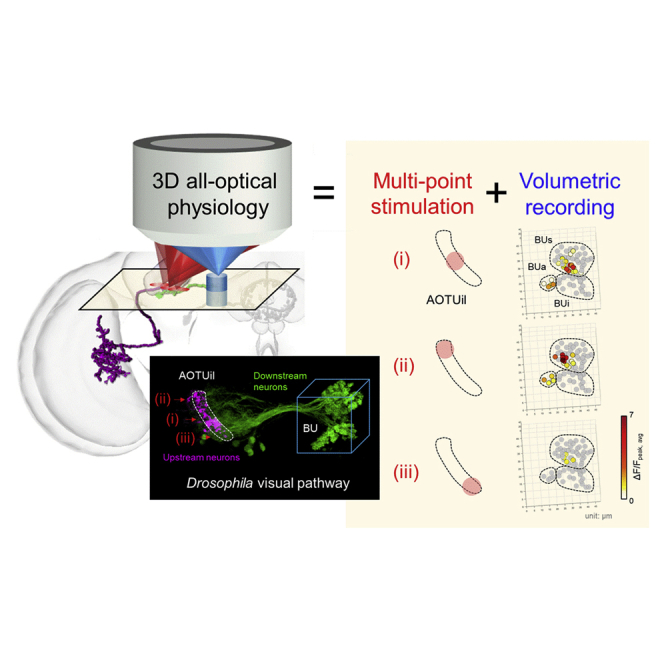
Summary
All-optical physiology (AOP) manipulates and reports neuronal activities with light, allowing for interrogation of neuronal functional connections with high spatiotemporal resolution. However, contemporary high-speed AOP platforms are limited to single-depth or discrete multi-plane recordings that are not suitable for studying functional connections among densely packed small neurons, such as neurons in Drosophila brains. Here, we constructed a 3D AOP platform by incorporating single-photon point stimulation and two-photon high-speed volumetric recordings with a tunable acoustic gradient-index (TAG) lens. We demonstrated the platform effectiveness by studying the anterior visual pathway (AVP) of Drosophila. We achieved functional observation of spatiotemporal coding and the strengths of calcium-sensitive connections between anterior optic tubercle (AOTU) sub-compartments and >70 tightly assembled 2-μm bulb (BU) microglomeruli in 3D coordinates with a single trial. Our work aids the establishment of in vivo 3D functional connectomes in neuron-dense brain areas.
Subject Areas: Neuroscience, Techniques in Neuroscience, Biological Sciences Research Methodologies, Biological Sciences Tools
Graphical Abstract
Highlights
-
•
All-optical volumetric physiology = precise stimulation + fast volumetric recording
-
•
Precise single-photon point stimulation among genetically defined neurons
-
•
3D two-photon imaging by an acoustic gradient-index lens for dense neural structures
-
•
Observation of 3D functional connectivity in Drosophila anterior visual pathway
Neuroscience; Techniques in Neuroscience; Biological Sciences Research Methodologies; Biological Sciences Tools
Introduction
It is now generally accepted that the function of the brain is based on the emergent property of its complex and numerous neuronal connections; for example, 100 trillion synaptic connections exist in a human brain. Since the era of Cajal and Golgi, mapping the interconnections between neurons (i.e., connectomes) has been a pursuit lasting more than a century (Alivisatos et al., 2012, Lichtman and Sanes, 2008). Although anatomical analyses using electron or optical microscopy have impressively revealed the structural connections among neurons (Lin et al., 2013, Takemura et al., 2013), they do not provide functional information encoded in these connections, such as the spike rates and precise times of calcium signals, types of connections (excitatory or inhibitory), or interaction strengths of the connections (Bargmann and Marder, 2013, Emiliani et al., 2015). Nevertheless, these functional details are critical to correctly understand the connectome.
To investigate functional connectivity among neurons, it is essential to be able to stimulate specific upstream neurons and record the activities of multiple downstream neurons. Conventionally, electrophysiology, which stimulates and records neurons with electrodes, is the method of choice, and is associated with excellent temporal resolution. However, it is impractical for simultaneous stimulation/recording of multiple neurons in a small volume (Mancuso et al., 2011). Conversely, with the aid of optogenetic tools, all-optical physiology (AOP) (Emiliani et al., 2015), which stimulates and records neurons with light, offers a non-invasive method to investigate multiple neurons with sub-micrometer spatial resolution (Scanziani and Häusser, 2009); therefore, AOP provides an opportunity to study functional connectomes in living animals (Hampel and Seeds, 2017, Olsen and Wilson, 2008).
In an AOP system, stimulation and recording need to be optimized separately and combined appropriately. For optical stimulation, various strategies have been demonstrated (Packer et al., 2013, Papagiakoumou, 2013), including different illumination geometries, such as wide-field (Bovetti et al., 2017) and point-scan (Wilson et al., 2012), as well as different stimulation mechanisms, such as single- (Akerboom et al., 2013) and two-photon stimulation (Papagiakoumou et al., 2010). For illumination geometries, the wide-field scheme irradiates all neurons within a large field of view, and the point-scan selectively stimulates a few neurons within its small focal volume, which allows for the more precise manipulation of a specific subpopulation of genetically defined neurons (Petreanu et al., 2007, Wang et al., 2007). For stimulation mechanisms, although two-photon stimulation provides desirable optical sectioning capabilities, special optical strategies such as serial scanning (Rickgauer and Tank, 2009) and beam enlargement (Hernandez et al., 2016, Papagiakoumou et al., 2010, Pégard et al., 2017) are typically required because the tiny focus volume (sub-femtoliter) of the two-photon laser is not sufficient to target enough opsin proteins for action potential generation (Peron and Svoboda, 2011). On the other hand, single-photon stimulation has higher efficiency and requires much lower power density than two-photon stimulation (Oron et al., 2012). Even though single-photon lacks optical sectioning ability, when combined with precise genetic expression of opsin, it is capable of providing localized stimulation without out-of-focus artifact in some cases (Jiao et al., 2018, Judkewitz et al., 2009).
For optical recording strategies, optical sectioning is a key requirement to ensure clear observation in thick tissues, such as the brain (Mertz, 2011). Several techniques are known to provide optical sectioning, including confocal (Conchello and Lichtman, 2005), light-sheet (Ahrens et al., 2013), HiLo (Lim et al., 2008), and two-photon microscopy (Denk et al., 1990). Confocal microscopy is conventionally used to achieve sectioned observations by point-scanning and pinhole filtering and is based on single-photon excitation that typically utilizes short wavelengths in the visible spectrum; therefore, its imaging depth is limited to less than 100 μm (Helmchen and Denk, 2005). Light sheet and HiLo microscopy are relatively new techniques that provide optical sectioning through orthogonal or structured illumination. However, owing to their wide-field detection schemes, scattering/aberration in deep tissues results in pixel cross talk (Yang and Yuste, 2017); thus, these techniques are typically adopted for transparent animals, such as zebrafish (Ahrens et al., 2013, Jiao et al., 2018, Lauterbach et al., 2015, Lim et al., 2011, Truong et al., 2011). Among the recording techniques, two-photon microscopy that relies on near-infrared excitation and point-scan detection could be a promising tool for deep-tissue imaging in highly scattering samples (Packer et al., 2015, Rickgauer et al., 2014).
Nevertheless, although two-photon optical sectioning allows for the observation of single layers deep inside brain tissues, it also poses difficulty for the high-speed monitoring of three-dimensionally (3D) distributed neural networks whose response times are in the order of milliseconds to seconds. Recently, researchers have developed various fast two-photon volumetric recording methods (Ji et al., 2016, Yang and Yuste, 2017) based on spatial light modulators (SLMs) (Yang et al., 2016), temporal focusing (Schrödel et al., 2013), piezo-controlled objectives (Göbel et al., 2007), acousto-optic deflectors (Reddy et al., 2008), electrically tunable lenses (ETLs) (Grewe et al., 2011), and ultrasound lenses (Dean and Fiolka, 2014, Duocastella et al., 2012, Duocastella et al., 2017, Olivier et al., 2009, Piazza et al., 2018, Zong et al., 2015). However, to the best of our knowledge, only multi-plane recording based on ETLs has been demonstrated in combination with optical stimulation (Dal Maschio et al., 2017, Mardinly et al., 2018, Yang et al., 2018). According to the results of previous studies, ETL requires a few milliseconds to shift the imaging plane; therefore, the axial sampling density is limited to a few separated layers. Such imaging density is sufficient to study the sparse distribution of large neurons, such as those in the hippocampus of mouse brains; however, for compactly distributed small neurons, such as neurons in Drosophila, high-density volume sampling that utilizes the full spatial resolution along the axial plane is necessary to allow for large-volume functional recording without the loss of subtle connection information.
In the current study, we adopted a high-speed, tunable acoustic gradient-index (TAG) lens (Hsu et al., 2017, Kong et al., 2015, Olivier et al., 2009, Zong et al., 2015) with an axial scanning speed that exceeded 100 kHz to achieve two-photon full-volumetric recordings with submicron/micron lateral/axial spatial resolution. Combined with an efficient single-photon point-scanning stimulation through a separate scanner, we constructed an AOP platform with precise stimulation and fast volumetric imaging. The system was applied to study functional connections in the anterior visual pathway (AVP) of Drosophila (Omoto et al., 2017). We chose Drosophila for connectomic mapping because their brains are complex (i.e., they contain 105 neurons) but they are also small enough to be completely mapped by optical microscopy with sub-cellular resolution. Furthermore, the genetic toolbox is more complete with Drosophila as compared with that of other animal models, and a comprehensive connectivity map based on in vitro structural registration of more than 30,000 cells has been established (Chiang et al., 2011), which serves as an invaluable reference for the study of functional connectomes in vivo. Here, we demonstrated the stimulation and single-depth recording inside a living Drosophila brain. Then, we performed full-volumetric recording and stimulation on the same brain to observe neuronal responses with high spatiotemporal resolution. Finally, through the combination of precise stimulation of subpopulations of genetically defined neurons and 3D high-speed recording of a large volume of downstream neurons, this platform allowed for the in vivo investigation of functional coding in the Drosophila brain visual pathway.
Results
Two Neuronal Populations in the Drosophila Anterior Visual Pathway
The AVP delivers visual information from the medulla (MED) to the anterior optical tubercle (AOTU) and then to the bulb (BU) in the Drosophila brain. Here, we focused on two populations of neurons in the AVP. One population consisted of MT neurons (neurons connect the MED to the AOTU), and the other consisted of TB neurons (neurons connect the AOTU to the BU) (Figures S1A–S1A″). Previous work has established that the AOTU is composed of three sub-compartments (intermediate medial AOTU [AOTUim], intermediate lateral AOTU [AOTUil], lateral AOTU [AOTUl]; see Figure S1A’) where TB neuronal dendrites arborize. The BU is an aggregate of tens of microglomeruli (Figure S1A″) where TB neuronal axons terminate (Omoto et al., 2017, Seelig and Jayaraman, 2013, Shiozaki and Kazama, 2017, Sun et al., 2017, Timaeus et al., 2017). Each microglomerulus has a diameter around 2–4 μm. The whole BU is about 35 μm in depth and about 70 μm beneath the frontal surface. The BU can also be divided into three sub-compartments: the superior BU (BUs), the anterior BU (BUa), and the inferior BU (BUi) (Figure S1A’’). Our AOP tool is used to study how the visual signal is transmitted (i.e., topographic functional coding) through MT and TB neurons between AOTU sub-compartments and BU microglomeruli. Previous anatomical studies have shown that these two neuron populations are highly stereotyped (Omoto et al., 2017, Timaeus et al., 2017). To investigate their functional coding patterns, optogenetic tools, such as CsChrimson, were expressed in MTil neurons (MT neurons innervating AOTUil [Omoto et al., 2017]) using the LexA system for stimulation, and GCaMP6f was expressed in TB neurons using the GAL4 system for mapping responses. By specifically stimulating the MTil neurons and spatiotemporally recording the axonal response of the TB neurons, we were able to construct the functional connections between AOTUil and BU microglomeruli.
All-Optical Physiology Scheme 1: Single-Point Stimulation and Single-Section Recording
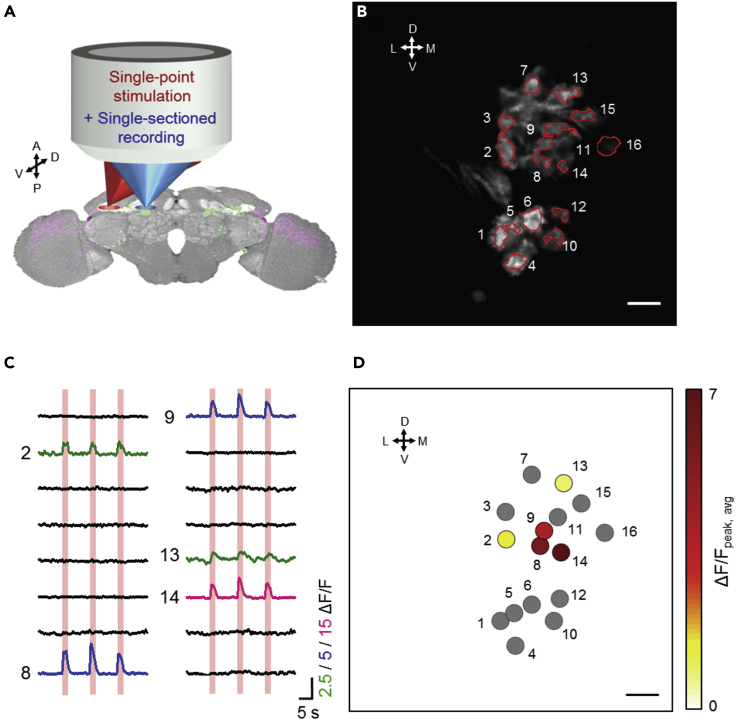
The concept of AOP within single-point stimulation and single-section recording is shown in Figure 1A. To achieve simultaneous stimulation and recording in different regions, we added a 638-nm stimulation beam with a customized, independent scanner beam path into a commercial 920-nm two-photon laser scanning microscope (see Figure S1B for setup). In AOP measurements, the stimulation spot is focused on the central AOTUil of MTil neurons with a 20% stimulation duty cycle (2 s on, 8 s off, repeated three times) with power <0.8 μW. The spot lateral size is 2 μm (Figure S1C) and axial size is ∼27 μm; 2 μm confinement of stimulation was verified in Figures S1D and S1E. Single optical sections of the TB neurons in the BU were recorded with a conventional 920-nm two-photon recording system with an average laser power of <2.8 mW to map the functional responses of microglomeruli. Figure 1B shows the distribution of microglomeruli at a single depth (∼70 μm beneath the frontal surface of the brain) that were observed with 0.5 μm lateral resolution and 2.75 μm axial section thickness (Figure S1F), which fit well with theoretical predictions (Zipfel et al., 2003). To determine the locations of microglomeruli, detailed BU structural images were acquired using low-speed scanning after functional imaging, and the structural images were next spatially registered to the functional images (Figures S2A–S2D, see Methods). After registration, the locations of individual microglomeruli were extracted automatically from low-speed images with the use of a home-built program based on fluorescence intensity (red circles in Figure 2B), whereas fiber structure and other neurons were excluded based on their location and morphology (Figures S2E and S2F, see Methods). Three repeated stimulations were applied, and the functional responses of all extracted microglomeruli are presented in Figure 1C. A generalized linear model (GLM [Miri et al., 2011]) was adopted to identify the stimulation-activated microglomeruli (Figure S2G, see Methods), whose response curves are shown in green/blue/red based on the level of changes in calcium concentrations (ΔF/F) (Figure 1C). After combining the spatiotemporal information, we constructed a 2D planar response strength map of activated microglomeruli (Figure 1D). Figure S1G verifies that sensory stimulation was avoided during optical stimulation.
Figure 1.
Single-Point Stimulation of the Intermediate-Lateral Anterior Optical Tubercle (AOTUil) with Single-Optical-Section Recording of the Bulb (BU) Microglomeruli
(A) Schematics of stimulation (red) and recording (blue) lasers whose focal points are independently controlled by two scanners in the all-optical physiology system.
(B) Single optical section (∼70 μm beneath the frontal surface of the brain) of microglomeruli of the BU. Red circles indicate the microglomeruli extracted by a home-built program based on neuronal fluorescence intensity and morphology. Detailed extraction procedure can be found in Figures S2E and S2F. Scale bar: 5 μm. Each extracted microglomeruli is labeled with a number from left to right.
(C) Neuronal temporal responses of the extracted microglomeruli with three repeated stimulations, which are represented by the red columns. The stimulation-activated microglomeruli are determined by the general linear model (GLM). Green/Blue/Red curves represent activated microglomeruli with ΔF/F scales equal to 2.5, 5, and 15, respectively, and black curves represent inactivated microglomeruli (scale same as blue curves).
(D) The spatial and functional response map of microglomeruli in a 2D plane. Inactivated microglomeruli are shown in gray, and the activated microglomeruli are presented as colors that represent their response strength (averaged peak ΔF/F of repeated stimulations). Scale bar: 5 μm.
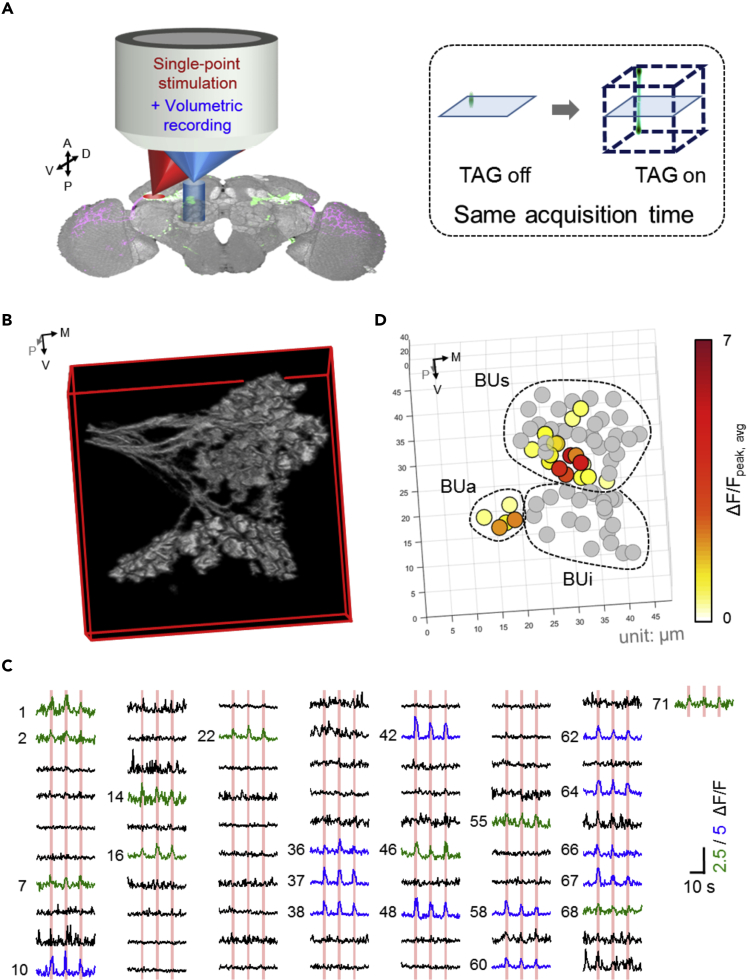
Figure 2.
Single-Point Optical Stimulation of Intermediate-Lateral Anterior Optical Tubercle (AOTUil) with Volumetric Recording of the Bulb (BU) Microglomeruli
(A) Left: all-optical physiology system combining a single-point stimulation beam (red) and a volumetric recording beam (blue). Right: the schematic of fast axial scan by a tunable acoustic gradient-index (TAG) lens that enables 3D recording with the same acquisition time as single-plane recording.
(B and C) (B) A full-volumetric image of microglomeruli of the BU (C) Temporal responses of each microglomerulus. The number-position correspondence can be found in Figure S3A. With general linear model (GLM) analysis, activated microglomeruli are shown as green/blue curves with ΔF/F scale equal to 2.5 and 5, respectively, and inactivated microglomeruli are shown as black curves (scale same as blue curves).
(D) The spatial and functional response map of microglomeruli in 3D. Inactivated microglomeruli are shown in gray, and activated microglomeruli are presented by color depending on their response strength. The subregions of BU (superior BU [BUs], inferior BU [BUi], and anterior BU [BUa]) are indicated by dashed circles.
Here we demonstrated simultaneous stimulation of MTil neurons at the center of the AOTUil and detailed single-section recording on the BU of TB neurons, which signifies that AOP with planar recording provides information regarding spatiotemporal functional connections and relative response strength. Nevertheless, similar to most neuronal structures in an intact brain, the microglomeruli of the BU are three-dimensionally distributed and extend to about a depth of 35 μm. Therefore, fast volumetric recording is required to perceive the complete spatiotemporal coding pattern in the AVP.
All-Optical Physiology Scheme 2: Single-Point Stimulation and Three-Dimensional Recording
The concept of single-point stimulation and volumetric recording is plotted in Figure 2A. Although stimulation was maintained at a single point, the recording plane was expanded into a 3D volume by introducing a high-speed axial scan lens (TAG lens, operating at 144 kHz scan rate) into the recording beam path without affecting the stimulation beam path (see Figure S1B for detailed setup information). With 1 m−1 dioptric power, the TAG lens provided an ∼40-μm axial recording range (Figure S1F), which was consistent with past theoretical studies (Duocastella et al., 2012, Hsu et al., 2017) and adequate to cover the whole BU structure. A 10-MHz data acquisition rate was used in the system to resolve the axial information acquired by the TAG lens. Considering that the 40-μm thickness was sampled twice during one period of TAG lens oscillation (144 kHz), 128 × 128 × 35 sampling voxels should have been obtained at ∼9 Hz. However, owing to the limited scan rate of the commercial microscope, the current volume rate was set to 2 Hz (see Methods), which is still fast enough to judge the neuronal calcium on/off response. The 35-optical-sectioned plane produced an axial digital resolution approaching 1 μm, which was better than the axial optical resolution (2.75 μm, Figure S1F). The power of the stimulation laser is maintained at the same level, whereas higher power is required for laser imaging in 3D recording since the voxel dwell time is much shorter compared with single-depth imaging. In principle, in 35-sectioned plane imaging, the power should increase to ∼6-fold (square root of 35) to achieve the same signal level as single-depth imaging. However, to avoid heating and photobleaching in a living brain, the power was controlled at < 8.2 mW, which is similar to other functional imaging works on GCaMP (Chen et al., 2013, Seelig and Jayaraman, 2013). Although the signal level of fast 3D imaging was not as high as single-plane imaging, with the aid of low-speed structural images (Figures S2A–S2D, see Methods), we can still establish structure-functional correlations of each neuron unit with high fidelity.
The results of AOP with volumetric recording are shown in Figures 2B–2D. Figure 2B shows complete 3D distribution of microglomeruli in the BU. The positions of 71 microglomeruli were extracted and labeled in a 3D coordinate system (see Figure S3A). Temporal responses of all the microglomeruli are presented in Figure 2C, where 22 stimulation-activated microglomeruli were quantitatively identified by the GLM and marked in green/blue colors based on response strength. The effectiveness of GLM analysis can be highlighted through microglomeruli #70 whose ΔF/F was large but not identified as activated because its responses did not temporally correlate with the repeated stimulations, resulting in a low statistical t-score (see Methods).
The relative response strength of activated microglomeruli are spatially color coded in Figure 2D. Three sub-compartments of the BU (BUs, BUa, BUi) are marked in dashed circles, indicating that most microglomeruli in the BUa and some microglomeruli in the BUs were activated following stimulation of MTil neurons. It is critical to note that the volumetric imaging capability enabled us to visualize and quantify the complete spatiotemporal response of a neural circuit connection.
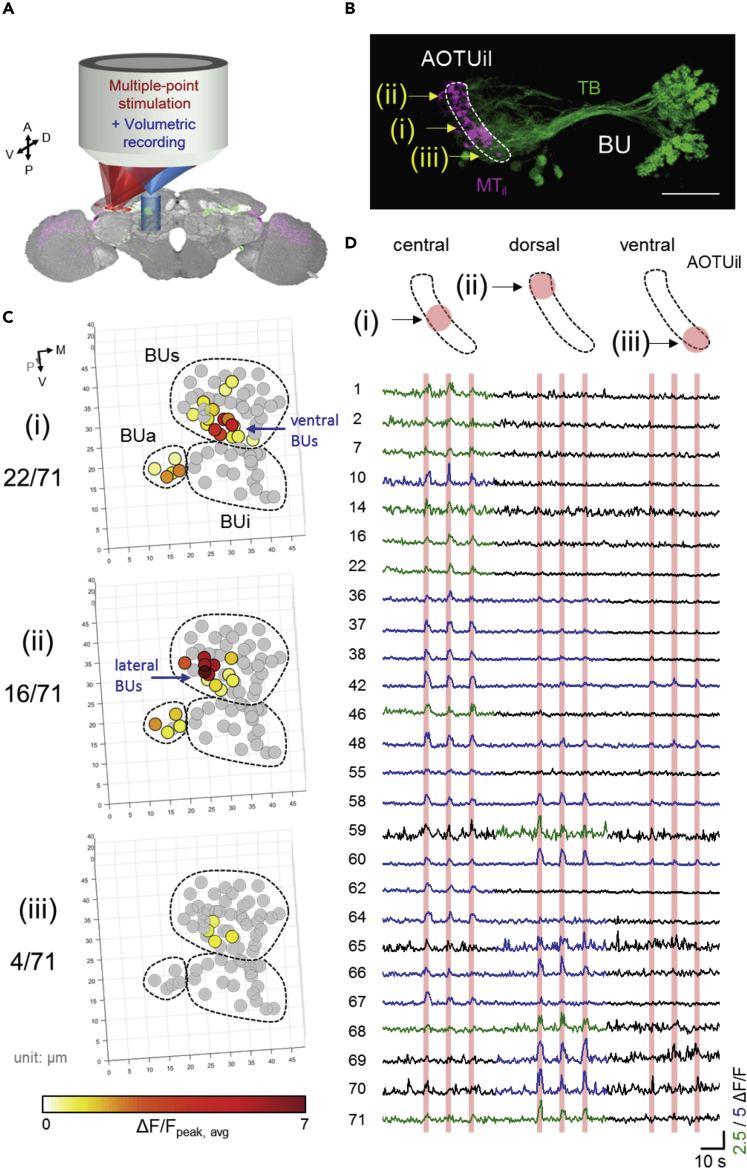
All-Optical Physiology Scheme 3: Multiple-Point Stimulation and Three-Dimensional Recording
In our system, point stimulation with an independent scanner provided the capability of position-dependent, precise stimulation on neurons. As shown by the conceptual illustration in Figure 3A, sequential point stimulation and volumetric recordings allowed for the complete mapping of a local functional connectome in the brain. In the previous two sections, the stimulation was directed only to the central AOTUil of MTil neurons (corresponding to region (i) in Figure 3B). Here we separately performed repeated stimulation on three regions of the AOTUil, including the (i) central, (ii) dorsal, and (iii) ventral AOTUil (Figure 3B), and recorded volumetric functional responses on the BU.
Figure 3.
Multiple-Point Optical Stimulation of the Intermediate-Lateral Anterior Optical Tubercle (AOTUil) with Volumetric Recording of the Bulb (BU) Microglomeruli
(A) Illustration of all-optical physiology with multiple-point stimulation (red) and volumetric recording (blue). The stimulation light is focused at different sites with volumetric recording at the downstream region.
(B) Stimulation strategy on upstream MTil neurons (neurons connecting medulla [MED] to AOTUil, magenta color) whose axon terminal AOTUil is marked with a dashed circle. Three sites of stimulation with equal spaces, including stimulation of the (i) central, (ii) dorsal, (iii) ventral AOTUil, are represented by yellow arrows. Scale bar: 20 μm.
(C) The volumetric functional response map of all microglomeruli in the BU corresponding to stimulation at the (i) central (same as Figure 2D), (ii) dorsal, and (iii) ventral AOTUil, respectively. Activated responses are determined by GLM and colored according to response strength. The numbers in the left show that 22, 16, and 4 of 71 microglomeruli were activated by stimulation of the central, dorsal, and ventral AOTUil, respectively. Microglomeruli located in the ventral and lateral BUs are indicated by the blue arrows.
(D) Temporal response of the microglomeruli that were activated by either stimulus (i, ii, or iii). Activated responses are marked in blue/green with ΔF/F scales equal to 2.5 and 5, respectively.
With whole volumetric recording and precise stimulation, we found that stimulation of each of the three regions activated a different number of microglomeruli with various strength distributions (the responses of all microglomeruli are presented in Figure S3B). The microglomeruli activated by each of the three stimulations are spatially represented in Figure 3C, color coded based on the strength of response. Stimulation at the (i) central and (ii) dorsal AOTUil activated many microglomeruli (22 and 16, respectively), whereas stimulation at the (iii) ventral AOTUil activated only four microglomeruli. This may be explained by the higher density of MTil axon terminals or higher amount of opsins expressed in the (i) central and (ii) dorsal AOTUil than in (iii) the ventral AOTUil (Figure 3B, magenta color).
We also observed the spatial distribution of activated microglomeruli. Stimulation at the (i) central and (ii) dorsal AOTUil activated microglomeruli in both BUa and in BUs (5 BUa and 17 BUs microglomeruli for (i); 4 BUa and 12 BUs microglomeruli for (ii)), whereas stimulation at (iii) the ventral AOTUil induced detectable Ca2+ signals only in BUs microglomeruli. Furthermore, we found that the spatial distribution of response strength following stimulation of the (i) central and (ii) dorsal AOTUil was different from that following the stimulation of the (iii) ventral AOTUil (i.e., ΔF/F in BUs). Specifically, in the presented fly with three-repeated stimulations, the relative response strength of ventral BUs was higher following stimulation of the (i) central AOTUil, whereas the relative response strength of lateral BUs was higher following stimulation of the (ii) dorsal AOTUil than the response strength of the ventral and lateral BUs following stimulation of the (iii) ventral AOTUil.
In Figure 3D, detailed responses from 26 selected microglomeruli that were activated by either of the three stimulations are shown. By comparing the changes in Ca2+ activity in the BU, these microglomeruli were further divided into four subgroups: (1) detectable activity change following stimulation at the (i) central AOTUil (Figure S3C); (2) detectable activity change following stimulation at the (ii) dorsal AOTUil (Figure S3D); (3) detectable activity change following stimulation at both the (i) central and (ii) dorsal AOTUil but not the (iii) ventral AOTUil (Figure S3E); (4) detectable activity change following stimulation of all three regions (Figure S3F). The results demonstrated that, with the capability of precise stimulation and 3D recording, we are able to identify important functional information including volumetric spatial distribution, temporal activation patterns, and response strengths of individual neuronal compartments. To further confirm the functional map we observed here, more replication of experiments on individual flies are necessary.
Discussion
The brain, including the neuronal circuits within it, is intrinsically 3D. However, current AOP platforms, which are based on conventional microscopic imaging tools, are mostly limited to 2D planar observations. In the current study, we established a 3D AOP platform combining precise stimulation and fast full-volumetric recording that offered 3D functional circuit interrogation in a living brain with high spatiotemporal resolution. In the following text, we compare the advantages and disadvantages between modern stimulation and recording methods and our system.
Currently, the most commonly adopted conventional method for the optogenetic stimulation of neurons is to illuminate the whole brain in a wide-field geometry with a light-emitting diode or a laser source (Bovetti et al., 2017). In wide-field illumination, neuron specificity is achieved via genetic labeling. More precise optical stimulation can be realized via scanning of a single-focus or multiple-focus illumination generated by digital micromirror devices or SLMs (Dal Maschio et al., 2017, Jiao et al., 2018, Mardinly et al., 2018, Yang et al., 2018). Here we adopted the former (single-focus scanning) because it is fully compatible with most commercial laser scanning microscopes and does not require extensive modifications (e.g., we only need to change the dichroic beam splitter inside the microscope to allow simultaneous stimulation and recording); conversely, it is technically difficult to implement multiple-focus illumination in an existing commercial system. In addition, there may be cross talk issues in the case of multiple stimulation foci. More importantly, we would like to note that quasi-simultaneous, multi-site stimulation can also be achieved by fast sequential scanning of the single focus in both lateral and axial directions by galvo mirrors (Wang et al., 2011) and ETL.
Both single-photon and two-photon setups have been extensively adopted for laser-scanning stimulation. In principle, the latter provides desirable optical sectioning and more penetration depth. However, in practice, optical sectioning with single-photon stimulation can be achieved with sparse and precise genetic labeling (Jiao et al., 2018, Judkewitz et al., 2009), which is one particular strength of using Drosophila. In this work, by only expressing opsin in MTil neurons (Figure S1A), we demonstrated precise single-photon stimulation on AOTU without artifacts from out-of-focus stimulation. Furthermore, in terms of depth, except for adopting special up-converting nanoparticles to achieve millimeter stimulation depth with a near-infrared laser (Chen et al., 2018), the current state-of-the-art two-photon stimulation extends only to a depth of 300 μm (Noguchi et al., 2011, Packer et al., 2015, Prakash et al., 2012, Ronzitti et al., 2017, Yang et al., 2018), whereas single-photon stimulation is capable of extending to a comparable depth of 250 μm in a living mouse (Ayling et al., 2009). In addition, because of the low two-photon stimulation efficiency, special optical strategies are typically required (Hernandez et al., 2016, Papagiakoumou et al., 2010, Pégard et al., 2017, Rickgauer and Tank, 2009). In contrast, the single-photon stimulation design that we adopted is less complex and requires less power density to achieve effective stimulation (Oron et al., 2012).
The volumetric recording capability was an important feature of our AOP platform. Compared with previous studies in microglomeruli with single-depth imaging (Omoto et al., 2017, Seelig and Jayaraman, 2013, Shiozaki and Kazama, 2017, Sun et al., 2017), fast volumetric imaging not only enables fast acquisition of 3D functional properties, but also allows for the comparison of the response between individual microglomeruli at different depths in a single trial.
In the current study, we adopted a TAG lens to achieve high-speed full volumetric recording, which is necessary for dense structures, such as the Drosophila BU. Recent studies employing AOP platforms have extended the recording region to three dimensions with fast acquisition speed, such as light-sheet microscopy, or integrated remote focusing and ETL into a laser scanning microscope. Light-sheet microscopy provides an excellent voxel throughput for whole brain imaging because of the camera-based detection strategy (Jiao et al., 2018). However, wide-field detection may suffer from cross talk between signals when applied to the opaque Drosophila brain, where scattering and aberration are strong (Hsu et al., 2019a). Conversely, remote focusing and ETL can be combined with scanning-based single-pixel detection and may be suitable for opaque samples. For remote focusing, by placing a movable light mirror after the auxiliary objective, the imaging plane can be tilted to align with an oblique neuron in 3D space (Anselmi et al., 2011) and can potentially achieve fast volumetric imaging with arbitrary trajectories (Botcherby et al., 2012). Fast multi-plane imaging can be achieved by refocusing the recording light at different depths for ETL (Dal Maschio et al., 2017, Mardinly et al., 2018, Yang et al., 2018). Nevertheless, the axial scan rates using the two approaches are limited so it is difficult to achieve full sampling. Specifically, the speed of remote focusing is limited by the inertia of optical elements., and the requirement of a few millisecond stabilization time of ETL also limits the recording to ∼ 3–10 discrete planes separated by 10 μm in depth. Although recording in trajectory or multiple planes with these methods (i.e., partial sampling) may be sufficient to obtain the dynamic response of large structures with sparse density (such as neuronal soma in a mouse brain), dense and full sampling is necessary for Drosophila BU microglomeruli, which are 2–4 μm in size, to avoid the loss of information. In our design, full 3D recording with femtoliter spatial resolution and sub-second temporal resolution is achieved with the aid of a TAG lens, unraveling the functional coding between densely packed BU microglomeruli and their upstream counterpart in the AVP.
For imaging opaque samples, other two-photon scanning-based, high-speed volumetric imaging strategies can also, potentially, be integrated in the AOP platform, such as random access microscopy (Katona et al., 2012, Reddy et al., 2008, Szalay et al., 2016), elongated point-spread function (PSF) (Lu et al., 2017, Song et al., 2017), and multiplexing techniques (Cheng et al., 2011, Yang et al., 2016). In the following text we discuss their respective advantages and disadvantages. First, random access microscopy allows high volume acquisition speeds from a few kHZ to tens of kHz, by moving focus quickly among several areas of interest in a sample with high-speed acousto-optic deflectors (Katona et al., 2012, Reddy et al., 2008, Szalay et al., 2016). However, the sparse sampling nature of this technique requires extremely high stability of brain, which is not easy for living animals. In addition, sparse sampling may result in information loss in dense structure in a dense tissue with highly diverse responses. Second, with elongated PSF (Lu et al., 2017, Song et al., 2017), video-rate (30 Hz) volumetric imaging can be achieved by simultaneously illuminating neurons at different depths and projecting the 3D morphology of neurons onto a 2D plane. However, when there are two or more neurons overlapped in the axial direction, information from different depths may be indistinguishable. Third, temporal multiplexing, i.e., scanning temporally delayed multiple beams at different depths, achieves a 60 Hz volume rate, whose voxel throughput can exceed the repetition rate of the imaging laser (Cheng et al., 2011). Nevertheless, the number of separated image planes is limited by laser pulse time interval divided by lifetime of fluorescent indicators. For example, with an 80-MHz pulsed laser (12.5-ns pulse interval) and fluorescence with 3-ns lifetime, only four planes are allowed with the multiplexing method. This is not enough for observation of dense structures such as BU microglomeruli. However, more imaging planes may be achieved by multiplexing techniques based on SLM, thus removing the limitations of the laser repetition rate or fluorescent lifetime. The number of planes is still limited by the total laser power, and only two imaging planes at 10-Hz acquisition speed have been demonstrated in a recent study (Yang et al., 2016). In short, for dense structures such as Drosophila BU, full 3D sampling with continuous axial imaging by using the TAG lens should be the most suitable technique among those currently available.
It is well known that TAG lens has been integrated into other fast volumetric systems in various configurations (Dean and Fiolka, 2014, Duocastella et al., 2012, Duocastella et al., 2014, Duocastella et al., 2017, Kong et al., 2015, Olivier et al., 2009, Piazza et al., 2018, Zong et al., 2015). In light-sheet microscopy, TAG lenses have been integrated in either the illumination (Dean and Fiolka, 2014, Zong et al., 2015) or the detection arm (Duocastella et al., 2017). By adding a TAG lens in the detection arm and combining acousto-optic deflectors in the illumination arm, inertia-free, i.e., all-acoustic-based, light-sheet microscopes have achieved outstanding volume rates up to 200 Hz (Duocastella et al., 2017). Yet, as mentioned earlier, wide-field detection in light-sheet microscopy makes the techniques more suitable for transparent samples such as C. elegans and paramecia. For imaging opaque adult Drosophila brains, combining TAG lenses with single-photon confocal or two-photon laser scanning microscopes should be a better choice (Duocastella et al., 2014, Kong et al., 2015, Olivier et al., 2009, Piazza et al., 2018). In our work, by integrating TAG lenses into a commercial two-photon laser scanning microscope, and adding a stimulation beam to the microscope, we demonstrated a combination of optical stimulation and TAG-lens-based fast volumetric imaging, i.e., volumetric AOP system, allowing 3D interrogation of neuronal dynamics in opaque and living Drosophila brains.
Our voxel throughput is now 10 Mvoxel/s. It is not the highest among current fast 3D imaging techniques. Tens of Mvoxel/s have been demonstrated by combining resonant scanning and multiplexing techniques (Cheng et al., 2011; Stirman et al., 2016), and tens to hundreds of Mvoxel/s have been achieved with light-sheet microscopy (Truong et al., 2011; Wolf et al., 2015; Zong et al., 2015). We have discussed the reasons why we did not adopt these techniques in this work in previous paragraphs.
In principle, the voxel speed of our TAG lens-based system can be enhanced to 80 MHz (equal to the repetition rate of the pulsed laser) (Kong et al., 2015). Nevertheless, there are two reasons that we chose the current voxel speed. First, higher sampling means higher axial digital resolution but not optical resolution. In our current setup, 10 MHz leads to an axial digital resolution of about 1 μm (35 layers in the range of 40 μm extension), which is smaller than the axial optical resolution (2.75 μm, Figure S1F). Therefore, increasing the sampling rate does not enhance image quality. If the axial extension range is beyond 100 μm, it would be necessary to use a higher sampling rate (Hsu et al., 2019a, Hsu et al., 2019b; Kong et al., 2015). Second, higher sampling rates reduce the signal-to-noise ratio. With an 80-MHz pulsed laser, fluorescent photons generated by eight pulses are integrated together as one voxel in our current system, leading to 10 Mvoxels/s data throughput with a reasonable signal-to-noise ratio.
Previous research has established that the Drosophila AVP plays a significant role in relaying spatial visual signals to the central brain for navigation behaviors (Omoto et al., 2017, Seelig and Jayaraman, 2013, Shiozaki and Kazama, 2017, Sun et al., 2017, Timaeus et al., 2017). The local topographic connections between the AOTU and BU sub-compartments (i.e., AOTUim, AOTUil, and AOTUl and BUs, BUi, and BUa, in the AVP) have been constructed based on anatomical methods, with one study demonstrating that the AOTUil connected with the BUa (Omoto et al., 2017) and another study reporting that the AOTUil is linked to the BUs (Timaeus et al., 2017). Now with our 3D AOP platform, we can investigate “function”-based connections among these compartments based on calcium activity in a living brain. The functional connections between the BU and AOTUil are presented in Figures 2 and 3. Figures 2 and 3 reveal that both dorsal and central AOTUil connect with BUs and BUa, but activation of ventral AOTUil only induced detectable transients in BUs. Although BUa and BUi do not display detectable Ca2+ signals following the activation of ventral and whole AOTUil, respectively, it should be noted that no Ca2+ response does not necessarily mean no connection. Sub-threshold excitatory or inhibitory functional connections may exist, but as long as they do not modulate intracellular calcium concentration, these connections are invisible in our current calcium-detection scheme. To further examine the connection between BU compartments and AOTUil, more studies involving electrophysiology or voltage-sensitive probes will be necessary.
Previous finding also indicates that there are smaller functional units, i.e., sub-regions, in AOTUil (Timaeus et al., 2017). However, currently no specific Gal4 drivers that target a single sub-region of the AOTUil for functional mapping is available. With precise stimulation of our 3D AOP platform, we can investigate the detailed functional connection pattern of AOTUil sub-regions and BU and link it to the structure-based map. The detailed calcium-dependent connection pattern of AOTUil sub-regions and BUs was distinguished in Figure 3. In terms of the number of activated microglomeruli, the result suggests that the central and ventral AOTUil activated more BUs microglomeruli than the dorsal AOTUil. We also identified the connection strength based on calcium activity (the ΔF/F level, whose peak is proportionally correlated with the number of action potential firings [Chen et al., 2013], indicating the strength of functional connection). The result reveals that following stimulation of the dorsal AOTUil, the ventral BUs showed a stronger connection than lateral BUs; conversely, following the stimulation of the central AOTUil, the lateral BUs showed stronger connection than ventral BUs, showing that the platform is a promising tool to provide detailed topographic coding of functional connectivity in the local AVP.
Previous studies have demonstrated that the topographical encoding of sensory information is a fundamental organization principle of neuronal circuits (Imai et al., 2010, Luo and Flanagan, 2007). In the past, the topographic analysis of neuronal diagrams of a circuit relied on the efforts of anatomical single-cell analyses (Fernandez et al., 2016, Omoto et al., 2017, Timaeus et al., 2017). However, our platform allows for functional, full-volume topographic dissection of neuronal circuits, paving the way toward single-trial, all-optical interrogation in living Drosophila.
This platform can also be used to study the functional connectome in other 3D neuronal structures because the axial range of volumetric recording can be tuned by the magnification of objectives and the optical power of TAG lenses (Duocastella et al., 2012). With fixed TAG lens optical power, lower magnification enables a larger axial recording range. For example, with 1 m−1 TAG lens optical power, 20× and 10× magnification of objectives lead to ∼160 and ∼640 μm axial recording range, respectively. Nevertheless, lower magnification typically means a lower numerical aperture (NA) of the objective, resulting in resolution degradation. Recently, objectives with large back apertures were introduced to allow for high NA with low magnification (for example, ZEISS W Plan-Apochromat 20×/1.0 objective). By combining TAG lenses with these objectives, high optical resolution with large recording volume is expected. However, even though the number of optically sectioned layers can be increased with a large axial extension range, the number of sectioned layers would still be limited by the repetition rate of the femtosecond laser and the signal-to-noise ratio of a high-speed data acquisition system.
Finally, although the current acquisition speed was only 2 volumes/s, a higher speed is achievable by changing the driving frequency of the TAG lens and the total scanning pixel number in the xy dimensions. For example, with a 500 kHz driving frequency (Zong et al., 2015) and 128×128 xy scanning pixels, a 31 Hz volume rate would be achieved. Note that, with higher driving frequency, the effective aperture of the TAG lens and signal-to-noise ratio in each z layer are both reduced. Volume acquisition rate can also be elevated to the kHz range using the ribbon-scanning strategy (Hsu et al., 2019b) and multiplex imaging (Yang et al., 2016).
In summary, we established a 3D AOP platform. Based on the combination of precise stimulation and fast full-volumetric imaging, 3D functional connectivity in Drosophila AVP was observed with high spatiotemporal resolution. These findings will aid in the non-invasive identification of 3D brain functional connectomes.
Limitations of the Study
There are three major limitations of our platform. First, the single-photon stimulation beam lacks optical-sectioning ability and out-of-focus stimulation may occur if opsin is not locally expressed. In such case, two-photon stimulation should be adopted. Second, the speed of shifting stimulation plane is limited in this work since we changed the stimulation plane by moving a lens in the telescope with a traditional translational stage. To achieve a higher shifting speed, an ETL with millisecond shifting speed can be adopted into the system. Third, the throughput in our recording system is ultimately limited by the repetition rate of the two-photon pulsed laser (i.e., 80 MHz). The throughput is also limited by the signal-to-noise ratio of the fluorescence and the detection system. To achieve higher throughput, pulsed laser with higher repetition rate, fluorescence with higher contrast and PMT with higher sensitivity should be adopted.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Yu-Chieh Lin, Ke Su, and Yung-Hsin Shih of NTHU and Yuan-Yao Lin of National Sun Yat-Sen University for initial test of the high-speed volumetric TAG recording. This work was financially supported by the Brain Research Center from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan and Ministry of Science and Technology (MOST) in Taiwan. S.-W.C., Y.-Y.L., and S.-C.W. received funding from MOST under grant MOST-108-2321-B-002-004, MOST-105-2628-M-002-010-MY4, MOST-107-2221-E-007-036-MY2, MOST-106-2221-E-007-059, and MOST104-2218-E-007-022-MY2. S.-W.C. also acknowledges the generous support from the Foundation for the Advancement of Outstanding Scholarship during 2015–2018.
Author Contributions
C.H., C.-Y.T., K.-P.Y. performed the experiments. C.-Y.T. prepared the fly strains and immunostaining sample. C.H. and K.-P.Y. built the optical part of the AOP platform. W.-K.C. assisted on mechatronic control of stimulation system, and K.-J.H. constructed data acquisition system of fast 3D imaging. C.-C.H. and S.-C.W. designed a program for high-speed 3D image reconstruction. C.H. characterized optical performance of the system and worked together with C.-C.H. and S.-C.W. on image analysis. C.H. and C.-Y.T. drafted the manuscript, polished by S.-W.C. and all co-authors. Y.-Y.L., A.-S.C., S.-W.C. initiated and conceived the project.
Declaration of Interests
The authors declare no competing interests.
Published: December 20, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.11.011.
Contributor Information
Yen-Yin Lin, Email: yylin@life.nthu.edu.tw.
Ann-Shyn Chiang, Email: aschiang@life.nthu.edu.tw.
Shi-Wei Chu, Email: swchu@phys.ntu.edu.tw.
Data and Code Availability
The raw data of Figures 1, 2, 3 and S1A as well as the code for 3D functional image analysis using the generalized linear model is available on Mendeley Data. doi: https://doi.org/10.17632/5t5kb6gc35.1.
Supplemental Information
References
- Ahrens M.B., Orger M.B., Robson D.N., Li J.M., Keller P.J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- Akerboom J., Carreras Calderón N., Tian L., Wabnig S., Prigge M., Tolö J., Gordus A., Orger M.B., Severi K.E., Macklin J.J. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alivisatos A.P., Chun M., Church G.M., Greenspan R.J., Roukes M.L., Yuste R. The brain activity map project and the challenge of functional connectomics. Neuron. 2012;74:970–974. doi: 10.1016/j.neuron.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmi F., Ventalon C., Bègue A., Ogden D., Emiliani V. Three-dimensional imaging and photostimulation by remote-focusing and holographic light patterning. Proc. Natl. Acad. Sci. U S A. 2011;108:19504–19509. doi: 10.1073/pnas.1109111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayling O.G., Harrison T.C., Boyd J.D., Goroshkov A., Murphy T.H. Automated light-based mapping of motor cortex by photoactivation of channelrhodopsin-2 transgenic mice. Nat. Methods. 2009;6:219–224. doi: 10.1038/nmeth.1303. [DOI] [PubMed] [Google Scholar]
- Bargmann C.I., Marder E. From the connectome to brain function. Nat. Methods. 2013;10:483–490. doi: 10.1038/nmeth.2451. [DOI] [PubMed] [Google Scholar]
- Botcherby E.J., Smith C.W., Kohl M.M., Débarre D., Booth M.J., Juškaitis R., Paulsen O., Wilson T. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc. Natl. Acad. Sci. U S A. 2012;109:2919–2924. doi: 10.1073/pnas.1111662109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovetti S., Moretti C., Zucca S., Dal Maschio M., Bonifazi P., Fellin T. Simultaneous high-speed imaging and optogenetic inhibition in the intact mouse brain. Sci. Rep. 2017;7:40041. doi: 10.1038/srep40041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Weitemier A.Z., Zeng X., He L., Wang X., Tao Y., Huang A.J., Hashimotodani Y., Kano M., Iwasaki H. Near-infrared deep brain stimulation via upconversion nanoparticle–mediated optogenetics. Science. 2018;359:679–684. doi: 10.1126/science.aaq1144. [DOI] [PubMed] [Google Scholar]
- Chen T.-W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V. Ultra-sensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A., Gonçalves J.T., Golshani P., Arisaka K., Portera-Cailliau C. Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nat. Methods. 2011;8:139–142. doi: 10.1038/nmeth.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang A.-S., Lin C.-Y., Chuang C.-C., Chang H.-M., Hsieh C.-H., Yeh C.-W., Shih C.-T., Wu J.-J., Wang G.-T., Chen Y.-C. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Conchello J.-A., Lichtman J.W. Optical sectioning microscopy. Nat. Methods. 2005;2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- Dal Maschio M., Donovan J.C., Helmbrecht T.O., Baier H. Linking neurons to network function and behavior by two-photon holographic optogenetics and volumetric imaging. Neuron. 2017;94:774–789. doi: 10.1016/j.neuron.2017.04.034. [DOI] [PubMed] [Google Scholar]
- Dean K.M., Fiolka R. Uniform and scalable light-sheets generated by extended focusing. Opt. Express. 2014;22:26141–26152. doi: 10.1364/OE.22.026141. [DOI] [PubMed] [Google Scholar]
- Denk W., Strickler J.H., Webb W.W. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Duocastella M., Sancataldo G., Saggau P., Ramoino P., Bianchini P., Diaspro A. Fast inertia-free volumetric light-sheet microscope. ACS Photon. 2017;4:1797–1804. [Google Scholar]
- Duocastella M., Sun B., Arnold C.B. Simultaneous imaging of multiple focal planes for three-dimensional microscopy using ultra-high-speed adaptive optics. J. Biomed. Opt. 2012;17:050505. doi: 10.1117/1.JBO.17.5.050505. [DOI] [PubMed] [Google Scholar]
- Duocastella M., Vicidomini G., Diaspro A. Simultaneous multiplane confocal microscopy using acoustic tunable lenses. Opt. Express. 2014;22:19293–19301. doi: 10.1364/OE.22.019293. [DOI] [PubMed] [Google Scholar]
- Emiliani V., Cohen A.E., Deisseroth K., Häusser M. All-optical interrogation of neural circuits. J. Neurosci. 2015;35:13917–13926. doi: 10.1523/JNEUROSCI.2916-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez D.C., Chang Y.-T., Hattar S., Chen S.-K. Architecture of retinal projections to the central circadian pacemaker. Proc. Natl. Acad. Sci. U S A. 2016;113:6047–6052. doi: 10.1073/pnas.1523629113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel W., Kampa B.M., Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat. Methods. 2007;4:73–79. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- Grewe B.F., Voigt F.F., van’t Hoff M., Helmchen F. Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens. Biomed. Opt. Express. 2011;2:2035–2046. doi: 10.1364/BOE.2.002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel S., Seeds A.M. Targeted manipulation of neuronal activity in behaving adult flies. In: Çelik A., Wernet M.F., editors. Decoding Neural Circuit Structure and Function. Springer; 2017. pp. 191–222. [Google Scholar]
- Helmchen F., Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Hernandez O., Papagiakoumou E., Tanese D., Fidelin K., Wyart C., Emiliani V. Three-dimensional spatiotemporal focusing of holographic patterns. Nat. Commun. 2016;7:11928. doi: 10.1038/ncomms11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.-J., Li K.-Y., Lin Y.-Y., Chiang A.-S., Chu S.-W. Optimizing depth-of-field extension in optical sectioning microscopy techniques using a fast focus-tunable lens. Opt. Express. 2017;25:16783–16794. doi: 10.1364/OE.25.016783. [DOI] [PubMed] [Google Scholar]
- Hsu K.-J., Lin Y.-Y., Chiang A.-S., Chu S.-W. Optical properties of adult Drosophila brains in one-, two-, and three-photon microscopy. Biomed. Opt. Express. 2019;10:1627–1637. doi: 10.1364/BOE.10.001627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.-J., Lin Y.-Y., Lin Y.-Y., Su K., Feng K.-L., Wu S.-C., Lin Y.-C., Chiang A.-S., Chu S.-W. Millisecond two-photon optical ribbon imaging for small-animal functional connectome study. Opt. Lett. 2019;44:3190–3193. doi: 10.1364/OL.44.003190. [DOI] [PubMed] [Google Scholar]
- Imai T., Sakano H., Vosshall L.B. Topographic mapping—the olfactory system. Cold Spring Harb. Perspect. Biol. 2010;2:a001776. doi: 10.1101/cshperspect.a001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N., Freeman J., Smith S.L. Technologies for imaging neural activity in large volumes. Nat. Neurosci. 2016;19:1154–1164. doi: 10.1038/nn.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Z.-F., Shang C.-F., Wang Y.-F., Yang Z., Yang C., Li F.-N., Xie J.-Z., Pan J.-W., Fu L., Du J.-L. All-optical imaging and manipulation of whole-brain neuronal activities in behaving larval zebrafish. Biomed. Opt. Express. 2018;9:6154–6169. doi: 10.1364/BOE.9.006154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judkewitz B., Rizzi M., Kitamura K., Häusser M. Targeted single-cell electroporation of mammalian neurons in vivo. Nat. Protoc. 2009;4:862–869. doi: 10.1038/nprot.2009.56. [DOI] [PubMed] [Google Scholar]
- Katona G., Szalay G., Maák P., Kaszás A., Veress M., Hillier D., Chiovini B., Vizi E.S., Roska B., Rózsa B. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods. 2012;9:201–208. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- Kong L., Tang J., Little J.P., Yu Y., Lämmermann T., Lin C.P., Germain R.N., Cui M. Continuous volumetric imaging via an optical phase-locked ultrasound lens. Nat. Methods. 2015;12:759–762. doi: 10.1038/nmeth.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach M.A., Ronzitti E., Sternberg J.R., Wyart C., Emiliani V. Fast calcium imaging with optical sectioning via HiLo microscopy. PLoS One. 2015;10:e0143681. doi: 10.1371/journal.pone.0143681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman J.W., Sanes J.R. Ome sweet ome: what can the genome tell us about the connectome? Curr. Opin. Neurobiol. 2008;18:346–353. doi: 10.1016/j.conb.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D., Chu K.K., Mertz J. Wide-field fluorescence sectioning with hybrid speckle and uniform-illumination microscopy. Opt. Lett. 2008;33:1819–1821. doi: 10.1364/ol.33.001819. [DOI] [PubMed] [Google Scholar]
- Lim D., Ford T.N., Chu K.K., Mertz J. Optically sectioned in vivo imaging with speckle illumination HiLo microscopy. J. Biomed. Opt. 2011;16:016014. doi: 10.1117/1.3528656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H.-H., Chu L.-A., Fu T.-F., Dickson B.J., Chiang A.-S. Parallel neural pathways mediate CO2 avoidance responses in Drosophila. Science. 2013;340:1338–1341. doi: 10.1126/science.1236693. [DOI] [PubMed] [Google Scholar]
- Lu R., Sun W., Liang Y., Kerlin A., Bierfeld J., Seelig J.D., Wilson D.E., Scholl B., Mohar B., Tanimoto M. Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat. Neurosci. 2017;20:620–680. doi: 10.1038/nn.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Flanagan J.G. Development of continuous and discrete neural maps. Neuron. 2007;56:284–300. doi: 10.1016/j.neuron.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Mancuso J.J., Kim J., Lee S., Tsuda S., Chow N.B., Augustine G.J. Optogenetic probing of functional brain circuitry. Exp. Physiol. 2011;96:26–33. doi: 10.1113/expphysiol.2010.055731. [DOI] [PubMed] [Google Scholar]
- Mardinly A.R., Oldenburg I.A., Pégard N.C., Sridharan S., Lyall E., Chesnov K., Brohawn S.G., Waller L., Adesnik H. Precise multimodal optical control of neural ensemble activity. Nat. Neurosci. 2018;21:881–893. doi: 10.1038/s41593-018-0139-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. Optical sectioning microscopy with planar or structured illumination. Nat. Methods. 2011;8:811–819. doi: 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- Miri A., Daie K., Burdine R.D., Aksay E., Tank D.W. Regression-based identification of behavior-encoding neurons during large-scale optical imaging of neural activity at cellular resolution. J. Neurophysiol. 2011;105:964–980. doi: 10.1152/jn.00702.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi J., Nagaoka A., Watanabe S., Ellis-Davies G.C., Kitamura K., Kano M., Matsuzaki M., Kasai H. In vivo two-photon uncaging of glutamate revealing the structure–function relationships of dendritic spines in the neocortex of adult mice. J. Physiol. 2011;589:2447–2457. doi: 10.1113/jphysiol.2011.207100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier N., Mermillod-Blondin A., Arnold C.B., Beaurepaire E. Two-photon microscopy with simultaneous standard and extended depth of field using a tunable acoustic gradient-index lens. Opt. Lett. 2009;34:1684–1686. doi: 10.1364/ol.34.001684. [DOI] [PubMed] [Google Scholar]
- Olsen S.R., Wilson R.I. Cracking neural circuits in a tiny brain: new approaches for understanding the neural circuitry of Drosophila. Trends Neurosci. 2008;31:512–520. doi: 10.1016/j.tins.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto J.J., Keleş M.F., Nguyen B.-C.M., Bolanos C., Lovick J.K., Frye M.A., Hartenstein V. Visual input to the Drosophila central complex by developmentally and functionally distinct neuronal populations. Curr. Biol. 2017;27:1098–1110. doi: 10.1016/j.cub.2017.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron D., Papagiakoumou E., Anselmi F., Emiliani V. Two-photon optogenetics. Prog. Brain Res. 2012;196:119–143. doi: 10.1016/B978-0-444-59426-6.00007-0. [DOI] [PubMed] [Google Scholar]
- Packer A.M., Roska B., Häusser M. Targeting neurons and photons for optogenetics. Nat. Neurosci. 2013;16:805–815. doi: 10.1038/nn.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer A.M., Russell L.E., Dalgleish H.W., Häusser M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods. 2015;12:140–146. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiakoumou E. Optical developments for optogenetics. Biol. Cell. 2013;105:443–464. doi: 10.1111/boc.201200087. [DOI] [PubMed] [Google Scholar]
- Papagiakoumou E., Anselmi F., Bègue A., de Sars V., Glückstad J., Isacoff E.Y., Emiliani V. Scanless two-photon excitation of channelrhodopsin-2. Nat. Methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pégard N.C., Mardinly A.R., Oldenburg I.A., Sridharan S., Waller L., Adesnik H. Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT) Nat. Commun. 2017;8:1228. doi: 10.1038/s41467-017-01031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peron S., Svoboda K. From cudgel to scalpel: toward precise neural control with optogenetics. Nat. Methods. 2011;8:30–34. doi: 10.1038/nmeth.f.325. [DOI] [PubMed] [Google Scholar]
- Petreanu L., Huber D., Sobczyk A., Svoboda K. Channelrhodopsin-2–assisted circuit mapping of long-range callosal projections. Nat. Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Piazza S., Bianchini P., Sheppard C., Diaspro A., Duocastella M. Enhanced volumetric imaging in 2-photon microscopy via acoustic lens beam shaping. J. Biophotonics. 2018;11:1–11. doi: 10.1002/jbio.201700050. [DOI] [PubMed] [Google Scholar]
- Prakash R., Yizhar O., Grewe B., Ramakrishnan C., Wang N., Goshen I., Packer A.M., Peterka D.S., Yuste R., Schnitzer M.J. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat. Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G.D., Kelleher K., Fink R., Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat. Neurosci. 2008;11:713–720. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer J.P., Deisseroth K., Tank D.W. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat. Neurosci. 2014;17:1816–1824. doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer J.P., Tank D.W. Two-photon excitation of channelrhodopsin-2 at saturation. Proc. Natl. Acad. Sci. U S A. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzitti E., Ventalon C., Canepari M., Forget B.C., Papagiakoumou E., Emiliani V. Recent advances in patterned photostimulation for optogenetics. J. Opt. 2017;19:113001. [Google Scholar]
- Scanziani M., Häusser M. Electrophysiology in the age of light. Nature. 2009;461:930–939. doi: 10.1038/nature08540. [DOI] [PubMed] [Google Scholar]
- Schrödel T., Prevedel R., Aumayr K., Zimmer M., Vaziri A. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nat. Methods. 2013;10:1013–1020. doi: 10.1038/nmeth.2637. [DOI] [PubMed] [Google Scholar]
- Seelig J.D., Jayaraman V. Feature detection and orientation tuning in the Drosophila central complex. Nature. 2013;503:262–266. doi: 10.1038/nature12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki H.M., Kazama H. Parallel encoding of recent visual experience and self-motion during navigation in Drosophila. Nat. Neurosci. 2017;20:1395–1403. doi: 10.1038/nn.4628. [DOI] [PubMed] [Google Scholar]
- Song A., Charles A.S., Koay S.A., Gauthier J.L., Thiberge S.Y., Pillow J.W., Tank D.W. Volumetric two-photon imaging of neurons using stereoscopy (vTwINS) Nat. Methods. 2017;14:420–426. doi: 10.1038/nmeth.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirman J.N., Smith I.T., Kudenov M.W., Smith S.L. Wide field-of-view, multi-region, two-photon imaging of neuronal activity in the mammalian brain. Nat. Biotechnol. 2016;34:857–862. doi: 10.1038/nbt.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Nern A., Franconville R., Dana H., Schreiter E.R., Looger L.L., Svoboda K., Kim D.S., Hermundstad A.M., Jayaraman V. Neural signatures of dynamic stimulus selection in Drosophila. Nat. Neurosci. 2017;20:1104–1113. doi: 10.1038/nn.4581. [DOI] [PubMed] [Google Scholar]
- Szalay G., Judák L., Katona G., Ócsai K., Juhász G., Veress M., Szadai Z., Fehér A., Tompa T., Chiovini B. Fast 3D imaging of spine, dendritic, and neuronal assemblies in behaving animals. Neuron. 2016;92:723–738. doi: 10.1016/j.neuron.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura S.-Y., Bharioke A., Lu Z., Nern A., Vitaladevuni S., Rivlin P.K., Katz W.T., Olbris D.J., Plaza S.M., Winston P. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timaeus L., Geid L., Hummel T. A topographic visual pathway into the central brain of Drosophila. bioRxiv. 2017:183707. [Google Scholar]
- Truong T.V., Supatto W., Koos D.S., Choi J.M., Fraser S.E. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- Wang H., Peca J., Matsuzaki M., Matsuzaki K., Noguchi J., Qiu L., Wang D., Zhang F., Boyden E., Deisseroth K. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc. Natl. Acad. Sci. U S A. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liu Y., Li Y., Guo Y., Song P., Zhang X., Zeng S., Wang Z. Precise spatiotemporal control of optogenetic activation using an acousto-optic device. PLoS One. 2011;6:e28468. doi: 10.1371/journal.pone.0028468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.R., Runyan C.A., Wang F.L., Sur M. Division and subtraction by distinct cortical inhibitory networks in vivo. Nature. 2012;488:343–348. doi: 10.1038/nature11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Supatto W., Debrégeas G., Mahou P., Kruglik S.G., Sintes J.-M., Beaurepaire E., Candelier R. Whole-brain functional imaging with two-photon light-sheet microscopy. Nat. Methods. 2015;12:379–380. doi: 10.1038/nmeth.3371. [DOI] [PubMed] [Google Scholar]
- Yang W., Carrillo-Reid L., Bando Y., Peterka D.S., Yuste R. Simultaneous two-photon imaging and two-photon optogenetics of cortical circuits in three dimensions. Elife. 2018;7:e32671. doi: 10.7554/eLife.32671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Miller J.-E.K., Carrillo-Reid L., Pnevmatikakis E., Paninski L., Yuste R., Peterka D.S. Simultaneous multi-plane imaging of neural circuits. Neuron. 2016;89:269–284. doi: 10.1016/j.neuron.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Yuste R. In vivo imaging of neural activity. Nat. Methods. 2017;14:349–359. doi: 10.1038/nmeth.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel W.R., Williams R.M., Webb W.W. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- Zong W., Zhao J., Chen X., Lin Y., Ren H., Zhang Y., Fan M., Zhou Z., Cheng H., Sun Y. Large-field high-resolution two-photon digital scanned light-sheet microscopy. Cell Res. 2015;25:254–257. doi: 10.1038/cr.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of Figures 1, 2, 3 and S1A as well as the code for 3D functional image analysis using the generalized linear model is available on Mendeley Data. doi: https://doi.org/10.17632/5t5kb6gc35.1.