Abstract
Sulfite resistance is an important oenological trait for wine yeasts because this compound is used during winemaking as a microbial inhibitor and antioxidant. The molecular mechanisms by which Saccharomyces cerevisiae responds and tolerates SO2 have been mainly focused on the sulfite efflux pump encoded by SSU1. Different chromosomal rearrangements in the regulatory region of this gene have been correlated with improved sulfite tolerance. However, other molecular factors must contribute to this trait because the SSU1 gene activity does not always fit with sulfite tolerance. An interesting approach to shed light onto this issue could be found by Lage et al. (2019). These authors have combined transcriptomic and genome-wide analysis to describe how the poorly characterized transcription factor Com2 controls, directly or indirectly, the expression of more than 80% of the genes activated by SO2. Additionally, large-scale phenotyping revealed the identification of 50 Com2-targets contributing to the protection against SO2. This information is very interesting for gaining knowledge regarding this important industrial trait.
Keywords: Saccharomyces cerevisiae, wine, sulfur dioxide, Com2
Microorganisms utilize a great variety of genetic strategies to adapt to natural and human-made environments. The wine strains of S. cerevisiae are a clear example of highly specialized microorganisms that have evolved to use the different ecological niches provided by human activity. The specific genetic characteristics of the wine yeast strains are a consequence of the process of domestication [1–6]. Thus, in the age of high throughput sequencing technologies and omics data, a current challenge is to unveil the molecular determinants underlying a specific trait of industrial interest. This knowledge will be of paramount importance for selection and genetic improvement of the industrial strains.
Sulfite (SO32−), which is produced by dissolution of sulfur dioxide (SO2) in water, is used during winemaking as a microbial inhibitor and antioxidant. Therefore, sulfite resistance is a desired trait for wine yeast strains [7]. S. cerevisiae cells have different mechanisms to deal with the stress produced by sulfites including the increase in the production of acetaldehyde, which binds to SO32−, the regulation of the sulfite uptake pathway, and sulfite efflux through a plasma membrane pump encoded by the SSU1 gene [8]. Indeed, wine S. cerevisiae strains are considerably more tolerant to SO2 than laboratory strains and the main molecular mechanism connected with this higher resistant phenotype results from a higher transcription of the SSU1 gene. Among this group of strains, three chromosomal rearrangements (VIIItXVI, XVtXVI and Inv-XVI) have been described to up-regulate SSU1 expression and thereby increasing sulfite tolerance [9–12]. In all cases, the chromosomal rearrangement involves the SSU1 promoter and leads to its transcriptional up-regulation. Surprisingly, SSU1 transcript levels are not responsive to SO2 [9, 10, 12], indicating that the different levels of resistance in S. cerevisiae wine strains are, in general, explained by the basal transcript levels of SSU1, except for the 71B strain that harbors a sulfur-inducible SSU1 gene that may have gained a new regulatory system [13]. However, in some cases SSU1 mRNA levels did not correlate with sulfite tolerance probably due to the contribution of other factors to yeast sulfite resistance. Thus, a key question remains to be addressed in the field: Which are the molecular mechanisms that explain yeast sulfite tolerance? This question is of paramount importance for understanding much better the regulation network between gene activity and metabolic response. However, this information is also crucial for the wine industry, which is leading a process of sulfite reduction in their wines
In this scenario, Lage et al., (2019) [14] have recently demonstrated the involvement of Com2 as the main transcriptional factor regulating genes that impart SO2 resistance. The results clearly highlight Com2 as the main player in the reprogramming of yeast genomic expression under SO2 stress, regulating approximately 80% of the SO2-activated genes. Moreover, the involvement in SO2 tolerance of genes regulated by Com2 was validated by a large-scale phenotyping of the corresponding mutants, proving the susceptibility of these mutants to SO2. Figure 1 shows a summary of the main mechanisms involved in SO2 resistance in S. cerevisiae.
Figure 1. FIGURE 1: Schematic representation of the main mechanisms involved in sulfites resistance in the yeast Saccharomyces cerevisiae.
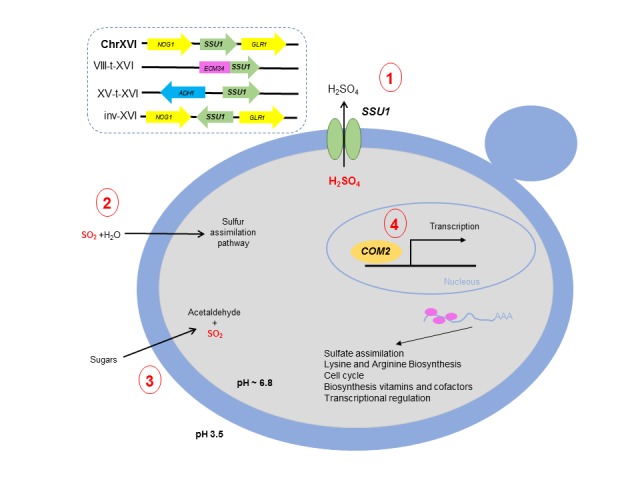
(1) Efflux of SO2 mediated by SSU1 including the different chromosomal rearrangements described so far; (2) Incorporation into the sulfur assimilation pathway; (3) Acetaldehyde production and (4) Com2 regulon.
COM2 encodes an orphan homologue of the environmental stress-responsive transcription factors Msn2 and Msn4 [15] and, the genes regulated by Msn2 were also found to be regulated by Com2, suggesting some overlap between them. In a phenotypic assay with and without sulfite, the strain lacking the Com2 gene was more sensitive to sulfite than its wild-type counterpart (BY4741). The transcriptomic profiling of BY4741 and BY4741_com2? cells revealed dramatic changes in both strains especially after exposure to SO2 (0.5 mM), although this effect was markedly different in the two strains. A higher number of genes were differentially expressed in the wild type compared to the com2? strain, indicating that these SO2-responsive genes present in the wild type and without activation in the mutant strain could be considered as Com2 targets. No significant effect of Com2 was found in the absence of SO2. The functional categories obtained after analyzing these genes are old known from the transcriptomic analysis performed to date and are mainly related to nitrogen, sulfur and some aminoacids metabolism [16]. In this line, authors have shown that supplementation of the medium with arginine and lysine somehow alleviated SO2 toxicity. A plausible explanation could be that exposure to SO2 can lead to a depletion of intracellular lysine and arginine or, in the case of arginine, by contributing to the integrity of the cell wall and the plasma membrane [17].
However, as mentioned above, the exposition to SO2 during hundreds of years and thousands of generations have caused S. cerevisiae wine strains to be more tolerant to this compound than the laboratory strains. They have evolved to employ various anthropic niches or environments during the so-called “unaware domestication” process [1, 3, 18]. Their genomes present genetic polymorphisms with different evolutionary consequences all of which help wine yeast genomes to adapt [19, 20]. Thus, the main limitation of this study is the use of a single laboratory strain genetic background to investigate a phenotype of industrial relevance in the wine industry. Thus, the remaining question is: How much is the contribution of the Com2 in a wine yeast background adapted to sulfite stress? The key role of this gene in the laboratory strains is undeniable but we have to be careful when transferring this information from one background to another and, therefore, more studies are needed to determine its possible application in the industry.
Funding Statement
This work was supported by the Spanish Government through “Ministerio de Ciencia, Innovación y Universidades” (MICINN) and “Fondo Europeo de Desarrollo Regional” (FEDER) funds (grant number PCIN-2015-143, AGL2016-77503-C3-1-R) to JMG. This study has been carried out in the context of the European Project ERA-IB “YeastTempTation”.
REFERENCES
- 1.Legras J, Galeote V, Bigey F, Camarasa C, Marsit S, Nidelet T, Sanchez I, Couloux A, Guy J, Franco-duarte R, Marcet-houben M, Gabaldon T, Schuller D, Sampaio JP. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol Biol Evol. 2018;35(May):1712–1727. doi: 10.1093/molbev/msy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O 'kelly MJT, Van Oudenaarden A, Barton DBH, Bailes E, Nguyen Ba AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almeida P, Barbosa R, Zalar P, Imanishi Y, Shimizu K, Turchetti B, Legras JL, Serra M, Dequin S, Couloux A, Guy J, Bensasson D, Gonçalves P, Sampaio JP. A population genomics insight into the Mediterranean origins of wine yeast domestication. Mol Ecol. 2015;24(21):5412–5427. doi: 10.1111/mec.13341. [DOI] [PubMed] [Google Scholar]
- 4.Gonçalves M, Pontes A, Almeida P, Barbosa R, Serra M, Libkind D, Hutzler M, Gonçalves P, Sampaio JP. Distinct Domestication Trajectories in Top-Fermenting Beer Yeasts and Wine Yeasts. Curr Biol. 2016;26(20):2750–2761. doi: 10.1016/j.cub.2016.08.040. [DOI] [PubMed] [Google Scholar]
- 5.Borneman AR, Forgan AH, Kolouchova R, Fraser JA, Schmidt SA. Whole Genome Comparison Reveals High Levels of Inbreeding and Strain Redundancy Across the Spectrum of Commercial Wine Strains of Saccharomyces cerevisiae. G3. 2016;6(4):957–971. doi: 10.1534/g3.115.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gayevskiy V, Lee S, Goddard MR. European derived Saccharomyces cerevisiae colonisation of New Zealand vineyards aided by humans. FEMS Yeast Res. 2016;16(7):1–12. doi: 10.1093/femsyr/fow091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Divol B, Du Toit M, Duckitt E. Surviving in the presence of sulphur dioxide: Strategies developed by wine yeasts. Appl Microbiol Biotechnol. 2012;95(3):601–613. doi: 10.1007/s00253-012-4186-x. [DOI] [PubMed] [Google Scholar]
- 8.Casalone E, Colella CM, Daly S, Gallori E, Moriani L, Polsinelli M. Mechanism of resistance to sulphite in Saccharomyces cerevisiae. Curr Genet. 1992;22(6):435–440. doi: 10.1007/BF00326407. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Ortín JE, Querol A, Puig S, Barrio E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002;12(10):1533–9. doi: 10.1101/gr.436602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmer A, Durand C, Loira N, Durrens P, Sherman DJ, Marullo P. QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS One. 2014;9(1):e86298. doi: 10.1371/journal.pone.0086298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuasa N, Nakagawa Y, Hayakawa M, Iimura Y. Distribution of the sulfite resistance gene SSU1-R and the variation in its promoter region in wine yeasts. J Biosci Bioeng. 2004;98(5):394–397. doi: 10.1016/S1389-1723(04)00303-2. [DOI] [PubMed] [Google Scholar]
- 12.García-Ríos E, Nuévalos M, Barrio E, Puig S, Guillamón JM. A new chromosomal rearrangement improves the adaptation of wine yeasts to sulfite. Environ Microbiol. 2019;21(5):1771–1781. doi: 10.1111/1462-2920.14586. [DOI] [PubMed] [Google Scholar]
- 13.Nardi T, Corich V, Giacomini A, Blondin B. A sulphite-inducible form of the sulphite efflux gene SSU1 in a Saccharomyces cerevisiae wine yeast. Microbiology. 2010;156(6):1686–1696. doi: 10.1099/mic.0.036723-0. [DOI] [PubMed] [Google Scholar]
- 14.Lage P, Sampaio-marques B, Ludovico P, Mira NP, Mendes-ferreira A. Transcriptomic and chemogenomic analyses unveil the essential role of Com2-regulon in response and tolerance of Saccharomyces cerevisiae to stress induced by sulfur dioxide. Microbial Cell. 2019;6(11):509–523. doi: 10.15698/mic2019.11.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nadai C, Treu L, Campanaro S, Giacomini A, Corich V. Different mechanisms of resistance modulate sulfite tolerance in wine yeasts. Appl Microbiol Biotechnol. 2016;100(2):797–813. doi: 10.1007/s00253-015-7169-x. [DOI] [PubMed] [Google Scholar]
- 17.Cheng Y, Du Z, Zhu H, Guo X, He X. Protective Effects of Arginine on Saccharomyces cerevisiae Against Ethanol Stress. Sci Rep. 2016;6:1–12. doi: 10.1038/srep31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter J, De Chiara M, Friedrich A, Yue J-X, Pflieger D, Bergstrom A, Sigwalt A, Barré B, Freel K, Llored A, Cruaud C, Labadie K, Aury JM, Istace B, Lebrigand K, Barbry P, Engelen S, Lemainque A, Liti G, Schacherer J. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339–344. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sicard D, Legras JL. Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011;334(3):229–36. doi: 10.1016/j.crvi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Bisson LF. Geographic origin and diversity of wine strains of Saccharomyces. Am J Enol Vitic. 2012;63(2):165–176. doi: 10.5344/ajev.2012.11083. [DOI] [Google Scholar]


