Abstract
Treatment of head and neck cancer has been slow to change with epidermal growth factor receptor (EGFR) inhibitors, PD1 inhibitors, and taxane-/plant-alkaloid-derived chemotherapies being the only therapies approved by the U.S. Food and Drug Administration (FDA) in the last 10 years for the treatment of head and neck cancers. Head and neck cancer is a relatively rare cancer compared to breast or lung cancers. However, it is possible that existing therapies for more common solid tumors or for the treatment of other diseases could also prove effective against oral cancers. Many therapies have molecular targets that could be appropriate in oral cancer as well as the cancer in which the drug gained initial FDA approval. Also, there may be targets in oral cancer for which existing FDA-approved drugs could be applied. This study describes informatics methods that use machine learning to identify influential gene targets in patients receiving platinum-based chemotherapy, non-platinum-based chemotherapy, and genes influential in both groups of patients. This analysis yielded 6 small molecules that had a high Tanimoto similarity (>50%) to ligands binding genes shown to be highly influential in determining treatment response in oral cancer patients. In addition to influencing treatment response, these genes were also found to act as gene hubs connected to more than 100 other genes in pathways enriched with genes determined to be influential in treatment response by a random forest classifier with 20 000 trees trying 320 variables at each tree node. This analysis validates the use of multiple informatics methods to identify small molecules that have a greater likelihood of efficacy in a given cancer of interest.
Keywords: Chemical informatics, oral cancer, virtual screening, traditional chinese medicine, random forest, machine learning, pathway analysis
Introduction
Head and neck cancers can be defined as cancers in the upper airway and/or digestive tract found in oral cavity and laryngeal, pharyngeal, oropharyngeal, and hypo-pharyngeal tissues. Such cancers make up 3% of cancers diagnosed each year.1 Head and neck cancer incidence has declined from 25 cases per 100 000 in the 1990s to 15 cases per 100 000 in the present day.2 While the decrease in head and neck cancer incidence may be due to a drop in tobacco use,3,4 the mortality associated with head and neck cancers has not changed significantly in the last 20 years. Treatment of head and neck cancer has also been slow to change with epidermal growth factor receptor (EGFR) inhibitors, PD1 inhibitors, and taxane-/plant-alkaloid-derived chemotherapies being the only therapies approved by the U.S. Food and Drug Administration (FDA) in the last 10 years for the treatment of head and neck cancers. Head and neck cancer is a relatively rare cancer compared to breast or lung cancers. However, it is possible that existing therapies for more common solid tumors could also prove effective in oral cancers. Many therapies have molecular targets that could be appropriate in oral cancer as well as the cancer in which the drug gained initial FDA approval. Also, there may be targets in oral cancer for which existing FDA-approved drugs could be applied. This study describes informatics methods that use machine learning to identify influential gene targets in patients receiving platinum-based chemotherapy, non-platinum-based chemotherapy, and genes influential in both groups of patients.
Drugs approved by the FDA for oral cancer are methotrexate, cetuximab, pembrolizumab, nivolumab, and docetaxel. These therapies are combined to be used in conjunction with platinum-based chemotherapies such as cisplatin or carboplatin unless those therapies are contraindicated due to comorbidities such as renal disease. The small number of new oral cancer drugs could be attributed in part to the low overall burden of oral cancer in comparison with other cancers. The current timeline for FDA approval of a novel small molecule or biologic is 10 years or more. Repurposing existing FDA-approved drugs is a popular method used to shorten this process to 3 to 4 years. Cetuximab is an EGFR inhibitor that has been shown to decrease the rate of progression of oral cancer used in conjunction with Cisplatin. Pembrolizumab and nivolumab are both PD1 inhibitors that used T-cells to attack cancer while it progresses. These therapies use the body’s immune system as another treatment modality to reduce the burden of oral cancer. Current literature provides support for the role of ligand channel gating, hedgehog signaling,5-7 NOTCH, B-WICH,8 inflammasome,9 WNT,10 and Calcineurin pathways in cancer.11-13 The role of these pathways and targeting specific genes within them has been pursued in other cancers, but have with few exceptions, not yet been examined in oral cancer. Possible gains from targeting these pathways would be initiating immune response, targeting cancer metabolism, targeting signaling for metastasis, and targeting inflammation pathways that may drive progression. If there is a synergistic effect to attacking multiple hallmarks of cancer simultaneously, then the net gain to the patient would be a net gain in overall post diagnosis survival time for the oral cancer patient.
Identifying a means by which drugs may be prioritized for further screening and validation for a specific cancer type would be desirable. Databases linking genes, the proteins they express, ligands corresponding to those proteins, and structural data that can be analyzed all exist in varying forms or completeness across different publicly available databases. This study describes how integration of analyses of these databases can be used to select gene targets in a specific cancer and how therapies can be prioritized for screening based on existing structural information for the ligands associated with genes and the proteins they express.
There are several hurdles to the analysis of high-dimensional genomic data using traditional regression analyses. Random forest analysis is a machine learning approach that is less hindered by datasets with large predictor to observation ratios. In this study, we apply random forests analysis to gene expression data to identify those genes and pathways that are most predictive of post diagnosis survival across treatment strata. This is the first application of this approach to head and neck cancer patient data in The Cancer Genome Atlas (TCGA). National Comprehensive Cancer Network (NCCN)14 guidelines recommend that node-positive patients with tumors of clinical stage 3 and greater receive chemotherapy. Platinum-based chemotherapy with radiation and surgery is the current standard of care recommended to these patients. Patients that do not receive chemotherapy recommendations by NCCN are node negative with clinical stage 2 and lower. Following standards set by NCCN guidance on treatment, this study chose to identify patients receiving and not receiving platinum-based chemotherapy as separate groups. Analysis of influential genes in each group will improve knowledge of possible mechanisms driving treatment response for early stage and more advanced tumors.
Random forest is a machine learning approach to identifying the most important predictors in high dimensional datasets.15,16 This approach is uniquely suited for classification of observations in datasets where P (predictors) are greater than N (number of observations). Random forest randomly selects predictors from a large group of predictors and then applies those predictors to a decision tree predicting overall survival. Random forest does not pay a statistical penalty when the number of observations is small. Instead the strength and limitation of this method is its reliance on computational intensity. That is, as the number of decision trees in a random forest increase, so does classification accuracy. Accuracy is also dependent on the number of predictors tried at decision tree nodes. As node size and forest size increase, so does forest classification accuracy. However, there is a rate of diminishing returns in the accuracy gained from each tree added to a forest. Therefore, computational time and cost must be factored into all random forest analysis plans to measure project feasibility. Random forest has been successfully applied to predicting cancer diagnosis and treatment response for a variety of cancers.17-21 For this study, we have selected to apply random forest analysis to the gene expression values of oral cavity cancer patients to identify the upregulated pathways most predictive of improved treatment response across gender and environmental exposure subgroups like alcohol and tobacco. RNAseq data are inherently high dimensional, applying typical regression models to such data can be costly as large sample sizes are required to identify even moderate effect. Identifying gene interactions can be even more costly in terms of the required statistical power. Stratified pathway analysis via random forest methods has been shown to be successful in identifying single influential genes (within the context of larger pathways) that are predictive of overall survival with limited sample size.22 This approach has not yet been applied to identification of influential genes and gene interactions within oral cancer patients stratified specifically by treatment. In this way, the importance of pathways and genes of interest can be compared across strata to assess which subgroups may be most sensitized to changes in gene expression within a given pathway.
Methods
This study focuses on the identification of the role of gene expression in oral cavity cancer patients and applying machine learning approaches like random forest to determine genes that are important in influencing treatment response. Reference ligands known to bind to proteins expressed by genes deemed influential by random forest can be sent through a virtual screening pipeline to identify small molecules with greater likelihood of acting as protein agonists/antagonists. Ligands that have a strong shape similarity to known binding ligands have greater potential for success in high-throughput screening endeavors. As shape similarity alone is insufficient in identifying new drug leads, all leads will be validated with existing literature, and those leads without previous biological validation will be presented as such.
By using a stratified random forest analysis, we will be able to rank genes within the strata of chemotherapy treatment status. This approach will allow for the identification of those top ranked genes that are unique to each stratum. This will be done by identifying common and unique genes between sets of genes influencing the treatment response in patients receiving platinum-based chemotherapy and those that do not. The result will be the identification of oral cavity cancer pathways influencing treatment response which will inform researchers on mechanisms driving treatment response in specific groups such as late-stage, node-positive patients who are more likely to receive chemotherapy treatment. This analysis will illustrate and support existing studies showing the strength of machine learning methods as an alternative method in identifying gene expression values influencing treatment response. This study is focused not only on the predictive power of an aggregated panel of gene expression values, but rather to integrate random forest with chemical informatics and thus describe methods to shorten the pursuit of novel therapies treating cancers with relatively lower incidence.
Retrieval of Public Data
This study used clinical and genetic data obtained from TCGA. Genetic data included raw counts per million (CPM) of RNA sequence expression values for 523 patients posted to TCGA. Of these 523 patients, 313 were diagnosed with Oral Squamous Cell Carcinoma (OSCC). Oral squamous cell carcinoma patients included tongue, buccal mucosa, alveolar ridge, general oral cavity, and soft palate tissues. Of these 313 patients, 267 were included based on complete survival time. Of these 267, 109 received either Carboplatin, Oxaliplatin, or Cisplatin, while 158 patients received a treatment other than the platinum-based chemotherapy treatment regimen. All tissue samples were collected prior to start of treatment. Clinical data on tumor stage, necrosis, size, and nuclei were also retrieved from TCGA. We obtained demographic data on ethnicity, race, and gender from TCGA clinical files. In addition, this dataset had information on environmental exposures like tobacco history (ever/never smoke) and number of alcohol containing drinks in a day (greater than 2 drinks consumed per day, 2 drinks or less consumed per day). Overall survival time in months was extracted as a measure of treatment response.
Machine Learning Methods
Description of random forest approach
Stratified pathway analysis considers important covariates in data analysis. In this analysis of head and neck cancer gene expression data, this study used information regarding patient age, sex, alcohol, and smoking exposures. In the first round of analysis, the sample of 267 OSCC patients was organized into 109 and 158 subsamples based on whether patients received platinum or non-platinum-based chemotherapy, respectively. For each group, this study built a random forest15 to predict survival time based on the gene expression levels within each subgroup.
To better communicate the function of random forests, understanding of a decision tree construction is needed. A decision tree is constructed by the following steps:
Step A: A bootstrap sample was taken from the original sample.
Step B: A decision tree is grown for each bootstrap sample.
Step C: At each tree node, a predetermined number of predictors were applied randomly to create branches within the tree.
Step D: A branch is formed using the predictor from step C.
Step E: Steps C and D are repeated until the end of every tree branch contains samples above or below the same survival threshold or contain only one sample.
Random forest result measurements
Random forests build many decision trees to comprise a forest. Each tree is put together by using a random bootstrap sample of the original data and applying a random number of predictors at each node of a decision tree. The SRCRandomForest R package23 employed in this study sets aside half of the data to be used for validation purposes to measure the accuracy of the random forest model. The SRCRandomForest package was chosen for this study because it applies a multivariate Cox regression model to produce each decision tree within each Forest. In this way, censored and non-censored data can be considered when carrying out this analysis. In this way, the model can measure the degree of influence of each gene on patient survival. The P values yielded through this analysis are defined as the “proportion of cross-validation errors smaller than the cross-validation errors obtained from 500 iterations of random forest runs of randomly permuted labels of patients.” This list of genes can be used to identify pathways that are enriched with the influential genes identified through random forest at odds that would be greater than can be attributed to chance alone with a P value of .05. This analysis will present pathways common to, and unique to, each chemotherapy treatment strata. Such analyses may identify plausible biological mechanisms that enhance understanding of observed differences in survival. To reiterate, the focus of this study is not to pursue a diagnostic tool but to identify those gene expression values exerting a strong influence on treatment response. This study adopted random forest as the machine learning method of choice due to its superior interpretability and scalability. Random forest and many applications incorporating random forest have been developed more than 30 years. Random forest was of interest to this study as an application developed specifically for the use of survival data had been developed as an open-source R package. Although random forest has been shown to be less efficient than extreme gradient boosting applications, the long history of this approach has allowed it to evolve to produce an application well suited toward achieving the analytic goals of this study.
Random forest tuning parameters
Our random forest model used 20 000 tree forests for a forest size, with 320 variables tried at each node in each decision tree; 20 000 trees were the point at which we could identify no significant increase in our ability to predict patient survival; 320 tries at each node in each decision tree were double the recommended number of tries given by the author of our R software package RandomForestSRC. The author H. Ishwaran et al23 refers to a generally accepted practice of using the “square root of the total number of predictors as a starting point for the number of variables tried at each node.” It was for this reason that we applied 20 000 trees and 320 variables tried at each node in each decision tree for every stratum in our analysis. This approach was applied to our entire final sample of 267 OSCC patients. We then divided this sample by whether a patient received platinum-based chemotherapy or not. All random forest analyses were carried out on 64 computational nodes in the University of California High Performance Computational Cluster. Random forest analysis was made more efficient by running analysis in conjunction with the R package parallel. The output of each group’s analysis produces a list ranking each gene. This analysis identified common and unique pathways between the entire dataset and each chemotherapy treatment group. We then identified the unique and common genes between chemotherapy groups. This analysis will allow us to observe the difference in gene importance and corresponding pathways in relation to overall survival.
Description of virtual screening approaches
Using chemical informatics techniques, ZINC drug database24 of 1379 FDA-approved drugs (FDA) and ZINC Traditional Chinese Medicine (TCM) database of 39 894 small molecules can be used to apply three-dimensional chemical informatics approaches to the identification of small molecules that are the best candidates for inhibition of proteins expressed by those genes influencing treatment response. Reference ligands for each protein are obtained from the Royal Chemistry Society Protein Database25 and then virtually screened against FDA and TCM small molecule libraries. Molecular shape overlay is an approach for measurement of the similarity of one molecule in comparison with another. A Tanimoto coefficient is used to measure the degree of similarity between 2 molecules.
The Tanimoto coefficient between 2 points, a and b, with k dimensions is calculated as follows
The Tanimoto similarity only applies to binary variables, for binary variables the Tanimoto coefficient ranges from 0 to 1 (where 1 is the highest possible similarity).26
A goal of this study is to perform searches of 2 small molecule databases FDA and TCM using a maximum common substructure measurement of Tanimoto similarity from the R Rcpi package27 that has been shown to perform robustly across a variety of molecule types.
Pathway analysis
Table 1 provides information on those genes that are significantly enriched within our gene set beyond what would be expected by chance alone. The significance of enrichment is calculated as the odds of randomly selecting the number of genes in the submitted set of genes by randomly selecting from 20 530 genes more than 100 times. The false discovery rate (FDR) reported by the Pathway Reactome application used for this study represents an adjustment for multiple comparisons across all pathways. Benjamini–Hochberg FDR is calculated as the P value ranking (smallest being 1 and all following having greater or equal rank dependent on size of P value) divided by the number of tests performed and multiplied by the significance criterion. In this study, .05 is the criterion used to measure significance. The FDR can be interpreted as the proportion of tests within a set of tests that falsely rejects the null hypothesis. If an FDR is .5, then 50% of those pathways identified will falsely reject the null hypothesis. It is important to note that the FDR calculation used by Pathway Reactome defaults to perform a large number of analyses/tests. In addition, there are many pathways examining similar genes and gene types. Unfortunately, this thorough examination strategy also inflates the number of analyses and causes the FDR to become overly conservative.
Table 1.
Top pathways enriched with genes influencing platinum-based chemotherapy treatment response in oral cancer.
| Pathway name | P value | FDR | Influential genes enriched in pathway |
|---|---|---|---|
| Signaling by Hedgehog | 6.25E-05 | 0.002 | ARRB1, ARRB2, KIF7, ADCY6, PSMA7, ADCY5, PSMB6, TUBB6, PSMC6, PSME4, PSME1, PSME2, CDON |
| Molecules associated with elastic fibers | 3.43E-04 | 0.006 | ELN, FN1, FBLN1, LTBP3, BMP7 |
| CLEC7A/inflammasome pathway | 9.93E-04 | 0.01 | IL1B, UBE2D4, ITPR2, ITPR3, PSMA7, PSMB6, PSMC6, IL1B, PSME4, PSME1, PSME2, TAB2, IKBKG, CALM1, CARD11 |
| Phase 0—rapid depolarization | .001 | 0.01 | CAMK2B, CAMK2D, CACNB3, CAMK2A, CACNA2D2, CALM1 |
| Adrenaline, noradrenaline inhibits insulin secretion | .001 | 0.01 | CACNB3, GNG2, CACNA2D2, ADCY6, ADCY5 |
| Signaling by NOTCH1 in cancer | .006 | 0.03 | HDAC5, HDAC1, EP300, CCNC, TBL1X |
| LGI-ADAM interactions | .0 | 0.08 | LGI2, ADAM11 |
| SeMet incorporation into proteins | .05 | 0.10 | QARS |
| Presynaptic depolarization and calcium channel opening | .05 | 0.10 | CACNB3, CACNA2D2 |
Abbreviations: FDR, false discovery rate.
To identify those genes that are most likely to be connected to influential pathways, 2 filters were applied to gene selection. First, the gene had to be in the top 5% of influential genes identified via random forest. Second, if the gene was not within the top 5% of genes, it could still be included within the analysis if it was within the top 40% of genes and was known to be connected via past experimental studies supporting gene network connections found in the Cytoscape/Pathway Reactome plugin database. Gene topology can be accessed by uploading topology from a given gene in Cytoscape and merging it with that gene’s corresponding random forest importance values. Gene topology refers to the connectedness of a gene (supported by experimental results referenced in cytoscape) to other genes within a Cytoscape network. Thus, the selection of genes included those ranked as the most important by random forest (top 5%) or of moderate importance and high topology (shown through past experiment or literature search to be connected to 50 genes or more). For each set of analyses, 1000 influential genes were selected and 100 high topology genes were selected. In this way, integration of Cytoscape network analysis with random forest results allowed for identification of pathways significantly enriched with the genes identified by the random forest model. For a complete overview of the analyses in this study please reference the online Supplemental Materials.
Results
Top pathways
This section will describe pathways uniquely influential in patient response to platinum-based chemotherapy and influential in response to non-platinum-based therapy. Influential pathways shared by both platinum-based chemotherapy users and non-users will also be presented. An influential pathway will be defined as a pathway that is significantly enriched with genes that were in the list of top 1000 (5%) of most influential genes yielded by random forest analysis for platinum-based chemotherapy users, non-platinum-based chemotherapy users, and for those pathways enriched with genes shared in common in lists of top 1000 genes for platinum-based chemotherapy users and non-users. The top 5% of genes were conservatively selected to produce greater certainty of the link between highly ranked genes, the pathways in which they were enriched, and the link between treatment response and significant pathways identified through gene enrichment analysis. Top pathways could also be enriched with those genes in the top 40% of important genes identified by random forest if they also had a high amount of connectedness (a gene was connected to 25 gene nodes in a gene network) reported by the Cytoscape/Reactome application.
Pathways significantly enriched with genes identified as the most influential (top 5%) in predicting treatment response for platinum-based chemotherapy users were those related to calcium channel gating, hedgehog signaling, histone acetylation, elastic fiber production, tRNA acetylation, hexokinase deficiency, inhibition of adenylate cyclase, and CLEC7A inflammasome pathway. It has been reported that calcium channel gating has been associated with multiple cancers.28-30 There have also been recent studies evaluating the benefit of targeting histone deacetylation pathways in oral cancer.31-33 The hedgehog signaling pathway has also been shown to signal progression in other cancers “Hh signaling has been shown to regulate the self-renewal of CSCs in breast, glioma and multiple myeloma, and more convincingly in the maintenance of chronic myelogenous leukemia (CML) stem cells.”34-39 All significant pathways for platinum-based chemotherapy users are in Table 1.
Significant pathways for patients not using platinum-based chemotherapy were those related to B-WICH complex, TP53 pathway, fibroblast growth factor receptor (FGFR) pathway, potassium channel gating, and RNA polymerase chain elongation pathways and their epigenetic regulation. TP53 and FGFR pathways represent the expression of canonical oncogenes which have been shown to be cancer drivers and associated with the production of all cancers.40-43 The B-WICH complex has been found to be linked to maturation of invadopodium in breast cancer and has been suggested as both a biomarker and target for cancer invasiveness.8,44 Potassium channel gating has also shown to be a potential target for head and neck cancers due to this pathways association with immune response and treatment response.45-49 RNA polymerase chain elongation and its role in transcription is a logical contributor to cancer progression and differentiation; however, the lack of specificity makes this a difficult pathway to target specifically in cancer cells. All significant pathways for non-platinum-based chemotherapy users are in Table 2.
Table 2.
Top pathways enriched with genes influencing treatment response in oral cancer patients not receiving platinum-based chemotherapy.
| Pathway identifier | Pathway name | P value | FDR | Influential genes enriched in pathway |
|---|---|---|---|---|
| R-HSA-5250924 | B-WICH complex positively regulates rRNA expression | 3.21E−12 | 1.70E−10 | HIST1H2BM; H2AFJ; H2AFZ; HIST1H2AJ; HIST1H2BK; H3F3A; POLR1C; H2AFV; HIST2H3C; HIST2H2BE |
| R-HSA-5250913 | Positive epigenetic regulation of rRNA expression | 8.82E−11 | 3.00E−09 |
HIST1H2BM; H2AFJ; H2AFZ; HIST1H2AJ;
HIST1H2BK; H3F3A; POLR1C; H2AFV; HIST2H3C; HIST2H2BE |
| R-HSA-1296065 | Inwardly rectifying K+ channels | 2.40E−03 | 9.61E−03 | GNG2; KCNJ14; GNB3 |
| R-HSA-1839130 | Signaling by activated point mutants of FGFR3 | 2.78E−03 | 1.07E−02 | FGFR3 |
| R-HSA-5655332 | Signaling by FGFR3 in disease | 3.91E−03 | 1.17E−02 | KRAS; FGFR3 |
| R-HSA-8853338 | Signaling by FGFR3 point mutants in cancer | 3.91E−03 | 1.17E−02 | KRAS; FGFR3 |
| R-HSA-2033514 | FGFR3 mutant receptor activation | 8.05E−03 | 2.42E−02 | FGFR3 |
| R-HSA-5654227 | Phospholipase C-mediated cascade; FGFR3 | 3.48E−02 | 6.96E−02 | FGFR3 |
| R-HSA-6803211 | TP53 Regulates Transcription of Death Receptors and Ligands | 3.48E−02 | 6.96E−02 | TNFRSF10D |
| R-HSA-2033515 | t(4; 14) translocations of FGFR3 | 4.73E−02 | 9.46E-02 | FGFR3 |
| R-HSA-5619109 | Defective SLC6A2 causes orthostatic intolerance (OI) | 4.73E−02 | 9.46E−02 | SLC6A5 |
| R-HSA-432030 | Transport of glycerol from adipocytes to the liver by Aquaporins | 4.73E−02 | 9.46E−02 | AQP7 |
| R-HSA-1226099 | Signaling by FGFR in disease | 5.37E−02 | 9.73E−02 | KRAS; FGFR3 |
Abbreviations: FDR, false discovery rate.
For those pathways enriched with genes shared by users and non-users of platinum-based chemotherapy, it was found that pathways related to g-protein beta folding, nuclear factor of activated T-cells (NFAT) activation, and repression of Wnt pathway genes were enriched with genes influential in treatment response in both groups of patients. Repression of Wnt pathway genes may be done through targeting the sonic hedgehog pathway as previously outlined or through more direct means which have been researched in multiple other cancers.50-52 Nuclear factor of activated T-cells proteins have been found to be associated with cancer progression in blood and solid tumors; however, the literature is mixed as to whether NFAT pathways are viable targets for treatment.53-55 These pathways influencing treatment response in both users and non-users of platinum-based chemotherapy can be seen in Table 3.
Table 3.
Top common pathways enriched with genes influencing platinum-based chemotherapy treatment response in all oral cancer patients.
| Pathway name | P value | FDR | Influential genes enriched in pathway |
|---|---|---|---|
| Signaling by WNT | 6.66E−15 | 5.21E−12 | HIST1H2BM; HIST1H2BK; CAMK2A; ITPR2; LRP6; PPP3CA; PPP3CB; GNG2; PSMB3; PPP2R1A; PSMD2; PSMB1; PSMD1; SOST; SOX6; BCL9L; SKP1; CSNK2A1; HIST1H2AJ; WNT5A; H2AFV; PPP2R5D; WNT16; RNF146; PSMC3; PSME4 |
| CLEC7A (Dectin-1) signaling | 8.51E−08 | 7.32E−06 | PPP3CA; PPP3CB; PSMC3; PSMB3; PSMD2; PSMB1; PSME4; ITPR2; PSMD1; BCL10; MALT1; SKP1 |
| Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding | 3.31E−05 | 4.77E−04 | GNG2; CSNK2A1; CCT8; RGS6; CCT6B; CCT4 |
| Calcineurin activates NFAT | .014851 | 0.032508 | PPP3CA; PPP3CB |
Abbreviations: FDR, false discovery rate.
Important genes and biological implications
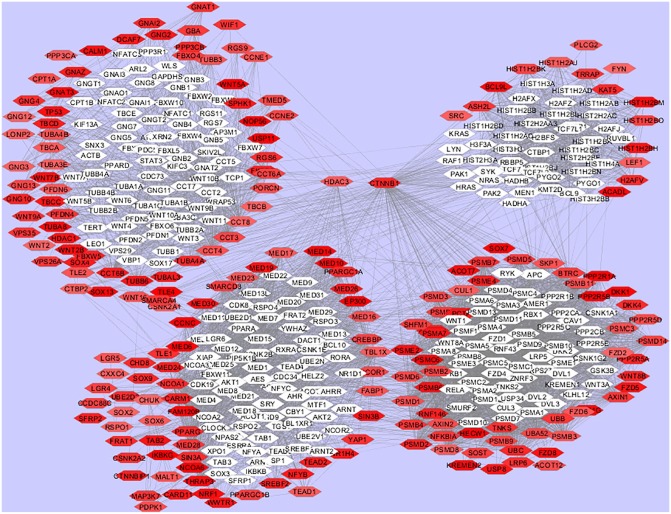
A visualization of pathways overlapping between users and non-users of platinum-based chemotherapy highlights the importance of several genes in a way that random forest analysis alone could not. By visualizing the 4 common pathways, it becomes possible to identify not only highly influential genes but also those genes that have the highest degree of connectivity to influential genes. Using annotation built into Cytoscape,56,57 we can also identify existing small molecules used in cancer therapy that are not yet commonly used in oral cancer, and we can also observe those genes previously found to be associated with oral cancer. Genes found to be influential in oral cancer for patients receiving platinum-based chemotherapy with existing literature supporting the targeting of these genes in cancer were INSR, BRAF, and PSMB7 which are targeted by Ceritinib (regorafenib and dabrafenib) and bortezomib, respectively. These drugs are not currently FDA approved for treatment in oral cancer. Genes found to be influential in oral cancer for patients not receiving platinum-based chemotherapy with existing FDA-approved chemotherapy drugs targeting the products of said genes are FGFR3, EGFR, PRKAA2, CSNK2A1, INSR, MET, CAMK2A, PSMB5, and PSMB1. There are multiple chemotherapy drugs targeting these pathways with 14 different drugs targeting EGFR alone. It should be noted that EGFR is a gene pathway being targeted in current oral cancer treatment. Sarafenib Tosylate, Pazopanib Hydrochloride, and Vadetanib all target the FGFR pathway specifically. Sunitinib Malate is unique in that it has been found to act on 4 different genes that were found by random forest to be influential in treatment response: FGFR3, CDNK2A1, PRKAA2, and CAMK2A. Again, we see that Certinib acts on a gene that is influential in both patient treatment groups, and that gene is INSR. Bortezomib, Carfilzobib, and Ixazomib all act on PSMB5, which is an influential gene in both Platinum and non-platinum-based therapy. PSMB5 and PSMB1 are both found to be within the top 5% of influential genes in random forest analysis and are genes that are significantly enriched within the sonic hedgehog pathway. Chemotherapy drugs and their relationship to genes in common influential pathways between users and non-users of platinum-based chemotherapy are visualized in Figure 1 for platinum-based chemotherapy users and Figure 2 for non-platinum-based chemotherapy users.
Figure 1.
Network visualization of pathways enriched with genes influencing platinum-based treatment response in oral cancer.
Hexagon shapes are genes. Dark red genes are of greater influence based on random forest analysis results (within top 5% of influential genes), white genes do not fall within the criteria of being in the top 5% of influential genes. All 4 common pathways enriched in patients receiving platinum and non-platinum therapy (described in Table 3) were merged into one pathway using Cytoscape. Genes were clustered according to the Reactome Pathway Plugin available via Cytoscape.
Figure 2.
Network visualization of pathways enriched with genes influencing non-platinum-based treatment response in oral cancer.
Hexagon shapes are genes. Dark red genes are of greater influence based on random forest analysis results (within top 5% of influential genes), white genes do not fall within the criteria of being in the top 5% of influential genes. All 4 common pathways enriched in patients receiving platinum and non-platinum therapy (described in Table 3) were merged into one pathway using Cytoscape. Genes were clustered according to the Reactome Pathway Plugin available via Cytoscape. Loss of ring structure is indicative of differences in influence of genes between patients receiving platinum and non-platinum therapies.
In addition to analysis of the intersection of existing cancer drugs and genes deemed influential by random forest, this study also looked at the intersection between gene topology within a pathway and random forest influence. This analysis identified CTNNB1, PLCG2, SHC1, UBA52, UBB, UBC, and HDAC3 as genes that meet filters of belonging to one of the 4 common enriched pathways, being a gene that is one of the top 5% of influential genes listed by random forest analysis and being connected to more than 100 genes within the 4 interconnected pathways (Figures 1 and 2). CTNNB1 mutations have been found to be predictive of lung and other thoracic cancers,58-62 and PLCG2 and calmodulin knockdown have been shown to induce paclitaxel sensitivity in cervical cancer tumors. This may prove of use to oral cancer patients, which may be assigned to paclitaxel or other taxol regimen.63,64 SHC1 has been shown to be a regulator of EGFR function and thus a potential target for multiple cancer types where EGFR is a key driver.65-67 Ubiquitin genes UBA52, UBB, and UBC have been shown to be associated with several cancers and research is currently being pursued in targeting ubiquitin ligases to improve treatment response.68-72 Histone-deacetylase genes specifically HDAC3 is shown to be a hub to several genes that are influential in platinum-based chemotherapy response genes that have also been associated with metastatic invasion in breast and pancreatic cancer (Figure 1). Inhibition of HDAC3 was shown to impact signaling to cancer stem cells. This gene has been shown to be a regulator of apoptosis control exerted by TP53.73-76 These genes were not shown to currently have any antibody or small molecule therapies targeting their action. High-level topology (more than 100 connected genes) and high random forest importance ranking should provide impetus for further research into the targeting of gene action in oral cancer.
Chemical informatics analysis of drug targets and leads
For those genes meeting topology and random forest filters, the known ligands of proteins expressed by each gene were identified though the RCSB protein data bank. Structural files of ligands were downloaded as .sdf files and uploaded into the chemical informatics R package Rcpi. Once loaded, each ligand had to undergo virtual screening against all FDA-approved drugs to identify existing FDA-approved drugs that may prove efficacious as therapeutic agents. Only those molecules with a Tanimoto similarity score >50% were included in results. A TCM small molecule database was also used as biologic-derived small molecules are known to provide better shape overlay when screened against other biologic small molecules. In addition, the molecules in the TCM database have been shown to be generally safe in people by merit of its long historical use in human populations. For CTNNB1, several ligands were identified via RCSB PDB (2s)-3-{[{[(2s)-2,3-dihydroxypropyl]aoxy}(hydroxy)phosphoryl]oxy}-2-[(6e)-hexadec-6-enoyloxy]propyl (8e)-octadec-8-enoate was the single ligand associated with CTNNB1 that was used for virtual screening against the FDA and TCM libraries. Unfortunately, neither library yielded a small molecule candidate with greater than a 50% Tanimoto score.
The ligand of PLCG2 did yield several interesting drug leads in both the FDA-approved library and the TCM library. Fludarabine has been tested in oral cancer cell lines and found to be effective in inducing cell apoptosis.77 Ganciclovir an HIV drug has also been tested in oral cancer and found to have a clinical effect on cell differentiation.78 Entecovir and didanosine are drugs used in the treatment of HepB infection and HIV and have not yet been tested on oral cancer. Small molecules in the TCM database meeting shape overlay filters were [4-[2,6-dimethylmorpholin-4-yl)sulfonylphenyl]-[4-(2-phenoxyethyl)piperazin-1-yl]-methanone which has not yet been used on oral cancer cell lines. There was overlap between drug leads for ligands of UBB and PLCG2. This is due to the structural similarity between cytosine and guanine ligands used as reference molecules for similarity matching. Drug leads for UBB included Cytarabine (Cancer), Fludarabine (Cancer), Azacitidine (myelodysplastic syndrome), Gemcitabine (Cancer), and Lamivudine (HIV). Gemcitabine is unique in that it is the only drug of those listed, which has been approved by the FDA for use in oral cancer patients. For HDAC3, there were no matches exceeding a Tanimoto score threshold of 50% of the reference molecule when using a library of FDA-approved drugs. The TCM database did yield a match with an extract from Mallotus philippensis, a member of the Euphorbiacae plant family. An extract of this plant known as Rottlerin has been found to inhibit growth of colon cancer and breast cancer cells.79,80
Quercetin and Diosmetin were other phenols found in citrus that were also identified as matches meeting Tanimoto thresholds. There are no reports of the effect of quercetin or diosmetin in oral cancer. It should be noted that Quercetin and Rottlerin have been noted in literature as promiscuous ligands that are often found in natural product in silico screenings.81 A recent study reported quercetin as the number one natural product in terms of number of occurrences within the database.82 The aforementioned studies do show that Rottlerin is related to the metastatic potential and viability of colorectal cancer cells.79 Caution and validation of results with existing literature or carefully designed follow-up experiments should always be pursued to justify the results of promising in silico analyses. These analyses enhance knowledge of genes influencing treatment response in oral cancer. This study describes how pathway, network, and chemical informatics analysis can be paired with literature review to identify drug leads for oral cancer treatment. Reference ligands associated with influential genes in the Royal Chemistry Society Protein Database are listed along with drug leads and their corresponding Tanimoto similarity scores in Table 4.
Table 4.
Drug leads identified in FDA-approved and Traditional Chinese Medicine database.
| Reference ligand | RCSB-linked protein/gene | Drug candidates FDA (disease treated) | TCM candidates | Tanimoto score (FDA), (TCM) |
|---|---|---|---|---|
| (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE | CTNNB1 | No candidates found | No candidates found | <50% |
| 5′-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE | PLCG2 | Fludarabine (lung cancer), inosine (multiple sclerosis), ganciclovir (HIV), didanosine (HIV), entecovir (HepB, HIV) | [4-(2,6-dimethylmorpholin-4-yl)sulfonylphenyl]-[4-(2-phenoxyethyl)piperazin-1-yl]-methanone | (60%, 59%, 56%, 53%, 51%), (55%) |
| CYTOSINE ARABINOSE-5′-PHOSPHATE | UBB | Cytarabine (cancer), fludarabine (cancer), azacitidine(myelodysplastic syndrome), gemcitabine (cancer), lamivudine (HIV) | No candidates found | (80%, 73%, 72%, 69%, 63%) |
| d-MYO-INOSITOL-1,4,5,6-TETRAKISPHOSPHATE | HDAC3 | No candidates | Mallotophillipen-D, quercetin, diosmetin | (NA), (77%, 77%, 77%) |
Abbreviations: TCM, Traditional Chinese Medicine.
Discussion
A machine learning approach known as random forest was used to identify genes influencing oral cancer treatment response specific to the platinum-based chemotherapy treatment type and the non-platinum-based chemotherapy treatment type. This article emphasizes the benefits of integrating the results of this line of analyses with pathway, network, and chemical informatics analysis to identify promising gene targets and drug leads. Biological plausibility of these findings was highlighted with a review of existing literature supporting the findings for pathways, genes, and small molecules that our reported approach identified as influential in oral cancer. The results of this work identify pathways influencing treatment response in platinum-based chemotherapy users, non-users, and those common to both users and non-users. Network analysis via Cytoscape allowed for the identification of those influential genes within each treatment modality group within the context of interconnected gene networks. The utility of random forest was underscored in that in addition to pathways it also provides a rank to each gene in its influence on treatment response. This approach is a low-cost method of prioritizing gene targets and drug leads. These methods are validated in that the genes identified have been shown to be associated with cancer progression in oral cancers and other cancers. Several drug leads identified were also shown to be effective in inhibiting oral cancer cells and were reported to be in different phases of the drug approval pipeline.
A possible criticism of the method outlined in this study is that there is uncertainty in the degree of trust that should be extended to random forest measures of gene influence and the inference of importance to the pathways in which “influential” genes reside. To further such criticisms, a point could be made that the Tanimoto threshold of >50% similarity could be perceived as low and the 50% difference in the molecules compared may prevent activity and may also be shown to have toxicity for a given disease state. Given such uncertainties, it may seem that the evidence supporting these methods is tenuous. This study recognizes these criticisms; however, the counterpoints must be made that gene influence is not observed in a single sample of the data, but rather in more than 20 000 permuted samples of the data in which the top-ranked genes were found to be more influential than thousands of other genes. The computational intensity provided in this study (20 000 trees and 320 tries at each node of each decision tree) provides justification of the trust provided for each gene influence value. In respect to the results yielded by chemical informatics analysis, it is important to note that the outlined chemical informatics method was able to identify gemcitabine as a drug that has been approved for use in oral cancer by the FDA. This method also identified Fludarabine and Ganciclovir which have both been reported as providing significant reduction in oral cancer cell line progression and viability.
Drug leads were identified in both FDA and TCM libraries, and the benefit in expanding the number of libraries is that it increases the probability of finding a match meeting the Tanimoto threshold of >50%. The negative aspect of adding libraries is that if computational resources are not planned for accordingly, then the amount of time required to screen against each reference molecule will scale upward with library size. The tools used in this study were all open source and freely available, and a limitation to the adoption of this pipeline is that tools and their dependencies are distributed across different R repositories that may or may not be kept up to date. Combining these tools into a single package that allows for the identification of both gene targets and drug leads may enhance the pace of drug discovery pipelines. We have shown in this study that random forest is well suited to datasets with small observations and high number of features. Gene targets that have been shown (through literature review) to be associated with treatment response and cancer progression were identified through this study’s use of random forest analysis. Stratifying this analysis by the type of chemotherapy received allows for interpretation of influential genes and pathways within the context of treatment. Indeed, the lack of overlap in the importance of genes from one treatment modality to another highlights the idea that the gene expression patterns influencing platinum-based treatment response differ from those gene expression patterns influencing non-platinum treatment response.
It is likely that there is bias inherent to the stratification of patients by chemotherapy treatment type. Chemotherapy treatment is associated with clinical variables like clinical stage, tumor size, and tumor grade, as well as gender and socioeconomic quintile.83-85 This study attempted to address these confounders by including them within the bag of randomly selected features available for construction of the random forest model. By integrating chemical informatics analyses, random forest results can be translated into lists of drug leads for each target gene. This method identified drug leads that have already entered or passed phase 3 trials. Our review of identified drug leads and comparison with existing annotations show that the chemical informatics methods described can identify small molecules with therapeutic potential. This study provides the impetus for further exploration of the role of the identified small molecules in oral cancer treatment response and the targeting of those genes identified as most influential by our series of analyses. This study also serves as a model for researchers identifying gene targets in rare cancers where the number of cases is limited.
Supplemental Material
Supplemental material, Supplemental_material for Identification of Targetable Pathways in Oral Cancer Patients via Random Forest and Chemical Informatics by John Schomberg in Cancer Informatics
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of conflicting interests:The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: JS is employed in bioinformatics research at Afecta Pharmaceuticals and the platform Pharmetrx.ai which is a branch of Afecta Pharmaceuticals.
Author Contributions: JS was the sole contributor to manuscript composition, experimental design, analysis, and production of all tables and figures in this manuscript.
ORCID iD: John Schomberg  https://orcid.org/0000-0001-5175-5911
https://orcid.org/0000-0001-5175-5911
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2. Cooper JS, Porter K, Mallin K, et al. National cancer database report on cancer of the head and neck: 10-year update. Head Neck. 2009;31:748-758. doi: 10.1002/hed.21022. [DOI] [PubMed] [Google Scholar]
- 3. Sinha P, Logan HL, Mendenhall WM. Human papillomavirus, smoking, and head and neck cancer. Am J Otolaryngol. 2012;33:130-136. doi: 10.1016/j.amjoto.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers. Cancer. 2007;110:1429-1435. doi: 10.1002/cncr.22963. [DOI] [PubMed] [Google Scholar]
- 5. Kelleher FC. Hedgehog signaling and therapeutics in pancreatic cancer. Carcinogenesis. 2011;32:445-451. doi: 10.1093/carcin/bgq280. [DOI] [PubMed] [Google Scholar]
- 6. Saqui-Salces M, Merchant JL. Hedgehog signaling and gastrointestinal cancer. Biochim Biophys Acta. 2010;1803:786-795. doi: 10.1016/j.bbamcr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang J, Hui C. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801-812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia E, Ragazzini C, Yu X, et al. WIP and WICH/WIRE coordinately control invadopodium formation and maturation in human breast cancer cell invasion. Sci Rep. 2016;6:23590. doi: 10.1038/srep23590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo B, Fu S, Zhang J, Liu B, Li Z. Targeting inflammasome/IL-1 pathways for cancer immunotherapy. Sci Rep. 2016;6:36107. doi: 10.1038/srep36107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aminuddin A, Ng PY. Promising druggable target in head and neck squamous cell carcinoma: Wnt signaling. Front Pharmacol. 2016;7:244. doi: 10.3389/fphar.2016.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baek KH, Zaslavsky A, Lynch RC, et al. Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature. 2009;459:1126-1130. doi: 10.1038/nature08062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peuker K, Muff S, Wang J, et al. Epithelial calcineurin controls microbiota-dependent intestinal tumor development. Nat Med. 2016;22:506-515. doi: 10.1038/nm.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao X, Wang Q, Yang S, et al. Quercetin inhibits angiogenesis by targeting calcineurin in the xenograft model of human breast cancer. Eur J Pharmacol. 2016;781:60-68. doi: 10.1016/j.ejphar.2016.03.063. [DOI] [PubMed] [Google Scholar]
- 14. National Comprehensive Cancer Network (NCCN). Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Head and Neck. Version 1.2016; 2016:1-174. https://oralcancerfoundation.org/wp-content/uploads/2016/09/head-and-neck.pdf. [Google Scholar]
- 15. Breiman L. Random forests. Mach Learn. 2001;45:5-32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 16. Qi Y. Random forest for bioinformatics. In: Zhang C, Ma Y, eds. Ensemble Machine Learning. Boston, MA: Springer; 2012:307-323. doi: 10.1007/978-1-4419-9326-7_11. [DOI] [Google Scholar]
- 17. Chen X, Wang L, Ishwaran H. An integrative pathway-based clinical-genomic model for cancer survival prediction. Stat Probab Lett. 2010;80:1313-1319. doi: 10.1016/j.spl.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mutanga O, Adam E, Cho MA. High density biomass estimation for wetland vegetation using worldview-2 imagery and random forest regression algorithm. Int J Appl Earth Obs Geoinf. 2012;18:399-406. doi: 10.1016/j.jag.2012.03.012. [DOI] [Google Scholar]
- 19. Tong W, Xie Q, Hong H, et al. Using decision forest to classify prostate cancer samples on the basis of SELDI-TOF MS data: assessing chance correlation and prediction confidence. Environ Health Perspect. 2004;112:1622-1627. doi: 10.1289/txg.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen XW, Liu M. Prediction of protein-protein interactions using random decision forest framework. Bioinformatics. 2005;21:4394-4400. doi: 10.1093/bioinformatics/bti721. [DOI] [PubMed] [Google Scholar]
- 21. Phillips M, Cataneo RN, Ditkoff BA, et al. Prediction of breast cancer using volatile biomarkers in the breath. Breast Cancer Res Treat. 2006;99:19-21. doi: 10.1007/s10549-006-9176-1. [DOI] [PubMed] [Google Scholar]
- 22. Pang H, Zhao H. Stratified pathway analysis to identify gene sets associated with oral contraceptive use and breast cancer. Cancer Inform. 2014;13:73-78. doi: 10.4137/CIN.S13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS. Random survival forests. Ann Appl Stat. 2008;2:841-860. doi: 10.1214/08-AOAS169. [DOI] [Google Scholar]
- 24. Irwin JJ, Shoichet BK. ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177-182. doi: 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams A, Tkachenko V. The Royal Society of Chemistry and the delivery of chemistry data repositories for the community. J Comput Aided Mol Des. 2014;28:1023-1030. doi: 10.1007/s10822-014-9784-5. [DOI] [PubMed] [Google Scholar]
- 26. Bajusz D, Racz A, Heberger K. Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations. J Cheminform. 2015;7:20. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao DS, Xiao N, Xu QS, Chen AF. Rcpi: R/Bioconductor package to generate various descriptors of proteins, compounds and their interactions. Bioinformatics. 2015;31:279-281. doi: 10.1093/bioinformatics/btu624. [DOI] [PubMed] [Google Scholar]
- 28. Monteith GR, McAndrew D, Faddy HM, Roberts-Thomson SJ. Calcium and cancer: targeting Ca2+ transport. Nat Rev Cancer. 2007;7:519-530. doi: 10.1038/nrc2171. [DOI] [PubMed] [Google Scholar]
- 29. Pahor M, Guralnik JM, Salive ME, Corti MC, Carbonin P, Havlik RJ. Do calcium channel blockers increase the risk of cancer? Am J Hypertens. 1996;9:695-699. doi: 10.1016/0895-7061(96)00186-0. [DOI] [PubMed] [Google Scholar]
- 30. Pahor M, Guralnik JM, Ferrucci L, et al. Calcium-channel blockade and incidence of cancer in aged populations. Lancet. 1996;348:493-497. doi: 10.1016/S0140-6736(96)04277-8. [DOI] [PubMed] [Google Scholar]
- 31. Giudice FS, Pinto DS, Jr, Nor JE, Squarize CH, Castilho RM. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS ONE. 2013;8:e58672. doi: 10.1371/journal.pone.0058672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Erlich RB, Kherrouche Z, Rickwood D, et al. Preclinical evaluation of dual PI3K-mTOR inhibitors and histone deacetylase inhibitors in head and neck squamous cell carcinoma. Br J Cancer. 2012;106:107-115. doi: 10.1038/bjc.2011.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haigentz M, Jr, Kim M, Sarta C, et al. Phase II trial of the histone deacetylase inhibitor romidepsin in patients with recurrent/metastatic head and neck cancer. Oral Oncol. 2012;48:1281-1288. doi: 10.1016/j.oraloncology.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349-354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 35. Gulino A, Ferretti E, De Smaele E. Hedgehog signalling in colon cancer and stem cells. EMBO Mol Med. 2009;1:300-302. doi: 10.1002/emmm.200900042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313-317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 37. Shaw G, Price AM, Ktori E, et al. Hedgehog signalling in androgen independent prostate cancer. Eur Urol. 2008;54:1333-1343. doi: 10.1016/j.eururo.2008.01.070. [DOI] [PubMed] [Google Scholar]
- 38. Kasper M, Jaks V, Fiaschi M, Toftgård R. Hedgehog signalling in breast cancer. Carcinogenesis. 2009;30:903-911. doi: 10.1093/carcin/bgp048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851-856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parikh N, Hilsenbeck S, Creighton CJ, et al. Effects of TP53 mutational status on gene expression patterns across 10 human cancer types. J Pathol. 2014;232:522-533. doi: 10.1002/path.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC. Targeting FGFR signaling in cancer. Clin Cancer Res. 2015;21:2684-2694. doi: 10.1158/1078-0432.CCR-14-2329. [DOI] [PubMed] [Google Scholar]
- 42. Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20:3299-3309. doi: 10.1158/1078-0432.CCR-13-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cerliani JP, Vanzulli SI, Pinero CP, et al. Associated expressions of FGFR-2 and FGFR-3: from mouse mammary gland physiology to human breast cancer. Breast Cancer Res Treat. 2012;133:997-1008. doi: 10.1007/s10549-011-1883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sadeghifar F, Bohm S, Vintermist A, Ostlund Farrants AK. The B-WICH chromatin-remodelling complex regulates RNA polymerase III transcription by promoting Max-dependent c-Myc binding. Nucleic Acids Res. 2015;43:4477-4490. doi: 10.1093/nar/gkv312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang X, Jan LY. Targeting potassium channels in cancer. J Cell Biol. 2014;206:151-162. doi: 10.1083/jcb.201404136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bonnet S, Archer SL, Allalunis-Turner J, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37-51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 47. Chang K-W, Yuan T-C, Fang K-P, et al. The increase of voltage-gated potassium channel Kv3.4 mRNA expression in oral squamous cell carcinoma. J Oral Pathol Med. 2003;32:606-611. [DOI] [PubMed] [Google Scholar]
- 48. Leanza L, Venturini E, Kadow S, Carpinteiro A, Gulbins E, Becker KA. Targeting a mitochondrial potassium channel to fight cancer. Cell Calcium. 2015;58:131-138. doi: 10.1016/j.ceca.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 49. Lew T-S, Chang C-S, Fang K-P, Chen C-Y, Chen C-H, Lin S-C. The involvement of K(v)3.4 voltage-gated potassium channel in the growth of an oral squamous cell carcinoma cell line. J Oral Pathol Med. 2004;33:543-549. doi: 10.1111/j.1600-0714.2004.00236.x. [DOI] [PubMed] [Google Scholar]
- 50. Pannone G, Bufo P, Santoro A, et al. WNT pathway in oral cancer: epigenetic inactivation of WNT-inhibitors. Oncol Rep. 2010;24:1035-1041. doi: 10.3892/or.2010.1035. [DOI] [PubMed] [Google Scholar]
- 51. Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2012;13:11-26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 52. Barker N, Clevers H. Mining the Wnt pathway for cancer therapeutics. Nat Rev Drug Discov. 2006;5:997-1014. doi: 10.1038/nrd2154. [DOI] [PubMed] [Google Scholar]
- 53. Qin JJ, Nag S, Wang W, et al. NFAT as cancer target: mission possible. Biochim Biophys Acta. 2014;1846:297-311. doi: 10.1016/j.bbcan.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mancini M, Toker A. NFAT proteins: emerging roles in cancer progression. Nat Rev Cancer. 2009;9:810-820. doi: 10.1038/nrc2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645-656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 56. Yeung N, Cline MS, Kuchinsky A, Smoot ME, Bader GD. Exploring biological networks with cytoscape software. Curr Protoc Bioinformatics. 2008;8:13. doi: 10.1002/0471250953.bi0813s23. [DOI] [PubMed] [Google Scholar]
- 57. Manual CU. Cytoscape user manual. Syst Biol (Stevenage). 2011;163:18-28. doi: 10.1111/j.1476-5381.2010.01178.x. [DOI] [Google Scholar]
- 58. Watanabe K, Biesinger J, Salmans ML, et al. Integrative ChIP-seq/microarray analysis identifies a CTNNB1 target signature enriched in intestinal stem cells and colon cancer. PLoS ONE. 2014;9:e92317. doi: 10.1371/journal.pone.0092317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Geyer FC, Lacroix-Triki M, Savage K, et al. Β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24:209-231. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 60. Morikawa T, Kuchiba A, Lochhead P, et al. Prospective analysis of body mass index, physical activity, and colorectal cancer risk associated with β-catenin (CTNNB1) status. Cancer Res. 2013;73:1600-1610. doi: 10.1158/0008-5472.CAN-12-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74-83. doi: 10.1016/j.ygeno.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 62. Hirata H, Hinoda Y, Ueno K, Shahryari V, Tabatabai L, Dahiya R. MicroRNA-1826 targets VEGFC, beta-catenin (CTNNB1) and MEK1 (MAP2K1) in human bladder cancer. Carcinogenesis. 2012;33:41-48. doi: 10.1093/carcin/bgr239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Recurrent BTK and PLCG2 mutations confer ibrutinib resistance. Cancer Discov. 2014;4:866. doi: 10.1158/2159-8290.CD-RW2014-128. [DOI] [Google Scholar]
- 64. Stanislaus A, Bakhtiar A, Salleh D, et al. Knockdown of PLC-gamma-2 and calmodulin 1 genes sensitizes human cervical adenocarcinoma cells to doxorubicin and paclitaxel. Cancer Cell Int. 2012;12:30. doi: 10.1186/1475-2867-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Choi KY, Cho YJ, Kim JS, Ahn YH, Hong SH. SHC1 sensitizes cancer cells to the 8-Cl-cAMP treatment. Biochem Biophys Res Commun. 2015;463:673-678. doi: 10.1016/j.bbrc.2015.05.123. [DOI] [PubMed] [Google Scholar]
- 66. Zheng Y, Zhang C, Croucher DR, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166-171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. SHC1 temporally regulates EGFR signaling. Cancer Discov. 2013;3. doi: 10.1158/2159-8290.CD-RW2013-154. [DOI] [Google Scholar]
- 68. Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006; 6:776-788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 69. Kirkin V, Dikic I. Ubiquitin networks in cancer. Curr Opin Genet Dev. 2011;21:21-28. doi: 10.1016/j.gde.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 70. Ohta T, Fukuda M. Ubiquitin and breast cancer. Oncogene. 2004;23:2079-2088. doi: 10.1038/sj.onc.1207371. [DOI] [PubMed] [Google Scholar]
- 71. Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Ther. 2003;2:623-629. doi: 10.4161/cbt.2.6.677. [DOI] [PubMed] [Google Scholar]
- 72. Shi D, Grossman SR. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol Ther. 2010;10:737-747. doi: 10.4161/cbt.10.8.13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sebban S, Farago M, Gashai D, et al. Vav1 fine tunes p53 control of apoptosis versus proliferation in breast cancer. PLoS ONE. 2013;8:e54321. doi: 10.1371/journal.pone.0054321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ilan L, Katzav S. Human Vav1 expression in hematopoietic and cancer cell lines is regulated by c-Myb and by CpG methylation. PLoS ONE. 2012;7:e29939. doi: 10.1371/journal.pone.0029939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Razidlo GL, Schroeder B, Chen J, Billadeau DD, McNiven MA. Vav1 as a central regulator of invadopodia assembly. Curr Biol. 2014;24:86-93. doi: 10.1016/j.cub.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Razidlo GL, Magnine C, Sletten AC, et al. Targeting pancreatic cancer metastasis by inhibition of Vav1, a driver of tumor cell invasion. Cancer Res. 2015;75:2907-2915. doi: 10.1158/0008-5472.CAN-14-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nitsche M, Christiansen H, Hermann RM, et al. The combined effect of fludarabine monophosphate and radiation as well as gemcitabine and radiation on squamous carcinoma tumor cell lines in vitro. Int J Radiat Biol. 2008;84:643-657. doi: 10.1080/09553000802241754. [DOI] [PubMed] [Google Scholar]
- 78. Neves SS, Sarmento-Ribeiro AB, Simoes SP, Pedroso de Lima MC. Transfection of oral cancer cells mediated by transferrin-associated lipoplexes: mechanisms of cell death induced by herpes simplex virus thymidine kinase/ganciclovir therapy. Biochim Biophys Acta. 2006;1758:1703-1712. doi: 10.1016/j.bbamem.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 79. Juneja M, Kobelt D, Walther W, et al. Statin and rottlerin small-molecule inhibitors restrict colon cancer progression and metastasis via MACC1. Plos Biol. 2017;15:e2000784. doi: 10.1371/journal.pbio.2000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yin X, Zhang Y, Su J, et al. Rottlerin exerts its anti-tumor activity through inhibition of Skp2 in breast cancer cells. Oncotarget. 2016;7:66512-66524. doi: 10.18632/oncotarget.11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Soltoff SP. Rottlerin: an inappropriate and ineffective inhibitor of PKCδ. Trends Pharmacol Sci. 2007;28:453-458. doi: 10.1016/j.tips.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 82. Bisson J, McAlpine JB, Friesen JB, Chen SN, Graham J, Pauli GF. Can invalid bioactives undermine natural product-based drug discovery. J Med Chem. 2016;59:1671-1690. doi: 10.1021/acs.jmedchem.5b01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Saloura V, Langerman A, Rudra S, Chin R, Cohen EEW. Multidisciplinary care of the patient with head and neck cancer. Surg Oncol Clin N Am. 2013;22:179-215. doi: 10.1016/j.soc.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 84. Marur S, Forastiere AA. Head and neck squamous cell carcinoma: update on epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2016;91:386-396. doi: 10.1016/j.mayocp.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 85. Mesía Nin R, Pastor Borgoñón M, Cruz Hernández JJ, Isla Casado D. SEOM clinical guidelines for the treatment of head and neck cancer. Clin Transl Oncol. 2010;12:742-748. doi: 10.1007/s12094-010-0589-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental_material for Identification of Targetable Pathways in Oral Cancer Patients via Random Forest and Chemical Informatics by John Schomberg in Cancer Informatics