Abstract
This study aimed at detecting Staphylococcus aureus from white coats of college students and characterizing antimicrobial susceptibility and biofilm production. Bacterial samples (n = 300) were obtained from white coats of 100 college students from August 2015 to March 2017 S. aureus was isolated and it´s resistance profile was assessed by antimicrobial disk-diffusion technique, screening for methicillin-resistant Staphylococcus aureus (MRSA), detection of mecA gene by PCR, and determination of staphylococcal cassette chromosome mec (SCCmec) by multiplex PCR. Congo red agar (CRA) and icaA and icaD genes by PCR were used for biofilm characterization. S. aureus was identified in 45.0% of samples. Resistance of S. aureus sample to antimicrobial was seen for penicillin (72.59%), erythromycin (51.85%), cefoxitin (20.74%), oxacillin (17.04%), clindamycin (14.81%) and levofloxacin (5.18%). MRSA was detected in 53.3% of the samples with SCCmec I (52.8%), SCCmec III (25%) and SCCmec IV (11.1%). Biofilm production was observed in 94.0% S. aureus samples. These data show that biosafety measures need to be enhanced in order to prevent dissemination of multiresistant and highly adhesive bacteria across other university settings, relatives, and close persons.
Keywords: S. aureus, MRSA, Biofilm, White coats
Introduction
Personal protective equipment (PPE), such as white coats, is used as biosafety measure, mainly recommended to healthcare professionals working in unwholesome environments with variable risk. [1]. Commensal microorganisms, most commonly Staphylococcus aureus, can be easily disseminated through white coats [2]. This bacteria is commonly found in nasal mucosa and can become pathogenic, triggering infections such as skin boils and pimples, cellulitis, bacteremia, pneumonia, osteomyelitis, and acute endocarditis, among others [3].
Inappropriate white coat use is an important way for transmission of methicillin-resistant S. aureus (MRSA) [4]. MRSA infections are difficult to treat due to it´s resistance to all beta lactam antimicrobials, leaving few alternatives for treatment [5].
The assessment of S. aureus susceptibility to methicillin is key to proper infections treatments without unnecessary use of vancomycin, since this agent can lead to several therapeutic complications, in spite of being a first-line and less expensive antimicrobial for MRSA infections [6]. Although vancomycin has been used since 1958, hospital samples have already show reduced susceptibility of S. aureus to vancomycin (vancomycin-intermediate Staphylococcus aureus-VISA) [7].
In addition to antimicrobial resistance, S. aureus has the ability to produce a complex molecular structure named biofilm in both biotic and abiotic surfaces, such as white coats. Biofilm promotes adhesion to substrate and protects the microorganism from host immune response and antimicrobial action through an extracellular matrix composed by proteins, carbohydrates and deoxyribonucleic acid (DNA) [8]. Biofilm can thus be associated to bacteria cells adhesion to the fabric white coats of professionals who use that PPE.
College students frequently wear white coats as mandatory PPE during pedagogical activities in hospitals and laboratories settings. Moreover, some students also wear coats in improper settings like libraries, cafeteria, and even during their journey back home [1]. According to Muhadi et al. [9], it is very common to find white coats laying down on chairs and outside laboratory and hospital settings, potentially causing pathogen dissemination.
Considering that multiresistant bacteria can be carried across different settings, it is especially important to evaluate the susceptibility of S. aureus from health students’ lab coats to antimicrobials when they wear such PPE item in different settings while attending classes. The characterization of S. aureus isolated from lab coats can help preventing these microorganisms from spreading through different university settings. This is why this study aimed at detecting S. aureus from white coats worn by college students and characterizing this microorganism in terms of antimicrobial susceptibility and biofilm production.
Methods
The present study is a cross-sectional trial, registered and approved by the Universidade do Oeste Paulista Committee for Ethics in Research, Presidente Prudente, Brazil. Participants were Biomedical Sciences students from the third semester on, i.e. when they started clinical practice. Participants answered a questionnaire regarding personal information related to white coat use: semester of the course, gender, laboratories they attended during the period of study, coat cleaning frequency and purpose of this PPE use.
Bacterial samples were obtained from white coats of college students from August 2015 to March 2017. Samples were collected by rolling a sterile swab moistened with sterile saline solution (0.85%) through three areas: collar, pocket and sleeve. Immediately upon collection, samples were forwarded to the Universidade do Oeste Paulista Microbiology Laboratory, Presidente Prudente, Brazil, where they were kept for 24 h at 37 °C in brain–heart infusion (BHI) broth for growth and subsequently seeded in mannitol salt agar for selection of Staphylococcus samples. After growth on agar plates, colony samples were submitted to GRAM staining and catalase and coagulase test for differentiation of S. aureus and coagulase-negative staphylococci (CoNS). Samples obtained were frozen at − 70 °C in nutrient broth with 10% glycerol.
Antimicrobial sensitivity was tested by drug diffusion in agar using disks infused with oxacillin, cefoxitin, penicillin, clindamycin, erythromycin, and levofloxacin according to Clinical Laboratory Standards Institute criteria [10].
Screening method with oxacillin (6 μg/ml) and NaCl (4%)-enriched Mueller–Hinton Agar was used to identify MRSA. Inoculums were standardized according to Pereira et al. [11] and MRSA detection was assessed by growth of at least one colony on the agar surface.
Staphylococcus aureus were seeded in Congo red agar (CRA); following incubation at 37 °C for 24–48 h, biofilm-producing colonies stained in black, whereas isolated staining in red to burgundy were considered non producers [12].
Bacterial DNA extraction was performed by using the Illustra tissue and cells prep genomic mini spin kit (GE Healthcare, Little Chalfont, UK), according to manufacturer´s instructions, following an adapted protocol described by Pereira et al. [13]. DNA quality and concentration were measured by assessing the 260/280 nm absorbance ratio (Gen5, Epoch, Bio Tek, Winooski, VT). The obtained DNA was stored at − 20 °C up to processing.
Detection of mecA gene was performed by conventional PCR reactions according to the protocol described by Murakami et al. [14]. Strains of S. aureus ATCC 33591 (positive control) and ATCC 25923 (negative control) were included in all reactions. Staphylococcal Cassette Chromosome mec (SCCmec) typing in MRSA strains was performed by multiplex PCR as described by Oliveira et al. [15]. PCR reactions for detection of biofilm formation-related genes (icaA e icaD) were performed according to Arciola et al. [16]. Effectiveness of all amplified reactions was monitored by electrophoresis in 2% agarose gel stained with ethidium bromide.
Statistical analyses were made through R program (3.3.2 version) and it‘s statistical package. Fisher’s exact test (table 2 × 2) was used to assess antimicrobial resistance and susceptibility. Sensitivity and specificity were evaluated by comparing the phenotypic methods used and PCR for the detection of MRSA and genes involved in biofilm synthesis [17]. Agreement between phenotypic and genotypic tests was evaluated using the kappa index [18].
Results
A total of 300 samples from sleeve, pocket, and collar of 100 white coats were included in this study. S. aureus was detected in 135 samples (45%), whereas coagulase-negative staphylococci, bacilli, and unidentified microorganisms were found in 20, 69 and 76 samples, respectively. S. aureus detection rate was significantly higher (p < 0.001). S. aureus was most often isolated from collar (n = 53, 39%), followed by sleeve (n = 45, 33%) and pocket samples (n = 37, 28%).
Table 1 shows the results of disk-diffusion assessment of microbial resistance for 135 S. aureus isolates. MRSA was identified by screening test on 44 samples (32.6%) as shown Table 2.
Table 1.
Antimicrobial susceptibility in S. aureus by disk-diffusion technique
| Antimicrobial | Resistant |
|---|---|
| N (%) | |
| Pen | 21 (15.5) |
| Ery | 3 (2.22) |
| Cef/Lev | 1 (0.74) |
| Pen/Clin | 3 (2.22) |
| Pen/Cef | 1 (0.74) |
| Pen/Ery | 34 (25.18) |
| Ery/Oxa/Clin | 1 (0.74) |
| Pen/Oxa/Cef | 5 (3.7) |
| Pen/Cef/Lev | 1 (0.74) |
| Pen/Ery/Oxa | 4 (2.96) |
| Pen/Ery/Cef | 8 (5.92) |
| Pen/Ery/Clin | 7 (5.18) |
| Pen/Oxa/Cef/Clin | 1 (0.74) |
| Pen/Ery/Oxa/Cef | 3 (2.22) |
| Pen/Ery/Oxa/Clin | 1 (0.74) |
| Pen/Ery/Oxa/Cef/Clin | 3 (2.22) |
| Pen/Ery/Oxa/Cef/Lev | 3 (2.22) |
| Pen/Ery/Cef/Clin/Lev | 1 (0.74) |
| Pen/Ery/Oxa/Clin/Lev | 1 (0.74) |
| Pen/Ery/Oxa/Cef/Clin/Lev | 1 (0.74) |
| Sensitivea | 32 (23.70) |
Pen penicillin, Ery erythromycin, Oxa oxacillin, Cef cefoxitin, Clin clindamycin, Lev levofloxacin, N number of S. aureus samples
aS. aureus sensitive to all antimicrobials
Table 2.
Sensibility and specificity to oxacillin in S. aureus samples by phenotypic and genotypic methods
| Phenotypic methods | mecA | Sensibility | Specificity | Kappa coefficient | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| N = 72 | N = 63 | % | % | k (interpretation) | |
| Oxacillin (10 µg) | 13 | 10 | 18.0 | 84.1 |
0.02 Non-concordance |
| Cefoxitin (30 µg) | 17 | 11 | 23.6 | 82.5 |
0.07 Non-concordance |
| Screeninga | 26 | 18 | 36.1 | 82.5 |
0.35 Satisfactory concordance |
N: number of S. aureus; k: Kappa coefficient
aOxacillin (6 μg/ml) and NaCl (4%)-enriched Mueller–Hinton Agar
All isolates were examined for mecA gene detection in order to confirm MRSA phenotypic results from screening and disk-diffusion with oxacillin and cefoxitin. Positivity for mecA gene was observed in 72 S. aureus samples (53.3%). Sensitivity and specificity results from phenotypic tests in comparison with mecA gene and Kappa’s concordance analysis are shown in Table 2.
Isolates positive for mecA gene were examined by multiplex PCR technique for SCCmec typing. Thirty-eight (52.8%) S. aureus samples presented SCCmec I, 18 (25.0%) SCCmec III, and 8 (11.1%) SCCmec IV. Typing was not possible in eight (11.1%) S. aureus samples. Table 3 shows susceptibility of these strains to other antimicrobials tested.
Table 3.
Determination of antimicrobial resistance by MRSA, according to SCCmec types
| Antimicrobial | N | SCCmec | |||
|---|---|---|---|---|---|
| Type I (%) | Type III (%) | Type IV (%) | Untyped (%) | ||
| Erythromycin | 38 | 50 | 30 | 8 | 12 |
| Clindamycin | 10 | 50 | 40 | 10 | 0 |
| Penicillin | 50 | 54 | 26 | 8 | 12 |
| Levofloxacin | 3 | 100 | 0 | 0 | 0 |
Of all identified S. aureus, 127 (94.0%) were biofilm producers as shown by black colonies on Cong red agar surface. Of all biofilm-producing isolates, 42 (33.0%) were MRSA.
Results regarding biofilm formation-related genes showed 109 (80.7%) strains positive for icaA gene and 127 (94.0%) for icaD gene. In 105 samples (77.8%) both genes were concomitantly detected; and 99 samples (73.3%) were shown to be positive on CRA. Sensibility and specificity of CRA regarding icaA was 94% and 7.7%, respectively. On the other hand, sensitivity was 94% and specificity 0% for icaD. There was minimum agreement between CRA and icaA gene (k = 0.03), and no agreement whatsoever between CRA and icaD gene (k = − 0.06).
Data on inside and outside university white coat use showed that S. aureus was isolated in 64 (77.1%) white coat samples from female participants (n = 83) and in 13 (76.5%) white coat samples from male participants (n = 17).
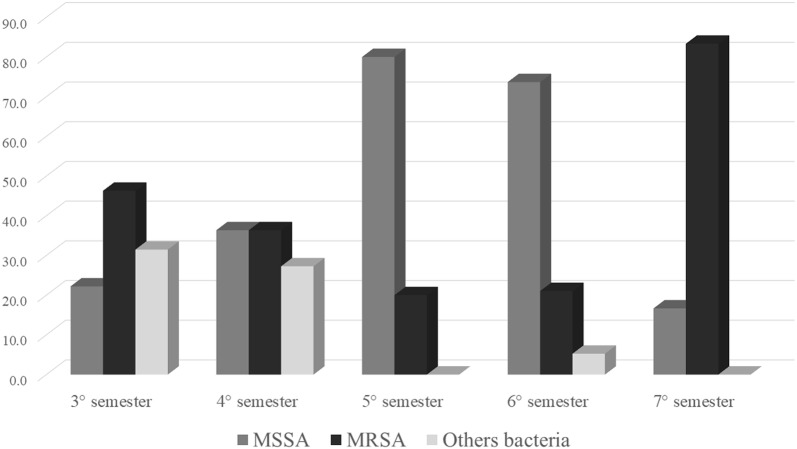
Colonization frequency by Methicillin-sensitive S. aureus (MSSA) and MRSA according to the different current course periods of participants is shown in Fig. 1. Highest rates of all three microorganisms are seen in samples from 3rd semester students.
Fig. 1.
Colonization frequency of white coats by S. aureus and MRSA according to the different current course period of the participants
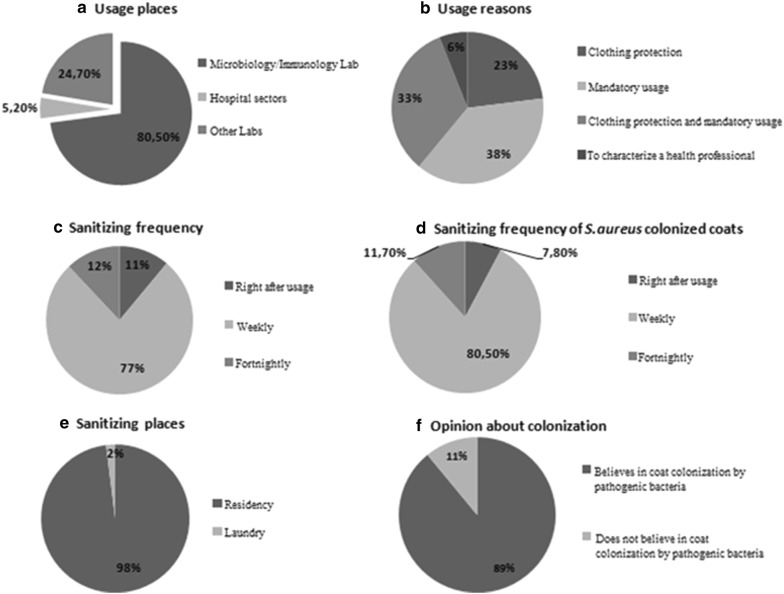
Figure 2 shows results regarding coat usage. Most students stated that wearing a white coat aims at protecting clothes and is a mandatory PPE item at Microbiology and Immunology lab, settings where white coats are mostly worn by students. Moreover, 98% of the students perform sanitization procedures at home, and about 80% do it weekly, regardless of S. aureus colonization (Fig. 2). Interesting, almost 90% of the students are aware that this PPE item is potentially colonized by bacterial pathogens.
Fig. 2.
Questions on coat usage by students of the Biomedicine course. a Usage places. b Usage reasons. c Sanitizing frequency. d Sanitizing frequency of S. aureus colonized coats. e Sanitizing places. f Opinion about colonization
Discussion
White coats are PPE items widely used by health professionals and students during their academic education. According to Brazilian law (n°. 14.446), the use of this item outside workplace is strictly forbidden, violators being subject to severe financial penalties. Wearing a white coat in improper places can lead to risk of pathogen transmission. This study identified a 45.0% contamination rate of college students´ white coats with S. aureus, a pathogen playing a major role in community-acquired infections.
The rate of S. aureus isolates found herein is higher than in other studies. Fenalte et al. [19] have detected this microorganism in 19.8% of samples from coats worn by nursing professionals in a midsize hospital, while Margarido et al. [20] identified S. aureus in 31.6% of samples from nursing students. The data presented in this study highlight S. aureus colonization in coats worn by students less exposed to hospital environment than those in other health science courses.
Since genotypic techniques are considered gold standard for MRSA detection, the high rate of such strains found in students´ coats is a major concern, as other studies have shown lower rates in white coat samples from health professionals with higher exposure to hospital environment. Treakle et al. [21] have detected 23% of S. aureus isolated from 149 samples from coats worn by health professionals, and 6 were identified as MRSA. Additionally, Fenalte et al. [19] described a 4.7% rate of MRSA in isolates from coats worn by nursing technicians. Low sensitivity and specificity of phenotypic methods in the present study reflect the difficulty in detecting MRSA, thus highlighting the importance of using molecular methods to detect these strains.
Another alarming result from the present work refers to SCCmec typing in MRSA, which showed high rates of types I and III, the most prevalent types in hospitals settings. A study with 62 MRSA isolated from six healthy professionals and 56 patients has shown SCCmec III as the most frequent type (76%), followed by SCCmec IV (11.2%), SCCmec I (4.8%) and SCCmec V (3.2%). In this same study, the authors point out that SCCmec I was isolated in blood culture only, whereas the SCCmec IV and V were associated to wounds and urine samples and SCCmec III was isolated from all analyzed clinical materials [22].
More than 50% of MRSA isolates with SCCmec I showed resistance to erythromycin, clindamycin, penicillin, and levofloxacin. Resistance to erythromycin, clindamycin and penicillin was also observed in MRSA with SCCmec III and IV. One of the main mechanisms of acquiring genes related to antimicrobial resistance involves acquisition of mobile genetic elements, mainly among larger SCCmec. Deletion events are also very common as a mechanism of acquiring genes related to antimicrobial resistance, as a MRSA strain can be self-modified to MSSA or lose antimicrobial resistance genes [22].
Ninety-four percent of S. aureus detected were biofilm producers, indicating that production of this polysaccharide is an important mechanism providing closer adhesion to coat fabrics. Adhesion to coat surface can be related to the polysaccharide intercellular adhesin (PIA), mediated by icaA and icaD genes and different mechanisms. According to Fredheim et al. [23], extreme conditions such as heat, pressure, and environmental chemical can influence biofilm formation. Furthermore, certain Staphylococcus species can develop alternatives mechanisms, such as PIA-independent biofilm formation.
Congo Red Agar test showed a 94% sensitivity rate for icaA and icaD genes. Although CRA method has shown low specificity compared to icaA and icaD genes, as well minimal agreement with icaA gene and no agreement with icaD gene, these results reflect the importance of studying other biofilm production mechanisms, since there are other genes, like bap, encoding biofilm-associated protein [24].
Although 87% of students with S. aureus-colonized coats are aware of the potential of this PPE item to carry bacterial pathogens, it seems, in view of the high colonization rates on white coats herein detected, that biosafety measures need to be reinforced. As for coat sanitization, 80.5% of coats colonized by S. aureus are sanitized weekly and 97.4% of the students reported performing the procedure at home. Silva et al. [25] point out that health professionals should be trained for better knowledge of measures involving precaution, proper use, and adequate maintenance of their white coats.
Students should therefore receive proper education regarding biosafety procedures, especially after classes and lab research activities. Ideally, white coats should be disposable, but since this practical is not viable it is mandatory that their use follow rigorous sanitization measures, including separate laundering with powder detergent followed by drying and ironing, which can prevent the possibility of bacterial pathogen dissemination, according to Margarido et al. [20]. These authors collected samples from white coats sanitized according to standard procedures, i.e. laundering with powder detergent, drying, and ironing, and could demonstrate absence of bacterial growth in 100% of samples. The present study shows that biosafety measures need to be enhanced in order to prevent dissemination of multiresistant and highly adhesive bacteria across other university settings, relatives, and close persons.
Acknowledgements
We thank the São Paulo State Research Foundation for Scientific and Technological Development (FAPESP; Grant 2016/12252-2 and 2016/15809-8) for supporting research development.
Abbreviations
- PPE
personal protective equipment
- MRSA
methicillin-resistant S. aureus
- MSSA
methicillin-sensitive S. aureus
- VISA
vancomycin-intermediate Staphylococcus aureus
- BHI
brain heart infusion
- CoNS
coagulase-negative staphylococci
- CRA
Congo Red Agar
- SCCmec
staphylococcal cassette chromosome mec
Authors’ contributions
IRB: Responsible for performing microbiological and antimicrobial resistance tests. ACLP: Responsible for performing microbiological and biofilm production tests. BSS: Responsible for performing microbiological and biofilm production tests. JCCA: Responsible for performing microbiological and antimicrobial resistance tests. YCOB: Responsible for performing microbiological and antimicrobial resistance tests. MVPR: Responsible for correcting the manuscript. GM: Responsible for correcting the manuscript. JP: Responsible for the translation of the manuscript. ACC: Responsible for correcting the manuscript. LKW: Responsible for correcting the manuscript. VCP: Responsible for conceiving the idea, data analysis, and manuscript writing. All authors read and approved the final manuscript.
Funding
São Paulo State Research Foundation for Scientific and Technological Development (FAPESP; Grant 2016/12252-2 and 2016/15809-8).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The present study is a cross-sectional study, registered and approved by Universidade do Oeste Paulista Committee for Ethics in Research, Presidente Prudente, Brazil. (CAAE: 47871015.0.0000.5515).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Banu A, Anand M, Nagi N. White coats as a vehicle for bacterial dissemination. J Clin Diagn Res. 2012;6:1381–1384. doi: 10.7860/JCDR/2012/4286.2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goyal S, Khot SC, Ramachandran V, Shah KP, Musher DM. Bacterial contamination of medical providers’ white coats and surgical scrubs: a systematic review. Am J Infect Control. 2019 doi: 10.1016/j.ajic.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 3.Santos AL dos, Santos DO, Freitas CC de, Ferreira BLA, Afonso IF, Rodrigues CR, et al. Staphylococcus aureus: visitando uma cepa de importância hospitalar. J Bras Patol e Med Lab [Internet]. SBPC, SBP, SBC; 2007;43:413–23. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1676-24442007000600005&lng=pt&nrm=iso&tlng=pt. Accessed 22 Nov 2016.
- 4.Carvalho CMRS, Madeira MZ de A, Tapety FI, Alves ELM, Martins M do C de C, Brito JNP de O. Aspectos de biossegurança relacionados ao uso do jaleco pelos profissionais de saúde: uma revisão da literatura. Texto Context - Enferm [Internet]. 2009;18. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S0104-07072009000200020&lng=pt&nrm=iso&tlng=pt.
- 5.Stefani S, Varaldo PE. Epidemiology of methicillin-resistant staphylococci in Europe. Clin Microbiol Infect. 2003;9:1179–1186. doi: 10.1111/j.1469-0691.2003.00698.x. [DOI] [PubMed] [Google Scholar]
- 6.Broekema NM, Van Tam T, Monson TA, Marshall SA, Warshauer DM. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA-mediated resistance in staphylococcus aureus in a large-scale study. J Clin Microbiol. 2009;47:217–219. doi: 10.1128/JCM.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palazzo ICV, Araujo MLC, Darini ALC. First report of vancomycin-resistant staphylococci isolated from healthy carriers in Brazil. J Clin Microbiol. 2005;43:179–185. doi: 10.1128/JCM.43.1.179-185.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fey PD, Climo MW, Archer GL. Determination of the chromosomal relationship between mecA and gyrA in methicillin-resistant coagulase-negative staphylococci. Antimicrob Agents Chemother. 1998;42:306–312. doi: 10.1128/aac.42.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muhadi SA, Aznamshah NA, Jahanfar S. A cross sectional study of microbial contamination of medical students’ white coat. Malays J Microbiol. 2007;3:35–38. [Google Scholar]
- 10.(CLSI) Clinical and Laboratory Standards. Performance standards for antimicrobial susceptibility testing, document M100-S23. 2016.
- 11.Pereira VC, Martins A, de Souza Rugolo LMS, de Lourdes Ribeiro de Souza da Cunha M. Detection of Oxacillin Resistance in Staphylococcus aureus Isolated from the Neonatal and Pediatric Units of a Brazilian Teaching Hospital. Clin Med Pediatr [Internet]. 2009;3:23–31. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3676290&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed]
- 12.Freeman D, Falkiner F, Keane C. New method for detecting slime production by coagulase-negative staphylococci. J Clin Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira VC, Pinheiro L, Oliveira A, Martins KB, Flávio D, Riboli M, et al. Microbial pathogenesis expression of superantigens and the agr system in Staphylococcus epidermidis. Microb Pathog. 2018;115:19–24. doi: 10.1016/j.micpath.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira DC, Multiplex PCR. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Society. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of Staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39:2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curi PR. Metodologia e análise de pesquisa em Ciências Biológicas. Botucatu: Tipomic; 1997. [Google Scholar]
- 18.Landis J. Richard, Koch Gary G. The Measurement of Observer Agreement for Categorical Data. Biometrics. 1977;33(1):159. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 19.Fenalte MP, Gelatti LC. Contaminação de jalecos usados pela equipe de enfermagem. Online. 2013;4:67–76. [Google Scholar]
- 20.Margarido CA, Villas-Boas TM, Mota VS, Da Silva CKM, Poveda VDB. Microbial contamination of cuffs lab coats during health care. Rev Bras Enferm. 2014;67:127–132. doi: 10.5935/0034-7167.20140017. [DOI] [PubMed] [Google Scholar]
- 21.Treakle AM, Thom KA, Furuno JP, Strauss SM, Harris AD, Perencevich EN. Bacterial contamination of health care workers’ white coats. Am J Infect Control. 2009;37:101–105. doi: 10.1016/j.ajic.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodríguez-Noriega E, Seas C, Guzman-Blanco M, Mejía C, Alvarez C, Bavestrello L, Zurita J, Labarca J, Luna CM, Salles MJ, Gotuzzo E. Staphylococcal cassette chromosome mec types among methicillin-resistant Staphylococcus aureus in northern Iran. Jundishapur J Microbiol. 2016;9:e33933. doi: 10.5812/jjm.33933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fredheim EGA, Klingenberg C, Rohde H, Frankenberger S, Gaustad P, Flægstad T, et al. Biofilm formation by Staphylococcus haemolyticus. J Clin Microbiol. 2009;47:1172–1180. doi: 10.1128/JCM.01891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain Z, Stoakes L, Massey V, Diagre D, Fitzgerald V, El Sayed S, et al. Correlation of oxacillin MIC with mecA gene carriage in coagulase- negative staphylococci. J Clin Microbiol. 2000;38:752–754. doi: 10.1128/jcm.38.2.752-754.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartels AS, Andrade AD, Neumann ESAM. Identificação de portadores de Staphylococcus enterotoxigênicos e avaliação da sensibilidade a antimicrobianos. Arq Bras Med Vet Zootec. 2009;61:1450–1453. doi: 10.1590/S0102-09352009000600029. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.