medium and then incubated for 20 min at room temperature. The mixture was then added to the cells. Six hours later, fresh medium replaced the spent culture medium, and the cells were cultured. The AMO-miR-1 sequence (5′-ATACATACTTCTTTACATTCCA-3′) was modified with locked nucleic acid, 2′-O-methyl, and phosphorothioate. The sequence of AMO-miR-1 was randomly scrambled, generating AMO-miR-1-scrambled (5′-CTGTGTTGATCCTG ATA-3′) with the same modifications as defined above. AMO-miR-1 and AMO-miR-1-scrambled were synthesized by Sangon Biotech (Shanghai, China). At different times after transfection, the cells were collected for later use.
Cell Viability Assay
Cell viability was monitored with 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) using a CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega, USA) according to the manufacturer’s protocol. Briefly, 20 μL of CellTiter 96 AQueous One Solution Reagent was added to the cells. The cells were incubated at 37°C for 3 h. Then, absorbance was measured at 490 nm using the iMark microplate reader (Bio-Rad, USA).
Transmission Electron Microscopy
To examine the mitochondrial morphology, cells were fixed with 2.5% (v/v) glutaraldehyde in phosphate buffer (0.14 M NaCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, pH 7.4). After centrifugation at 1,000 × g, the cells were postfixed with 1% OSO4 in phosphate buffer for 1–2 h. Then, the fixed cells were dehydrated in a graded ethanol series and embedded in Epon resin that was polymerized for 16 h at 60°C. Sections were mounted on copper grids and contrasted with uranyl acetate for 2–3 h at 42°C and then citrate for 20 min at room temperature. The samples were imaged with a FEI Tecnai G2 Spirit BioTWIN transmission electron microscope (Hillsboro, OR, USA) at an operating voltage of 120 kV.
Quantification of mRNA with Real-Time PCR
At 36 h after miR-1 transfection of cancer stem cells, total RNAs were extracted from the cells using an RNA isolation kit (Ambion, USA). Subsequently, the first-strand cDNA was synthesized by reverse transcription with a PrimeScript first-strand cDNA synthesis kit (Takara Bio, Japan). Quantitative real-time PCR was conducted with gene-specific primers (GAPDH [glyceraldehyde-3-phosphate dehydrogenase], 5′-GGACCTGACCTGCCGTCTAG-3′ and 5′-GTAGC CCAGGATGCCCTTGA-3′; COX1, 5′-GAGAAA TGAATGAGCCTACAGA-3′ and 5′-TACACCCTAGACCAAACTTACG-3′; ND1, 5′-CGATTCCGCTACGACCAACT-3′ and 5′-AG GTTTGAGGGGGAATGCTG-3′; ATP6, 5′-GGGCGCA GTGATTATAGGCT-3′ and 5′-TAAGGGGTGTAGGTGTGCCT-3′; GPD2, 5′-GGCTTCCAGATACCCTTCCTT-3′ and 5′-TGTT GATGTTCAGCGTGTATTAGAG-3′; MINOS1, 5′-AGGATTCTTCCCCTGCTAATA-3′ and 5′-GAACTGCTT CCACCTCGTAAT-3′; ND4, 5′-ACAAGCTCCATCTGC CTACG-3′ and 5′-GCTTCAGGGGGTTTGGATGA-3′). The reaction mixture contained 5 μL of SYBR Green PCR Master Mix (Takara Bio, Japan), 0.4 μL of 10 μM forward and reverse primers, and 1 μL of cDNA at a final volume of 10 μL. The PCR was carried out at 95°C for 5 min, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. GAPDH was used as an internal standard for normalization.
Mitochondrial Membrane Potential (mtΔΨ) Analysis
A mitochondrial membrane potential assay kit with JC-1 (Beyotime Institute of Biotechnology, China) was used to investigate mtΔΨ according to the manufacturer’s protocol. At 36 h after miR-1 transfection, cells were incubated with JC-1 dye for 20 min and then subjected to flow cytometry analysis. JC-1 aggregates formed red fluorescence in the cells with high mtΔΨ, while the JC-1 monomer formed green fluorescence in the cells with low mtΔΨ. The value of mtΔΨ was indicated as the ratio of red fluorescence intensity to green fluorescence intensity. All experiments were performed in triplicate.
Target Gene Prediction of miR-1
The putative target genes of miR-1 were predicted using the miRanda, TargetScan, and PicTar algorithms. The overlapped targets of three algorithms were the potential target genes of miR-1.
Dual-Luciferase Reporter Assay
The 3′ UTR of target gene was amplified with sequence-specific primers (GPD2, 5′-GAAGAGCTCGTCTGGGCAGTAAATCCA-3′ and 5′-GACCTCGAGAGAACTCATTTAGAATAAGG-3′; MINOS1, 5′-AGGCT CGAGCTGGAACCAAATCCAAGGAA-3′ and 5′-GGCGTCGACAAACTACATC GTCATCTCCT-3′). As controls, the 3′ UTRs of target genes of miR-1 were mutated by PCR using sequence-specific primers (GPD2, 5′-AAAGCGACTAAAA CTTTAAGGT-3′ and 5′-AGTTTTAGTCG CTTTTCTAGTGTTAAAAAAG-3′; MINOS1, 5′-TTGGAGCCATATGTACCCACTAGGGGAAGC-3′ and 5′-ACATAT GGCTCCAAAGTCATTCACCTAAAT-3′), generating a GDP2 3′ UTR mutant and MINOS1 3′ UTR mutant, respectively. The wild-type and mutant 3′ UTRs were cloned into the pmirGLO Dual-Luciferase miRNA target expression vector (Promega, USA). Then, 50 nM synthesized miR-1 or control miRNA was co-transfected with 2 μg of 3′ UTR or 3′ UTR mutant into melanoma stem cells using Lipofectamine 2000. At 36 h after transfection, the luciferase activity of cells was measured using the Dual-Luciferase reporter assay system (Promega, USA) according to the manufacturer’s protocol.
Co-localization of Protein and Mitochondria in Cells
Cells were plated in a glass-bottomed dish (In Vitro Scientific, USA) and cultured overnight. Then, the cells were incubated with 1 μM mitochondrial tracker (Yeasen Biotech, Shanghai, China) that can label mitochondria at 37°C for 30 min. After fixing in 4% paraformaldehyde for 15 min, the cells were permeabilized with 0.1% Triton X-100 for 20 min. Subsequently, the cells were incubated with a primary antibody against target protein at room temperature for 2 h, followed by incubation with a secondary antibody labeled with a fluorophore for 30 min. All antibodies used were purchased from Abcam (USA). The nuclei were labeled with DAPI for 5 min at room temperature. Images were captured with a laser-scanning microscope. The Pearson coefficient of the signals for co-localization was calculated using ImageJ software (https://imagej.en.softonic.com). The ImageJ program provided a Pearson’s correlation and overlap coefficient of co-localization that was a method to measure the degree of co-localization of objects in confocal dual-color images. The higher was the coefficient value, the better was the co-localization of two molecules.
RNAi Assay
To silence the expression of target genes in cancer stem cells, an RNAi assay was conducted using sequence-specific siRNAs (LRPPRC-siRNA, 5′-GGUGCCAGCAAG AUUCUUG-3′; GPD2-siRNA, 5′-GCAUUUCAGAACCAGUUAATT-3′; MINOS1-siRNA, 5′-CCAACUGUCAGCAUGAUUUTT-3′). The melanoma stem cells (1 × 105) were transfected with 50 nM siRNA with Lipofectamine 2000 (Invitrogen, USA). As a control, LRPPRC-siRNA-scrambled (5′-GUGUGGGAUACAACUUCCA-3′), GPD2-siRNA-scrambled (5′-GUAGUCAACCACGAGAAUAGT-3′), or MINOS1-siRNA-scrambled (5′-CGUACAGGCUGGAAGAGCUAT-3′) was included in the transfection. All siRNAs were synthesized by Shanghai GenePharma (Shanghai, China). At different times after transfection, the cells were harvested for later use.
Western Blot
The proteins were separated by SDS-PAGE and then electrotransferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, USA). After incubation in blocking solution (5% skim milk) for 2 h, the membrane was incubated with a primary antibody overnight at 4°C. Subsequently, alkaline phosphatase-conjugated secondary antibody (Roche, Switzerland) was added and incubated for 2 h at room temperature. The membrane was rinsed and then incubated with 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitroblue tetrazolium (NBT) substrate (Sangon, China) until the blot was visualized.
RNA Pulldown Assay
Cancer stem cells (5 × 106) were lysed using immunoprecipitation lysis buffer (Beyotime, China) containing 2 mM protease inhibitor. The cell lysate was incubated with streptavidin-conjugated Dynabeads (Life Technologies, USA) at 4°C for 1 h to remove the proteins non-specifically bound to beads. After centrifugation at 300 × g for 5 min, the cell lysate was incubated with biotin-labeled miR-1 (GenePharma, China) at 4°C overnight. Subsequently, the mixture was incubated with streptavidin-conjugated Dynabeads on a rotator for 2 h at 4°C. The beads were washed with lysis buffer and then boiled in protein loading buffer (Sangon, China). The proteins were separated on a 12% polyacrylamide gel and stained with Coomassie brilliant blue (Beyotime, China). The separated proteins were identified by mass spectrometry.
CLIP Assays
Cells were irradiated in a UV cross-linker for 400 mL/cm2 and then for 200 mJ/cm2. After centrifugation at 300 × g for 5 min, the cells were resuspended in immunoprecipitation lysis buffer (Beyotime, China), followed by incubation with DNase (Generay, Shanghai, China) at 37°C for 20 min. The lysate was centrifuged at 10,000 × g for 10 min to remove the cell debris and then precleared by incubation with protein A Dynabeads (Life Technologies, USA) at 4°C for 60 min. The supernatant was incubated with anti-LRPPRC IgG or goat anti-mouse IgG (Beyotime, China) at 4°C overnight, followed by incubation with protein A Dynabeads at 4°C for 2 h. After washes with PBST (0.5% Tween 20 in PBS), the beads were subjected to 12% SDS-PAGE with Coomassie staining. At the same time, the RNAs in the precipitated complex were extracted using a mirVanaP miRNA isolation kit (Ambion Thermo, USA) and analyzed by quantitative real-time PCR.
EMSAs
The RBD of LRPPRC protein or the mutant of LRPPRC protein (ΔLRPPRC) lacking RBD was cloned into pGEX-6p-2 vector downstream of GST using sequence-specific primers (5′-GGATCCACA GAACCTGATTTCCAGAA-3′ and 5′-GGCCTCGAGAG AAGAGTTTTCCCTCAA TT-3′). The recombinant GST-RBD protein was purified. To explore the binding of miR-1 to RBD of LRPPRC protein, 40 mM synthesized mature miR-1 (GenePharma, China) was incubated with GST-RBD in EMSA buffer (Beyotime Biotechnology, Shanghai, China) for 30 min at room temperature. GST alone was included in the incubation as a control. Subsequently, the mixture was separated on a 1% agarose gel at 100 V for 0.5 h. RNAs were stained with ethidium bromide. At the same time, the mixture was separated by SDS-PAGE, followed by staining with Coomassie brilliant blue to detect the proteins.
Isolation of Mitochondria
A mitochondria isolation kit (Beyotime Biotechnology, China) was used to isolate mitochondria. Melanoma stem cells were collected by centrifugation, resuspended in TE buffer (10 mM Tris-HCl [pH 7.4], 20 mM EDTA), and homogenized in a prechilled Dounce homogenizer. Mitochondria were sedimented at 13,000 × g for 10 min, washed once in the same buffer, and further purified by centrifugation at 40,000 × g for 1 h at 4°C on a sucrose gradient (17%, 31%, 42%, and 50%) in TE buffer.
Cell Cycle Assay
FACS analysis was used to examine the cell cycle of melanoma and breast cancer stem cells. Cell samples were fixed in ice-cold ethanol overnight. Then, the cells were treated with DNase-free RNase A (20 μg/mL) for 30 min. After centrifugation at 500 × g for 5 min, the cells were stained with propidium iodide (PI) (50 μg/mL). The fluorescence intensity of 1 × 105 cells was measured with a flow cytometer at an excitation wavelength of 488 nm.
Lentiviral Constructs Stably Overexpressing miR-1
miR-1 with 250 bp of flanking sequence was amplified from human genomic DNA using sequence-specific primers (5′-AACGGATCCGAGAGATGGATTCAGG GATGGA-3′ and 5′-ACAGAATTCTGTCTGGTGAGCACTTCCACCTGC-3′) and then cloned into the lentiviral vector pSMPUW-miR-Puro (Cell Biolabs, USA). Subsequently, the lentiviral vector expressing miR-1 was transfected into 293T cells using Lipofectamine 2000 reagent (Life Technologies, USA). Lentiviral vector alone was included in the transfection as a control. At 48 h after transfection, the viral particles were collected to infect melanoma stem cells. Melanoma stem cells overexpressing miR-1 were selected with 4 μg/mL puromycin.
Tumorigenicity in Nude Mice
The miR-1-overexpressing cells were collected at 6 × 106 cells/mL in physiological saline. Matrigel (Becton Dickinson) was added to the cell suspension at a ratio of 1:2. Then, 250 μL of cell suspension was subcutaneously injected into BALB/c nude mice to induce tumor growth. The tumor volume was examined weekly. Six weeks later, the nude mice were sacrificed and their tumor sizes were evaluated. Animal experiments were approved by the Animal Experiment Center of Zhejiang University, China. All methods were carried out in accordance with the approved guidelines.
Immunohistochemical Analysis
To examine proteins in solid tumors of mice by immunohistochemical staining, a 5-μm-thick section was placed on a precoated slide with 3-triethoxysilylpropylamine (Merck, Darmstadt, Germany). The slide was soaked in xylol for 1 h and washed in a series of decreasing alcohol concentrations. After deparaffinizing the tissue, antigen retrieval of the section was performed in a microwave for 5 min in TEC buffer (0.05 M Tris-HCl, 0.05 M EDTA, 0.02 M sodium citrate, pH 7.8), followed by peroxidase blocking. Then, the slide was incubated with a primary antibody for 12 h and a subsequent incubation with the biotinylated secondary antibody (Vector, Grünberg, Germany) for 30 min. The slide was stained with diaminobenzidine (DAB) (Sigma, USA) for 10 min to label proteins and then counterstained with hematoxylin to label nuclei.
Statistical Analysis
All assays were biologically repeated three times. Numerical data were analyzed using a one-way ANOVA. The statistical significance between two treatments was determined by Student’s t test.
Author Contributions
X.Z.: conceptualization, funding acquisition, project administration, supervision, writing, and editing. S.Z.: data curation, validation, formal analysis, methodology, validation, resources, software, and writing. C.L.: data curation, methodology, validation, and resources.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by China Ocean Mineral Resources R&D Association (DY135-B-04) and the National Key Research and Development Program of China (2018YFC0310703).
References
- 1.Rycaj K., Tang D.G. Cell-of-origin of cancer versus cancer stem cells: assays and interpretations. Cancer Res. 2015;75:4003–4011. doi: 10.1158/0008-5472.CAN-15-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takebe N., Harris P.J., Warren R.Q., Ivy S.P. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat. Rev. Clin. Oncol. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 3.de Almeida R.A., Fraczek M.G., Parker S., Delneri D., O’Keefe R.T. Non-coding RNAs and disease: the classical ncRNAs make a comeback. Biochem. Soc. Trans. 2016;44:1073–1078. doi: 10.1042/BST20160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman M.A., Hammond S.M. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Vito C., Riggi N., Cornaz S., Suvà M.L., Baumer K., Provero P., Stamenkovic I. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell. 2012;21:807–821. doi: 10.1016/j.ccr.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Bu P., Wang L., Chen K.Y., Srinivasan T., Murthy P.K., Tung K.L., Varanko A.K., Chen H.J., Ai Y., King S. A miR-34a-Numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18:189–202. doi: 10.1016/j.stem.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang W.L., Jiang J.K., Yang S.H., Huang T.S., Lan H.Y., Teng H.W., Yang C.Y., Tsai Y.P., Lin C.H., Wang H.W., Yang M.H. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014;16:268–280. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- 8.Cui Y., Huang T., Zhang X. RNA editing of microRNA prevents RNA-induced silencing complex recognition of target mRNA. Open Biol. 2015;5:150126. doi: 10.1098/rsob.150126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Ren Q., Huang X., Cui Y., Sun J., Wang W., Zhang X. Two white spot syndrome virus MicroRNAs target the Dorsal gene to promote virus infection in Marsupenaeus japonicus shrimp. J. Virol. 2017;91:e02261-16. doi: 10.1128/JVI.02261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shu L., Zhang X. Shrimp miR-12 suppresses white spot syndrome virus infection by synchronously triggering antiviral phagocytosis and apoptosis pathways. Front. Immunol. 2017;8:855–860. doi: 10.3389/fimmu.2017.00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King I.N., Yartseva V., Salas D., Kumar A., Heidersbach A., Ando D.M., Stallings N.R., Elliott J.L., Srivastava D., Ivey K.N. The RNA-binding protein TDP-43 selectively disrupts microRNA-1/206 incorporation into the RNA-induced silencing complex. J. Biol. Chem. 2014;289:14263–14271. doi: 10.1074/jbc.M114.561902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eiring A.M., Harb J.G., Neviani P., Garton C., Oaks J.J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C.J. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen A.K., Sengupta P., Waki K., Van Engelenburg S.B., Ochiya T., Ablan S.D., Freed E.O., Lippincott-Schwartz J. MicroRNA binding to the HIV-1 Gag protein inhibits Gag assembly and virus production. Proc. Natl. Acad. Sci. USA. 2014;111:E2676–E2683. doi: 10.1073/pnas.1408037111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C., Zhang S., Wang Q., Zhang X. Tumor suppressor miR-1 inhibits tumor growth and metastasis by simultaneously targeting multiple genes. Oncotarget. 2017;8:42043–42060. doi: 10.18632/oncotarget.14927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han C., Yu Z., Duan Z., Kan Q. Role of microRNA-1 in human cancer and its therapeutic potentials. BioMed Res. Int. 2014;2014:428371. doi: 10.1155/2014/428371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang Y.S., Chen W.Y., Yin J.J., Sheppard-Tillman H., Huang J., Liu Y.N. EGF receptor promotes prostate cancer bone metastasis by downregulating miR-1 and activating TWIST1. Cancer Res. 2015;75:3077–3086. doi: 10.1158/0008-5472.CAN-14-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y.N., Yin J.J., Abou-Kheir W., Hynes P.G., Casey O.M., Fang L., Yi M., Stephens R.M., Seng V., Sheppard-Tillman H. miR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296–306. doi: 10.1038/onc.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paumard P., Vaillier J., Coulary B., Schaeffer J., Soubannier V., Mueller D.M., Brèthes D., di Rago J.P., Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh A., Happel C., Manna S.K., Acquaah-Mensah G., Carrerero J., Kumar S., Nasipuri P., Krausz K.W., Wakabayashi N., Dewi R. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J. Clin. Invest. 2013;123:2921–2934. doi: 10.1172/JCI66353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alkhaja A.K., Jans D.C., Nikolov M., Vukotic M., Lytovchenko O., Ludewig F., Schliebs W., Riedel D., Urlaub H., Jakobs S., Deckers M. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol. Biol. Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valadi A., Granath K., Gustafsson L., Adler L. Distinct intracellular localization of Gpd1p and Gpd2p, the two yeast isoforms of NAD+-dependent glycerol-3-phosphate dehydrogenase, explains their different contributions to redox-driven glycerol production. J. Biol. Chem. 2004;279:39677–39685. doi: 10.1074/jbc.M403310200. [DOI] [PubMed] [Google Scholar]
- 23.Mourier A., Ruzzenente B., Brandt T., Kühlbrandt W., Larsson N.G. Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet. 2014;23:2580–2592. doi: 10.1093/hmg/ddt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mili S., Piñol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA-binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 2003;23:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang A., Qin S., Schulte B.A., Ethier S.P., Tew K.D., Wang G.Y. MYC inhibition depletes cancer stem-like cells in triple-negative breast cancer. Cancer Res. 2017;77:6641–6650. doi: 10.1158/0008-5472.CAN-16-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank N.Y., Schatton T., Frank M.H. The therapeutic promise of the cancer stem cell concept. J. Clin. Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viale A., Corti D., Draetta G.F. Tumors and mitochondrial respiration: a neglected connection. Cancer Res. 2015;75:3685–3686. doi: 10.1158/0008-5472.CAN-15-0491. [DOI] [PubMed] [Google Scholar]
- 28.Viale A., Pettazzoni P., Lyssiotis C.A., Ying H., Sánchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez F., Lim J.H., Chim H., Bhalla K., Girnun G., Pierce K., Clish C.B., Granter S.R., Widlund H.R., Spiegelman B.M., Puigserver P. PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G., Frederick D.T., Wu L., Wei Z., Krepler C., Srinivasan S., Chae Y.C., Xu X., Choi H., Dimwamwa E. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Invest. 2016;126:1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagadinou E.D., Sach A., Callahan K., Rossi R.M., Neering S.J., Minhajuddin M., Ashton J.M., Pei S., Grose V., O’Dwyer K.M. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12:329–341. doi: 10.1016/j.stem.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janzer A., German N.J., Gonzalez-Herrera K.N., Asara J.M., Haigis M.C., Struhl K. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl. Acad. Sci. USA. 2014;111:10574–10579. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jans D.C., Wurm C.A., Riedel D., Wenzel D., Stagge F., Deckers M., Rehling P., Jakobs S. STED super-resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc. Natl. Acad. Sci. USA. 2013;110:8936–8941. doi: 10.1073/pnas.1301820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daoud H., Gruchy N., Constans J.M., Moussaoui E., Saumureau S., Bayou N., Amy M., Védrine S., Vu P.Y., Rötig A. Haploinsufficiency of the GPD2 gene in a patient with nonsyndromic mental retardation. Hum. Genet. 2009;124:649–658. doi: 10.1007/s00439-008-0588-3. [DOI] [PubMed] [Google Scholar]
- 35.Ruzzenente B., Metodiev M.D., Wredenberg A., Bratic A., Park C.B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–456. doi: 10.1038/emboj.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y., Huang J., Zhao X., Zhou J., Yan Y. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]