Abstract
Background
Ochratoxin A (OTA) is a widespread mycotoxin and induces liver inflammation to human and various species of animals. The intestinal microbiota has critical importance in liver inflammation; however, it remains to know whether intestinal microbiota mediates the liver inflammation induced by OTA. Here, we treated ducklings with oral gavage of OTA (235 μg/kg body weight) for 2 weeks. Then, the microbiota in the cecum and liver were analyzed with 16S rRNA sequencing, and the inflammation in the liver was analyzed. To explore the role of intestinal microbiota in OTA-induced liver inflammation, intestinal microbiota was cleared with antibiotics and fecal microbiota transplantation was conducted.
Results
Here, we find that OTA treatment in ducks altered the intestinal microbiota composition and structure [e.g., increasing the relative abundance of lipopolysaccharides (LPS)-producing Bacteroides], and induced the accumulation of LPS and inflammation in the liver. Intriguingly, in antibiotic-treated ducks, OTA failed to induce these alterations in the liver. Notably, with the fecal microbiota transplantation (FMT) program, in which ducks were colonized with intestinal microbiota from control or OTA-treated ducks, we elucidated the involvement of intestinal microbiota, especially Bacteroides, in liver inflammation induced by OTA.
Conclusions
These results highlight the role of gut microbiota in OTA-induced liver inflammation and open a new window for novel preventative or therapeutic intervention for mycotoxicosis.
Keywords: Ochratoxin A, Intestinal microbiota, LPS, Liver inflammation, Fecal microbiota transplantation
Background
Mycotoxins are secondary metabolites produced by fungal genera (e.g., Aspergillus, Penicillium, and Fusarium) and are the most common natural food contaminants in human and animal diets, such as cereals and animal forages [1–3]. Ochratoxin A (OTA), the most prevalent and relevant fungal toxin produced by Aspergillus species and Penicillium species [4], is found to be one of the most common contaminants in cereals, coffee, wine, dried fruits and nuts, meat products [5], herbal medicines [6–8], food coloring agents [9], and even in bottled water [10]. OTA induces diverse toxic effects in host, including carcinogenic [11], hepatotoxic [12], nephrotoxic [13], and immunotoxic [14, 15]. OTA is metabolized and accumulated mainly in the liver and kidney; thus, the liver and kidney are the key target organs for OTA to exert its toxic effects [16, 17]. Previous studies have found that OTA induces inflammation and even cancer in the liver [12, 18–20]. Notably, OTA induces inflammation through the toll-like receptor (TLR)-4/myeloid differentiation factor (MyD) 88 signaling pathway [21].
Indeed, the absorption rate of OTA varies from animals to human (e.g., 66% in pigs, 56% in rabbits, and 40% in chicken) [22]. Intestinal barrier is the first line of host defense against encroaching commensal bacteria, invading enteric pathogens and natural toxins [23]. Numerous studies have shown that OTA disrupts intestinal barrier function, thereby inducing extraintestinal organ (e.g., liver) inflammation [24, 25]. Intestinal microbiota highly shapes the intestinal barrier function and the physiological function of extraintestinal organs [26]. Interestingly, the recent investigations showed that intestinal dysbiosis is tightly associated with the onset of hepatic inflammation and injury [27, 28]. Notably, OTA treatment alters intestinal microbiota in rats by changing the relative abundance of Bacteroidaceae and Lactobacillaceae [29]. However, whether OTA-induced liver inflammation involving in intestinal microbiota remains largely unknown.
Therefore, this study was conducted to explore the underlying mechanism of intestinal microbiota and bacterial translocation in the liver inflammation induced by OTA in ducks. The ducklings were used in this study since infants and young animals are more sensitive to OTA than matures due to their incomplete development of organs [30, 31], especially for duckling which serves as the most sensitive species by oral gavage OTA [32–35].
Results
Oral OTA gavage alters cecum microbiota composition and promotes cecum LPS biosynthesis in ducks
To explore the effects of oral OTA gavage on 21-day ducks, OTA residue, feed intake, final weight, weight gain, and feed/gain ratio were monitored during the experiment. The OTA residue was found in different organs, including the kidney, liver, muscle, and intestinal tissues (Additional file 1: Figure S1A). OTA had little effects on the growth performance (Additional file 1: Figure S1B–E), and showed little effects on the relative weight of organs, except the liver (Additional file 1: Figure S1F–H).
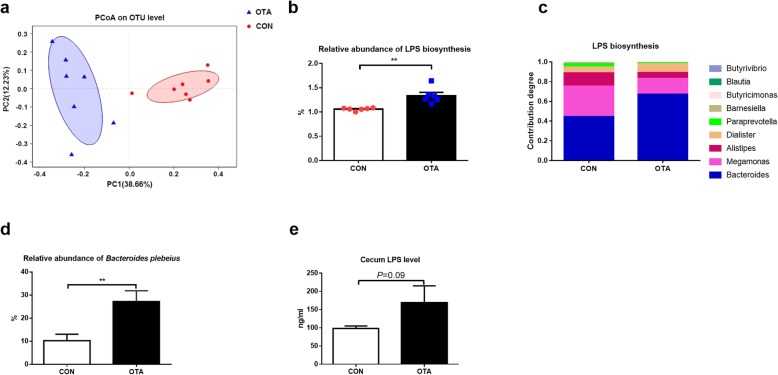
To explore the effect of OTA on intestinal microbiota, cecum microbiota of ducks was analyzed by sequencing the cecum bacterial 16S rRNA V3 + V4 region and metagenomics. PCoA analysis showed a clear separation between the cecum microbiota of ducks in CON and OTA group (Fig. 1a), demonstrating a strong effect of OTA on cecum microbiota. OTA also significantly reduced the richness (ACE index) and diversity (Shannon index) of cecum microbiota (Additional file 2: Figure S2A). Besides the difference in diversity, OTA increased the relative abundance of Bacteroidetes in the phylum (Wilcoxon rank-sum test, P = 0.01; Additional file 2: Figure S2B) and Bacteroides in the genus (Wilcoxon rank-sum test, P < 0.05; Additional file 2: Figure S2C). Metagenomic results showed that OTA increased gene and gene family with LPS biosynthesis (P < 0.01, Fig. 1b). With the analysis of contribution capacity of different strains to LPS biosynthesis by metagenomic sequencing, Bacteroides displayed the greatest contribution to LPS biosynthesis (Fig. 1c). Notably, the relative abundance of Bacteroides plebeius was higher in OTA group than those in CON group (Fig. 1d, Additional file 2: Figure S2D). To test the LPS biosynthesis ability, the LPS levels in the cecum were determined. The LPS level in OTA-treated ducks showed 1.5-fold higher than those ducks without OTA treatment (Fig. 1e). Collectively, OTA induces dysbiosis of the intestinal microbiota, especially increasing LPS-producing Bacteroides.
Fig. 1.
The composition and metagenomic function prediction of cecum microbiota after oral OTA treatment in ducks. a Principal component analysis of microbial communities in the cecum from CON and OTA. Plots are based on Bray-Curtis dissimilarities of 16S-based OTUs (n = 7). b Genes and gene families associated with LPS biosynthesis in different groups (n = 6, data shown as mean with SEM). c The contribution of different bacterial classes to LPS biosynthesis (n = 6). d Effect of OTA on the relative abundance of Bacteroides plebeius (n = 7). e The effect of OTA on cecum LPS level in ducks (n = 6, data shown as mean with SEM). For b, d, and e, data were analyzed with unpaired t test, **P < 0.01
OTA alters microbiota in the liver of ducks
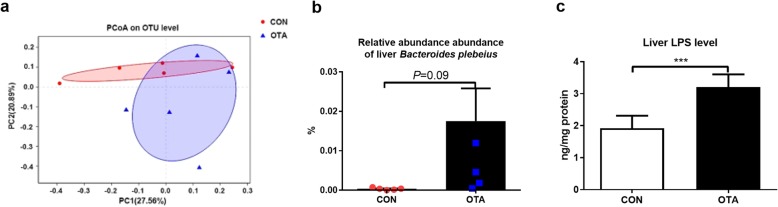
Besides its effect on cecum microbiota, OTA also lowered the mRNA expression and protein abundance of tight junction proteins (TJP1 and Occludin) (Additional file 3: Figure S3), suggesting that OTA enhances the cecum permeability, which is a widely accepted conclusion from previous studies [24, 25]. Based on these results, we hypothesized that LPS-producing Bacteroides may enter the liver through the leaky gut after OTA treatment. To explore this possibility, the microbiota in the liver was analyzed. PCoA analysis showed the difference about the liver microbiota in the CON and OTA groups (Fig. 2a). Although OTA had no effect on richness and diversity of liver microbiota in ducks (Additional file 4: Figure S4A), OTA significantly increased the relative abundance of Bacteroidetes in the phylum level (Student’s t test, P=0.02), the relative abundance of Bacteroides (Student’s t test, P < 0.01) at the genus level, and the relative abundance of Bacteroides plebeius at the species level (Fig. 2b and Additional file 4: Figure S4B–D). Interestingly, we also found that OTA treatment significantly increased the level of LPS in the liver (Fig. 2c). Collectively, OTA increases the relative abundance of LPS-producing Bacteroides and the level of LPS in the liver of ducks.
Fig. 2.
The composition of liver microbiota after oral OTA treatment in ducks. a Principal component analysis of microbial communities in the liver from CON and OTA (n = 5). b Effect of OTA on the relative abundance of Bacteroides plebeius. c Effect of OTA on liver LPS level (n = 5, mean with SEM). Data in b and c were analyzed with unpaired t test, ***P < 0.001
Oral OTA promotes liver inflammation
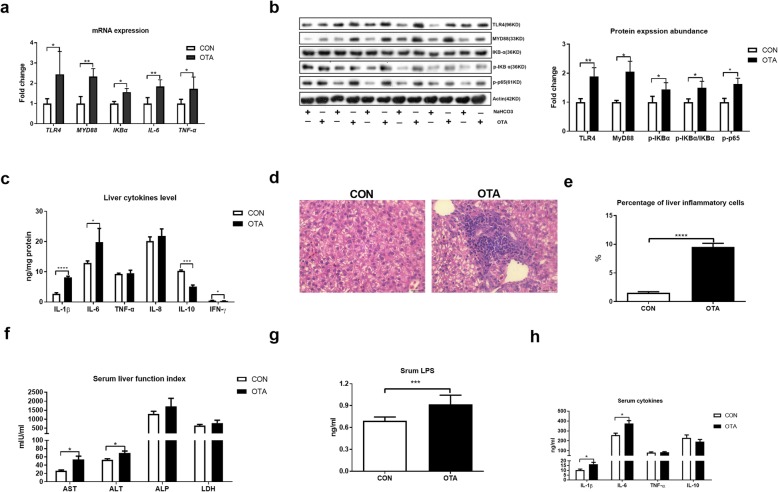
Based on the higher relative weight of the liver and level of LPS in the liver after OTA treatment, we then analyzed the inflammatory responses in the liver. The activation of the TLR-4 signaling pathway was activated after OTA treatment based on the higher mRNA expression and protein abundance of TLR4 Myd88 and p-p65 and higher ratio of p-IKBα/IKBα (Fig. 3a, b). OTA treatment also promoted the mRNA expression and secretion of inflammatory cytokines, including IL-1β and IL-6 (Fig. 3a, c). Notably, the level of anti-inflammation cytokine IL-10 was lowered after OTA treatment (Fig. 3c). Histological analysis also indicated that OTA induced inflammation in the liver based on extensive inflammatory cell infiltration in OTA-treated liver (Fig. 3d, e). The inflammation in the liver was also supported by the evidence that OTA treatment increased the activities of serum AST and ALT and the levels of serum LPS, IL-1β, and IL-6 (Fig. 3f, h). Together, OTA promotes liver inflammation in ducks.
Fig. 3.
OTA promotes liver inflammation. a Relative mRNA expressions of TLR4, MYD88, IKBα, IL-6, and TNF-α in the liver after OTA oral gavage (n = 6, mean with SEM). b Relative protein abundances of TLR4, MYD88, p-IKBα, p-IKBα/IKBα, and p-p65 in the liver after OTA oral gavage (n = 6, mean with SEM). c Effect of OTA on levels of liver cytokines, including IL-1β, IL-6, TNF-α, IL-8, IL-10, and IFN-γ (n = 6, mean with SEM). d Representative images of H&E-stained liver sections in CON and OTA group (magnification × 400, scale bar 100 μm, n = 6). e Statistical analysis of the percentage of inflammatory cells in different groups shown in d (n = 6, mean with SEM). f Effect of OTA on serum levels of AST, ALT, ALP, and LDH (n = 6, mean with SEM). g Serum LPS level with or without OTA treatment (n = 6, mean with SEM). h Effect of OTA on serum levels of IL-1β, IL-6, TNF-α, and IL-10 (n = 6, mean with SEM). Data in a, b, c, e, f, g, and h were analyzed with unpaired t test, *P < 0.05; **P < 0.01, *** P < 0.001, P<0.0001
OTA has little effect on cecum microbiome in antibiotics-treated ducks
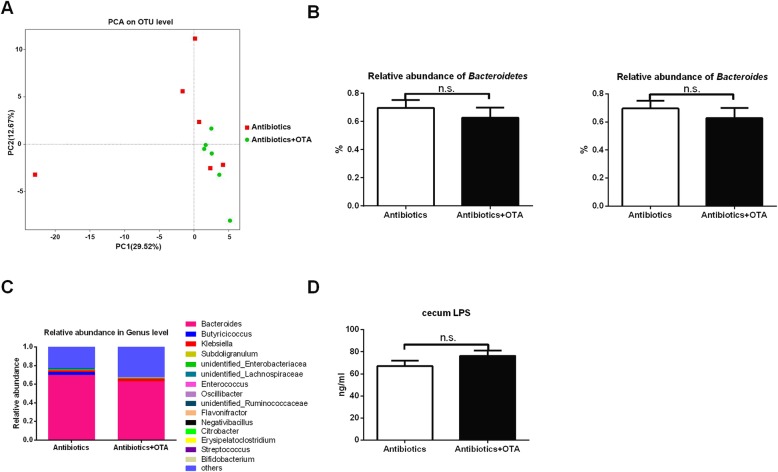
We have shown that OTA induces dysbiosis of intestinal microbiota and liver inflammation in ducks; thus, we hypothesized that OTA promotes the liver inflammation through intestinal microbiota. To clear the intestinal microbiota, ducks were treated with antibiotic mixtures, including streptomycin, ampicillin, and neomycin. Antibiotic treatment showed little effect on OTA residue in ducks, and OTA had little effects on growth performance, organ indexes, and relative weight and length of the intestine in antibiotics-treated ducks (Additional file 5: Figure S5A–H). Notably, OTA had little effects on the relative weight of the liver in antibiotics-treated ducks (Additional file 5: Figure S5F). PCoA analysis showed the similarity of cecum microbiota between two groups (Fig. 4a), and the diversity of cecum microbiota between two groups were similar (Additional file 6: Figure S6A), suggesting OTA has little effect on intestinal microbiota in antibiotics-treated ducks.
Fig. 4.
OTA has little effect on microbiome composition and LPS level in the cecum after antibiotics treatment. a PCoA of microbiota communities in the cecum between two groups (n = 6). b The relative abundance of Bacteroidetes (at the phylum level) and Bacteroides (at the genus level) (n = 6, mean with SEM). c Relative abundance of top 15 genus in each group (n = 6). d Effect of OTA on cecum LPS level in antibiotics-treated ducks (n = 6, mean with SEM). Data in b and d were analyzed with unpaired t test; n.s., not significant
Antibiotic treatment extremely decreased the absolute abundance of Bacteroidetes (Additional file 6: Figure S6B, P < 0.0001) and Bacteroides (Additional file 6: Figure S6B, P < 0.001) compared with ducks without antibiotic treatment, while OTA showed little effect on the relative and absolute abundance of Bacteroidetes and Bacteroides in antibiotics-treated ducks (Fig. 4b and Additional file 6: Figure S6B). Moreover, OTA had little effect on the composition of cecum bacteria at the phylum, genus, and species level (Fig. 4c and Additional file 6: Figure S6C–D). Likewise, OTA had little effect on the cecum LPS level (Fig. 4d). These results indicated that OTA has little effect on cecum microbiota in antibiotics-treated ducks.
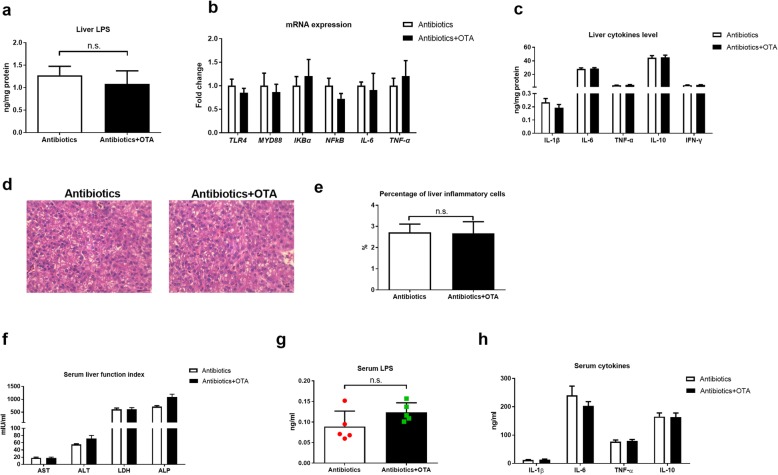
OTA failed to promote liver inflammation in antibiotics-treated ducks
Besides the results that OTA had little effects on cecum microbiome in antibiotics-treated ducklings, OTA had little effects on the mRNA expression and protein abundance of TJP1 and Occludin in antibiotics-treated ducks (Additional file 7: Figure S7). Notably, OTA had little effects on the levels of LPS in liver and the liver inflammation in antibiotics-treated ducks, including mRNA expression of TLR4 and Myd88, secretion of inflammatory cytokines, and inflammatory cell infiltration (Fig. 5a–e). OTA also had little effects on the activities of serum AST and ALT and levels of serum LPS and inflammatory cytokines (Fig. 5f–h). Together, OTA fails to promote liver inflammation in antibiotics-treated ducks.
Fig. 5.
OTA fails to promote liver inflammation in antibiotics-treated ducks. a Liver LPS level in different groups (n = 6, mean with SEM). b Relative mRNA expressions of TLR4, MYD88, IKBα, IL-6, and TNF-α in the liver of antibiotics-treated ducks with or without OTA (n = 6, mean with SEM). c Levels of IL-1β, IL-6, TNF-α, IL-8, IL-10, and IFN-γ in the liver of antibiotics-treated ducks with or without OTA (n = 6, mean with SEM). d Representative images of H&E-stained liver sections (magnification × 400; scale bar 100 μm; n = 6). e Statistical analysis of the percentage of inflammatory cells in different groups shown in d (n = 6, mean with SEM). f Effect of OTA on serum levels of AST, ALT, ALP and LDH in antibiotics-treated ducks (n = 6, mean with SEM). g Serum LPS level with or without OTA treatment in antibiotics-treated ducks (n = 6, mean with SEM). h Effect of OTA on serum levels of IL-1β, IL-6, TNF-α, and IL-10 (n = 6, mean with SEM). Data were analyzed with unpaired t test. n.s., not significant
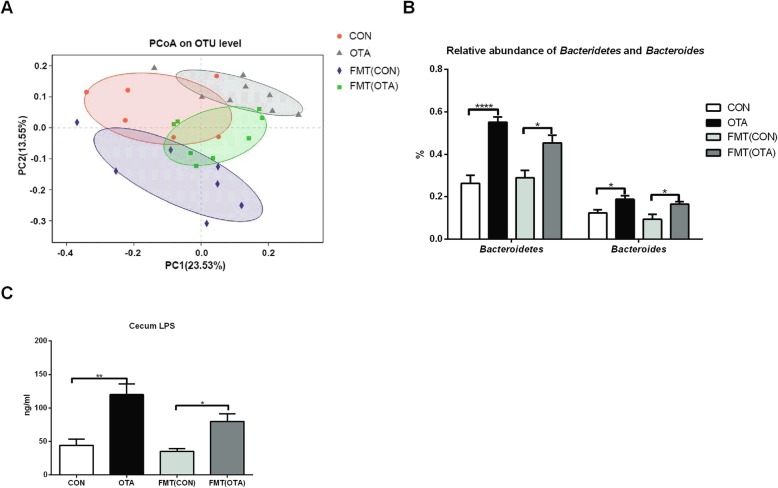
OTA-originated microbiota promotes the accumulation of LPS-producing Bacteroides and LPS in the cecum
To further verify the role of gut microbiota in OTA-induced liver inflammation, we transplanted fecal microbiota from oral OTA-treated ducks into antibiotics-treated ducks. As a control, the fecal microbiota from normal ducks was also transplanted into antibiotics-treated ducks. After OTA-originated microbiota transplantation, the load of OTA in the cecum was much lower than those with OTA treatment, and it was hardly detected in the liver (Additional file 1: Figure S1A, Additional file 5: Figure S5A and Additional file 8: Figure S8A). Although OTA-originated microbiota had little effect on the bodyweight of recipient ducks, it significantly enhanced the relative weight of the liver (Additional file 8: Figure S8B–C). Considering the OTA residue in OTA-originated fecal microbiota, we analyzed the OTA residue in OTA-originated microbiota. Recipient duck after OTA-originated microbiota transplantation received OTA at a dosage of 2 ng/kg body weight. Notably, OTA failed to induce liver inflammation in ducks even with a dosage of 60 μg/kg body weight (Additional file 8: Figure S8D), ruling out the possibility that the liver inflammation in recipient ducks is from OTA in OTA-originated fecal microbiota. Interestingly, OTA-originated microbiota significantly altered the microbiota in the cecum and enhanced the relative abundance of Bactroidetes and Bacteroides, as well as the level of LPS (Fig. 6a–c, Additional file 9: Figure S9). These results suggest that OTA-originated microbiota promotes the accumulation of LPS-producing Bacteroides and LPS in the cecum.
Fig. 6.
Gut microbiota composition and LPS level after intestinal microbiota transplantation. a Principal-coordinate analysis (PCoA) of cecum microbiota for transplant donors and recipients. Each circle represents a group. b Relative abundance of Bactroidetes and Bacteroides for transplant donors and recipients (n = 7, Mean with SEM). c Cecum LPS levels for different groups (n = 5, mean with SEM). Data in b and c were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test, *P < 0.05, **P < 0.01, ****P < 0.0001. FMT (CON): ducks received the CON group fecal microbiota. FMT (OTA): ducks received the OTA group fecal microbiota
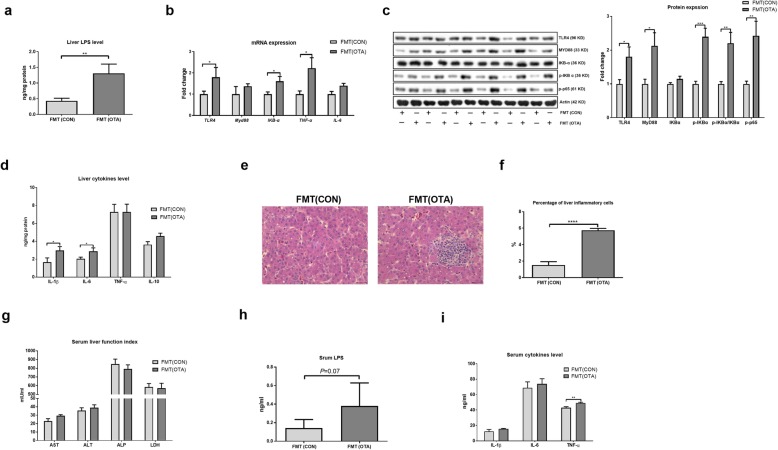
OTA-originated microbiota induces liver inflammation
To explore the role of gut microbiota in OTA-induced liver inflammation, the expression of tight junctions in the cecum and liver inflammation was analyzed. OTA-originated microbiota lowered the mRNA expression and protein abundance of TJP1 and Occludin in recipient ducks (Additional file 10: Figure S10). Interestingly, OTA-originated microbiota promoted the liver level of LPS and liver inflammation in recipient ducks, including mRNA expression of TLR4 and TNF-α, protein abundance of TLR4, Myd88 and p-p65, ratio of p-IKBα/IKBα, secretion of IL-1β and IL-6, and inflammatory cell infiltration (Fig. 7a–f). OTA-originated microbiota also enhanced the levels of LPS and TNF-α in the serum (Fig. 7g–i). Together, OTA-originated microbiota induces liver inflammation in recipient ducks.
Fig. 7.
OTA-originated microbiota induces liver inflammation. a Liver LPS level in FMT (CON) and FMT (OTA) ducks. b Relative mRNA expressions of TLR4, MYD88, IKBα, IL-6, and TNF-α in the liver after FMT (n = 6, mean with SEM). c Relative protein abundance of TLR4, MYD88, p-IKBα, p-IKBα/IKBα, and p-p65 in the liver after FMT (n = 6, mean with SEM). d Effect of FMT on the liver levels of IL-1β, IL-6, TNF-α, and IL-10 (n = 6, mean with SEM). e Representative H&E-stained liver sections. f Statistical analysis of the percentage of inflammatory cells in different groups shown in e (n = 6, mean with SEM). g Serum levels of AST, ALT, ALP, and LDH in different groups (n = 6, mean with SEM). h Serum LPS level in different FMT groups (n = 6, mean with SEM). i Serum levels of IL-1β, IL-6, and TNF-α after FMT (n = 6, mean with SEM). FMT (CON): ducks received the CON group fecal microbiota. FMT (OTA): ducks received the OTA group fecal microbiota. Data were analyzed with unpaired t test, n = 6. *P < 0.05; **P < 0.01, *** P < 0.001, ****P<0.0001
Discussion
OTA is a major food-contaminating mycotoxin and shows nephrotoxic, hepatotoxic, teratogenic, immunotoxic, and carcinogenic effects within phylogenetically distant organisms from animals to human [36]. Intestinal microbiota represents a crucial bridge between environmental substances and host health. Indeed, in addition to the important roles in maintaining gastrointestinal tract homeostasis, intestinal microbiota also affects multitudinous physiological functions of other visceral organs (e.g., brain, liver, and lung) and is involved in mediating the pathogenesis of different diseases in these organs [37–42]. Although previous investigations have demonstrated that OTA reduces the relative abundance of beneficial microbes (e.g., Lactobacillus and Bifidobacteria) [43], shapes the diversity of the intestinal microbiota, and increases the fecal total facultative anaerobes in rats [44], whether OTA-induced injuries in extraintestinal organs (e.g., liver and kidney) through intestinal microbiota remains to be revealed.
We found that OTA alters the intestinal microbiota composition in ducks, including the decrease in the richness and diversity of microbiota, and the relative abundance of Firmicutes, as well as the increase of the relative abundance of Bacteroidetes and Bacteroides. The relative abundance of Bacteroides accounts for 32.69% of the bacterial abundance in the normal ducks, but up to 52.99% after OTA treatment. Guo et al [44] also found that OTA decreases the within-subject diversity of the intestinal microbiota and induces changes in functional genes of gut microbiota, like signal transduction, carbohydrate transport, and amino acid transport system in OTA-treated rats. These results indicate that OTA changes intestinal microbiota composition in various animal species. Given intestinal microbiota metabolism has various physiological functions in the intestine and extraintestinal organs, thus, it is interesting to investigate whether OTA affects the intestinal metabolites (e.g., acetate, propionate, and butyrate) in the future.
Although the Bacteroides (Gram-negative bacteria) is often associated with the leanness and other desirable health traits [45, 46], some of its strains (e.g., Bacteroides fragilis, Bacteroides vulgatus, and Bacteroides dorei) have been linked to abdominal infections, metabolic disease, and inflammation [47, 48]. LPS or endotoxin is the main component of Gram-negative bacteria and induces inflammatory responses after across the mucosa [49–51]. After translocation in the portal circulation through enterohepatic recycling, LPS or the translocated microbiota, such as L. monocytogenes, contributes to inflammation through the engagement of various pattern-recognizing receptors (PRRs), including TLR4 or activation of NF-κB [52, 53]. Mechanistically, TLR4 (special receptor for LPS) leads to a cascade of the phosphorylation of mitogen-activated protein kinases (MAPKs), activation of NF-κB signaling, and expressions of various inflammatory cytokines, chemokines, and adhesion molecules [52].
A leaky gut permits the passage of microbial products, such as LPS, across the mucosa, which manifests as a moderate increase in the plasma LPS concentration [54, 55]. Indeed, when the gut barrier is compromised, microorganisms and microorganism-derived molecules can translocate to the liver, causing inflammation and hepatic injury [56]. Previous studies also have demonstrated that OTA drives intestinal barrier dysfunction and induces inflammation [57, 58]. In this study, we found that OTA treatment triggers intestinal barrier dysfunction and increases subsequent systemic LPS levels and inflammatory responses in the liver of ducks. Besides, studies have shown that anaerobic bacteria do not readily translocate, whereas aerobic Gram-negative bacteria translocate easily and even across an intact intestinal epithelium barrier [59, 60]. We detected Bacteroides, which were dominated in intestinal microbiota, still occupied the main status in the liver microbiome of the OTA group, indicating that they were probably from the intestinal microbiota by a leaky gut. Given the findings that OTA increases the relative abundance of gene and gene family associating with LPS biosynthesis, and that there is a positive correlation between LPS biosynthesis and Bacteroides; thus, OTA promotes liver inflammation may through intestinal LPS-producing Bacteroides. It is worth mentioning that OTA increases the relative weight of the liver [57, 61, 62]. Interestingly, in the current study, we also find that OTA treatment increases the liver weight of ducks. The possible explanations include (1) as the main target organ for OTA, OTA causes hepatocyte excessive proliferation, swelling, and hyperemia [63]; (2) OTA induces compensatory increment of connective tissue of liver [64]; and (3) OTA induces the infiltration of immune cells during the inflammation. It is interesting to investigate some opening questions, like the underlying mechanisms by which OTA promotes the relative percentage of LPS-producing Bacteroides.
Notably, OTA shows little effect on the accumulation of LPS in the cecum, serum, and liver and the activation of the TLR4 signaling pathway, as well as the liver inflammation in antibiotics-treated ducks. Additionally, in order to explore the contribution of the intestinal microbiome to OTA-induced liver inflammation, we carried out the fecal microbiota transplantation (FMT) experiment. Microbial transplantation provides an excellent way to demonstrate the role of gut microbiota in host defense mechanisms under a similar genetic background [65]. The previous study has also shown that FMT has a very high success rate in altering metabolic phenotypes, curing diseases, and affecting the host immune status [66–69]. In our study, antibiotics-treated ducks colonized with the fecal microbiota from OTA-treated ducks show similar phenotypes (cecum microbiota composition, cecum LPS, intestinal permeability, and liver inflammation) with OTA-treated ducks. Altogether, based on these results, we conclude that the gut microbiota, especially LPS-producing Bacteroides, contributes to the liver inflammation. Indeed, the liver is the target of xenobiotics and performs xenobiotic metabolism [70]; thus, it is meaningful to investigate whether OTA affects liver biosynthetic (e.g., total protein and prothrombin) and metabolic (e.g., lipid metabolism) capacity directly and/or indirectly through intestinal microbiota. Likewise, it is also interesting to study whether the pathogenesis of OTA-induced disease in other organs (e.g., brain and kidney injury) depends on intestinal microbiota.
Conclusions
Collectively, our study has identified a new mechanism linking the intestinal microbiota to OTA-induced liver inflammation. OTA alters cecum microbiota diversity and composition, leading to an increase in Gram-negative bacterial-derived LPS and entrance into the blood and liver through defective intestinal barrier, and ultimately facilitates the development of liver inflammation in ducks.
Methods
Animal and diet
All animal experiments were conducted according to the guidelines of Guangdong Province on the Review of Welfare and Ethics of Laboratory Animals, and approved by the Guangdong Province Administration Office of Laboratory Animals (GPAOLA). Male one-day-old Peking ducklings were purchased from Guiliu Poultry Co., Ltd. (Foshan, China). Animals were housed in stainless steel cages (five ducklings/cage) with free access to water and food, and kept under controlled room (temperature, maintained at 33 ± 1 °C for the first 3 days and then reduced by 2.5 ± 0.5 °C per week to a final temperature of 26 °C; relative humidity, 45%–60%; lighting, 24 h lighting with 10 Lux). Ducklings were fed a corn-soybean meal basal diet formulated to meet the nutritional requirements for starter ducks (National Research Council, 1994).
OTA oral gavage for ducklings
One-day-old male Peking ducklings were divided randomly into CON and OTA group with 15 replicates per group. On day 8, ducklings in the OTA group were challenged once a day for 2 weeks by i.g gavage OTA (Pribolab, Qingdao, China) in doses of 235 μg/kg body weight with 0.1 M sodium bicarbonate. This dosage was selected according to the previous study, which could increase the relative weight of the liver, result in a clear sign of enterotoxity, and increase levels of the pro-inflammatory cytokines in duck jejunal mucosa [57]. After 2 weeks of OTA oral gavage, ducks were executed by bloodletting of the jugular vein (at 09:00). Individual blood samples were collected from the jugular vein, and serum samples were separated by centrifugation of blood at 1200×g for 10 min at 4 °C and stored at − 30 °C for analysis. For conventional analysis, the entire liver, duodenum, jejunum, ileum, cecum, and rectum were collected and weighed. The middle part of liver samples (about 1–2 cm) was collected after the phosphate-buffered saline (PBS; pH = 7.2–7.4) washing. The liver was fixed in fresh 4% paraformaldehyde for paraffin embedding. For molecular biological analysis, parts of the liver and cecal mucosa were snap-frozen in liquid nitrogen for mRNA and protein extraction for qRT-PCR and Western blotting. Cecum digesta were collected and stored at − 80 °C until further analyses. The body weights of the animals were regularly monitored during the treatment period.
OTA oral gavage for antibiotics-treated ducklings
One-day-old Peking male ducklings were divided randomly into antibiotics and antibiotics + OTA groups with 15 replicates per group. All ducklings received a basic diet and drinking water containing streptomycin (1 g/L, Sigma), ampicillin (1 g/L, Sigma), and neomycin (1 g/L, Sigma) to clear intestinal bacteria, and drinking water was prepared daily [71]. The ducklings in the antibiotics + OTA group were treated similarly to the OTA group. After 2 weeks of OTA oral gavage, ducks were executed by bloodletting of the jugular vein (at 09:00) to collect the serum, liver, cecum mucosa, and cecum digesta for further analyses. The body weights of the animals were regularly monitored during the treatment period.
Fecal microbiota transplantation
One-day-old Peking male ducklings were divided randomly into CON, OTA, FMT (CON), and FMT(OTA) groups with 15 ducklings per group (n = 15). FMT(CON) and FMT(OTA) groups received a basal diet and drinking water containing streptomycin (1 g/L), ampicillin (1 g/L), and neomycin (0.5 g/L) for 2 weeks to remove indigenous gut microorganisms. After this treatment, the antibiotics-containing water was replaced with the regular water, and the microbiota-depleted ducklings were transplanted with donor microbiota. For fecal microbiota transplantation, 10 g fresh fecal samples were collected from the CON and OTA groups and resuspended in 50 ml sterile anaerobic saline, vortexed for 3 min and allowed to settle by gravity for 2 min. Transplant into recipient ducklings was achieved by gavage with 10 ml/kg body weight of the supernatant from the fecal sample once a day for 2 weeks. Then, ducks per group were executed by bloodletting of the jugular vein (at 09:00) to collect the serum, liver, cecum mucosa, and cecum digesta for further analysis. The body weights of the animals were regularly monitored during the treatment period.
Histopathology
After embedding in paraffin blocks, formalin-fixed liver tissues were sectioned to 6 μm thicknesses on a microtome (LeicaRM2235, Leica, Nussloch, Germany). The sections were placed on silicon-coated glass slides (Leica Biosystem, Richmond, IL, USA), dried, deparaffinized with xylene, and rehydrated in decreasing ethanol series. The sections were stained with 2.5% hematoxylin (Merck, Darmstadt, Germany) followed by counterstaining with 0.5% eosin (Sigma Aldrich, USA). Stained tissue sections were examined under an Olympus BX61 light microscope (Olympus, Tokyo, Japan) with × 200 magnification. Images were captured (scale bar=100 µm) using an Olympus DP70 digital camera (Olympus, Japan).
Biochemical analysis
The organs were pretreated with 70% methanol, and the residual OTA was measured by an ELISA Kit (RNM98008, REAGEN, USA). The activity of serum aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), and lactate dehydrogenase (LDH) and the levels of cytokines, including IL-1β, TNF-α, IL-6, and IL-10 in the liver and serum, were measured with ELISA Kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer’s instructions. LPS levels were measured in the cecum, liver, and serum using ELISA Kit (Cloud-Clone Crop., Houston, USA).
Western blotting for protein expression
Whole protein, including Occludin and TJP-1, which are responsible for maintaining the intestinal barrier function [72], and TLR4 and NF-κB signaling pathway, which are highly crucial for modulating inflammatory responses [52], from the liver and cecum mucous were lysed by RIPA lysis buffer (LifeTechnologies Inc., USA) supplemented with protease inhibitor cocktail (Roche, USA). The protein concentration in the tissue lysate was measured with BCA. Proteins were loaded onto the SDS-PAGE gel (BioRad) and electrophoresed and analyzed by WB using antibodies against TLR4 (BA1717, Boster, Wuhan, China), MYD88 (abs135682, Absin, Shanghai, China), IKBα (D120138, Sangon Biotech, Shanghai, China), p-IKBα (D151548, Sangon Biotech, Shanghai, China), p-p65 (HZ4902812, TW reagent, Shanghai, China), TJP-1 (mAb13663, Cell Signaling, USA), Occludin (ab167161, Abcam, USA), and actin (60008, Proteintech, USA). The bands were detected using the chemiluminescence kits (Amersham Biosciences, UK). Chemiluminescence was recorded with an Image Station 440CF, and results were analyzed with the 1DImage Software (Kodak Digital Science, Rochester, NY, USA).
16S rRNA amplicon sequencing, data processing, and analysis
DNA was extracted from the feces and liver using the E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to the protocol for isolation of DNA. Illumina MiSeq sequencing and general data analyses were performed by a commercial company (Majorbio Bio-Pharm Technology, Shanghai, China). Because of initially low bacterial DNA concentrations in some samples, a nested PCR was applied to increase specificity and amplicon yield [73, 74]. The V3–V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′-ACT CCT ACG GGA GGC AGC AG-3′) and 806R (5′-GGA CTA CHV GGG TWT CTA AT-3′) by thermocycler PCR system (GeneAmp 9700, ABI, USA). The PCR reactions were conducted using the following program: 3 min of denaturation at 95 °C, 27 cycles of 30 s at 95 °C, 30s for annealing at 55 °C, 45 s for elongation at 72 °C, and a final extension at 72 °C for 10 min. PCR reactions were performed in triplicate 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. The resulted PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, USA) according to the manufacturer’s protocol. Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Raw fastq files were demultiplexed, quality-filtered by Trimmomatic, and merged by FLASH with the following criteria: (a) The reads were truncated at any site receiving an average quality score < 20 over a 50-bp sliding window. (ii) Primers were exactly matched allowing two nucleotide mismatching, and reads containing ambiguous bases were removed. (iii) Sequences whose overlap longer than 10 bp were merged according to their overlap sequence. Operational taxonomic units (OTUs) were clustered with 97% similarity [75] cutoff using UPARSE (version 7.1 http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16S rRNA database using a confidence threshold of 70%.
Shotgun metagenomics of cecum microbiota
A total of 12 samples (CON1–6, OTA1–6) were selected for shotgun metagenomics sequencing. Using Nextra XT protocols (Illumina), individual libraries were sequenced on the Miseq platform. 3′ and 5′ ends were stripped using SeqPrep (https://github.com/jstjohn/SeqPrep). Low-quality reads (length < 50 bp or with a quality value < 20 or having N bases) were removed by Sickle (https://github.com/najoshi/sickle). Reads were aligned to the Anas platyrhynchos (mallard) (GenBank assembly accession: GCA_000355885.1 and RefSeq assembly accession: GCF_000355885.1) by BWA (http://bio-bwa.sourceforge.net) and any hit associated with the reads and their mated reads were removed. De bruijn-graph-based assembler SOAPdenovo (http://soap.genomics.org.cn, version 1.06) was employed to assemble short reads. K-mers, varying from 1/3~2/3 of reads length, were tested for each sample. Scaffolds with a length over 500 bp were retained for statistical tests; we evaluated the quality and quantity of scaffolds generated by each assembly and finally chose the best K-mer which yielded the minimum scaffold number and the maximum value of N50 and N90. Then, scaffolds with a length of over 500 bp were extracted and broken into contigs without gaps. Contigs were used for further gene prediction and annotation.
Gene prediction, taxonomy, and functional annotation
Open reading frames (ORFs) from each metagenomic sample were predicted using MetaGene (http://metagene.cb.k.u-tokyo.ac.jp/). The predicted ORFs with length being or over 100 bp were retrieved and translated to amino acid sequences using the NCBI translation table (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi?chapter=tgencodes#SG1). All sequences from gene sets with a 95% sequence identity (90% coverage) were clustered as the non-redundant gene catalog by the CD-HIT (http://www.bioinformatics.org/cd-hit/). Reads after quality control were mapped to the representative genes with 95% identity using SOAP aligner (http://soap.genomics.org.cn/), and gene abundance in each sample was evaluated. BLASTP (version 2.2.28+, http://blast.ncbi.nlm.nih.gov/Blast.cgi) was employed for taxonomic annotations by aligning non-redundant gene catalogs against NCBI NR database with e-value cutoff of 1e−5. The cluster of orthologous groups of proteins (COG) for the ORF annotation was performed using BLASTP against eggNOG database (v4.5) with an e-value cutoff of 1e−5. The KEGG pathway annotation was conducted using BLASTP search (version 2.2.28+) against the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/keeg/) with an e-value cutoff of 1e−5.
Transcriptional analysis
Total RNA was isolated from liquid nitrogen-frozen liver and cecum mucus using the Quick-RNA™ MiniPrep Plus (Zymo, USA) according to the manufacturer’s instructions. The synthesis of the first strand (cDNA) was performed using oligo (dT) 20 and Superscript II reverse transcriptase (Takara, Japan). The transcriptional analysis of the related genes were performed using the following primers: TJP1: 5′- TCA GCG AGA TGA ACG AGC C-3′, 5′- TCT GAA GGC TCT GAC CTC TGG-3′, OCLN: 5′- GCT GGG CTA CAA CTA CGG GT-3′, 5′- TAC GCC AAC ACG GTG CTG-3′, TLR4: 5′- TTA ACT GCC AAT TTG CTC C-3′, 5′- CCG GTT TCC ACC AAT ACT A-3′, MYD88: 5′- 5'-GAAGAGGAAGCAGCAGCAA-3', 5'-TGAACCGCAGGATACTTGG-3', IKB-α: 5′- CGT GTC TCC ATT TGG CAT CT-3′, 5′- GCC CTG GTA GGT CAC TTT GT-3′, TNF-α: 5′- ACA GCC TAT GCC AAC AAG-3′, 5′- TAC AGG AAG GGC AAC TCA-3′, IL-6: 5′- AAA GCA TCT GGC AAC GAC-3′, 5′- AAT AGC GAA CAG CCC TCA-3′. β-Actin (5′- TAC GCC AAC ACG GTG CTG-3′, 5′- GAT TCA TCA TAC TCC TGC TTG-3′) was used as an internal control to normalize target gene transcriptional levels.
Statistical analysis
The data are expressed as mean ± SEM and analyzed using GraphPad Prism 6.0 (GraphPad Software). Significant differences between the two groups were evaluated by two-tailed unpaired Student’s t test or Mann-Whitney U test for samples that were not normally distributed. Significant differences among three or more groups were evaluated by one-way ANOVA with Bonferroni’s multiple comparisons test. The level of significance was set at P < 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Supplementary information
Additional file 1: Figure S1. OTA residue and effect of OTA on growth performance and organ index in ducks. a: OTA residue in different organs (n = 6, Mean with SEM). The different letters above the column show significant differences. b-e: Effect of OTA on growth performance of ducks from day 1 to day 21. f: Effect of OTA on relative organ weight of 21 d ducks (n=6, Mean with SEM). g: Effect of OTA on relative length of intestine on day 21 ducks (n=6, Mean with SEM). h: Effect of OTA on relative weight of intestine on day 21 ducks. Data in a were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test, while data in b-h were analyzed by unpaired t test, *P <0 .05.
Additional file 2: Figure S2. OTA alters cecum microbial diversity and composition. a: Alpha diversity of cecum microbiota in different groups (n=7). Data were analyzed by student’s t test, *** P<0.001. b: Relative abundance of bacteria at Phylum level. OTUs with an occurrence lower than 1% are not represented (n=7). c: Relative abundance of top 15 Genus in each group (n=7). d: Relative abundance of top 15 Species in each group (n = 7).
Additional file 3: Figure S3. OTA lowers intestinal abundance of tight junctions. a: Effect of OTA on relative mRNA expressions of TJP1 and Occludin in the cecum of 21d ducks (n=6, Mean with SEM). Data were analyzed by unpaired t test. b: Effect of OTA on the protein abundance of TJP1 and Occludin in the cecum (n=6, Mean with SEM). Data were analyzed by unpaired t test. (n=6). *P<0 .05, **P<0 .01, *** P<0.001.
Additional file 4: Figure S4. OTA alters liver microbial composition. a: Alpha diversity of liver microbiota in different groups (n=5). Data were analyzed by student’s t test, n.s., not significant. b: Relative abundance of bacteria at Phylum level. OTUs with an occurrence lower than 1% are not represented (n=5). c: Relative abundance of top 15 Genus in each group (n=5). d: Relative abundance of top 15 Species in each group (n=5).
Additional file 5: Figure S5. OTA residue, effect of OTA on growth performance and organ index in antibiotics-treated ducks. a: OTA residue in different organs (n=6, Mean with SEM). Data were analyzed by the one-way ANOVA followed by Dunnett multiple comparisons (n=6, Mean with SEM). The different letters above the column show significant differences. b-e: Effect of OTA on growth performance of antibiotics-treated ducks from day 1 to day 21. f: Effect of OTA on relative organ weight of antibiotics-treated ducks (n=6, Mean with SEM). g: Effect of OTA on relative length of intestine on antibiotics-treated ducks (n=6, Mean with SEM). h: Effect of OTA on relative weight of intestine on antibiotics-treated ducks. Data in b-h were analyzed by unpaired t test.
Additional file 6: Figure S6. OTA has little effect on cecum microbiota diversity and composition in antibiotics-treated ducks. a: Alpha diversity of liver microbiota in different groups (n=6, Mean with SEM). b: The OTU numbers of Bacteroidetes (left) and Bacteroides (right) in different groups (CON, n=10; Antibiotics, n=6; Anti+OTA, n=6, mean with SEM). c: Relative abundance of top 7 bacteria at Phylum level in antibiotics-treated ducks. d: Relative abundance of top 15 bacteria at species level in antibiotics treated ducks. Data in a were analyzed by unpaired t test; while data in b were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test. n.s., not significant; ****P<0.0001.
Additional file 7: Figure S7. OTA shows little effect on intestinal abundance of tight junctions in antibiotics-treated ducks. a: Effect of OTA on relative mRNA expressions of TJP1 and Occludin in the cecum of antibiotics-treated ducks (n=6, Mean with SEM). Data were analyzed by unpaired t test. b: Effect of OTA on the protein abundance of TJP1 and Occludin in the cecum (n=6, Mean with SEM). Data were analyzed by unpaired t test. (n=6). *P<0 .05.
Additional file 8: Figure S8. OTA residue, growth performance and organ index after intestinal microbiota transplantation. a: OTA residue in the cecum digesta and liver of FMT (OTA) ducks (n=6, Mean with SEM). b-c: Final weight and relative weight of liver in FMT (CON) and FMT (OTA) ducks (n=9, mean with SEM). d: Levels of pro-inflammatory cytokines in duck liver after administration of OTA at dosage of 0, 15, 30 or 60 μg/kg (n=6, mean with SEM). Data in a, b and c were analyzed by unpaired t test; while data in d were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test. *P<0 .05. FMT (CON), ducks received CON group fecal microbiota; FMT (OTA), ducks received OTA group fecal microbiota.
Additional file 9: Figure S9. The cecum microbial diversity and composition of transplant donors and recipients. a: Alpha diversity of intestinal microbiome in transplant donors and recipients. b: Relative abundance of bacteria at Phylum level. OTUs with an occurrence lower than 1% are not represented (n=7). c: Relative abundance of top 15 Genus in each group (n=7). d: Relative abundance of top 15 Species in each group (n=7). Data in a and b were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test *P<0.05. FMT (CON): ducks received CON group fecal microbiota. FMT (OTA): ducks received OTA group fecal microbiota.
Additional file 10: Figure S10. OTA-originated microbiota lowers intestinal abundance of tight junctions. a: Relative mRNA expression of TJP1 and Occludin in different groups (n=6). b: Relative protein abundance of TJP1 and Occludin (n=6). Data were analyzed by unpaired t test, n=6, mean with SEM. *P<0 .05, **** P<0.0001. FMT (CON): ducks received CON group fecal microbiota. FMT (OTA): ducks received OTA group fecal microbiota.
Acknowledgments
The authors would like to acknowledge Prof. Yulong Yin (Institute of Subtropical Agriculture, the Chinese Academy of Sciences) for reading the manuscript and providing helpful comments.
Abbreviations
- ALP
Alkaline phosphatase
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- FMT
Fecal microbiota transplantation
- LDH
Lactate dehydrogenase
- LPS
Lipopolysaccharides
- MAPKs
Mitogen-activated protein kinases
- OTA
Ochratoxin A
- PRRs
Pattern-recognizing receptors
- TLR4
Toll-like receptor 4
Authors’ contributions
WW, WR, and LY designed the study. WW, SZ, and YX acquired the data and performed experiments. HW, DR, TZ, and YZ helped conduct the animal experiments. HZ, MZ, and HY advised on the data analysis. WW, SZ, YX, and WR wrote the manuscript. All authors read and approve the manuscript.
Funding
This study was sponsored by the National Key Research Program (2016YFD0500509-07), National Natural Science Foundation of China grants (31922079, 31872365, and 31790411), the China Agriculture Research System (CARS-42-15), the Special Fund for Agro-scientific Research in the Public Interest, China (201303143), National Youth Fund Project of China (31501959), and Innovation Team Project in Universities of Guangdong Province (2017KCXTD002).
Availability of data and materials
All raw data of 16S rRNA amplicon sequencing and Shotgun metagenomics are available at Sequence Read Archive (Accession number SUB5539876).
Ethics approval and consent to participate
All animal experiments were conducted according to the guidelines of Guangdong Province on the Review of Welfare and Ethics of Laboratory Animals, and approved by the Guangdong Province Administration Office of Laboratory Animals (GPAOLA). Written informed consent was received from all participants.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wence Wang, Shuangshuang Zhai and Yaoyao Xia contributed equally to this work.
Contributor Information
Wenkai Ren, Email: renwenkai19@126.com.
Lin Yang, Email: ylin@scau.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s40168-019-0761-z.
References
- 1.Robert H, Payros D, Pinton P, Theodorou V, Mercier-Bonin M, Oswald IP. Impact of mycotoxins on the intestine: are mucus and microbiota new targets? J Toxicol Environ Health B Crit Rev. 2017;20:249–275. doi: 10.1080/10937404.2017.1326071. [DOI] [PubMed] [Google Scholar]
- 2.Wild CP, Gong YY. Mycotoxins and human disease: a largely ignored global health issue. Carcinogenesis. 2010;31:71–82. doi: 10.1093/carcin/bgp264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manizan AL, Oplatowska-Stachowiak M, Piro-Metayer I, Campbell K, Koffi-Nevry R, Elliott C, Akaki D, Montet D, Brabet C. Multi-mycotoxin determination in rice, maize and peanut products most consumed in Cote d’Ivoire by UHPLC-MS/MS. Food Control. 2018;87:22–30. doi: 10.1016/j.foodcont.2017.11.032. [DOI] [Google Scholar]
- 4.Liuzzi VC, Fanelli F, Tristezza M, Haidukowski M, Picardi E, Manzari C, Lionetti C, Grieco F, Logrieco AF, Thon MR. Transcriptional analysis of Acinetobacter sp. neg1 capable of degrading ochratoxin A. Front Microbiol. 2017;7:2162. doi: 10.3389/fmicb.2016.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fink-Gremmels J. Conclusions from the workshops on ochratoxin A in food: recent developments and significance, organized by ILSI Europe in Baden (Austria), 29 June–1 July 2005. Food Addit Contam. 2005;22:1–5. doi: 10.1080/02652030500358415. [DOI] [PubMed] [Google Scholar]
- 6.Chen AJ, Jiao X, Hu Y, Lu X, Gao W. Mycobiota and mycotoxins in traditional medicinal seeds from China. Toxins. 2015;7:3858–3875. doi: 10.3390/toxins7103858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shim WB, Ha KS, Kim MG, Kim JS, Chung DH. Evaluation of the transfer rate of ochratoxin a to decoctions of herbal medicines. Food Sci Biotechnol. 2014;23:2103–2108. doi: 10.1007/s10068-014-0286-y. [DOI] [Google Scholar]
- 8.Veprikova Z, Zachariasova M, Dzuman Z, Zachariasova A, Fenclova M, Slavikova P, Vaclavikova M, Mastovska K, Hengst D, Hajslova J. Mycotoxins in plant-based dietary supplements: hidden health risk for consumers. J Agric Food Chem. 2015;63:6633. doi: 10.1021/acs.jafc.5b02105. [DOI] [PubMed] [Google Scholar]
- 9.Michele S, Luca P, Lucia G, Rosanna Z, Francesco L. Food coloring agents and plant food supplements derived from Vitis vinifera: a new source of human exposure to ochratoxin A. J Agric Food Chem. 2015;63:3609–3614. doi: 10.1021/acs.jafc.5b00326. [DOI] [PubMed] [Google Scholar]
- 10.Mata AT, Ferreira JP, Oliveira BR, Batoréu MC, Crespo MT, Barreto PVJ, Bronze MR. Bottled water: analysis of mycotoxins by LC-MS/MS. Food Chem. 2015;176:455–464. doi: 10.1016/j.foodchem.2014.12.088. [DOI] [PubMed] [Google Scholar]
- 11.Polovic M, Dittmar S, Hennemeier I, Humpf H-U, Seliger B, Fornara P, Theil G, Azinovic P, Nolze A, Koehn M, et al. Identification of a novel lncRNA induced by the nephrotoxin ochratoxin A and expressed in human renal tumor tissue. Cell Mol Life Sci. 2018;75:2241–2256. doi: 10.1007/s00018-017-2731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madalena M, Sobral C, Faria MA, Cunha SC, Ferreira IMPLVO. Toxicological interactions between mycotoxins from ubiquitous fungi: impact on hepatic and intestinal human epithelial cells. Chemosphere. 2018;202:538–548. doi: 10.1016/j.chemosphere.2018.03.122. [DOI] [PubMed] [Google Scholar]
- 13.Vettorazzi A, Pastor L, Guruceaga E, Lopez de Cerain A. Sex-dependent gene expression after ochratoxin A insult in F344 rat kidney. Food Chem Toxicol. 2019;123:337–348. doi: 10.1016/j.fct.2018.10.057. [DOI] [PubMed] [Google Scholar]
- 14.Ladeira C, Frazzoli C, Orisakwe OE. Engaging one health for non-communicable diseases in Africa: perspective for Mycotoxins. Front Public Health. 2017;5:266. doi: 10.3389/fpubh.2017.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou L, Gan F, Zhou X, Zhou Y, Qian G, Liu Z, Huang K. Immunotoxicity of ochratoxin A and aflatoxin B1 in combination is associated with the nuclear factor kappa B signaling pathway in 3D4/21 cells. Chemosphere. 2018;199:718–727. doi: 10.1016/j.chemosphere.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Vettorazzi A, de Trocóniz IF, González-Peñas E, Arbillaga L, Corcuera L-A, Gil AG, de Cerain AL. Kidney and liver distribution of ochratoxin A in male and female F344 rats. Food Chem Toxicol. 2011;49:1935–1942. doi: 10.1016/j.fct.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Hort V, Nicolas M, Minvielle B, Maleix C, Desbourdes C, Hommet F, Dragacci S, Dervilly-Pinel G, Engel E, Guérin T. Ochratoxin A determination in swine muscle and liver from French conventional or organic farming production systems. J Chromatogr B. 2018;1092:131–137. doi: 10.1016/j.jchromb.2018.05.040. [DOI] [PubMed] [Google Scholar]
- 18.Al-Anati L, Essid E, Reinehr R, Petzinger E. Silibinin protects OTA-mediated TNF-alpha release from perfused rat livers and isolated rat Kupffer cells. Mol Nutr Food Res. 2009;53:460–466. doi: 10.1002/mnfr.200800110. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Arias CA, Crespo-Sempere A, Marin S, Sanchis V, Ramos AJ. Modulation of the xenobiotic transformation system and inflammatory response by ochratoxin A exposure using a co-culture system of Caco-2 and HepG2 cells. Food Chem Toxicol. 2015;86:245–252. doi: 10.1016/j.fct.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Pfohl-Leszkowicz A, Manderville RA. An update on direct genotoxicity as a molecular mechanism of ochratoxin A carcinogenicity. Chem Res Toxicol. 2012;25:252–262. doi: 10.1021/tx200430f. [DOI] [PubMed] [Google Scholar]
- 21.Xu H, Hao S, Gan F, Wang H, Xu J, Liu D, Huang K. In vitro immune toxicity of ochratoxin A in porcine alveolar macrophages: a role for the ROS-relative TLR4/MyD88 signaling pathway. Chem Biol Interact. 2017;272:107–116. doi: 10.1016/j.cbi.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 22.Galtier P, Alvinerie M, Charpenteau JL. The pharmacokinetic profiles of ochratoxin A in pigs, rabbits and chickens. Food Cosmet Toxicol. 1981;19:735–738. doi: 10.1016/0015-6264(81)90528-9. [DOI] [PubMed] [Google Scholar]
- 23.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14:9. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Li S, Wang J, Luo C, Zhao S, Zheng N. Modulation of intestinal epithelial permeability in differentiated Caco-2 cells exposed to aflatoxin M1 and ochratoxin A individually or collectively. Toxins. 2017;10:13. doi: 10.3390/toxins10010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Chen Y, Zhai N, Chen X, Gan F, Li H, Huang K. Ochratoxin A-induced apoptosis of IPEC-J2 cells through ROS-mediated mitochondrial permeability transition pore opening pathway. J Agric Food Chem. 2017;65:10630–10637. doi: 10.1021/acs.jafc.7b04434. [DOI] [PubMed] [Google Scholar]
- 26.Martens EC, Neumann M, Desai MS. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol. 2018;16:457–470. doi: 10.1038/s41579-018-0036-x. [DOI] [PubMed] [Google Scholar]
- 27.Liao L, Schneider KM, Galvez EJC, Frissen M, Marschall H-U, Su H, Hatting M, Wahlstrom A, Haybaeck J, Puchas P, et al. Intestinal dysbiosis augments liver disease progression via NLRP3 in a murine model of primary sclerosing cholangitis. Gut. 2019;68(8):1477–1492. doi: 10.1136/gutjnl-2018-316670. [DOI] [PubMed] [Google Scholar]
- 28.Ponziani FR, Zocco MA, Cerrito L, Gasbarrini A, Pompili M. Bacterial translocation in patients with liver cirrhosis: physiology, clinical consequences, and practical implications. Expert Rev Gastroenterol Hepatol. 2018;12:641–656. doi: 10.1080/17474124.2018.1481747. [DOI] [PubMed] [Google Scholar]
- 29.Guo Mingzhang, Huang Kunlun, Chen Siyuan, Qi Xiaozhe, He Xiaoyun, Cheng Wen-Hsing, Luo Yunbo, Xia Kai, Xu Wentao. Combination of Metagenomics and Culture-Based Methods to Study the Interaction Between Ochratoxin A and Gut Microbiota. Toxicological Sciences. 2014;141(1):314–323. doi: 10.1093/toxsci/kfu128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adetunji Modupeade C., Atanda Olusegun O., Ezekiel Chibundu N. Risk Assessment of Mycotoxins in Stored Maize Grains Consumed by Infants and Young Children in Nigeria. Children. 2017;4(7):58. doi: 10.3390/children4070058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherif SO, Salama EE, Abdel-Wahhab MA. Mycotoxins and child health: the need for health risk assessment. Int J Hyg Environ Health. 2009;212:347–368. doi: 10.1016/j.ijheh.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Van der Merwe K, Steyn P, Fourie L, Scott DB, Theron J. Ochratoxin A, a toxic metabolite produced by Aspergillus ochraceus Wilh. Nature. 1965;205:1112. doi: 10.1038/2051112a0. [DOI] [PubMed] [Google Scholar]
- 33.Purchase I, Theron J. The acute toxicity of ochratoxin A to rats. Food Cosmet Toxicol. 1968;6:479–483. doi: 10.1016/0015-6264(68)90138-7. [DOI] [PubMed] [Google Scholar]
- 34.Peckham JC, Doupnik B, Jones OH. Acute toxicity of ochratoxins A and B in chicks. Appl Environ Microbiol. 1971;21:492–494. doi: 10.1128/am.21.3.492-494.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prior M, Sisodia C, O’neil J. Acute oral ochratoxicosis in day-old white leghorns, turkeys and japanese quail. Poult Sci. 1976;55:786–790. doi: 10.3382/ps.0550786. [DOI] [PubMed] [Google Scholar]
- 36.Pfohl-Leszkowicz A, Manderville RA. Ochratoxin A: an overview on toxicity and carcinogenicity in animals and humans. Mol Nutr Food Res. 2007;51:61–99. doi: 10.1002/mnfr.200600137. [DOI] [PubMed] [Google Scholar]
- 37.Safari Z, Gerard P. The links between the gut microbiome and non-alcoholic fatty liver disease (NAFLD) Cell Mol Life Sci. 2019;76:1541–1558. doi: 10.1007/s00018-019-03011-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology. 2014;146:1513–1524. doi: 10.1053/j.gastro.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porras D, Nistal E, Martínez-Flórez S, Pisonero-Vaquero S, Olcoz JL, Jover R, González-Gallego J, García-Mediavilla MV, Sánchez-Campos S. Protective effect of quercetin on high-fat diet-induced non-alcoholic fatty liver disease in mice is mediated by modulating intestinal microbiota imbalance and related gut-liver axis activation. Free Radic Biol Med. 2017;102:188–202. doi: 10.1016/j.freeradbiomed.2016.11.037. [DOI] [PubMed] [Google Scholar]
- 40.Del Chierico F, Nobili V, Vernocchi P, Russo A, Stefanis CD, Gnani D, Furlanello C, Zandonà A, Paci P, Capuani G. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65:451–464. doi: 10.1002/hep.28572. [DOI] [PubMed] [Google Scholar]
- 41.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20:145. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dickson RP, Singer BH, Newstead MW, Falkowski NR, Erb-Downward JR, Standiford TJ, Huffnagle GB. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1:16113. doi: 10.1038/nmicrobiol.2016.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ouethrani M, Van de Wiele T, Verbeke E, Bruneau A, Carvalho M, Rabot S, Camel V. Metabolic fate of ochratoxin A as a coffee contaminant in a dynamic simulator of the human colon. Food Chem. 2013;141:3291–3300. doi: 10.1016/j.foodchem.2013.05.157. [DOI] [PubMed] [Google Scholar]
- 44.Guo M, Huang K, Chen S, Qi X, He X, Cheng W-H, Luo Y, Xia K, Xu W. Combination of metagenomics and culture-based methods to study the interaction between ochratoxin a and gut microbiota. Toxicol Sci. 2014;141:314–323. doi: 10.1093/toxsci/kfu128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 46.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1079–U1049. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Palma G, Nadal I, Medina M, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010;10:63. doi: 10.1186/1471-2180-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis-Richardson AG, Ardissone AN, Dias R, Simell V, Leonard MT, Kemppainen KM, Drew JC, Schatz D, Atkinson MA, Kolaczkowski B, et al. Bacteroides dorei dominates gut microbiome prior to autoimnnunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Liu Y, Li S, Jin Y, Zhao L, Zhao F, Feng J, Yan W, Wei Y. Altered gut microbiota after traumatic splenectomy is associated with endotoxemia. Emerg Microbes Infect. 2018;7(1):197. doi: 10.1038/s41426-018-0202-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Opal SM. The clinical relevance of endotoxin in human sepsis: a critical analysis. J Endotoxin Res. 2002;8:473–476. doi: 10.1177/09680519020080060401. [DOI] [PubMed] [Google Scholar]
- 51.Shamsollahi HR, Ghoochani M, Jaafari J, Moosavi A, Sillanpaa M, Alimohammadi M. Environmental exposure to endotoxin and its health outcomes: a systematic review. Ecotoxicol Environ Saf. 2019;174:236–244. doi: 10.1016/j.ecoenv.2019.02.046. [DOI] [PubMed] [Google Scholar]
- 52.Lu Y-C, Yeh W-C, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 53.Drolia R, Tenguria S, Durkes AC, Turner JR, Bhunia AK. Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe. 2018;23:S1931312818301331. doi: 10.1016/j.chom.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho Y-E, Kim D-K, Seo W, Gao B, Yoo S-H, Song B-J. Fructose promotes leaky gut, endotoxemia and liver fibrosis through CYP2E1-mediated oxidative and nitrative stress. Hepatology (Baltimore, Md). 2019. 10.1002/hep.30652. [DOI] [PMC free article] [PubMed]
- 55.Cho Y-E, Yu L-R, Abdelmegeed MA, Yoo S-H, Song B-J. Apoptosis of enterocytes and nitration of junctional complex proteins promote alcohol-induced gut leakiness and liver injury. J Hepatol. 2018;69:142–153. doi: 10.1016/j.jhep.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ekihiro S, Bernd S. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590:447–458. doi: 10.1113/jphysiol.2011.219691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruan D, Wang W, Lin C, Fouad A, Chen W, Xia W, Wang S, Luo X, Zhang W, Yan S. Effects of curcumin on performance, antioxidation, intestinal barrier and mitochondrial function in ducks fed corn contaminated with ochratoxin A. Animal. 2019;13:42–52. doi: 10.1017/S1751731118000678. [DOI] [PubMed] [Google Scholar]
- 58.Wang H, Zhai N, Chen Y, Fu C, Huang K. OTA induces intestinal epithelial barrier dysfunction and tight junction disruption in IPEC-J2 cells through ROS/Ca 2+ -mediated MLCK activation &z.star. Environ Pollut. 2018;242:106–112. doi: 10.1016/j.envpol.2018.06.062. [DOI] [PubMed] [Google Scholar]
- 59.Wells CL. Colonization and translocation of intestinal bacterial flora. Transplant Proc. 1996;28:2653–2656. [PubMed] [Google Scholar]
- 60.Steffen EK, Berg RD, Deitch EA. Comparison of translocation rates of various indigenous bacteria from the gastrointestinal tract to the mesenteric lymph node. J Infect Dis. 1988;157:1032–1038. doi: 10.1093/infdis/157.5.1032. [DOI] [PubMed] [Google Scholar]
- 61.Aoudia N, Callu P, Grosjean F, Larondelle Y. Effectiveness of mycotoxin sequestration activity of micronized wheat fibres on distribution of ochratoxin A in plasma, liver and kidney of piglets fed a naturally contaminated diet. Food Chem Toxicol. 2009;47:1485–1489. doi: 10.1016/j.fct.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Z, Gan F, Xue H, Liu Y, Huang D, Khan AZ, Chen X, Huang K. Nephropathy and hepatopathy in weaned piglets provoked by natural ochratoxin A and involved mechanisms. Exp Toxicol Pathol. 2015;68:205–213. doi: 10.1016/j.etp.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 63.Khatoon A, Abidin Z. An extensive review of experimental ochratoxicosis in poultry: I. growth and production parameters along with histopathological alterations. Worlds Poult Sci J. 2018;74:627–646. doi: 10.1017/S0043933918000685. [DOI] [Google Scholar]
- 64.Stoev SD, Daskalov H, Radic B, Domijan AM, Peraica M. Spontaneous mycotoxic nephropathy in Bulgarian chickens with unclarified mycotoxin aetiology. Vet Res. 2002;33:83–93. doi: 10.1051/vetres:2001008. [DOI] [PubMed] [Google Scholar]
- 65.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W, Ching JYL, Chan PKS, Chan MCW, Wu JCY, et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2018;67:634–643. doi: 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kreznar JH, Keller MP, Traeger LL, Rabaglia ME, Schueler KL, Stapleton DS, Zhao W, Vivas EI, Yandell BS, Broman AT, et al. Host genotype and gut microbiome modulate insulin secretion and diet-induced metabolic phenotypes. Cell Rep. 2017;18:1739–1750. doi: 10.1016/j.celrep.2017.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Schooneveld TC, Gross A, Kalil AC. Duodenal infusion of feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:2143. doi: 10.1056/NEJMc1303919. [DOI] [PubMed] [Google Scholar]
- 68.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smillie CS, Sauk J, Gevers D, Friedman J, Sung J, Youngster I, Hohmann EL, Staley C, Khoruts A, Sadowsky MJ. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe. 2018;23:229–240.e225. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sturgill MG, Lambert GH. Xenobiotic-induced hepatotoxicity: mechanisms of liver injury and methods of monitoring hepatic function. Clin Chem. 1997;43:1512–1526. [PubMed] [Google Scholar]
- 71.Ren W, Wang P, Yan J, Liu G, Zeng B, Hussain T, Peng C, Yin J, Li T, Wei H. Melatonin alleviates weanling stress in mice: involvement of intestinal microbiota. J Pineal Res. 2018;64:e12448. doi: 10.1111/jpi.12448. [DOI] [PubMed] [Google Scholar]
- 72.Pinton P, Nougayrede JP, Del Rio JC. The food contaminant deoxynivalenol, decreases intestinal barrier permeability and reduces claudin expression. Toxicol Appl Pharmacol. 2009;237:41–48. doi: 10.1016/j.taap.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Berg M, Stenuit B, Ho J, Wang A, Parke C, Knight M, Alvarez-Cohen L, Shapira M. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J. 2016;10:1998. doi: 10.1038/ismej.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;8(7):R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. OTA residue and effect of OTA on growth performance and organ index in ducks. a: OTA residue in different organs (n = 6, Mean with SEM). The different letters above the column show significant differences. b-e: Effect of OTA on growth performance of ducks from day 1 to day 21. f: Effect of OTA on relative organ weight of 21 d ducks (n=6, Mean with SEM). g: Effect of OTA on relative length of intestine on day 21 ducks (n=6, Mean with SEM). h: Effect of OTA on relative weight of intestine on day 21 ducks. Data in a were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test, while data in b-h were analyzed by unpaired t test, *P <0 .05.
Additional file 2: Figure S2. OTA alters cecum microbial diversity and composition. a: Alpha diversity of cecum microbiota in different groups (n=7). Data were analyzed by student’s t test, *** P<0.001. b: Relative abundance of bacteria at Phylum level. OTUs with an occurrence lower than 1% are not represented (n=7). c: Relative abundance of top 15 Genus in each group (n=7). d: Relative abundance of top 15 Species in each group (n = 7).
Additional file 3: Figure S3. OTA lowers intestinal abundance of tight junctions. a: Effect of OTA on relative mRNA expressions of TJP1 and Occludin in the cecum of 21d ducks (n=6, Mean with SEM). Data were analyzed by unpaired t test. b: Effect of OTA on the protein abundance of TJP1 and Occludin in the cecum (n=6, Mean with SEM). Data were analyzed by unpaired t test. (n=6). *P<0 .05, **P<0 .01, *** P<0.001.
Additional file 4: Figure S4. OTA alters liver microbial composition. a: Alpha diversity of liver microbiota in different groups (n=5). Data were analyzed by student’s t test, n.s., not significant. b: Relative abundance of bacteria at Phylum level. OTUs with an occurrence lower than 1% are not represented (n=5). c: Relative abundance of top 15 Genus in each group (n=5). d: Relative abundance of top 15 Species in each group (n=5).
Additional file 5: Figure S5. OTA residue, effect of OTA on growth performance and organ index in antibiotics-treated ducks. a: OTA residue in different organs (n=6, Mean with SEM). Data were analyzed by the one-way ANOVA followed by Dunnett multiple comparisons (n=6, Mean with SEM). The different letters above the column show significant differences. b-e: Effect of OTA on growth performance of antibiotics-treated ducks from day 1 to day 21. f: Effect of OTA on relative organ weight of antibiotics-treated ducks (n=6, Mean with SEM). g: Effect of OTA on relative length of intestine on antibiotics-treated ducks (n=6, Mean with SEM). h: Effect of OTA on relative weight of intestine on antibiotics-treated ducks. Data in b-h were analyzed by unpaired t test.
Additional file 6: Figure S6. OTA has little effect on cecum microbiota diversity and composition in antibiotics-treated ducks. a: Alpha diversity of liver microbiota in different groups (n=6, Mean with SEM). b: The OTU numbers of Bacteroidetes (left) and Bacteroides (right) in different groups (CON, n=10; Antibiotics, n=6; Anti+OTA, n=6, mean with SEM). c: Relative abundance of top 7 bacteria at Phylum level in antibiotics-treated ducks. d: Relative abundance of top 15 bacteria at species level in antibiotics treated ducks. Data in a were analyzed by unpaired t test; while data in b were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test. n.s., not significant; ****P<0.0001.
Additional file 7: Figure S7. OTA shows little effect on intestinal abundance of tight junctions in antibiotics-treated ducks. a: Effect of OTA on relative mRNA expressions of TJP1 and Occludin in the cecum of antibiotics-treated ducks (n=6, Mean with SEM). Data were analyzed by unpaired t test. b: Effect of OTA on the protein abundance of TJP1 and Occludin in the cecum (n=6, Mean with SEM). Data were analyzed by unpaired t test. (n=6). *P<0 .05.
Additional file 8: Figure S8. OTA residue, growth performance and organ index after intestinal microbiota transplantation. a: OTA residue in the cecum digesta and liver of FMT (OTA) ducks (n=6, Mean with SEM). b-c: Final weight and relative weight of liver in FMT (CON) and FMT (OTA) ducks (n=9, mean with SEM). d: Levels of pro-inflammatory cytokines in duck liver after administration of OTA at dosage of 0, 15, 30 or 60 μg/kg (n=6, mean with SEM). Data in a, b and c were analyzed by unpaired t test; while data in d were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test. *P<0 .05. FMT (CON), ducks received CON group fecal microbiota; FMT (OTA), ducks received OTA group fecal microbiota.
Additional file 9: Figure S9. The cecum microbial diversity and composition of transplant donors and recipients. a: Alpha diversity of intestinal microbiome in transplant donors and recipients. b: Relative abundance of bacteria at Phylum level. OTUs with an occurrence lower than 1% are not represented (n=7). c: Relative abundance of top 15 Genus in each group (n=7). d: Relative abundance of top 15 Species in each group (n=7). Data in a and b were analyzed by one-way ANOVA with Bonferroni’s multiple comparisons test *P<0.05. FMT (CON): ducks received CON group fecal microbiota. FMT (OTA): ducks received OTA group fecal microbiota.
Additional file 10: Figure S10. OTA-originated microbiota lowers intestinal abundance of tight junctions. a: Relative mRNA expression of TJP1 and Occludin in different groups (n=6). b: Relative protein abundance of TJP1 and Occludin (n=6). Data were analyzed by unpaired t test, n=6, mean with SEM. *P<0 .05, **** P<0.0001. FMT (CON): ducks received CON group fecal microbiota. FMT (OTA): ducks received OTA group fecal microbiota.
Data Availability Statement
All raw data of 16S rRNA amplicon sequencing and Shotgun metagenomics are available at Sequence Read Archive (Accession number SUB5539876).