Short abstract
Ebola virus high lethality relies on its ability to efficiently bypass the host innate antiviral response, which senses the viral dsRNA through the RIG-I receptor and induces type I interferon α/β production. In the bypassing action, the Ebola virus protein VP35 plays a pivotal role at multiple levels of the RIG-I cascade, masking the viral 5′-triphosphorylated dsRNA from RIG-I, and interacting with other cascade components. The VP35 type I interferon inhibition is exerted by the C-terminal domain, while the N-terminal domain, containing a coiled-coil region, is primarily required for oligomerization. However, mutations at key VP35 residues L90/93/107A (VP35-3m) in the coiled-coil region were reported to affect oligomerization and reduce type I interferon antagonism, indicating a possible but unclear role of homo-oligomerization on VP35 interaction with the RIG-I pathway components. In this work, we investigated the VP35 dimerization thermodynamics and its contribution to type I interferon antagonism by computational and biological methods. Focusing on the coiled-coil region, we combined coarse-grained and all-atom simulations on wild type VP35 and VP35-3m homo-dimerization. According to our results, wild type VP35 coiled-coil is able to self-assemble into dimers, while VP35-3m coiled-coil shows poor propensity to even dimerize. Free-energy calculations confirmed the key role of L90, L93 and L107 in stabilizing the coiled-coil homo-dimeric structure. In vitro type I interferon antagonism studies, using full-length wild type VP35 and VP35-3m, revealed that VP35 homo-dimerization is an essential preliminary step for dsRNA binding, which appears to be the main factor of the VP35 RIG-I cascade inhibition, while it is not essential to block the other steps.
Keywords: Models/projections, mutations, Filoviridae
Introduction
Ebola virus (EBOV) is the etiological agent of one of the deadliest diseases caused by infective agents, with many symptoms including a severe, mostly lethal, hemorrhagic fever in humans.1 Among the six species included in the genus Ebolavirus, Zaire, Sudan, Reston, Tai Forest, Bundibugyo and Bombali,2,3 Zaire EBOV has generally been associated with the highest fatality rates; in the past, this strain has shown a mortality rate often approaching 90%,4 whereas the estimate of case fatality based on 2014–2015 outbreak was less than 60%.5 Because of the severity of EBOV disease, the high mortality rates and the potential use as a bioterrorist agent, EBOV remains an urgent-to-care hazard to global human health. Despite a deep effort, up to date, acute case management is essentially supportive,6 approved specific treatments and licensed vaccination are still lacking,7–9 although several compounds and vaccines are under investigation.10
The EBOV genome is ∼19 kilobases in length and contains seven genes (3′ NP VP35 VP40 GP VP30 VP24 L 5′) encoding two soluble non-structural glycoproteins (sGP and ssGP) and seven structural proteins, NP (nucleoprotein), VP35 (polymerase cofactor), VP40 (matrix protein), GP (glycoprotein), VP30 (transcription activator), VP24 (secondary matrix protein) and the RNA-dependent RNA polymerase L.11 The virulence and lethality of this virus is due to the profound suppression of the innate immune system, together with a strong production of pro-inflammatory cytokines and a profound immunosuppression associated with a peripheral T lymphocyte apoptosis.1,12,13 The ability of EBOV to avoid and inhibit the innate immune response in the early stages of infection is due to the viral multifunctional proteins, VP24 and VP35,14,15 which are the main determinants of virulence16–18 and thus attractive drug targets.19–24
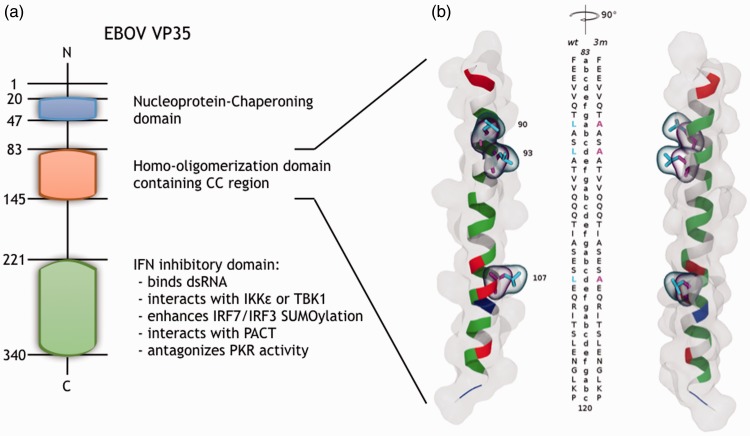
VP35 is a polymerase cofactor essential for EBOV replication, and it also carries out many other functions necessary for virus infection through which it is able to influence the virus interaction with the host: suppression of RNA silencing,25 blocking the stress granule assembly through interaction with its multiple components,26 and suppression of the type I interferon (IFN)-α/β production in a dose-dependent mode.27,28 VP35 exerts its IFN production inhibitory function through different mechanisms (Figure 1(a)): (i) binding to dsRNA through the C-terminal IFN inhibitory domain (IID) and preventing RIG-I-like receptor activity;29 (ii) directly interacting with either IKKε or TBK1, blocking kinases binding and phosphorylation of the critical transcription factors IRF3 and IRF7 responsible for IFN-α/β promoter activation;30,31 (iii) acting downstream of IKKε and TBK1, enhancing the SUMOylation of IRF7 and IRF3;32 (iv) interacting with PACT, activator of RIG-I ATPase activity and RIG-I signaling, consequently disabling it to interact with the RIG-I carboxy-terminal domain and impairing PACT dependent induction of IFN-α/β gene expression;33 (v) inhibiting the activation of the IFN-stimulated genes interfering with the dsRNA-activated kinase PKR regulated pathway.34,35
Figure 1.
EBOV VP35 overview. (a) Sequence position and corresponding known function(s) of the protein domains: the NP-chaperoning domain (navy), the homo-olimerization domain (pale red) and the IID (green); the amino acid numbering, the N- and C-terminals are noted. (b) CC region model super-imposition of Zaire EBOV VP35-wt and VP35-3m. Structures (image pair with a stereo angle of 90°) are shown as cartoon colored by residue type (non-polar, polar, basic and acidic side chains, respectively, in white, green, blue and red) and as surface representation. The mutated amino acids (90, 93 and 107) are highlighted in cyan (VP35-wt) and magenta (VP35-3m) licorice. The two sequences are aligned along the structure with the associated CC heptad-register.
VP35 consists of an N-terminal domain of a C-terminal domain (Figure 1(a)): the former domain, containing a coiled-coil (CC) region, is required for its oligomerization and responsible for the interaction with NP:18,36,37 the latter domain, also called IID, is required for binding the viral polymerase L and for IFN inhibition.27,38 The high-resolution structure of the Zaire EBOV VP35 C-terminal domain has been solved showing that the dsRNA is bound to an IID dimer,39,40 while its N-terminal half, and in particular the oligomerization domain, including the CC motif, has been crystallized only very recently.41 Such a CC complex, which was suggested to involve the region between residues 82 and 118 for Zaire EBOV (Figure 1),15 has been structurally characterized by Zinzula et al. as a bipartite parallel helix bundles composed by the proposed residues (trimer/tetramer).41 The CC is a widely spread α-helical protein motif42 involved in different processes, including protein oligomerization.43 The typical CC structure consists of two or more amphipathic α-helices having a heptad-repeating amino acid pattern (abcdefg, Figure 1) that arrange each-other in a twisted fashion forming the so-called “knobs-into-holes” packing.44 It is noteworthy that also the closely related Marburg virus encodes a similar domain in its VP3536; however, Marburg VP35 differs in efficiency from the EBOV counterpart.45 In addition, the CC domain of Marburg virus VP35 is a whole heptad shorter than the one of EBOV and it has been recently solved in a trimeric state,46 differently from the one of EBOV VP35 that has been resolved crystallographically in both oligomeric forms, trimer and tetramer,41 while in all the other viruses of the genus Ebolavirus, VP35 oligomerization domain in solution exists exclusively as homo-tetramer.41 In the same framework, even more recently, further insights into the parallel orientation and tetrameric arrangement of the EBOV VP35 N-terminus were gained by means of a multidisciplinary structural approach.47
It has been shown that the presence of VP35 carrying IID mutations that directly suppress dsRNA interaction leads to non-virulent EBOV variants.17 While the full structure of EBOV VP35 has not been determined yet, it has been reported that VP35 oligomeric form facilitates IFN antagonism,15 but it has not been determined whether the oligomerization is required for the complete VP35 inhibition of all the different steps through which it exerts its IFN antagonism.
We recently modeled Zaire EBOV VP35 CC domain and reported the homo-oligomerization profile of the full-length recombinant viral protein48 leaving open the question of the dimer as the possible fundamental oligomeric unit to originate also trimer, tetramer or even higher orders VP35 aggregates.49 Reid et al.15 reported that the substitution of leucine residues at positions 90, 93 and 107 in a and d positions of the predicted CC domain (Figure 1) with three alanines affected VP35 IFN antagonism, potentially abolishing oligomerization. Note that the study of Zinzula et al. has further pointed out the importance of the same three inner-core leucines by biophysical assays on a Leu-Ala variant.41 In the present work, to improve our understanding on the microscopic details of the interactions and to assess the role of dimerization in the process, we combined computational and biochemical methods. By means of molecular dynamics (MD) simulations, we compared the CC domains of the wild type VP35 (VP35-wt) and the L90/93/107A VP35 variant (VP35-3m) to characterize their differences during the dimerization process. Experimentally, we gained insights into the role of EBOV VP35 N-terminal CC domain, performing in vitro comparison of the properties of the full-length VP35-wt and VP35-3m through dsRNA binding biochemical assay with recombinant proteins and luciferase gene reporter assay to outline the functional contribution of dimerization on IFN antagonism.
Materials and methods
MD simulations
The homo-dimerization process of the CC domains of Zaire EBOV VP35-wt and VP35-3m was investigated using a combined coarse-grained (CG)50 and all-atom (AA)51 approach as previously described in details.48 Briefly, the protocol of these simulations was made up of a 1 µs CG MD, whose final conformation was first back-mapped to AA and then simulated for additional 100 ns. Both the CG and AA MD simulations to study the dimerization of the VP35 N-terminal CC forming domain were performed using the GROMACS 2016 package.52 The protein was represented by CG Martini models and simulated using Martini force field version 2.2. The starting configuration for the VP35-wt CG MD simulations was taken from an in-house simulation of the VP35 N-terminal CC domain modeled48 via a knowledge-based approach on the Nipah P protein (PDB id: 4N5B). The VP35-3m CG simulation was performed after the in silico mutation of the L90, L93 and L107 into alanine. The VP35-wt and VP35-3m models were converted to a CG Martini representation starting from the atomistic structures applying Martini22 force-field (http://cgmartini.nl/images/tools/martinize/python3/martinize-2.6/martinize.py). Two monomer (VP35-wt or VP35-3m) helices were parallelly placed at a distance of at least 15 Å for the dimerization simulations. The CG model was subjected to energy minimization in vacuum and solvated. Successively, steepest-descent energy minimization, equilibration with position restraints on protein backbone and finally 1 µs long production runs were performed. All the simulation parameters were set to the recommended values for proteins CG simulation (http://cgmartini.nl/index.php/tutorials-general-introduction-gmx5/proteins-gmx5). We simulated 12 independent replicas for VP35-wt and VP35-3m, for a total CG simulation time of 24 µs. The starting structures for the following independent AA simulations for both systems were taken from the final conformations of the CG simulations that reached and maintained five heptad-repeats of CC dimeric structure (heptads-register in Figure 1). We used the SOCKET program53 to verify the effective sampling of this particular conformation during the CG replicas (resulting CC timeline in Figure 2). In order to make the CG structures suitable to start the AA simulations, we back-mapped them to an atomistic detailed description (http://cgmartini.nl/images/tools/backward/initram-v5.sh). The resulting models based on the AMBER99sb*ILDN force-field were used to set up the systems as previously reported:48 (i) steepest descent minimization in vacuum; (ii) solvation in a 0.15 M NaCl solution with TIP3P water molecules and re-optimized monovalent ions in a rhombic dodecahedron box; (iii) steepest descent minimization with ions and solvent; (iv) thermalization at 300 K in NVT ensemble; (v) pressure equilibration at 1 atm and (vi) 200 ns production run in NPT ensemble.
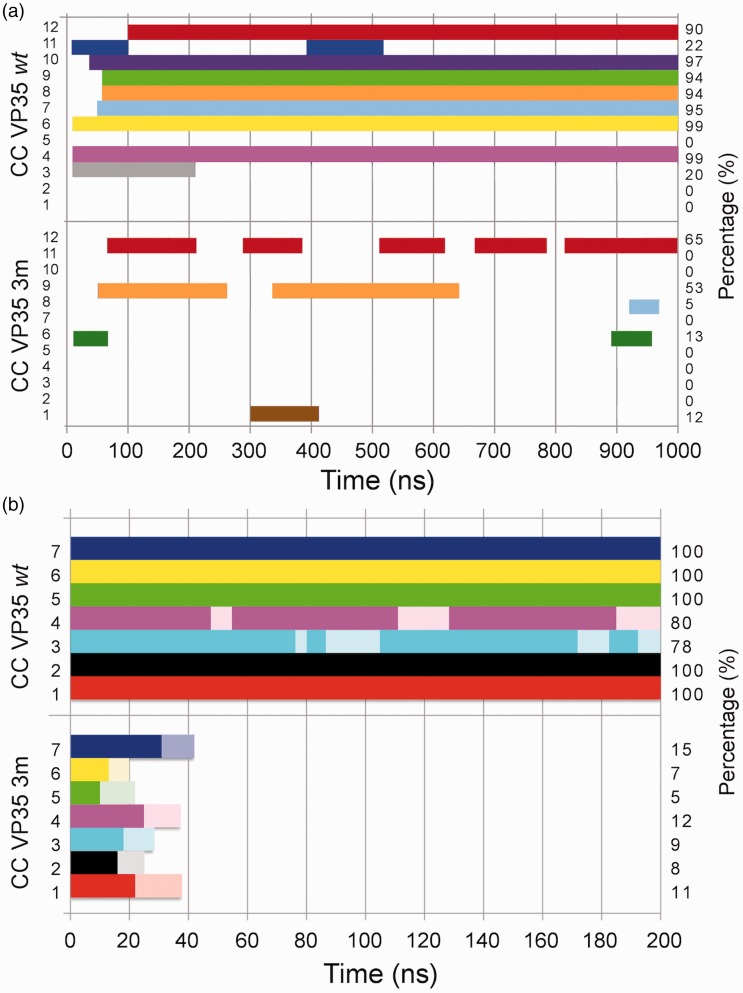
Figure 2.
Final conformations for VP35-wt (top) and VP35-3m (bottom) variants from 12 independent replicas of CG simulations. Dimers resulting in a proper CC configuration are shown in green boxes, improper CC and uncoiled dimers are in yellow boxes, disordered/anti-parallel dimers are in red boxes. The two monomers are shown in red and blue beads with the mutated residues in cyan and pink, respectively; yellow terminal prolines give the relative monomer orientation (parallel/anti-parallel).
The SOCKET program53 was employed to determine the presence of CC conformations in the resulting trajectories. For each VP35-wt and VP35-3m CG trajectory, the analysis was performed on 1000 equidistantly picked snapshots (every 1 ns) properly back-mapped to an AA model in order to be suitable for SOCKET, using the default cut-off (7 Å).
The dimerization free-energy between the two VP35-wt and VP35-3m CC domain monomers was evaluated by means of the molecular mechanics-generalized Born surface area (MM-GBSA) post-processing method.54,55 For this aim, the MMPBSA program56 was used setting the implicit solvent model (igb = 8), intrinsic radii (mbondi2) and 0.15 M as ionic strength. This analysis allows to decompose the binding free energy within the framework of the MM-GBSA using both the pairwise and per-residue schemes.56 The free energies were calculated on 1500 frames (saved every 20 ps or 10,000 steps), extracted from a stable part of each production trajectory. The contribution from solute conformational entropy (TΔS) was not included in the evaluation of the free energy57 considering its computational demand.
Plasmid construction and cloning
EBOV VP35 gene cloning in pET45b(+) vector (Novagen) and production of pET45b-EBOV-VP35 plasmid for the expression of recombinant N-terminal His6-tag-VP35 protein (rVP35) was performed as previously described.48 Cloning of the EBOV VP35 gene (Zaire ebolavirus, Yambuku-Mayinga, GenBank: NC_002549.1) in pcDNA3 plasmid (Invitrogen) and production of pcDNA3-EBOV-VP35 plasmid for the transfection of rVP35 was performed as previously described in Cannas et al.21
To construct the EBOV VP35-3m procariotic expression plasmid in order to introduce three-point mutations (Leu 90 to Ala, Leu 93 to and Leu 107 to Ala) into the pET45b-EBOV-VP35 plasmid, a series of three consecutive site-directed mutagenesis was performed using the QuickChange Lightning Site-Directed Mutagenesis kit (Agilent technologies), manufacturer instructions, using the following primers:
L90A forward 5′-GAGGAGGTAGTACAAACA GCGGCTTCATTGGCTACT GTTGTGC-3′;
L90A reverse 5′-GCACAACAGTAGCCAATGAAGCCGCTGTTTGTAC TACCTCCTC-3′;
L107A forward 5′-CCATCGCATCAGAATCAGCAGAACAACGCATTA CGAGTCTTGAG-3′;
L107A reverse 5′-CTCAAGACTCGTAATGCGTTGTTCTGCTGATTCT GATGCGATGG-3′;
L93A forward 5′-GTACAAACAGCGGCTTCAGCGGCTACTGTTGTGCA ACAACAAACC-3′;
L93A reverse 5′-GGTTTGTTGTTGCACAACAGTAGCCGCTGAAGCCGC TGTTTGTAC-3′.
The obtained plasmid encodes for the triple mutant EBOV-VP35-L90A/L93A/L107A recombinant protein (rVP35-3m). The PCR reaction was carried out in a final volume of 50 µl of reaction mixture containing: 100 ng of pET45b-EBOV-VP35 plasmid, 125 ng of each primer. The PCR cycle consisted of: an initial denaturation at 95°C for 2′ (2 minutes), 18 cycles of denaturation at 94°C for 20″ (20 seconds), annealing at 60°C for 10″ (10 seconds), extension at 68°C for 3′30″ (3 minutes and 30 seconds) and a final extension at 68°C for 5′. XL10-Gold® Ultracompetent Cells were transformed with PCR products following suggested heat shock protocol – 30′ (30 minutes) on ice, 30″ (30 seconds) at 42°C, 2′ (2 minutes) on ice – and cultured. Plasmids was extracted and sequenced step by step for control.
For the construction of EBOV VP35-3m mammalian expression plasmid starting from pcDNA3-EBOV-VP35 plasmid21 a site-directed mutagenesis was performed using Q5® Site-Directed Mutagenesis Kit in order to obtain new insert that express the rVP35-3m in a mammalian cellular system, using the following primers:
forward 5′-ACCATCGCATCAGAATCAGCAGAACAACGCATTACGAGTCTTGAGAATG-3′;
reverse 5′-GTTGTTGCACAACAGTAGCCGCTGAAGCCGCTGTTTGTACTACCTCCTC-3′.
PCR reaction was carried out in a final volume of 25 µl of a mixture containing: pcDNA3-EBOV-VP35 plasmid (25 ng), each primer (0.5 µM) and Q5 Hot Start High-Fidelity 2X Master Mix (12.5 µl) provided within the kit. The PCR cycle consisted of: an initial denaturation at 98°C for 30″ (30 seconds), 25 cycles of denaturation at 98°C for 10″ (10 seconds), annealing at 59°C for 30″ (30 seconds), extension at 72°C for 2′30″ (2.30 minutes) and a final extension at 72°C for 2′ (2 minutes). After PCR, following steps were performed: the kinase, ligase and Dpn-I (KLD) treatment that allows phosphorylation, circularization and template removal in a single 5′ step and the subsequent transformation of 50 µl of NEB-5α competent cells with 5 µl of KLD products, following suggested heat shock protocol – 30′ (30 minutes) on ice, 30″ (30 seconds) at 42°C, 5′ (5 minutes) on ice – and cultured. The plasmid was extracted and sequenced.
Protein expression, purification, denaturation and refolding
BL21AI Escherichia coli were transformed with pET45b-EBOV-VP35 and pET45b-EBOV-VP35-3m plasmids. Recombinant proteins were expressed as previously reported.58 Bacterial pellets were re-suspended in 5 ml/g of lysis buffer (100 mM sodium phosphate pH 8.0, 500 mM NaCl, 1 mM 2-mercaptoethanol, 10% glycerol, 20 mM imidazole, 6 M urea), incubated on ice for 15′ minutes and sonicated on ice. The cell lysate was centrifuged at 32,000×g at 4°C for 45′ minutes. Supernatant was loaded (0.5 ml/min) to an IMAC Econo-Column® (Biorad) prepared with 3 ml of Ni-Sepharose High Performance (GE Healthcare), connected with a BioLogic LP Chromatographic System (Biorad) and pre-equilibrated in binding buffer (100 mM sodium phosphate pH 8.0, 300 mM NaCl, 1 mM 2-mercaptoethanol, 10% glycerol, 20 mM imidazole, 6 M urea). Urea was removed by a decreasing gradient from 6 to 0 M with a flow rate of 0.5 ml/min for around 130 column volumes. The column was washed with Washing Buffer (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 1 mM 2-mercaptoethanol, 10% glycerol) with increasing concentrations of imidazole (70 mM – 150 mM – 250 mM). Proteins were eluted in Washing Buffer 1 M imidazole. Eluted fractions were analyzed by 12% SDS–PAGE. Fractions containing rVP35 protein were gathered and dialyzed against Dialysis buffer (50 mM sodium phosphate pH 8.0, 800 mM NaCl, 1 mM 2-mercaptoethanol, 10% glycerol). Afterwards, protein concentration was determined with the Protein Quantification Kit-Rapid (Sigma Aldrich).
Preparation of dsRNA substrates
dsRNA of 500 bp was produced by in vitro transcription using the T7 MEGAscript® RNAi kit (Ambion) from the linearized DNA provided with the kit as template, according to manufacturer’s instructions. In vitro transcribed (IVT) oligomers were purified from transcription reaction with Quick Spin G25 columns (Roche), and quantified by spectrophotometry.
The fluorescent and non-fluorescent dsRNA oligomers of 30 bp were prepared by annealing from three different ssRNAs (Metabion International AG (Germany)):
5′-ppp-cccuuuccuccuuccuuuuguuccucuccc-3′;
5′-gggagaggaacaaaaggaaggaggaaaggg-3′;
5′-Fluorescein-gggagaggaacaaaaggaaggaggaaaggg-3′.
Annealing was conducted in order to have one end of each dsRNA with triphosphate. The integrity of DNA templates, IVT dsRNAs and synthetic dsRNA molecules was assessed by agarose-gel electrophoresis.
EBOV VP35 RNA binding assay
Seven hundred nanograms of rVP35 were added in each well of a 96-well Pierce® Nickel Coated Plate (Thermo Fisher) resulting in a final volume of 200 µl coating buffer (50 mM Sodium Phosphate pH 7.5, 150 mM NaCl) and incubated for 30′ (30 minutes) at 4°C, 120 r/min, then washed twice with 200 µl of Washing Buffer (50 mM Sodium Phosphate pH 7.5, 150 mM NaCl, 0.03% Tween-20). Then, 7.5 nM of 30 bp 5′-fluorescein-dsRNA was added in 100 µl reaction buffer (50 mM Sodium Phosphate pH 7.5, 100 mM NaCl, 20 mM MgCl2, 0.03% Tween-20) and the plate was incubated for 60′ (60 minutes) at 37°C, 200 r/min. Unbound dsRNA was removed by two washes with 200 µl of a reaction buffer and then fluorescence signals of samples were read with PerkinElmer Victor3™ at 490/528 nm (excitation/emission wavelength). All experiments were repeated three times in duplicate. The bound dsRNA has been quantified (in femtomoles) interpolating the obtained values in a 30 bp 5′-fluorescein-dsRNA calibration curve.
EBOV VP35 luciferase gene-reporter inhibition assay
The luciferase gene-reporter assay was performed using pcDNA3-EBOV-VP35 and pcDNA3-EBOV-VP35-3m plasmids as previously described.28 A549 cells (5 × 104 per well in a 48-well plate) were transfected using T-Pro P-Fect Transfection Reagent (T-Pro Biotechnology, Twin Helix) with the construct pGL(IFN-β) luc. For the evaluation of the IFN-β production inhibition mediated by VP35, cells were co-transfected with different amounts the pcDNA3-ZEBOV-VP35wt or pcDNA3-ZEBOV-VP353m using pcDNA3 as empty vector control at the maximum transfected concentration (250 ng). Twenty-four hours after transfection, cells were additionally transfected with Influenza virus A/PR/8/34 (H1N1) RNA and incubated for further 6 h at 37°C with 5% CO2. Next, cells were harvested with Luciferase Assay Buffer (50 mM Na-MES pH 7.8, 50 mM Tris-HCl pH 7.8, 1 mM dithiothreitol, 0.2% Triton X-100). The crude cell lysates were clarified by centrifugation and 50 µl of cleared lysates were added to 50 µl of luciferase assay buffer (125 mM Na-MES pH 7.8, 125 mM Tris-HCl pH 7.8, 25 mM magnesium acetate, 2.5 mg/mL ATP) in a white 96-well plate (OptiPlate, PerkinElmer). Immediately after addition of 50 µl of 1 mM D-luciferin into each well, the luminescent signal was measured in Victor3 luminometer (Perkin Elmer). The measured signal is normalized as the percentage of increase of pGL IFN-β luc activation versus unstimulated control. Each assay was carried out in triplicate.
Results and discussion
Starting from our model of Zaire EBOV VP35 CC domain,47 we investigated the impact of substituting with alanines the leucines 90, 93 and 107, key residues sitting at the a and d positions of the heptad-repeat framework of the VP35 CC domain. These mutations were designed to disrupt the CC structure and were showed to impair VP35 oligomerization15 or to promote the formation of aberrant oligomeric species.41 In order to understand the molecular mechanism thereof and characterize quantitatively the structural and thermodynamical importance of these mutations, we performed MD dimerization simulations of VP35-wt and VP35-3m. After investigation of the oligomerization propensity analysis of the CC domain investigated by Ramaswamy et al.,48 we designed new sets of simulations to draw attention to the differences between the VP35-wt and the VP35-3m systems in the formation and stabilization of the dimers. It should be mentioned that these simulations were performed in the absence of a X-ray structure of the EBOV VP35 N-terminal domain in its trimeric form, which was resolved only recently.41 Thus, we were forced to perform the whole computational study described here using the in-house built in silico model of the monomer48 as starting structure. Our choice is validated by the very low root-mean-square deviation (0.4 Å) of our model with respect to the structure of a single VP35 monomer taken from the recently resolved and published structure (PDB id: 6GBO).41
The results of CG MD oligomerization simulations for VP35-wt and VP35-3m clearly showed a different behavior in the dynamics of the two systems. Indeed, the 12 replicas for each variant indicated a marked reduction in the VP35-3m propensity to form dimers when compared with that of VP35-wt (Figure 2). Via SOCKET,53 we assessed whether the final configuration of each replica for the two systems was in a CC arrangement or not. In VP35-wt simulations, 7/12 replicas were found in a perfect parallel CC dimer, 4/12 in a not-coiled dimer and only one formed an anti-parallel dimer that should not be able to bind dsRNA. At opposite, in VP35-3m simulations, only 1/12 simulations led to the formation of a parallel CC structure, 4/12 formed a dimer without coiling and 7/12 replicas either did not even properly dimerized or formed an anti-parallel dimer, likely not functional for the dsRNA binding (Figure 2). A more quantitative analysis of the trajectories was performed with SOCKET, which revealed the whole time-line evolution of the CG simulations. As it can be noticed in Figure 3(a), VP35-wt clearly showed a high propensity to form stable CC dimers. In contrast, only one VP35-3m system formed transient CC structures over the whole simulation length.
Figure 3.
Coiled-coil timeline for the CG (a) and AA (b) simulations. (a) Twelve replicas for VP35-wt (top) and VP35-3m (bottom) variants (numbered on the left side of the box) of CG simulations. (b) Seven AA replicas for both variants (VP35-wt top, VP35-3m bottom). The occurrence of the CC conformation over time is evaluated by means of SOCKET.53 The time-percentage is indicated on the right border of the box. In the presence of CC fraying of the terminal three residues at the 5′- and/or 3′-end, the bars are pale-colored.
In order to investigate, at higher accuracy, the behavior and the stability of the single VP35-3m variant displaying an acceptable propensity to dimerize, this structure was back-mapped to an AA representation and simulated for 200 ns (as described in the “Materials and methods” section). As a control, also the seven VP35-wt CC dimers were back-mapped to an AA representation and simulated. In order to collect the same statistics for both the systems, the single VP35-3m CC structure was simulated in seven independent replicas using different starting velocities.
The analysis performed with SOCKET on these AA simulations showed that 5/7 VP35-wt dimers maintained the CC conformation over the whole length of the simulation (Figures 3(b), upper panel, and Figure 4, left panel), while the remaining two systems sampled some transient fraying of the last three residues on the 5′- and/or 3′-end. Most importantly, none of the VP35-3m AA MD simulations displayed a stable CC conformation (Figure 3(b), lower panel). Indeed, during the simulation, the two VP35-3m monomers partially lost their helical structure too (Figure 4, right panel). We can reasonably exclude that this behavior could be due to issues associated with back-mapping from CG to AA models, because the same procedure when applied to a single VP35-3m monomer lead to a stable structure over more than 100 ns of AA MD simulation (data not shown). Furthermore, to avoid any possible bias derived from the use of the initial conformation resulted from the CG dynamics, we also tested different protocols; we restrained the Cα or the backbone atoms during annealing cycles applying a force constant of 0.1 or 1.0 (kcal/mol)/Å2 on the back-mapped VP35-3m dimeric structure to generate different starting conformations. However, also in these additional 50 ns-long simulations (three replicas for each setting) the 3 m dimers lost their coiling and at least partially the helicity of the monomers, as in the original simulations (data not shown). We therefore can reasonably suggest that the instability of the 3 m system is due to the presence of three alanine residues in place of leucines at crucial positions of the heptads, presumably disfavoring a stable “knob-into-hole” arrangement and thus leading to CC disruption.
Figure 4.
Representative final structures from the AA MD simulations. The systems, VP35-wt (left) in perfect CC conformation and VP35-3m (right) as disordered oligomer, are shown in cartoon representation with the three mutated residues highlighted as licorice.
To determine the hinges of the CC interaction, also clarifying in this framework the role of the mutations, an estimation of the contribution given by the single residues to the dimerization process is required. To this aim, we performed free energy calculations over the equilibrium trajectories of the five stable AA VP35-wt simulations, using the MM-GBSA method,54,55 evaluating both the pairwise and the per-residue contributions:56 the former measures the contribution of every single inter-monomer interaction (Table 1), whereas the latter ranks the contribution of each residue towards CC formation (Figure 5).
Table 1.
Pairwise free-energy decomposition.
| Residue pair | Average pairwise free-energy (ΔG) (kcal/mol) |
|---|---|
| LEU90|LEU90 | –1.9±0.2 |
| LEU93|LEU93 | –1.9±0.1 |
| GLN100|GLN100 | –1.9±0.4 |
| GLN100|SER104 | –2.2±0.7 |
| LEU107|LEU107 | –1.9±0.1 |
| ARG110|ILE111 | –2.1±0.3 |
| ARG110|GLU115 | –3.0±1.3 |
| GLU115|ARG110 | –3.2±1.2 |
Note: For the listed interactions (first and second amino acid of each pair belong to monomers 1 and 2, respectively) the pairwise ΔG weighted mean value and the corresponding standard deviation from the VP35-wt simulations are reported. Only the residue pairs contributing by more than 1.5 kcal/mol to the dimerization free-energy (in each replica) are listed; the pairs involving leucine 90, 93, 107 are highlighted in red.
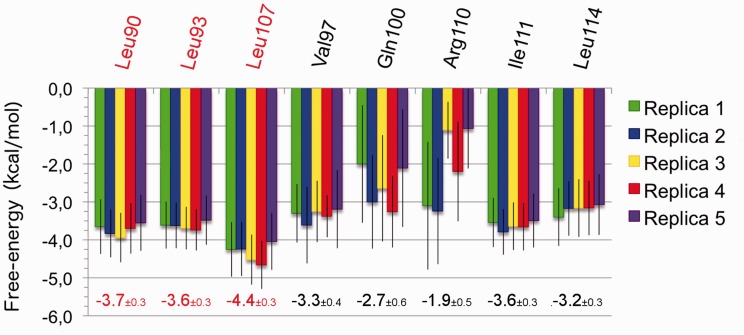
Figure 5.
Per-residue-based free-energies for VP35-wt complexes. The residues contributing to the total free-energy with at least 3.0 kcal/mol in at least one replica are included. The value resulting from the weighted mean and the associated standard deviation of the five replicas are reported emphasizing in red the contributions from leucine 90, 93 and 107.
The pairwise decomposition (Table 1) reveals that six out of eight pairs mostly contributing to stabilization of the dimer represent interactions between hydrophobic and apolar residues at a and d positions in the heptad-repeats (Figure 1). This finding is in agreement with the literature,43 confirming the key role of the amino acids at these positions also for the VP35 CC domain dimerization. In particular, our analysis quantitatively assessed their importance for the thermodynamic stability as well as for the structural complementarity at the interface between the two monomers. The interactions between R110 and E115, respectively, at positions g and e seem to be contributing most to the stability of the system, although the standard deviation of their free-energy contribution resulted higher than those of apolar interactions (Table 1). These two charged residues created a pair of crucial salt bridges tightly keeping the two monomers together in our reduced model. In association with the previously mentioned strong interactions involving positions a and d, this pair of non-covalent interactions creates a cooperative effect that prevents the unzipping of the dimer.59 These outcomes concerning R110 and E115 supported and complemented the experimental evidences of these residues as key for the stabilization of the oligomerization domain and the modulation between the CC oligomeric states in EBOV.41 Additionally, as the interactions between the L90, L93 and L107 of one monomer with their corresponding counterpart in the other monomer appear as those mostly contributing to the dimer stabilization, we can confirm the importance of these residues in the global framework of the dimerization process.
The per-residue decomposition reveals the effect of each amino acid on the stability of the complex by summing all its interactions with the others. The residues contributing more to the dimerization free-energy turned out to be L90, L93, V97, Q100, L107, R110, I111, L114 (Figure 5). The presence of the 90, 93 and 107 leucines among the list of amino acids giving the major contribution to the free-energy of binding further confirmed their importance for the stability of the VP35 CC domain dimer. Thus, their absence in VP35-3m, due to the contemporary mutation to alanines, could be the reason behind the impossibility of all the seven AA simulations to maintain a CC conformation. Although aware of the approximation associated with this evaluation, it can be inferred that the mutation of L107 might have a larger impact than the single mutations of L90 and L93, as the per-residue contribution of L107 is larger than those of the other two leucines.
Having computationally assessed the relevance of the three Leu residues in the dimerization process, we proceeded to explore experimentally which is the role of dimerization in the VP35 inhibition of IFN production. Firstly, we investigated if the presence of the three mutations in the full-length rVP35 could influence the protein binding to dsRNA. Implementing the previously developed protocols to determine VP35-dsRNA binding activity using full-length EBOV VP35 His-tagged VP35 purified in bacterial system,58,60 we compared rVP35-wt and rVP35-3m binding to dsRNA to test if the 3 m retains this function or not. The point mutations were introduced on the pET45b(+)-EBOV-VP35 plasmid, and the full-length protein was expressed and purified. We then performed three experiments of binding at a 7.5 nM fixed concentration of ligand. The results demonstrated that rVP35-3m showed almost abolished dsRNA binding ability as compared to VP35-wt (Figure 6(a)) (p-value = 0.0024), showing that the disruption of the CC functional structure compromises the ability of the full-length protein to bind the 3ppp-dsRNA even in presence of an unaltered IID. These data reveal that the IID requires a functional CC domain to exert its dsRNA binding function that can probably occur only in a dimeric form in agreement with structural studies.40
Figure 6.
Biochemical assays. (a) Comparison between rVP35-wt (red column) and rVP35-3m (gray column) ability to bind to dsRNA, performed with a nickel-coated plate assay; 700 ng/well of rVP35 or vVP35 3 m were incubated with 7.5 nM of 30 bp 5′-fluorescein-dsRNA and incubate for 60′. Unbound dsRNA was removed and fluorescence signals of samples were read. All experiments were repeated three times in duplicate. The bound dsRNA has been quantified (in femtomoles) interpolating the obtained values in calibration curve. (b) Comparison of the inhibitory effect of EBOV VP35-wt and VP35-3m in the luciferase reporter gene assay. A549 cells were co-transfected with pGL interferon β (IFN-β) luc plus different amounts of the pcDNA3-ZEBOV-VP35wt (red column) or pcDNA3-ZEBOV-VP353m (gray column), using pcDNA3 as empty vector control (EV) (black column). Twenty-four hours after transfection, cells were additionally transfected with influenza A virus (IAV) RNA. Six hours after transfection, cells were lysed and luciferase activity was measured. Results show the percentage of luciferase expression over the unstimulated control. Significant at: p-value < 0.05 (*); p-value <0.01 (**).
However, a question was still open: provided that the sole presence of VP35 IID is able to affect the IFN activation,15 we asked whether an altered CC oligomerization in presence of an unaltered IID could affect the VP35-mediated inhibition of the IFN production. Thus, we investigated the impact of VP35-3m in a luciferase gene reporter cellular assay. Starting from pcDNA3-EBOV-VP35 plasmid, we performed a site-directed mutagenesis to obtain an insert expressing EBOV VP35-3m in a mammalian cellular system. In order to compare the IFN inhibitory ability of rVP35 and rVP35-3m, cells were transfected with different amounts (100, 150, 200, 250 ng) of the mammalian expression vectors pcDNA3-EBOV-VP35 and pcDNA3-EBOV-VP35-3m. The result showed that the VP35-3m still maintains the ability to inhibit the RIG-I signaling cascade but to a lower extent with respect to VP35-wt, and this difference is higher at lower amount of transfected plasmid (Figure 6(b)), hence at lower level of protein expressed.
Interestingly, the loss of IFN inhibitory activity, observed at low cellular VP35-3m concentrations, was recovered almost at wt-levels as the intracellular levels of EBOV VP35-3m increased, suggesting that mutations affecting CC oligomerization do not impair the other VP35 interactions related to IFN inhibition. These results suggest that oligomerization may not be required for the VP35 effect on the other components of the RIG-I pathway that may be exerted by protein–protein interactions with unaltered portions of the VP35-3m. At higher VP35 cytosolic concentration, such interactions with the components of the RIG-I pathway may play a major role compensating the loss of dsRNA binding capacity. Although it remains to be determined if during viral infection, the VP35 cytoplasmatic levels will be sufficient to exert this effect. Finally, it is worth to note that it has been recently reported that the presence of VP35 of filoviral origins in the genomes of the Myotis genus of bats.61,62 Consistently with our results, such bat VP35s were shown to lack the dsRNA binding capacity and have a substantially decrease ability to inhibit the RIG-I mediated IFN activation with respect to EBOV VP35 that could be somehow restored at high protein concentrations.61
In the general framework of the characterization of the EBOV VP35 different multimeric forms and in the light of the two just published structural works,41,47 the possibility that in the presence of dsRNA this crucial virulence factor could be prompt to preliminary assemble as a dimer is still an open debate. The data of the present work as well as the crystal structures of the VP35 IID dimer in complex with dsRNA39,49,63 seem to confirm this alternative.
Conclusions
The impairment of RIG-I pathway is due to the capacity of VP35 to counteract the cascade at different levels by interacting with different targets. Given the complexity of the system, it is difficult to dissect the impact of structural impairments on the overall VP35 inhibition of IFN production. In silico and in vitro results demonstrated that L90, L93 and L107, previously shown to affect VP35 oligomerization, exert a crucial role on dimerization, which is an important preliminary step for VP35-dsRNA binding in the presence of unaltered VP35 IID and thus for VP35 inhibitory function on IFN production. Remarkably, according to our outcome, VP35 oligomerization may not be required for the VP35 interaction with the other components of the RIG-I pathway, compensating the loss of VP35-dsRNA binding ability in a concentration-dependent manner. Although relevant, this finding deserves to be investigated in a full-replicant EBOV variant carrying a dimerization-deficient VP35 to fully assess the importance of the dsRNA masking in the EBOV VP35 elusion of IFN activation.
Authors’ contribution
FDP performed the simulations and analyzed the data together with VKR and under the supervision of AVV. GLD and AF carried out the experiments, AC and EF interpreted the biochemical assay outcomes. AVV, ET and PR devised, conceived and designed the project. The manuscript was written through contributions of all authors and all authors have given approval to the final version of the manuscript.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: A Corona, F Di Palma, E Fanunza and VK Ramaswamy were supported by the Research Grant RAS LR 07/2007 grant no. CRP-78711/F72I15000900002 awarded by the Regione Autonoma della Sardegna (RAS, PO Sardegna FSE 2007–2013, L.R. 7/2007 “Promozione della Ricerca scientifica e dell’innovazione tecnologica in Sardegna”).
ORCID iD
Paolo Ruggerone https://orcid.org/0000-0003-0825-0824
References
- 1.Mahanty S, Bray M. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect Dis 2004; 4: 487–498. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn JH, Becker S, Ebihara H, et al. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch Virol 2011; 155: 2083–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein T, Anthony SJ, Gbakima A, et al. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 2018; 3: 1084–1089. Erratum in: Nat Microbiol 2018; 3: 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. Lancet 2011; 377: 849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaner J, Schaack S. Understanding Ebola: The 2014 epidemic. Global Health 2016; 53. DOI:10.1186/s12992-016-0194-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malvy D, McElroy AK, de Clerck H, et al. Ebola virus disease. Lancet 2019; 393: 936–948. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds P, Marzi A. Ebola and Marburg virus vaccines. Virus Genes 2017; 53: 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuler J, Hudson ML, Schwartz D, et al. A systematic review of computational drug discovery, development, and repurposing for ebola virus disease treatment. Molecules 2017; 22: 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fanunza E, Frau A, Corona A, et al. Antiviral agents against Ebola virus infection: repositioning old drugs and finding novel small molecules. In: Botta M (ed.) Annual reports in medicinal chemistry. Neglected diseases: extensive space for modern drug discovery. Vol. 51, 2018, pp.135–173. Elsevier, Amsterdam: The Netherlands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J. Ebola outbreak continues despite powerful vaccine. Science 2019; 364: 223. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez A, Kiley MP, Holloway BP, et al. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res 1993; 29: 215–240. [DOI] [PubMed] [Google Scholar]
- 12.Mohamadzadeh M. Potential factors induced by filoviruses that lead to immune supression. Curr Mol Med 2009; 9: 174–178. [DOI] [PubMed] [Google Scholar]
- 13.Zinzula L, Tramontano E. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: Hide, mask, hit. Antiviral Res 2013; 100: 615–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid SP, Leung LW, Hartman AL, et al. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J Virol 2006; 80: 5156–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reid SP, Ca WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the Ebolavirus VP35 protein. J Virol 2005; 341: 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebihara H, Takada A, Kobasa D, et al. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog 2006; 2: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prins KC, Delpeut S, Leung DW, et al. Mutations abrogating VP35 interaction with double-stranded RNA render Ebola virus avirulent in guinea pigs. J Virol 2010; 84: 3004–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung DW, Prins KC, Basler CF, et al. Ebolavirus VP35 is a multifunctional virulence factor. Virulence 2010; 1: 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanunza E, Frau A, Sgarbanti M, et al. Development and validation of a novel dual luciferase reporter gene assay to quantify Ebola virus VP24 inhibition of IFN signaling. Viruses 2018; 10: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fanunza E, Frau A, Corona A, et al. Insights into Ebola virus VP35 and VP24 interferon inhibitory functions and their initial exploitation as drug targets. Infect Disord Drug Targets. Epub ahead of print 23 November 2018. DOI: 10.2174/1871526519666181123145540. [DOI] [PubMed]
- 21.Daino GL, Frau A, Sanna C, et al. Identification of myricetin as an Ebola virus VP35-double-stranded RNA interaction inhibitor through a novel fluorescence-based assay. Biochemistry 2018; 57: 6367–6378. [DOI] [PubMed] [Google Scholar]
- 22.Darapaneni V. Virion protein 24 of Ebola virus as a potential drug target. Am J Curr Microbiol 2014; 3: 14–22. [Google Scholar]
- 23.Seesuay W, Jittavisutthikul S, Sae-Lim N, et al. Human transbodies that interfere with the functions of Ebola virus VP35 protein in genome replication and transcription and innate immune antagonism. Emerg Microbes Infect 2018; 7: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iversen PL, Warren TK, Wells JB, et al. Discovery and early development of AVI-7537 and AVI-7288 for the treatment of Ebola virus and Marburg virus infections. Viruses 2012; 4: 2806–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haasnoot J, De Vries W, Geutjes EJ, et al. The ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 2007; e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Sage V, Cinti A, McCarthy S, et al. Ebola virus VP35 blocks stress granule assembly. Virology 2017; 502: 73–83. [DOI] [PubMed] [Google Scholar]
- 27.Cárdenas WB, Loo Y-M, Gale M, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol 2006; 80: 5168–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannas V, Daino GL, Corona A, et al. A luciferase reporter gene assay to measure Ebola virus viral protein 35 – associated inhibition of double-stranded RNA – stimulated, retinoic acid – inducible gene 1 – mediated induction of interferon β. J Infect Dis 2015; 212: S277–S281. [DOI] [PubMed] [Google Scholar]
- 29.Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza a virus. Virology 2004; 328: 177–184. [DOI] [PubMed] [Google Scholar]
- 30.Basler CF, Mikulasova A, Martinez-Sobrido L, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol 2003; 77: 7945–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prins KC, Ca WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKK ε and TBK-1. J Virol 2009; 83: 3069–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang TH, Kubota T, Matsuoka M, et al. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog 2009; 5: e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luthra P, Ramanan P, Mire CE, et al. Mutual antagonism between the Ebola virus VP35 protein and the RIG-I activator PACT determines infection outcome. Cell Host Microbe 2013; 14: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Z, Cerveny M, Yan Z, et al. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J Virol 2007; 81: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumann M, Gantke T, Muhlberger E. Ebola virus VP35 antagonizes PKR activity through Its C-terminal interferon inhibitory domain. J Virol 2009; 83: 8993–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Möller P, Pariente N, Klenk H-D, et al. Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J Virol 2005; 14876–14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirchdoerfer RN, Abelson DM, Li S, et al. Assembly of the Ebola virus nucleoprotein from a chaperoned VP35 complex. Cell Rep 2015; 12: 140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Groseth A, Charton JE, Sauerborn M, et al. The Ebola virus ribonucleoprotein complex: a novel VP30-L interaction identified. Virus Res 2009; 8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung DW, Ginder ND, Fulton DB, et al. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci 2009; 106: 411–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung DW, Prins KC, Borek DM, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol 2010; 17: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinzula L, István N, Massimiliano O, et al. Structures of Ebola and Reston virus VP35 oligomerization domains and comparative biophysical characterization in all Ebolavirus species. Structure 2019; 27: 39–54.e6. [DOI] [PubMed] [Google Scholar]
- 42.Moutevelis E, Woolfson DN. A periodic table of coiled-coil protein structures. J Mol Biol 2009; 726–732. [DOI] [PubMed] [Google Scholar]
- 43.Lupas AN, Gruber M. The structure of α-helical coiled coils. Adv Protein Chem 2005; 95–129. [DOI] [PubMed] [Google Scholar]
- 44.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 2001; 82–88. [DOI] [PubMed] [Google Scholar]
- 45.Edwards MR, Liu G, Mire CE, et al. Differential regulation of interferon responses by Ebola and Marburg virus VP35 proteins. Cell Rep 2016; 1632–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruhn JF, Kirchdoerfer RN, Urata SM, et al. Crystal structure of the Marburg Virus VP35 oligomerization domain. J Virol. 2016; e01085–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chanthamontri CK, Jordan DS, Wang W, et al. The Ebola viral protein 35 N-terminus is a parallel tetramer. Biochemistry 2019; 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaswamy VK, Di Palma F, Vargiu AV, et al. Insights into the homo-oligomerization properties of N-terminal coiled-coil domain of Ebola virus VP35 protein. Virus Res 2018; 247: 61–70. [DOI] [PubMed] [Google Scholar]
- 49.Kimberlin CR, Bornholdt ZA, Li S, et al. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci 2010; 107: 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingólfsson HI, Lopez CA, Uusitalo JJ, et al. The power of coarse graining in biomolecular simulations. Wires Comput Mol Sci 2014; 225–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuckerman ME. Statistical mechanics: theory and molecular simulation. Oxford: Oxford University Press, 2010. [Google Scholar]
- 52.Páll S, Abraham MJ, Kutzner C, et al. (2015) Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. In: Markidis S and Laure E (eds) Solving Software Challenges for Exascale. EASC 2014. Lecture Notes in Computer Science, vol 8759. Springer, Cham [Google Scholar]
- 53.Walshaw J, Woolfson DN. SOCKET: a program for identifying and analysing coiled-coil motifs within protein structures. J Mol Biol 2001; 1427–1450. [DOI] [PubMed] [Google Scholar]
- 54.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 2015; 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kollman PA, Massova I, Reyes C, et al. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res 2000; 889–897. [DOI] [PubMed] [Google Scholar]
- 56.Miller BR, McGee TD, Swails JM, et al. MMPBSA.py: An efficient program for end-state free energy calculations. J Chem Theory Comput 2012; 3314–3321. [DOI] [PubMed] [Google Scholar]
- 57.Hou T, Wang J, Li Y, et al. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model 2011; 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zinzula L, Esposito F, Mühlberger E, et al. Purification and functional characterization of the full length recombinant Ebola virus VP35 protein expressed in E. coli. Protein Expr Purif 2009; 66: 113–119. [DOI] [PubMed] [Google Scholar]
- 59.Walther TH, Gottselig C, Grage SL, et al. Folding and self-assembly of the TatA translocation pore based on a charge zipper mechanism. Cell 2013; 316–326. [DOI] [PubMed] [Google Scholar]
- 60.Zinzula L, Esposito F, Pala D, et al. DsRNA binding characterization of full length recombinant wild type and mutants Zaire ebolavirus VP35. Antiviral Res 2012; 93: 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edwards MR, Liu H, Shabman RS, et al. Conservation of structure and immune antagonist functions of filoviral VP35 homologs present in microbat genomes. Cell Rep 2018; 861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kondoh T, Manzoor R, Nao N, et al. Putative endogenous filovirus VP35-like protein potentially functions as an IFN antagonist but not a polymerase cofactor. PLoS One 2017; e018645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bale S, Julien J-P, Bornholdt ZA, et al. Ebolavirus VP35 Coats the backbone of double-stranded RNA for interferon antagonism. J Virol 2013; 10385–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]