Abstract
Since the advent of whole slide imaging, the utility of digitized slides for education in medical school and residency has been amply documented. Pathology departments at most major academic medical centers have made digitized slides available to pathology residents for study, even before the use of digitized slides for clinical purposes (i.e., primary diagnosis) has become commonplace. This article describes the experience of one academic medical center with the storage and indexing of large volumes of digitized slides. Our goal was to be able to retrieve scanned slides for a variety of educational applications and thereby maximize the heuristic value of the slides. This posed a formidable challenge in terms of development and deployment of an index system that would allow exemplary slides to be identified and retrieved irrespective of the purpose for which the slide was scanned. We used the structure inherent in Aperio's image management software (eSlide Manager) to build an educational database that allowed each image to be appended with a unique taxonomic identifier so that the individual files could be retrieved in a flexible and utilitarian manner.
Keywords: Diagnosis, eSlide manager, pathology, taxonomy
INTRODUCTION
Since the advent of whole slide imaging (WSI), the utility of digitized slides for education in medical school and residency has been amply documented. Entire histology and histopathology databases exist on digital platforms,[1,2,3] “unknown” slides are routinely made available to residents for review prior to conferences,[1] many major academic medical centers have made thematic slide collections digitally available for medical student[4,5,6] and resident[7,8,9] education, and continuing medical education seminars in pathology are frequently hosted on digital platforms. These applications have enhanced the educational value of histologic and pathologic slides by affording use that is unrestricted in time or place and by preventing loss of educational slide sets through breakage, misplacement, and theft. WSI has increased accessibility to valuable educational material by sharing slide collections that are otherwise locked away in individual pathologist's offices. Digitization of glass slides is contributing to the process of democratization of information, as seen in other fields, because the best and most illustrative material is less likely to be restricted to the most popular consultation and highly rated academic centers. At the same time, digital storage of educational slides has decreased laboratory cost associated with generation of educational “recuts” for individual teaching slide collections.
The experience at the University of Wisconsin-Madison Department of Pathology and Laboratory Medicine (UW) was similar to that of many other educational programs when it first began to scan slides with a WSI device. The initial impetus to whole slide scanning was to make “Unknowns” slides available to residents to preview from off-site for their weekly conference. The number of slides in the database rapidly increased, as did the number of ways in which the diagnosis was recorded with a slide. Despite the concerted effort of excellent chief residents, continuity in WSI slide management was lax. There were typographical errors and problems with standardized nomenclature, not to mention the haphazard placement of scanned images into random “interesting cases” folders. This made searching the WSI storage system to utilize the slides for other educational applications virtually impossible. For example, if an educator were interested in putting together a lesson on inflammatory bowel disease, potentially pertinent slides may have been added to the database with labels as diverse as “Crohn disease,” “toxic megacolon (UC),” “colovaginal fistula,” “cecal red patch,” “inflammatory bowel disease, NOS,” and “anal fissure,” to name a few. For all intents and purposes, the vast database that was rapidly accumulating was using up storage space with little to no utility past the “Unknowns” conference itself.
The acquisition of scanners and associated file storage software was an economic challenge willingly born by the department in the interest of resident education; but, as often occurs with adoption of new technologies, it was done without due consideration of the work and infrastructure required to fully reap the benefits of implementation. The assumption that residents on rotation and support staff would themselves set up and maintain a system without serious consideration of what that infrastructure would entail, led to confusion and expenditure of resources for little educational gain. In short, 2 years' worth of slides (approximately 1000 WSI s) are now taking up electronic storage space without any hope of retrieval of their educational value without an extensive, manual, and time-consuming investment in their recovery.
At the same time, faculty at the UW had extensive personal collections of educational cases that were mostly inaccessible to residents because the residents either did not know of the existence of the slide collections, or faculty were hesitant to loan them for fear of attrition of the valuable glass slides they had collected over decades of practice. The slides were often kept in an inchoate manner and laboriously accessed via reference to handwritten notes or, at best, an Excel spreadsheet that was not always kept up-to-date. Moreover, the physical slide boxes were apt to become disorganized as the slides were inadvertently shuffled during the process of their removal for microscopic examination. More serious yet, documentation associated with the slides was prone to be lost when the collections were loaned out for study.
The leadership of the Surgical Pathology division decided to develop a unified taxonomic structure based on standardized terminology to support the WSI database. Such an infrastructure was deemed necessary to derive maximal value from the educational database and to encourage faculty to scan their slide boxes to a permanent and readily accessible platform. The logic inherent in the Aperio eSlide Manager (previously Spectrum) (Leica Biosystems, Nussloch, Germany) software offered the ability to preserve the organization of the faculty's slide boxes around themes or lessons (e.g. “infections of the placenta,” “gastrointestinal curriculum,” or “pattern-based approach to soft tissue tumors”). To make the scanned slides searchable and recoverable across all folders within the database, we created a unique “taxonomic string” associated as a type of metadata with each glass slide. The taxonomic string contains the organ of origin, the pathologic process, and standardized disease nomenclature, as well as subclassification where appropriate (e.g. for a case of Crohn Disease the taxonomic string leading to the disorder's niche would be: small intestine → Reactive, including: Inflammatory, Autoimmune, Degenerative and Metabolic Disorders → Idiopathic inflammatory bowel disease → Crohn's disease).
The unique taxonomic string appended to each WSI file is searchable with the “advanced search” option in the Aperio eSlide Manager software. Examples of the same disease entity scanned into several different folders can be recovered via a search on any portion of the taxonomic string. This indexing system has greatly increased the utility of the educational database by allowing images to be rapidly recovered and accessed in a comprehensive manner. Less time is spent searching for didactic material, and access to illustrative examples for teaching and publication has been greatly facilitated.
Procedure
Slides are selected for scanning by residents and faculty for conferences, tumor boards, and other clinical and academic purposes. Files of whole scanned slides are stored on a server linked to our Aperio C2 WSI scanner and organized and viewed using Aperio eSlide Manager database software provided by Leica/Aperio Imaging systems. The image is displayed on a monitor via Aperio WebScope (platform independent web-based image viewer) or Aperio ImageScope (free downloadable image viewer from Aperio/Leica).
The taxonomic system was created by subspecialized pathology faculty at the UW. The classification and nomenclature of neoplastic diseases was adopted from the latest editions of the World Health Organization (WHO) Classification of Tumors Atlases (International Agency for Research on Cancer, Lyon, France). The sequence of nodes in the taxonomic strings was standardized by the lead author. The complete system consists of >5000 rows in an Excel spreadsheet, with each row containing a unique taxonomic string. The taxonomic strings were not intended to be dogmatic statements about the pathophysiologic disease processes, but to reflect a current classification and disease etiology that most experts would agree on. For the most part, the structure of the taxonomy reflects disease classifications found in most standard pathology textbooks. In some cases, the classification is a judgment call predicated on an incomplete understanding of the pathophysiologic process. For example, “juxta articular myxoma” is arguably neoplastic or inflammatory; our taxonomic string puts the entity into the “neoplastic” category with full recognition that the exact nature of the entity is currently unknown. The string in this case is merely a placeholder for the disease entity. Such judgment calls are surprisingly rare across the spreadsheet.
The first node in the taxonomy is the name of the organ or organ system. For practical purposes, some organs or tissues were grouped together, such as “peritoneum” with “omentum,” and “colon” with “rectum.” This resulted in a list of 42 organs. The next node consists of disease processes, such as neoplastic, inflammatory, infectious, genetic, and so forth. The choices in this node vary slightly across the organ systems. From this node, the taxonomic tree branches into variable subclassification lists, each of which themselves may have additional taxonomic “children.”
The original excel spreadsheet was put into a software parser written in C# (C Sharp) (Microsoft Corporation, Redmond, WA, USA). The parser creates a directed tree data structure and stores it in a Microsoft SQL Server database. Each individual taxonomic term of each taxonomic string is a node in the tree. Traversing the tree in any route from the root node to a leaf node produces a valid taxonomic string.
A web-based interface for the database was created in the C# language using the ASP.NET MVC framework. The interface provides a way to visually traverse the taxonomy as well as execute text-based searches that return taxonomic strings that contain the search terms. This interface was integrated with our existing web form for submitting slide scanning requests. Users submitting a scanning request associate a taxonomic string with each slide on the form by either selecting the appropriate terms from a series of drop-down menus, or by performing a text search and selecting one of the returned taxonomic strings. Because the user does not manually type out the taxonomic string, there is no possibility of clerical errors that would complicate future searches. The form will not let users submit slides without entering a valid taxonomic string for each one. In the event that the appropriate taxonomic string for a given slide is not present in the taxonomic database, the submitter would mark the slide as “unclassified.” This will allow the user to submit the slide scan request form, and an email will automatically be generated and sent to the curator of the taxonomic tree. The curator and the submitter can then craft the appropriate string and request that it be added to the taxonomic tree. This level of oversight was deemed necessary to ensure that users cannot create conflicting terms which would diminish the database's integrity and utility. A similar mechanism can be utilized if the user discovers an error in the existing taxonomic string at the time of submission or if the string needs to be updated to incorporate the evolving pathology nomenclature.
On receipt of a submitted slide scan request form, the submitted slides are scanned using the Aperio CS2 WSI and uploaded to the Aperio eSlide Manager server. The digital slides are then assigned to the appropriate lesson in Aperio eSlide Manager. The taxonomic string and other associated information submitted on the scanning request form are entered into the corresponding digital slide record in Aperio eSlide Manager. To avoid transcription errors, each taxonomic string in the request form has a simple “copy” button next to it. The technician performing the scan can simply click the “copy” button to import the taxonomic string to the computer's clipboard, so it can be pasted into Aperio eSlide Manager without need for typing. After all slides have been scanned and appropriately sorted in Aperio eSlide Manager, the request is marked as completed, and the submitter receives an automated email indicating that the slides are available for review in Aperio eSlide Manager.
RESULTS
The Aperio eSlide Manager software offers the ability to “nest” subfolders into folders in a hierarchical organization that is well suited for creating image groups around “lessons” similar to chapters in textbooks. In pathology, it is particularly suitable for storage and organization of slide sets, including organ-based study sets and case collections. For organ-based study sets, particular diagnostic entities can easily be stored and retrieved by following an intuitive pathway, such as Gynecologic pathology → Neoplastic Diseases of the Ovary → Germ cell tumors. However, case collections such as “Unknowns,” The College of American Pathologists' (CAP) Performance Improvement Program (PIP) slide sets (CAP, Chicago, IL, USA), or “Interesting cases on the gastroenterology service” are not recoverable by diagnosis (if only using the Aperio eSlide Manager) because they are not scanned into the database in a thematic manner. In other words, the organization inherent in the Aperio eSlide Manager software is well suited for the creation of “lessons” but does not allow searching across the “lessons” or folders for entities that may be present in more than one “lesson” folder. For example, “angiolipoma” may be present in a PIP collection, the soft tissue curriculum, resident weekly “unknowns,” and a collection of dermatopathology. If only using the Aperio eSlide Manager software, a user interested in seeing as many examples of angiolipoma as possible would have to search for that entity in every one of the lessons.
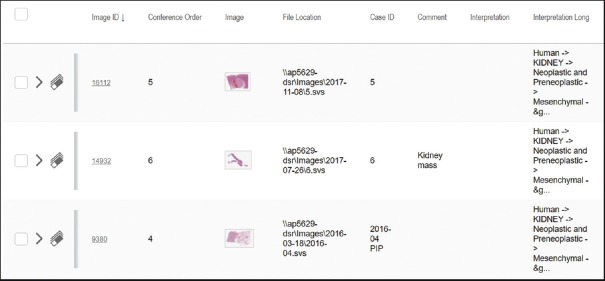
We have circumvented this problem of inaccessibility by appending a standardized taxonomic string to every image scanned into the Aperio database of educational slides. The taxonomic string contains the name of the organ, the pathophysiologic disease process, and the standard nomenclature for the diagnostic entity. In many cases, further granularity is achieved by addition of the grade of the disease process (e.g. primary biliary cholangitis Grades 1 through 4, or neuroendocrine tumor [NET] G1–G3). A search can be performed on any portion of the taxonomic string, across all image files within the educational database. In the example in Figure 1, the search for “angiomyolipoma” retrieved three images in two folders (Case ID column). These can now easily be viewed by clicking on the thumbnail of the image.
Figure 1.
Screenshot of search performed for “Angiomyolipoma” in the “Interpretation Long” field. This search found three images in two folders. The Case ID column identifies the two folders: the “Resident Unknowns” folder (indicated by numbers 5 and 6) and the College of American Pathologists' Performance Improvement Program folder. The full taxonomic string (in the Interpretation Long column) is only partly displayed due to settings in the Aperio eSlide Manager software
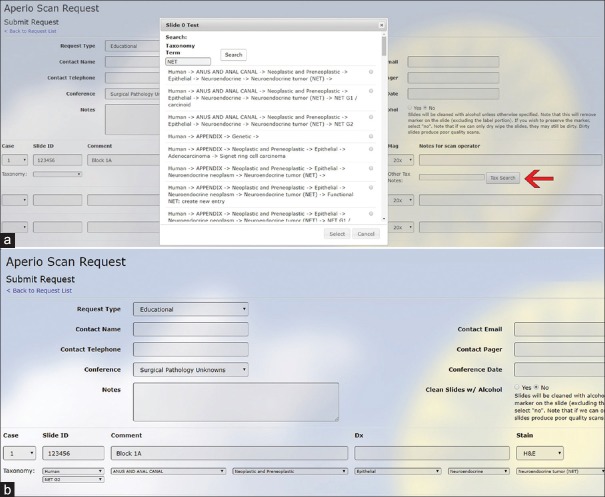
The program that was created to locate and append a taxonomic string to individual scanned slide files is easy to use. The appropriate taxonomic string can be retrieved in one of two ways. A unique term such as “angiolipoma,” “Crohn,” or an abbreviation such as “NET” can be entered into the search field. Our taxonomic program displays all the taxonomic strings in which this term occurs [Figure 2], and the user selects the appropriate string by clicking on the corresponding radio button. This will import the entire taxonomic string into the digital slide scan request form. Alternatively, to fill out the digital slide scan request form, the user can drill down on the appropriate taxonomic string by beginning with the most comprehensive term (the organ system) in the first drop-down menu and then selecting the next appropriate term in each of the subsequent boxes, representing nodes [Figure 3]. This pathway is more labor intensive, but easier for entities that share a name across many organ systems, such as “adenocarcinoma” or “Histoplasmosis.”
Figure 2.
Search function on the slide scan request form. (a) Clicking the “Tax Search” button (red arrow) on the Aperio Scan Request Form opens a search window. The search does not have complete fidelity, as it also pulls up nonrelated hits on that string, including, for example, “Genetic.” To see all the hits, one can use the scroll bar to move down the list. The string selected with the radio button will import the taxonomic string into the slide scan request form (b)
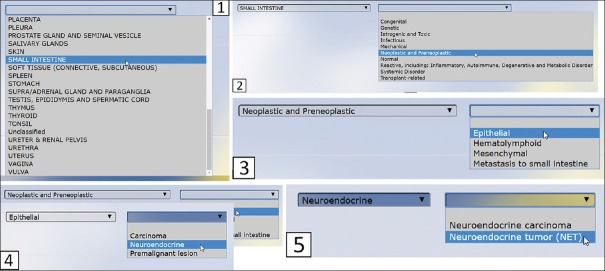
Figure 3.
Manual selection of nodes in slide scan request form. Selection of any of the options in the dropdown boxes prompts the choices available in successive nodes, as illustrated with the path for the taxonomic string for neuroendocrine tumor
The taxonomic string can also be appended to an already existing digital file from an Aperio scanner or other compatible source, i.e. previously digitized educational slides can retroactively be brought into the searchable database by adding the taxonomic string.
Because the taxonomic strings for each digital slide are stored in an associated text field in Aperio eSlide Manager, they are easily searchable across that field using Aperio eSlide Manager's built in “Advanced Search” feature. Using this feature, users can limit their search to the taxonomic string field and search based on a whole taxonomic string or just a portion of it. Aperio eSlide Manager will return each digital slide for which the taxonomic string matches the search term, regardless of which lesson it is assigned to. Each digital slide in the search result can then be viewed by the user by clicking on the slide's thumbnail. For example, if the user was preparing a presentation on neoplastic conditions in the ovary, a search for “ovary → neoplastic” would recover all ovarian neoplasms, benign and malignant, currently contained in the database. Alternatively, if the user were interested only in adult granulosa cell tumors, a search containing only the terms “adult granulosa” would retrieve only slides showing that entity.
DISCUSSION
The creation of WSI was a marvelous technological feat, but the day-to-day utility of files created from that technology remains limited by difficulties in systematic image storage and retrieval. The management of storage systems is a formidable informatics challenge. WSI files cannot even contribute to “big data” if they are not adequately tagged for retrieval. This project represents an attempt to manage the large digital WSI files in a robust and comprehensive manner while at the same time allowing flexibility in (a) construction of educational “lessons” using existing image management software and (b) revision of the taxonomy based on the inevitable evolution of classificatory nomenclature in pathology.
The taxonomic index developed by the Surgical Pathology Team at the UW amplifies the educational value of the large number of images stored in our digital slide database, by allowing the images to be easily retrieved by diagnosis or disease process. The utility of the scanned images is no longer limited to the lesson or slide collection to which it is restricted by the logic inherent in the native Aperio eSlide Manager software. The taxonomic string appended to each image allows every slide to be recovered by performing an advanced search on any segment of the taxonomic string. Rapid recall of slides is beneficial in two obvious situations: (1) a learner would like to see a selection of examples of a disease entity, such as hepatocellular carcinoma or neuroblastoma, to become familiar with its various histologic manifestations. Retrieval of slides from the Aperio eSlide Manager is much more simple and rapid relative to conventional retrieval through a search of the electronic medical record followed by manual retrieval of fragile material from the glass slide filing system. Residents can quickly pull up examples of such diseases while studying or when developing a differential diagnosis for a case in real time. (2) A pathologist needs still photomicrographs of a particular disease for a lecture or presentation. The Aperio software allows high-resolution still images to be screen captured from a digital slide and directly exported to a PowerPoint presentation. Recalling diagnostic entities via the taxonomic string while sitting at a desktop computer is much easier and quicker than hunting for slides even in one's own educational glass slide collections.
The taxonomic strings devised in this project are not intended to make an absolute, scientific, or philosophic statement about the nature of the diagnostic entity. Instead, the taxonomy represents an entirely utilitarian classification scheme. Each string reflects a current, generally acceptable representation of the organ, disease process, and contemporary nomenclature for every disease entity that might be encountered in pathology. The string is the metadata that allows each slide to be recalled from the database. The taxonomy was created with the full recognition that nomenclature changes as the understanding of disease processes change. The taxonomic strings can be changed. The search function identifies all the images appended with a string that needs correction, and in a matter of minutes, all the strings can be updated with the new nomenclature. With proper curation, slides do not lose their relevance simply because the name of the disease process has changed.
At the beginning of this project, we explored whether already-existing systems of disease nomenclature, such as SNOMED, International Classification of Diseases for Oncology (ICD-O), or ICD-10 codes, could be used for our purposes. We decided not to pursue these alternatives for a variety of reasons. (a) SNOMED and the various versions of ICD are created for epidemiologic and clinical research and for billing, and therefore, their decision points are not entirely consistent with those that are relevant to a pathologist. For example, SNOMED codes “cancer of the stomach” by its anatomic location in the stomach, which is largely irrelevant to the morphologic diagnosis, but does not differentiate between the various morphologic types of gastric cancer as defined by the WHO and as relevant to pathologic practice. To add this level of detail, the SNOMED codes would need to be augmented by ICD-O for neoplastic diseases. For nonneoplastic diseases, SNOMED codes do not always drill down on pathologic processes as the pathologist sees them. For example, “fistula” cannot be differentiated as to whether it occurs in the setting of radiologic damage or Crohn's disease. (b) Instructions for use of ICD-O, with its six layers of numeric identifiers, are described over 40 pages of the ICD-O manual (Fritz et al., ICD-O, 3rd ed.), i.e. it is far from intuitive. SNOMED is even less so. We wanted to create a system that was intuitive and reflected the manner in which pathologists approach disease classification. (c) Both SNOMED and ICD revisions are published periodically and affect all diagnoses and terminological strings, at once. Revision of the educational taxonomy to keep up with periodic, holistic, all-encompassing revisions would be much more cumbersome than the piecemeal revisions that can be accomplished on an as-needed basis with the UW taxonomy. In addition, these revisions lag behind changes in terminology as they occur in real-time, evidence-based clinical practice at an academic institution. (d) SNOMED and the ICD nomenclatures are redundant, which limits their usability for an archival database. For example, SNOMED contains entries for both “Crohn disease of the colon” and “Crohn disease of the large bowel.” A user interested in finding cases of Crohn disease of the colon would have to search either “Crohn” and retrieve cases of Crohn disease anywhere in the GI tract, or search both “…of the colon” and “…of the large bowel.” Such redundancy could only be circumvented by deleting one of the two entries.
The time and effort it would have taken to adapt SNOMED or any of the ICD classificatory systems to meet the needs of a purely educational database was deemed prohibitive at the outset of the project. The only existing classification system that was suitable for our purposes was the WHO classification of neoplastic diseases, which formed the basis of the classification of all neoplastic diseases in our taxonomy. The classification of nonneoplastic diseases was largely based on that present in well-known and reputable textbooks.
A considerable amount of discussion was generated over the issue of granularity. Where to break off the subclassification of the disease process was often a judgment call based on utilitarian considerations. For example, when it was first designed, the taxonomic string for NETs of the gastrointestinal tract ended at “NET”. The UW group scanned a large number of gastrointestinal tumors, including NET. Within a short amount of time, the collection of GI NET spanned several pages. We decided at this point to introduce grade (level of disease severity) as additional leaves in the taxonomic string to provide an additional layer of oversight to the slide holdings. On the other hand, we had very few examples of primary sclerosing cholangitis, so it did not make sense to include additional leaves for the various grades of this entity, although they were included in the original iteration of the taxonomy. If we ever have sufficient examples to warrant subclassifications based on disease severity, it would not be difficult for an experienced liver pathologist to review the cases and assign the grade, retrospectively.
Some slides found their way into educational collections because they represented a classic example of a particular histologic finding, such as koilocytes or skeinoid fibers. The taxonomic system was not able to handle such descriptors. A user interested in finding a classic example of such an entity would have to search by the diagnosis and review several slides to find the “best” representation of these findings. In the future, users could be invited to curate the collection by providing descriptors in the Comment field to expand the utility of the database.
Despite the fact that the taxonomic system is robust, with >5000 unique entries, the 1st year of its use saw modification of several of the taxonomic strings and creation of new ones. There were changes in nomenclature (e.g. the new WHO classification of hematopoietic tumors came out just after the taxonomy was completed), or recognition that a taxonomic string was too granular or not granular enough for practical purposes. Over time, as a wider range of entities was scanned into the database and the “bugs” worked out, these occurrences became less frequent. Nevertheless, the issue of curation of the database remains active. Any index system requires dedicated maintenance and modification. Adoption of the diagnostic taxonomy is therefore not a “one-time deal.” A content expert with interest in maintaining the database and a working understanding of its structure needs to oversee the regular upkeep of the taxonomic system, or the database will lose its value.
The issue of “backward compatibility” plagues any database of archival material, whether in a museum, a collection of Kodachromes or digital WSI slides. It is an unfortunate and not infrequent experience at academic medical centers to see whole-scale disposal of a large collection of educational materials, such as Kodachromes of autopsy or gross specimen photographs, simply because of a lack of workforce or interest to sort, label and archive – i.e. curate – the collection. This taxonomy grew out of the recognition that a worthwhile collection of educational cases needs to be curated to maintain viability and significance beyond the memory of the individuals who assemble the database. Without curation, servers rapidly fill with WSI files that immediately lose value even as they are scanned into the system.
Part of the duty of a curator is to make sure that the archived material will be retrievable even as terminology changes. With the UW taxonomy, it is a comparatively simple exercise to (a) identify WSI slides that are filed with outdated terminology by searching the WSI files for it (e.g. “goblet cell carcinoid”), (b) asking a pathologist to review the slides retrieved by the search for diagnostic accuracy and educational value, and (c) replacing the old term with the new (“goblet cell adenocarcinoma” in this example). It is important to recognize that although this seems labor intensive, terminology actually does not change all that frequently, the process is quick (once you put your mind to it), and does not require review of the entire 5000 line taxonomic database at one time, but is tailored to specifically the terminological changes as they are deemed relevant by the curator. An educational database is only relevant as long as the information contained within it is maintained current; for this reason, the issue of “version control” is not relevant. Since the database is not used for clinical purposes and is not attached to any patient information, treatment, or outcomes, the emphasis can be entirely on maintaining a robust, current, and easily searched database rather than maintaining past versions of the taxonomy.
Attaching a taxonomic string to a slide using the software described here is quick and easy. Submission of 20 slides for scanning using the slide scan request form takes less than 15 minutes, and appending the taxonomic string to the image file is a part of the routine slide scan process performed by the technician.
Nevertheless, the biggest limitation to the use of the taxonomy is “the human factor.” The work and time required to submit slides for scanning via the slide scan request form can be perceived as prohibitive. Furthermore, there may be general reluctance on the part of faculty to learn how to use yet another new computer-based application. Faculty and residents need individual encouragement to learn to use the system, as well as individual instruction in real-time applications to demonstrate the utility of the system (e.g. as they are putting together a PowerPoint presentation and want to illustrate it with a disease process that is not readily found in their personal collections).
This taxonomic system could be used to generate a searchable field in most databases and is not restricted to the Aperio eSlide Manager application that our department had on hand. If an individual or company had sufficient time and talent, it could be integrated into a different database's interface. In addition, attaching the taxonomic string to slides scanned for other purposes (e.g. interdisciplinary conferences) can rapidly increase the breadth and depth of cases easily accessible for educational purposes.
In an ideal workflow, educational images would be embedded in or otherwise linked to a patient's file in the laboratory information system (LIS), obviating the need to rescan a slide simply for educational purposes. Patient reports in most LIS systems can be searched for diagnosis, so theoretically, WSI images that are linked to a patient's report in the LIS could be retrieved with a keyword search. Currently, this approach to archiving educational cases has three drawbacks: (a) since surgical pathologic diagnoses are not necessarily entered with “discrete” terminology in surgical pathology reports, retrieval of cases for educational purposes would be constrained by lack of standardization of terminology. The WSI would still need to be appended with metadata, as described in this article. (b) A search of the LIS will retrieve entire cases, and the learner or educator would still have to scan through all WSI files attached to the case to identify the one(s) suitable for education, unless these are identified and “flagged” proactively. (c) Most pathology laboratories that have WSI capability do not (yet) have a robust interface with the LIS. (d) Ancillary documents such as narratives, indices, or lecture outlines will still need to be stored and referenced separately. Retrieval by case number, date of service, ICD code, or text string in the diagnostic field is not going to be as accurate as a thought-out taxonomic system. Further, creation of search functions needed to retrieve material for teaching is not in line with the primary mission of LIS software, which is patient care and revenue capture.
The database at the UW now contains >5000 slide files, all of which are appended with the taxonomic string. Residents are increasingly becoming familiar with the organization of the “lessons” in the Aperio eSlide Manager database and appreciate the range of educational materials it contains. The residents are also increasingly exploring the advanced search function and turning to the database to find illustrative examples of less common and rare entities that frequently appear in differential diagnoses but are otherwise not readily “at hand” to study. This is particularly useful during live review sessions with faculty, during which diverse topics of discussion arise and participants benefit from real-time teaching done from digital slides available at a moment's notice. An additional educational function is for residents preparing for pathology specialty boards. Since the slide files are organized into folders by organs and organ combinations, it is easy to navigate the folders and browse the images in a systematic review of topics. Many of the slides are accompanied by explanatory notes in the database, so residents can view the image as an “unknown” and then read the supplementary material to reinforce learning. As faculty and residents gain familiarity with the system, it is hoped that faculty will share their prodigious slide collections so that their educational potential can be made readily available to learners at all levels. An organized and accessible digital slide collection also contributes to greater resource stewardship and efficiency, as the need for individuals to request personal teaching recuts could decrease with the recognition that comparable specimens are available digitally.
CONCLUSION
The pace of digitization of pathology slides is increasing exponentially. Retrieval of the slides by diagnosis will be facilitated when these are linked to the LIS and searchable by free text searches. However, finding slides for educational purposes via this route will still be problematic and require upfront work to identify those of heuristic value. As this project shows, a taxonomic approach to select samples for educational and publication efforts can be built into the mundane workflow. Efforts such as that by the UW Surgical Pathology team will become progressively more important to manage the huge numbers of slides that are scanned by WSI into educational databases.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
This project could not have been completed without a heavy investment in time and effort by residents and subspecialty pathologists at the UW, specifically: Drs. Darya Buehler, Ali Chaudri, Michael Fritsch, Daniel Matson, Patrick Rush, Shahriar Salamat, James Stewart, Paul Weisman, and David Yang.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2019/10/1/33/270774
REFERENCES
- 1.Christensen PA, Lee NE, Thrall MJ, Powell SZ, Chevez-Barrios P, Long SW, et al. RecutClub.com: An open source, whole slide image-based pathology education system. J Pathol Inform. 2017;8:10. doi: 10.4103/jpi.jpi_72_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker EU, Reder NP, Glasser D, Henriksen J, Kilgore MR, Rendi MH, et al. NDER: A Novel web application for teaching histology to medical students. Acad Pathol. 2017;4:1–5. doi: 10.1177/2374289517691061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee LM, Goldman HM, Hortsch M. The virtual microscopy database-sharing digital microscope images for research and education. Anat Sci Educ. 2018;11:510–5. doi: 10.1002/ase.1774. [DOI] [PubMed] [Google Scholar]
- 4.Bertram CA, Firsching T, Klopfleisch R. Virtual microscopy in histopathology training: Changing student attitudes in 3 successive academic years. J Vet Med Educ Summ. 45:241–9. doi: 10.3138/jvme.1216-194r1. [DOI] [PubMed] [Google Scholar]
- 5.Gopalan V, Kasem K, Pillai S, Olveda D, Ariana A, Leung M, et al. Evaluation of multidisciplinary strategies and traditional approaches in teaching pathology in medical students. Pathol Int. 2018;68:459–66. doi: 10.1111/pin.12706. [DOI] [PubMed] [Google Scholar]
- 6.Kuo KH, Leo JM. Optical Versus Virtual Microscope for Medical Education: A Systematic Review. Anat Sci Educ. 2018 Nov 10; doi: 10.1002/ase.1844. doi: 10.1002/ase.1844. [Epub ahead of print] Review. PubMed PMID: 30414261. [DOI] [PubMed] [Google Scholar]
- 7.Wong M, Frye J, Kim S, Marchevsky AM. The use of screencasts with embedded whole-slide scans and hyperlinks to teach anatomic pathology in a supervised digital environment. J Pathol Inform. 2018;9:39. doi: 10.4103/jpi.jpi_44_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shahriari N, Grant-Kels J, Murphy MJ. Dermatopathology education in the era of modern technology. J Cutan Pathol. 2017;44:763–71. doi: 10.1111/cup.12980. [DOI] [PubMed] [Google Scholar]
- 9.Saco A, Bombi JA, Garcia A, Ramírez J, Ordi J. Current status of whole-slide imaging in education. Pathobiology. 2016;83:79–88. doi: 10.1159/000442391. [DOI] [PubMed] [Google Scholar]