Abstract
Background
Bistability and ability to switch between two stable states is the hallmark of cellular responses. Cellular signaling pathways often contain bistable switches that regulate the transmission of the extracellular information to the nucleus where important biological functions are executed.
Results
In this work we show how the method of Gröebner bases can be used to detect bistability and output switchability. The method of Gröebner bases can be seen as a multivariate, non-linear generalization of the Gaussian elimination for linear systems which conveniently seperates the variables and drastically simplifies the simultaneous solution of polynomial equations. A necessary condition for fixed-point state bistability is for the Gröbner basis to have three distinct solutions for the state. A sufficient condition is provided by the eigenvalues of the local Jacobians. We also introduce the concept of output switchability which is defined as the ability of an output of a bistable system to switch between two different stable steady-state values. It is shown that bistability does not necessarily guarantee switchability of every state variable of the system. We further show that, for a bistable system, the necessary conditions for output switchability can be derived using the Gröebner basis. The theoretical results are incorporated into an analysis procedure and applied to several systems including the AKT (Protein kinase B), RAS (Rat Sarcoma) and MAPK (Mitogen-activated protein kinase) signal transduction pathways. Results demonstrate that the Gröebner bases can be conveniently used to analyze biological switches by simultaneously detecting bistability and output switchability.
Conclusion
The Gröebner bases provides a novel methodology to analyze bistability. Results clarify the distinction between bistability and output switchability which is lacking in the literature. We have shown that theoretically, it is possible to have an output subspace of an n-dimensional bistable system where certain variables cannot switch. It is possible to construct such systems as we have done with two reaction networks.
Keywords: Bistability, Output switchability, The Gröebner bases, Univariate basis polynomial, Steady-state solutions, Bifurcation, Polynomial equations, Biomolecular reactions
Background
Bistable dynamical systems are frequently encountered in cellular processes. Information processing within cells is carried out by a complex network of switches and oscillators [1]. A bistable system is a system with two attractors. The system can switch between two distinct stable states without resting in intermediate states. Switch-like bistable responses have been observed in many applications including signal transduction [2–6], cell cycle control [7–13], learning and memory [14], growing bacterial biofilms [15], epileptic spike-wave discharges [16], neurons [17] and synaptic transmission [18]. Bistable switches have been designed synthetically as well. A genetic toggle switch in Escherichia coli has been constructed in [19]. The bistable switch forms an addressable cellular memory unit and has implications for biotechnology, biocomputing and gene therapy.
Considering its biological importance, significant research has been devoted to explaining the physical origin of bistability, to develop necessary conditions for its existence and to construct algorithms for its detection. In particular positive feedback and ultrasensitivity have been proposed as two necessary conditions for the physical appearance of bistability [20]. It is also well known that adding negative feedback to positive feedback can turn bistability into oscillations [21]. The theory of chemical reaction network (CRN) [22] proposes conditions for bistability by making use of the properties of a species-reaction graph. Angeli et al. [23] presented a graphical method to detect bistability for biological positive-feedback systems. Under some mild assumptions, if the open-loop response (when the positive feedback loop is opened) is monotone and has a sigmoidal shape, the system is guaranteed to be bistable for some values of the feedback gains. Finally, Wilhelm [24] proposed a smallest chemical reaction system with bistability. Two reactions constitute a positive feedback loop; a third reaction filters out small stimuli, and a fourth reaction prevents explosions. Analysis is based on the method of Instability Causing Structure Analysis (ICSA) which is based on the Jacobian of the reaction network. Recently a new method was proposed to study multistationarity and bistability of chemical reaction networks with few chemical complexes. The method uses polynomial systems with few distinct monomials and Gale duality [25].
The method of Gröebner bases was introduced by Buchberger in [26, 27] as a powerful computational tool to address fundamental questions in commutative algebra (polynomial ideal theory, algebraic geometry). Since its original inception, the method of Gröebner bases was applied to simplify the algorithmic solution of many difficult problems expressed in terms of multivariate polynomials. These include [28]: solving polynomial equations, coding theory, integer programming, partial differential equations, symbolic summation, graph theory and statistics. Today exploring its applicability to many diverse fields such as computational biology [29], chemical kinetics [30–32] and systems theory [33–35] is an active area of research.
The objective of this work is to explore the method of Gröebner bases to analyze the bistability and output switchability of biological signaling systems. The utility of the method is demonstrated using several examples from the cellular signaling systems literature. Next we give working definitions and examples of bistability and output switchability.
Bistability
Consider the dynamical system Sf expressed as DAE (Differential Algebraic Equations):
| 1 |
| 2 |
where Eq. (1) represents the dynamic mass balances with the nonnegative concentrations of species x and y. Eq. (2) is a set of algebraic constraints due to the species conservation laws. Substituting y from Eq. (2) into Eq. (1), one gets:
| 3 |
The steady-states of Sf are the solutions of 0 = f(x).
A bistable system is a system with two attractors. In general, a plethora of interesting possibilities with different attractors and separatrices can result in different types of bistabilities which coexist in the parameter space of interest. Some of the popular bistabilities are between two stable fixed points [10, 11, 14], between a stable fixed point and a limit-cycle oscillator [15–17]; or between two stable periodic orbits [36, 37]. In this paper, we adopt the following definition for bistability:
Definition 1: The dynamical system Sf is state bistable if it has three nonnegative distinct real steady-states (ss) for the state x, two of which are stable ( and one is unstable where the superscript S and U denote stable and unstable, respectively.
We refer to such bistability as fixed-point bistability to distinguish it from other types of bistabilities mentioned above. Each stable steady-state (or fixed-point) has its own basin of attraction (i.e. the set of initial conditions that asymptotically converge to that steady-state). These basins are separated with a boundary defined by a separatrix. Most often, the separatrix contains a steady-state that is an unstable saddle-point [11]. Upon perturbations in the medium reflected by the parameter changes in the model, other types of attractors like limit cycles can be born from these fixed points via Hopf bifurcation [36]. For example, it was shown in [38] that bistability is a necessary condition for the emergence of oscillations in the MAPK cascade signaling.
Throughout the paper bistability will mean fixed-point state bistability.
A one-dimensional example
Figure 1 shows a bistable system resulting from a one-dimensional ordinary differential equation There are three positive fixed points at x = 1, 2 and 3 . The unstable fixed point at x= 2 separates the basins of attraction of the stable fixed points. The trajectories starting from the initial conditions to the left of 2 approach the stable fixed point x = 1, and the trajectories starting from the initial conditions to the right of 2 approach the stable fixed point at x = 3.
Fig. 1.
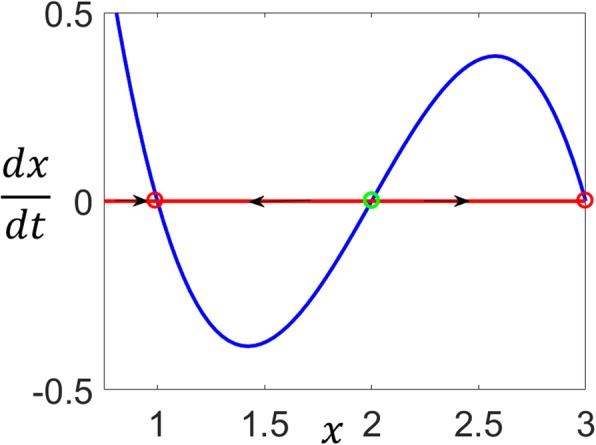
Bistability of a one-dimensional system. Two stable fixed points (red circles) separated by an unstable saddle-point (green circle)
A two-dimensional example
In [24] a smallest chemical reaction system with bistability was proposed. The model consists of the following two-component mass-action kinetic ODE system:
| 4 |
| 5 |
The system has three steady-states, two of which are stable at (0, 0), (6, 4.5) and an unstable steady-state which is a saddle-point at (2, 0.5). Figure 2 shows the phase plane with trajectories emanating from different initial conditions. Due to the saddle-point, the phase plane is divided into two basins of attraction which contain the trajectories approaching the two stable steady-states.
Fig. 2.
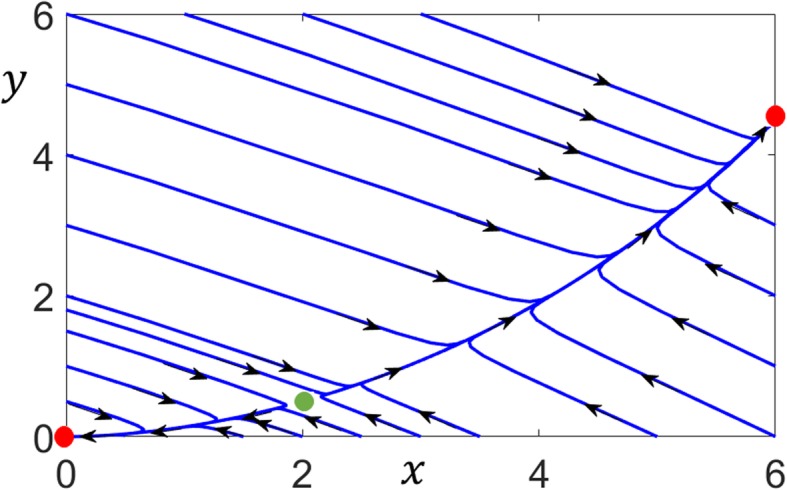
Phase-plane of a two-dimensional bistable system. The system has two stable fixed points (red circles) and an unstable saddle-point (green circle). Trajectories are separated into two basins of attraction of the stable steady-states
Bistability exits in models with higher dimensions n > 2 as well. For example, in [11] existence of a saddle point and two stable fixed points are highlighted with an apoptosis model that consists of 8 states. The authors present a local analysis to identify the saddle point that helps to understand the global properties of biological switches.
Output switchability
Many biological events are binary with certain variables switching on and off between active and inactive states to perform important biological functions. For a bistable dynamical system Sf, one is often interested in the switching response of the concentration of some species. Therefore, we include an output variable y in the model and the analysis. In general output y is taken to be any of the system states xi i = 1 : m. Without loss of generality, the concentration of the first species, or the first state variable x1, is defined as the output. The new dynamical system with output x1 is denoted by and expressed as:
Next, we introduce the following definition:
Definition 2: A bistable dynamical system is called output switchable if the steady-state output values x1, ss are different at the two stable steady-states of the state x i.e. at (.
Differentiation between state bistability (Definition 1) and output switchability (Definition 2) is not made in the literature. For a one-dimensional system with a single output, bistability implies output switchability. However, for higher dimensional systems, bistability does not necessarily guarantee switchability for every output variable or state variable xi i = 1 : m. Theoretically, it is possible to have an output subspace containing certain variables that do not switch.
Results
The method of Gröebner bases (see Methods section) is applied to several systems to detect biological switches.
Bistable systems with switchable outputs.
Example 1. In [24] a smallest bistable system is given that consists of the following four reactions:
The system is described by a two-component mass-action ODE system:
| 6 |
| 7 |
with k1 = 8, k2 = k3 = 1, k4 = 1.5.
Since the steady-states are determined by the solutions f1(x, y) = 0 and f2(x, y) = 0, the Gröbner basis is computed for these two polynomials using the reduced Gröebner basis program gbasis available in the Symbolic Math Toolbox of MATLAB:
| 8 |
| 9 |
Solving this triangular system and checking the eigenvalues of the Jacobians confirms that the system is bistable with three steady states:
Considering x as the output, the system is output switchable since g1(x) satisfies the necessary and sufficient conditions (Eqs. 59–61) for switchable outputs given in the Methods section. Plot of g1(x) with its three distinct roots is given in Fig. 3.
Fig. 3.
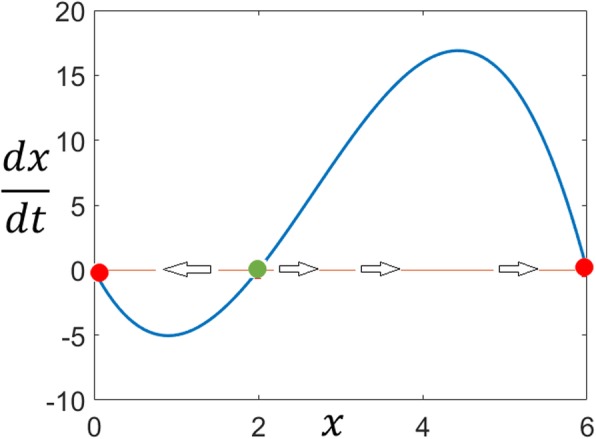
The univariate basis polynomial for Example 1. Two stable fixed points (red circles) separated by an unstable saddle-point (green circle)
Note that the inverse problem of given a “desirable” univariate basis polynomial such as (8), reconstruction of a corresponding reaction network is possible although this network is not unique in general. The cubic depletion term −x3 suggests a bilinear term (−xy) where y is proportional to x2 so that (−xy) = − x3 . This can be realized by the following set of reactions:
Steady-state mass balance for y gives The third reaction provides the cubic depletion rate . Without this reaction the system can not have three steady-states and bistability is not possible. The first two reactions also provide the quadratic production term x2 for the species x. Finally, a first order reaction:
gives the linear depletion rate −1.5x. Summing up all the terms yields
g1(x) = x2− which has the same solutions as the targeted univariate basis (8). Note that in [24] the above smallest bistable system is constructed in a similar way but without introducing the Gröebner basis.
In order to check if the output y is switchable, we change the lexicographic monomial order of the unknown variables (x, y) and recompute the Gröebner basis where the univariate polynomial is now a function of y:
| 10 |
| 11 |
Since g1(y) satisfies the conditions for switchable outputs (Eqs. (59–61)), the system is switchable in the output y as well.
Example 2. Consider the Edelstein reaction scheme analyzed in [29] and given in Fig. 4.
Fig. 4.
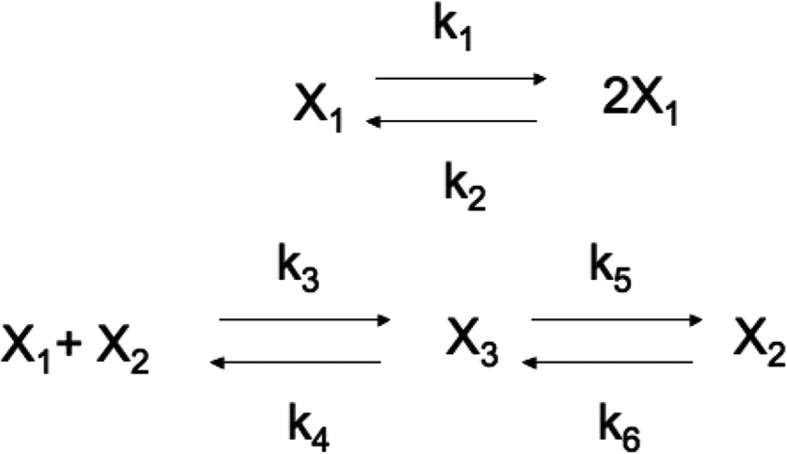
Edelstein Chemical Reaction Network [27]. Parameters: k1 = 8.5, k2 = k3 = k4 = k5 = 1, k6 = 0.2
The system is described by the following DAEs:
| 12 |
| 13 |
| 14 |
Substituting the parameter values (see Fig. 4) and eliminating x3, one gets:
| 15 |
| 16 |
The Gröbner basis is calculated as
| 17 |
| 18 |
The system is bistable with three steady states for the state x:
In the Methods section we derive that for a bistable system to have a switchable output, the rate of generation of the output must consist of a quadratic and a constant term, and the rate of depletion must consist of a cubic and a linear term. Considering x1 as the output in the current example, the system is output switchable since g1(x1) has quadratic and constant generation, and linear plus cubic depletion terms (see (17)), and it satisfies the necessary and sufficient conditions (Eqs. 59–61). The dynamic output responses of the original system (x1, f) and the univariate basis polynomial dynamical system () are compared in Fig. 5 for one initial condition. Trajectories converge to the same stable steady-state.
-
b.
Bistable systems with unswitchable outputs.
Fig. 5.
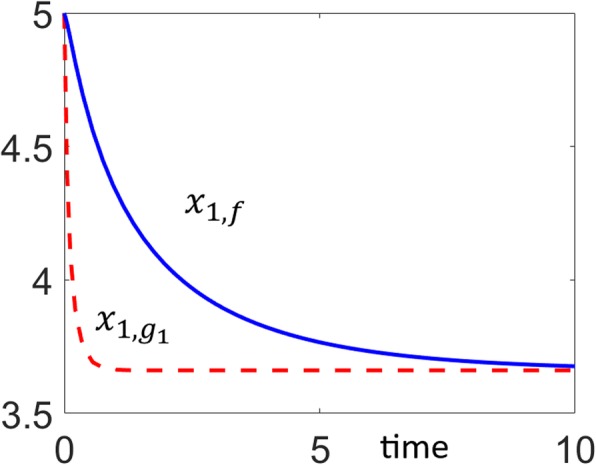
Comparison of the output responses of the original system (x1, f) and the univariate dynamical system (). Responses start from an initial condition and converge to one of the stable fixed points
It is difficult to find physical examples in the literature for bistable systems with unswitchable output(s). There are several reasons for this seemingly lacking data. First, only the switching variables are analyzed to show bistability; therefore, even if there are some outputs (or states) that do not switch, they are not reported. Second, it is possible that some outputs lose their switchability under abnormal conditions only (e.g. disease states due to mutations etc.) that create the right conditions for the emergence of unswitchable output(s). As a result, no distinction is made in the literature between bistability and output switchability. However, as we have presented above, the two concepts are not the same. In addition, at a practical level, for a complex system with many states, it is plausible for some output(s) of a bistable sytem to keep the same steady-state values as the system state switches from one stable steady-state to another. These special outputs can be acting as chaperons that change their values in transient only in order to help the other outputs to switch, and when they complete their tasks they return to their steady-states.
Example 3. The smallest bistable system whose output is not switchable is given by a two dimensional system:
which satisfies the following conditions at steady-state:
For example, the following ODEs meet the above conditions:
| 19 |
| 20 |
In this example we construct a reaction network that satisfies (19) and (20). Consider the reaction network shown in Fig. 6.
Fig. 6.
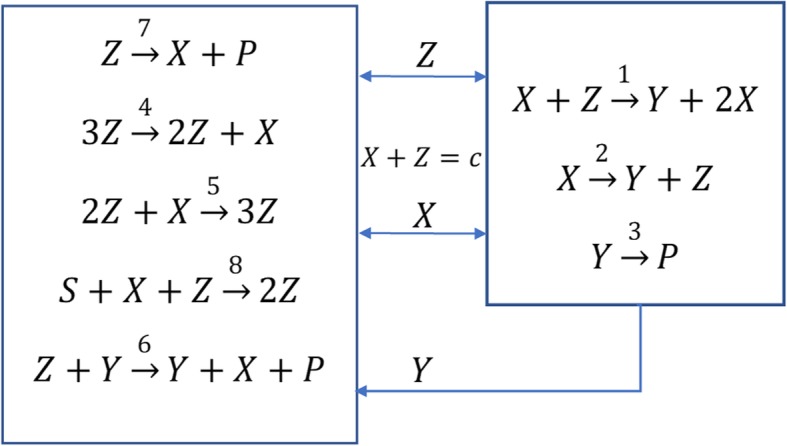
Example 3. X + Z = c (constant) is the conserved moiety
The conservation equations with mass action kinetics are given by two ODEs and one algebraic equation:
| 21 |
| 22 |
| 23 |
Using the values for the rate constants given in Table 1 and eliminating X via (23) gives:
Table 1.
Parameters for the reaction network of Example 3
| k1 | k2 | k3 | k4 | k5 | k6 | k7 | c | |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.1623 | 1 | 0.0156 | 1 | 0.0156 | 5.0983 | 0.1 | 5.1623 |
* constant concentration of the species S is lumped into
| 24 |
| 25 |
The Gröebner basis G of (f1, f2) is calculated using the GroebnerBasis program under Polynomial Algebra of MATHEMATICA and the triangular system of basis polyniomials is given by (compare with Eq. 58):
| 26 |
| 27 |
| 28 |
Solving this triangular system of equations yields three distinct steady-state solutions for the state = as shown in Fig. 7. Therefore, the necessary condition NC for bistability stated in the Methods section (see Eq. 58) is satisfied. Next, one proceeds with the calculation of the Jacobians to establish sufficiency. Both eigenvalues of the Jacobian are negative at steady-states indicating that these are the stable steady-states; one eigenvalue is positive for indicating that this is the unstable steady-state. Therefore, the system is bistable.
Fig. 7.
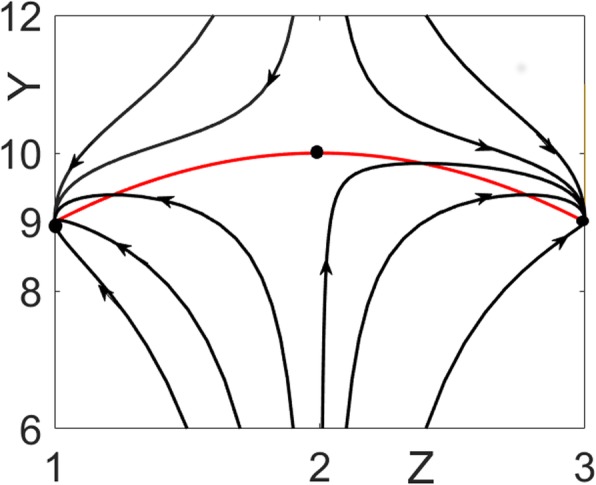
Bistable system with an output that does not switch at steady-state. Three steady-states are marked by black circles. The middle one at Z = 2, Y = 10 is the unstable steady-state. Trajectories starting from different initial conditions converge to the stable steady-state, where the output (Y) value does not change (i.e. Y = 9). The red curve is the production rate of Y which is the locus of steady-states for Y as a function of Z
g1(Y) = 0 has two solutions, one less than the total number of steady-states for the state. Therefore, one of the roots Y = 9 is necessarily repeated in the steady-state solutions for the state: = . Since the repeated roots belong to the stable steady-state solutions, the system is not switchable in the output Y. Figure 7 shows the state trajectories x(t) approaching the stable steady-states separated by the middle unstable fixed point at .
Figure 7 shows the state trajectories x(t) approaching the stable steady-states separated by the middle unstable fixed point at . However, unlike Z and X = c-Z, Y is not a switchable output since its value remains the same (equal to 9) at the stable steady-states.
The reason for Y not to be a switchable output can be physically explained as follows.
The mass balance for Y is determined by the following set of reactions (see Fig. 6):
and the constraint X + Z = c.
The first two reactions produce Y and the third reaction depletes Y. Conservation of Y is given by the following ODE:
| 29 |
The first term k1(c − Z)Z is the rate of production of Y by the first reaction, and the second term k2(c − Z) is the rate of production of Y by the second reaction. The last term k3 Y is the rate of consumption of Y which is equal to the sum of the two production rates at steady-state:
| 30 |
The total production rate of Y is maximum at the middle unstable steady-state as shown in Fig. 7. At the stable steady-state to the left of the maximum, the first production rate is greater than the second production rate k1(c − Z)Z > k2(c − Z) and the total production rate is 9. At the stable steady-state to the right of the maximum, the reverse is true i.e. k1(c − Z)Z < k2(c − Z) but the total production rate k3Y remains the same. Since k3 = 1 (see Table 1), k3Y = Y = 9 at the stable steady-states; thus, it cannot switch. The above result shows that, if an output species (Y) is produced by two reactions and the sum of the reactants is constant X + Z = c; then, for some values of the rate constants, the total production rate can remain the same at the stable steady-states leading to unswitchability of Y while both X and Z can switch.
Example 4. Consider the reaction network given in Fig. 8.
Fig. 8.
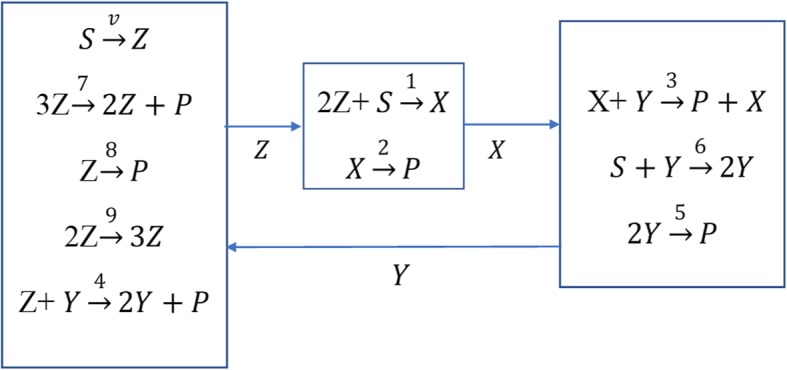
Reaction network for Example 4
The conservation equations with mass action kinetics are given by:
| 31 |
| 32 |
| 33 |
The parameter values are given in Table 2.
Table 2.
Parameters for the reaction network of Example 4
| k1 | k2 | k3 | k4 | k5 | k6 | k7 | k8 | k9 | v |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0.166 | 1 | 0.166 | 1.667 | 2 | 16 | 16 | 24 |
At steady-state Yf3(X, Y, Z) = 0 and one solution is Y = 0, but for this value of Y, there is only one real positive solution for Z at 6.56; therefore, the system cannot be bistable. Thus, we consider the other solutions that satisfy f3(X, Y, Z) = 0. If Y is designated as the output variable, the univariate polynomial in Y does not exist. Choosing Z as the output variable and using the lexicographic order (X, Y, Z), the Gröbner basis is computed for the polynomials f1(Y, Z), f2(X, Z), f3(X, Y, Z) and the triangular system of basis polyniomials is given by
| 34 |
| 35 |
| 36 |
The steady-state solutions for the state are easily computed: The system is bistable as determined by the Jacobians.
The univariate basis polynomial (34) satisfies the output switchability conditions; thus, the system is switchable in output Z. Due to (35) it is switchable in X as well. But it is not switchable in the output Y, since the solution Y = 18 determined from (36) is repeated in the stable steady-states. This is also shown in the bifurcation diagrams in Fig. 9.
-
c.
Cellular signaling pathways: AKT, RAS and MAPK signal transduction systems.
Fig. 9.
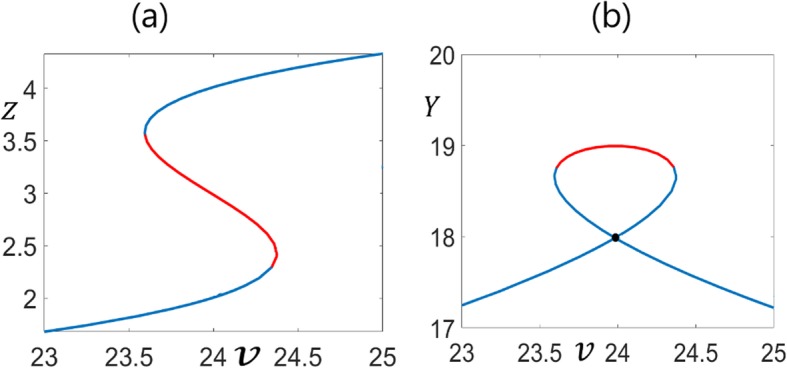
a. Bifurcation of Z with respect to v. Red and blue branches are the unstable and stable solutions. At v = 24, there are three distinct solutions for Z (stable, unstable, stable). b. Bifurcation of Y with respect to v. Red and blue branches are the unstable and stable solutions. At v = 24, there are two repeated stable solutions for Y at 18 (shown by black circle) and one unstable solution at 19
Example 5. AKT signaling pathway
AKT signaling pathway plays a key role in the most significant metabolic action of insulin, which is the glucose uptake. Insulin resistance can develop through impairments in the signaling events involved in the activation of AKT. We use the following minimal dimensionless two-state model which we have derived earlier [39] from the original model presented in [6]:
| 37 |
| 38 |
where the states are the dimensionless concentrations x1 = pAKT (active AKT) and x2= pIRS (insulin receptor substrate). The input is the amount of insulin represented by λ. The parameter values are taken from [39].
Since the Gröebner basis is defined for polynomials, the right-hand side of (37) is first converted to a rational polynomial function so that (37) and (38) can be expressed as:
= and = f2(x1, x2)
where both f1(x1, x2) and f2(x1, x2) are polynomials. Since the steady-states are determined by the solutions f1(x1, x2) = 0 and f2(x1, x2) = 0, the Gröbner basis is computed for these two polynomials using MATHEMATICA, and for the insulin level λ =0.4 it is given by:
| 39 |
| 40 |
Since g1(x1) given by (39) satisfies all the conditions for three distinct roots (Eqs. 59–61), the output x1 = pAKT is switchable, if the system is bistable. The necessary condition NC for bistability is satisfied since the triangular system (39)–(40) gives three steady-states:
=
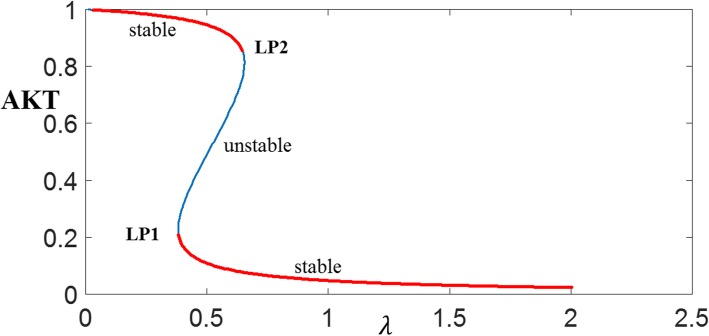
The sufficiency of bistability (i.e. checking the stability status of the three steady-states) is established by bifurcation analysis of eqs. (37)–(38) using XPPAUTO and is given in Fig. 10. The stable and unstable branches show that AKT is bistable for the range of λ between LP1 = 0.38 and λ = LP2 = 0.65. This confirms that the system is indeed bistable for λ = 0.4 and output x1 = pAKT is switchable.
Fig. 10.
Bifurcation diagram for AKT showing the stable and unstable branches. LP1 and LP2 are the turning points. The bistable region is for λ between LP1 = 0.38 and λ = LP2 = 0.65
In order for insulin to perform its function, AKT has to switch between its inactive and active states. Activated AKT (pAKT) enables the translocation of glucose transporter-4 (GLUT-4) from cytosol to the plasma membrane, thus glucose is taken into the cell.
The bistable behavior of pAKT is shown in Fig. 11. pAKT resides on either its active or inactive stable state depending on the initial condition.
Fig. 11.
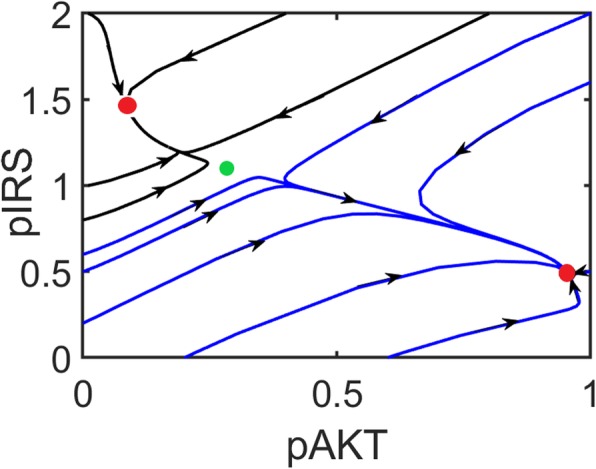
Bistability of the AKT signaling pathway. AKT system has two stable fixed points (red circles) and an unstable saddle-point (green circle). Trajectories are separated into two basins of attraction of the stable steady-states. λ = 0.4
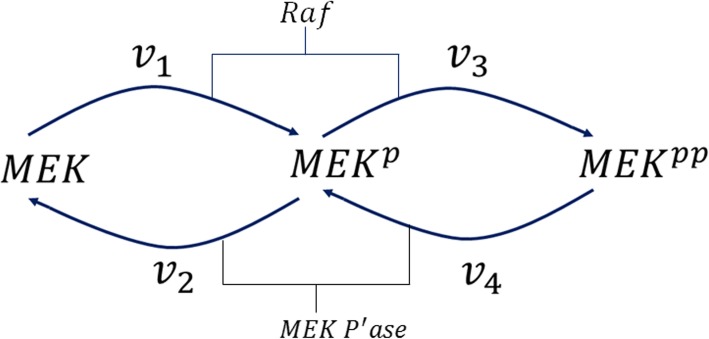
The MAPK cascade is an integral part of the ERK (Extracellular Signal-Regulated Kinase) signaling pathway which plays a key role in cell cycle control. In the first stage of the cascade, RAF (Rapidly Accelerated Fibrosarcoma) gets activated by RAS-GTP (Rat Sarcoma Guanosine Triphosphate), and it triggers the second stage where MEK (Mitogen activated protein kinase kinase) gets double phosphorylated [2, 38]. This is followed by the activation ERK in the last stage. Here we will focus on the second stage which is shown in Fig. 12.
Fig. 12.
Dual phosphorylation/dephosphorylation cycle of protein MEK
The two-site MAPK phosphorylation and dephosphorylation cycle with a distributive kinetic mechanism for the kinase and phosphatase possesses the necessary properties to exhibit bistable response [2, 40]. MEK and MEKp compete for the same kinase (RAF) for phosphorylation; MEKpp and MEKp compete for the same phosphatase (MEK P’ase) for dephosphorylation. Through this competition, MEK inhibits the production of MEKpp, and MEKpp inhibits the production of MEK. This double inhibition results in a positive feedback loop which leads to bistability under the right set of operating conditions or parameter values. Next we detect and confirm this bistability by using the method of Gröebner bases.
The model is taken from [40]:
| 41 |
| 42 |
| 43 |
, (respectively)
MT is the total concentration of MEK. The rates are given by:
with the parameters:
where c and p are the concentrations of RAF kinase and the MEK phosphatase, respectively. Pertinent data is listed in Table 3.
Table 3.
Parameter values for MAPK
| a1 | 0.0204 |
|---|---|
| a2 | 0.0493 |
| a3 | 0.0564 |
| a4 | 0.0326 |
| d1 | 10.386 |
| d2 | 2.716 |
| d3 | 10.088 |
| d4 | 0.813 |
| k1 | 7 |
| k2 | 11.13 |
| k3 | 3.57 |
| k4 | 1.13 |
| MT | 5128 nM |
| c | 0.6 nM |
| p | 1 nM |
First (41)–(43) are re-expressed as:
| 44 |
| 45 |
| 46 |
The Gröebner basis was obtained using MATHEMATICA:
which can be solved easily to give three solutions:
. Evaluation of the eigenvalues of the Jacobians shows that the system is bistable. Since the univariate basis polynomial g1(y) meets the conditions for three distinct roots, the system is switchable for the output
as well. In fact, all the states are switchable.
Example 7. RAS signaling
RAS, which is a small GTP (Guanosine Triphosphate) binding protein, serves as an important molecular switch in signaling pathways. For example, in ERK signaling pathway, RAS interacts with the ShC-Grb2-SOS complex, and it is transformed to its active conformation by exchanging GDP (Guanosine Diphosphate) for GTP. Active Ras-GTP starts the sequential phosphorylation of the MAPK pathway that consists of the RAf-MEK-ERK signaling cascade. Catalytic activation of RAS by the SOS (Son of Sevenless) complex Shc-Grb2-SOS while RAS-GTP is bound to its allosteric site creates a positive loop resulting in a bistable switching response of Ras-GTP [41]. The model is taken from [41] and it is converted to a dimensionless form. It consists of the following equations:
| 47 |
| 48 |
| 49 |
| 50 |
| 51 |
The variables are defined as follows: S is the Shc-Grb2-SOS complex; RT is RAS-GTP; RD is RAS-GDP, SRD and SRT are the complexes formed by the reactions; [.] denotes the concentration. The total concentration of S molecules is α, and the total concentration of RAS molecules is β. The values for the parameters are given in Table 4.
Table 4.
Parameter values for RAS
| β | 200 nM |
|---|---|
| α | 10 nM |
| k1 | 1.125e − 4 nM−1 s−1 |
| k2 | 1.0625e − 4nM−1 s−1 |
| k−1 | 3s−1 |
| k−2 | 0.4s−1 |
| 1.75s−1 | |
| 0.003s−1 | |
| 0.1s−1 | |
| RGAP | 0.1 nM |
| K3m | 2.7388e3 nM |
| K4m | 1.52304e4nM |
| K5m | 17.869nM |
The model has three steady-states, two of which are stable representing the active and inactive states of RAS, and a saddle point with a positive eigenvalue. Bistability is illustrated in Fig. 13. Trajectories first converge to the unstable manifold, and then they are attracted to either of the two steady-states.
Fig. 13.
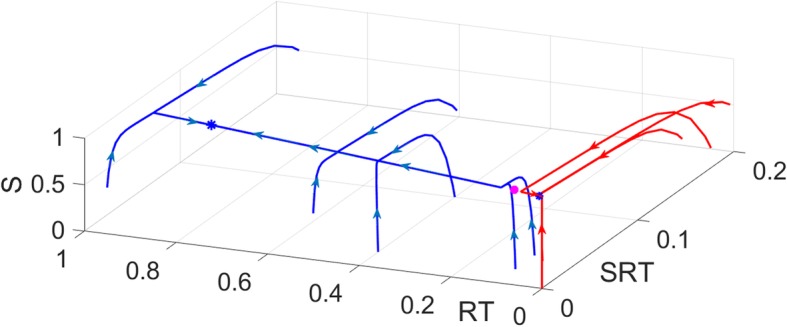
Bistability of a three-dimensional RAS model. The system has two stable fixed points (blue stars) and an unstable saddle-point (magenta circle). Trajectories reach the stable fixed points after following the unstable manifold of the saddle point. S is the Shc-Grb2-SOS complex. RT is RASGTP. SRT is the SOS-RASGTP complex
MATLAB computes the univariate basis polynomial as a cubic polynomial:
with three distinct roots, thus RT is a switchable output.
Discussion
We have developed a new method to detect and analyze biological switches by simultaneously treating bistability and output switchability using the Gröebner bases. As demonstrated by several examples, the proposed methodology offers the following:
First the method provides computational advantages due to its nice properties. Specifically, the method of Gröebner bases to solve polynomial systems can be seen as a multivariate, non-linear generalization of the Gaussian elimination for linear systems. Multistationarity is easily checked by solving a triangular set of equations which facilitates the root finding. Bistability is confirmed by local stability analysis using the Jacobian which is also straightforward.
It provides a theoretical framework and a systematic methodology that analyzes both bistability and output switchability simultaneously. Output switchability conditions follow immediately from the univariate Gröebner polynomial basis and are easy to check. We show by Examples 3 and 4 that some bistable systems can have outputs that do not switch their steady-states.
The univariate Gröebner basis polynomial provides useful biological insight which can help in the design of biological switches. A bistable dynamical system with output x, is output switchable, if its univariate basis polynomial g1(x) = − x3 + bx2 − cx + d with b > 0, c > 0, d ≥ 0 has three distinct nonnegative roots. This result provides some biological insight. The terms −x3 − cx represent the rate of depletion of species x, and the terms bx2 + d represent the rate of production of species x. This suggests that a biological switch for an output species x can be designed by constructing a reaction network (with its corresponding ODEs) whose univariate Gröbner basis polynomial in x has the above types of depletion and production terms. In fact Examples 1, 3 and 4 were constructed in this fashion.
Conclusions
We have presented a new method to detect biological switches by analyzing their bistability and output switchability properties. The methodology is based on the Gröebner bases. Conditions are established to make the connections between the Gröebner bases, bistability and output switchability. Various examples are given to elucidate the theoretical results. We show that the method can analyze bistability and output switchability while providing useful insight into the underlying mechanisms. The method is easy to apply since significant software such as MAPLE, MATLAB and MATHEMATICA exists to perform the Gröebner bases computation.
It goes without saying that high dimensionality can pose computational problems as in other methods. As a remedy, techniques such as lumping, network complexity reduction can be used to reduce the number of ODEs before the Gröebner basis calculation is carried out. In general the method can be applied to other types of polynomial differential equations derived from data instead of first principles. Such potential models include the nonlinear polynomial regression models.
Methods
Bistability and output switchability analysis is based on the Gröebner bases.
The Gröebner bases
Any finite set of multivariate polynomials F can be transformed by an algorithm (see Buchberger’s algorithm [26]) into another set of basis polynomials, called the Gröebner basis G. Many problems that are difficult to handle by the original set of polynomials can be easily solved by using the method of Gröebner bases due to its “nice” properties. Readily available computer software such as MAPLE, MATHEMATICA and MATLAB are equipped with the computational machinery of the Gröebner bases. The most basic definitions and properties of the Gröebner bases are presented in the Additional file 1. In this paper we explore the Gröebner bases within the context of bistability analysis. To this end, we state some of the useful properties of the Gröebner bases for solving polynomial equations.
Solution of polynomial equations by the Gröebner bases
The method of Gröebner bases to solve polynomial systems can be seen as a multivariate, non-linear generalization of the Gaussian elimination for linear systems [42].
The ideal I = < f1, f2, . . , fn> is the set of all possible linear combinations of fi ′ s where the coefficients are polynomials pi (Additional file 1). Since F = (f1, f2, , .., fn) and its Gröebner basis G generate the same ideal, they have the same solutions [42, 43]. The advantageous property of the Gröebner basis G is that it yields a triangular system which conveniently seperates the variables and drastically simplifies the calculation. This triangular system is like the reduced row echelon form obtained by pivoting in Gaussian elimination in the case of linear systems.
Consider the steady-state solutions of the dynamical system (Eqs. 4–5) which satisfy following set of polynomial equations:
| 52 |
| 53 |
The Gröebner basis G for these two polynomials with respect to the lexicographic ordering is given in a triangular form:
| 54 |
| 55 |
First the univariate basis polynomial g1(x) is easily solved for its roots x: (0, 6,2). Next these x values are substituted into the bivariate basis g2(x, y) to determine its corresponding roots y: (0,4.5,0.5). Thus, the solutions (x, y) of the original set of polynomials F are obtained: (0, 0), (6,4.5) and (2,0.5).
Detection of Bistability
Consider the dynamical system Sf given by:
| 56 |
where the steady-state solutions satisfy f(x) = 0, and they are denoted as
with m the number of solutions.
Let G = [g1(x1), g(x)] be the Gröebner basis for the ideal I = < f1, f2, …, fn>, where g1(x1) is the univariate basis polynomial, and g(x) is the vector of remaining polynomials arranged in triangular form:
| 57 |
A necessary condition for bistability (NC)
The dynamical system Sf is bistable only if the following set of equations have three distinct real nonnegative solutions for the state x:
| 58 |
Necessity follows from the working definition of bistability which requires three distinct steady-state solutions for the state vector x, and the fact that the steady-state solutions of Sf and the solutions of the Gröebner basis polynomials (58) are the same. Moreover, for a zero-dimensional ideal, the triangular structure in the Gröebner basis always exists [42]. For Gröbner bases, unlike other triangular systems, it is guaranteed that each partial solution can be extended to a full solution. This means that every solution x1 of the first polynomial can be extended to a solution (x1, x2) of the polynomials in x1 and x2, and each of these solutions can be further extended to a solution (x1, x2, x3) of the polynomials in x1, x2, x3, etc.
It is important to note that bistability cannot be detected by checking the number of solutions of the univariate basis polynomial g1(x1) alone but the whole basis must be considered. This follows from the fact that the number of solutions of g1(x1) = 0 can be less than the number of solutions for the state x.
Systems that fail to meet the necessary condition cannot be bistable; thus, they are easily eliminated from further consideration. However, satisfying the necessary condition does not guarantee bistability. Further tests should be applied to confirm it. The most common approach is to compute the eigenvalues of the Jacobian of f(x). The Jacobian matrix is obtained by linearizing the dynamical system Sf at its steady-states :
A steady-state of the dynamical system Sf is stable if all the eigenvalues of Jacobian matrix Ji have negative real parts. The steady-state is unstable if at least one of the eigenvalues has a positive real part [44]. Bistability can be easily ascertained by checking the stability status of each of the three distinct steady-states of Sf.
Detection of switchable outputs
A bistable dynamical system with output is output switchable, if its univariate basis polynomial satisfies the following conditions:
| 59 |
| 60 |
| 61 |
These conditions guarantee that g1(x1) has three distinct nonnegative roots so that, the output x1 can take different values when the state x changes between its stable steady-states. Existence of three distinct nonnegative roots can be shown as follows. According to the Descartes’ rule of signs, the maximum number of negative real roots of a polynomial f(x) is equal to the number of changes in sign of the coefficients of the terms of f(−x). When (60) is true, there are no sign changes in the coefficients of g1(−x1) in (59); thus, g1(x1) can have maximum three nonnegative roots. The inequality (61) is the cubic discriminant condition which guarantees that there are three distinct real roots. The general graph of g1(x1) satisfying conditions (59)–(61) is shown in Fig. 14.
Fig. 14.
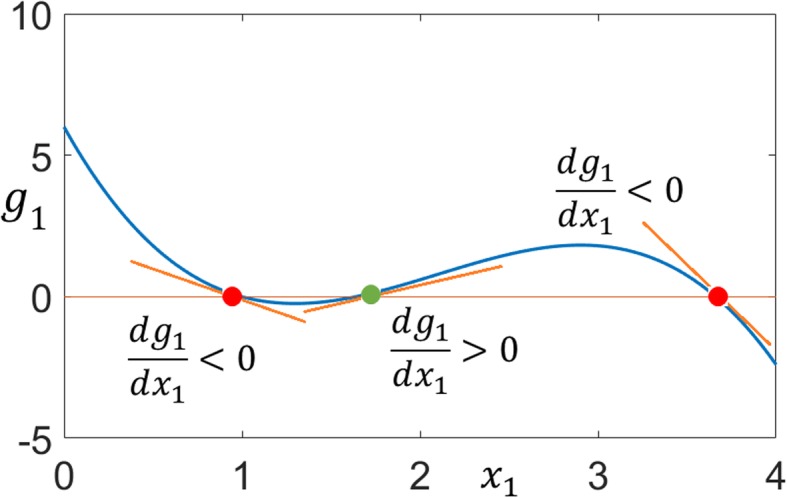
The univariate basis poynomial. Distinct roots are shown by the circles (red for stable; green for unstable). Slopes of the tangent lines (Jacobians) are negative and positive at the stable fixed points and saddle-point, respectively
If the univariate basis polynomial g1(x1) does not have three distinct roots; then, the bistable system is switchable in its output xi, only if one of the repeated solutions belongs to the unstable steady-state solution . This follows from the definition of output switchability which requires that the output values are different at the two stable steady-states.
It is possible that for an output variable of interest y, the univariate basis polynomial g1(y) may not exist. If this happens, by changing the lexicographic ordering, the triangular system of basis polyniomials (58) is calculated using a different univariate basis polynomial g1(xi) where xi ≠ y. In such cases, the bistable system is switchable in its output y, only if the solutions y obtained from the triangular system of basis polynomials are different at the two stable steady-states (.
The univariate basis polynomial dynamics
For a bistable system with a switchable output x1, we define the univariate basis polynomial dynamics describing the output as:
| 62 |
is bistable as seen from the signs of the local Jacobians depicted in Fig. 14. Note that the output trajectories of the original system are different in transient but both converge to the same stable steady-state, since they have the same output solutions due to the property of Gröbner basis. When the univariate basis polynomial has three distinct roots, it is always possible to construct the one-dimensional bistable system
It can be seen from Eq. (62) that |g1(x1)| is the imbalance between the rate at which the output species is generated and the rate at which it is depleted:
| 63 |
At the roots of g1(x1), the two rates equilibrate. Between the roots, the sign of (ggeneration − gdepletion) alternates as (+ − +) to create a bistable switching output response as shown in Fig. 15.
Fig. 15.
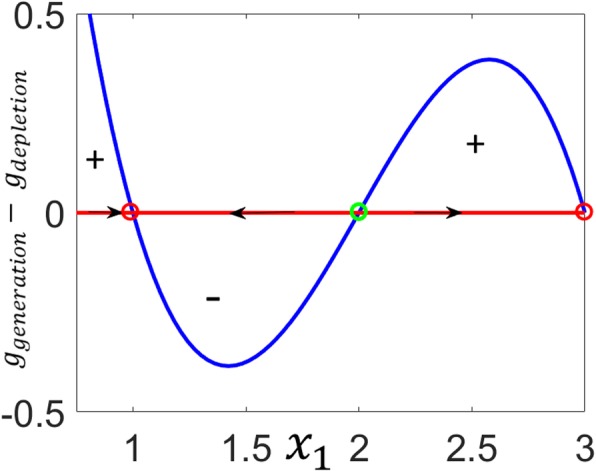
Bistable univariate dynamical system. Depending on the initial sign of ggeneration − gdepletion, trajectories approach either of the stable steady-states where. ggeneration = gdepletion.
According to (63) for a bistable system to have a switchable output x1, the rate of generation must consist of a quadratic and a constant term, and the rate of depletion must consist of a cubic and a linear term:
| 64 |
| 65 |
This suggests that a biological switch for an output species can be synthetically designed by constructing a reaction network (and its corresponding ODEs) who’s univariate Gröbner basis polynomial has the above types of depletion and production terms. Examples 1, 3 and 4 were constructed in this fashion.
Analysis procedure
We have incorporated the theoretical results into the following analysis procedure:
Given a Differential Algebraic System, compute its Gröbner basis. We have used MATLAB and MATHEMATICA for this purpose.
By solving the triangular system of eqs. (58), check if three distinct nonnegative solutions exist in some region of the state space. If it does, check the eigenvalues of the Jacobian at the three steady-states to decide if the system is bistable. If the system is bistable, proceed to the next step. If three distinct nonnegative solutions for the state do not exist, the system cannot be (fixed-point) bistable and stop.
For the bistable system, identify the output of interest y and compute the univariate basis polynomial in y i.e. g1(y) by reordering the variables, if necessary. Compute the roots of g1(y). If the number of roots is three, the bistable system is switchable in the output if these roots are all distinct. In all other cases where there are two repeated roots, one of the repeated roots must belong to the unstable steady-state solution . Otherwise the system is not switchable in output y.
In case the univariate basis polynomial g1(y) does not exist, solutions y are calculated using a different univariate basis polynomial g1(xi) where xi ≠ y. The bistable system is switchable in its output y, only if the solutions at two stable steady-states ( are distinct.
Supplementary information
Acknowledgements
The financial support through grant 117F123 by TÜBİTAK, The Scientific and Technological Research Council of Turkey, is gratefully acknowledged.
Abbreviations
- AKT
Protein kinase B
- CRN
Chemical Reaction Network
- DAE
Differential Algebraic Equations
- ERK
Extracellular Signal-Regulated Kinase
- GDP
Guanosine Diphosphate
- GLUT-4
Glucose transporter-4
- Grb2
Growth factor receptor-bound protein 2
- GTP
Guanosine Triphosphate
- ICSA
Instability Causing Structure Analysis
- IRS
Insulin receptor substrate
- LP
Limit Point
- MAPK
Mitogen activated protein kinase
- MEK
Mitogen activated protein kinase kinase
- NC
Necessary condition
- ODE
Ordinary Differential Equations
- pAKT
Phosphorylated AKT
- pIRS
Phosphorylated Insulin Receptor Substrate
- RAF
Rapidly Accelerated Fibrosarcoma
- RAS
Rat Sarcoma
- RAS-GTP
Active form of RAS
- RGAP
RAS GTPase Activating Proteins
- Shc
Src homology 2 domain containing
- SOS
Son of Sevenless
Authors’ contributions
YA is solely responsible for developing the methodology, collecting data, obtaining results and preparing the manuscript. The author read and approved the final manuscript.
Funding
The work was supported through grant 117F123 by TÜBİTAK, The Scientific and Technological Research Council of Turkey. TÜBİTAK did not play any role in the design of the study and collection, analysis, and interpretation of data, or in writing this manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this manuscript. Given the data, the Gröbner bases were calculated by calling the GroebnerBasis routine available in MATHEMATICA or the gbasis routine available in the Symbolic Math Toolbox of MATLAB. No special software is needed except to enter the data reported in the manuscript in MATLAB or MATHEMATICA.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The author declares that he has no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12859-019-3155-0.
References
- 1.Tyson JJ, Albert R, Goldbeter A, Ruoff AP, Sible J. Biological switches and clocks. J R Soc Interface. 2008;5:S1–S8. doi: 10.1098/rsif.2008.0179.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J Cell Biol. 2004;164:353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrell JE., Jr The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280(5365):895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 4.Arkun Y, Yasemi M. Dynamics and control of the ERK signaling pathway: sensitivity, bistability, and oscillations. PLoS One. 2018;13(4):e0195513. doi: 10.1371/journal.pone.0195513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giri L, Mutalik VK, Venkatesh KVA. Steady state analysis indicates that negative feedback regulation of PTP1B by Akt elicits bistability in insulin-stimulated GLUT4 translocation. Theor Biol Med Model. 2004;1:1–16. doi: 10.1186/1742-4682-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G. Singularity analysis of the AKT signaling pathway reveals connections between cancer and metabolic diseases. Phys Biol. 2010;7:046015. doi: 10.1088/1478-3975/7/4/046015. [DOI] [PubMed] [Google Scholar]
- 7.Eissing T, Conzelmann H, Gilles ED, Allgöwer F, Bullinger E, Scheurich P. Bistability analyses of a caspase activation model for receptor-induced apoptosis. J Biol Chem. 2004;279(35):36892–36897. doi: 10.1074/jbc.M404893200. [DOI] [PubMed] [Google Scholar]
- 8.Nakakuki T, Birtwistle MR, Saeki Y, Yumoto N, Ide K, Nagashima T, et al. Ligand-specific c-fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell. 2010;141(5):884–896. doi: 10.1016/j.cell.2010.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao G, Lee TJ, Mori S, Nevins JR, You L. A bistable Rb–E2F switch underlies the restriction point. Nat Cell Biol. 2008;10(4):476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]
- 10.Verdugo A, Vinod PK, Tyson JJ, Novak B. Molecular mechanisms creating bistable switches at cell cycle transitions. Open Biol. 2013;3:120179. doi: 10.1098/rsob.120179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trotta L, Sepulchre R, Bullinger E. Global analysis of dynamical decision-making models through local computation around the hidden saddle. PLoS One. 2012;7(3):e33110. doi: 10.1371/journal.pone.0033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T, Brazhnik P, Tyson JJ. Computational analysis of dynamical responses to the intrinsic pathway of programmed cell death. Biophys J. 2009;97:415–434. doi: 10.1016/j.bpj.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bala SI, Ahmad NMR. Comp Appl Math. 2018;37:266. doi: 10.1007/s40314-017-0467-4. [DOI] [Google Scholar]
- 14.Song H, Smolen P, Av-Ron E, Baxter DA, Byrne JH. Bifurcation of singularity analysis of a molecular network for the induction of long-term memory. Biophys J. 2006;9:2309–2325. doi: 10.1529/biophysj.105.074500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez-Corral R, Liu J, Suel G, Garcia-Ojalvo J. Bistable emergence of oscillations in structured cell populations. BioRxiv. 2018. 10.1101/276113.
- 16.Fan D, Liu S, Wang Q. Epileptic stimulus-induced epileptic spike-wave discharges in thalamocortical model with disinhibition. Sci Rep. 2016;6:1–21. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dovzhenok A, Kuznetsov AS. Exploring neuronal bistability at the depolarization block. PLoS One. 2012;7(8):e42811. doi: 10.1371/journal.pone.0042811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Byrne JH, Heidelberger R, Waxham MN. (eds)From Molecules to Networks. An Introduction to Cellular and Molecular Neuroscience: Academic. Academic Press, Elsevier; 2014.
- 19.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 20.Ferrell JE, Ha SH. Ultrasensitivity part II: multisite phosphorylation, stoichiometric inhibitors, and positive feedback. Trends Biochem Sci. 2014;39(11):556–569. doi: 10.1016/j.tibs.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas R. On the relation between the logical structure of systems and their ability to generate multiple steady states or sustained oscillations. Springer Ser Synergetics. 1981;9:180–193. doi: 10.1007/978-3-642-81703-8_24. [DOI] [Google Scholar]
- 22.Craciun G, Tang Yand Feinberg M. Understanding bistability in complex enzyme-driven reaction networks. Proc Natl Acad Sci. 2006;103(23):8697–8702. doi: 10.1073/pnas.0602767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angeli D, Ferrell JE, Sontag ED. Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc Natl Acad Sci U S A. 2004;101:1822–1827. doi: 10.1073/pnas.0308265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilhelm T. The smallest chemical reaction system with bistability. BMC Syst Biol. 2009;3:90. doi: 10.1186/1752-0509-3-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feliu E, Helmer M. Multistationarity and bistability for fewnomial chemical reaction networks. M Bull Math Biol. 2019;81:1089. doi: 10.1007/s11538018-00555-z. [DOI] [PubMed] [Google Scholar]
- 26.Buchberger B. An Algorithm for Finding the Bases Elements of the Residue Class Ring Modulo a Zero Dimensional Polynomial Ideal (German) Austria: University of Innsbruck; 1965. [Google Scholar]
- 27.Buchberger B. An Algorithmical Criterion for the Solvability of Algebraic Systems of Equations (German) Aequationes Math. 1970;4(3):374–383. doi: 10.1007/BF01844169. [DOI] [Google Scholar]
- 28.Buchberger B, Winkler F. editorsGröebner Bases and Applications, volume 251 of London MATHEMATICAl Society Series, Proc. of the International Conference “33 Years of Gröebner Bases”: Cambridge University Press. London Mathematical Society Lecture Note Series; 1998.
- 29.Martínez-Forero I, Pelá Ez-Ló Pez A, Villoslada P. Steady State Detection of Chemical Reaction Networks Using a Simplified Analytical Method. PLoS One. 2010;5(6):5. doi: 10.1371/journal.pone.0010823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minimair M, Barnett MP. Solving polynomial equations for chemical problems using Gröebner bases. Mol Phys. 2004;102:2521-535.
- 31.Mercedes PM, Dickenstein A, Shiu A, Conradi C. Chemical Reaction Systems with Toric Steady States. Bull Math Biol. 2012;74:1027–1065. doi: 10.1007/s11538-011-9685-x. [DOI] [PubMed] [Google Scholar]
- 32.Grimbs S, Arnolda A, Koseskac A, Kurths J, Selbiga J, Nikoloski Z. Spatiotemporal dynamics of the Calvin cycle: Multistationarity and symmetry breaking instabilities. BioSystems. 2011;103:212–223. doi: 10.1016/j.biosystems.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Calandrini GL, Paolini EE, Moiola JL. Gröebner bases for designing dynamical systems. Lat Am Appl Res. 2003;33:4. [Google Scholar]
- 34.Zhiping L, Xu L, Bose NK. A tutorial on Gröebner bases with applications in signals and systems. IEEE Trans Circuits Syst I. 2008;55(1):445–461. doi: 10.1109/TCSI.2007.914007. [DOI] [Google Scholar]
- 35.Wenz M, Wörn H. Solving the inverse kinematics problem symbolically by means of knowledge-based and linear algebra-based methods. 2007. pp. 1346–1353. [Google Scholar]
- 36.Guevara MR. Bifurcations Involving Fixed Points and Limit Cycles in Biological Systems. In: Beuter A, Glass L, Mackey MC, Titcombe MS, editors. Nonlinear Dynamics in Physiology and Medicine. Interdisciplinary Applied Mathematics. New York: Springer; 2003. [Google Scholar]
- 37.Abraham R, Shaw CD. Dynamics-the geometry of behavior: periodic behavior: Aerial Press. Basic Books; 1982.
- 38.Qiao L, Nachbar RB, Kevrekidis IG, Shvartsman SY. Bistability and oscillations in the Huang-Ferrell model of MAPK signaling. PLoS Comput Biol. 2007;3(9):1819–1826. doi: 10.1371/journal.pcbi.0030184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cizmeci D, Arkun Y. Regulatory networks and complex interactions between the insulin and angiotensin II signaling systems: Models and implications for hypertension and diabetes. PLoS One. 2013;8(12):e83640. doi: 10.1371/journal.pone.0083640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortega F, Garcés JL, Mas F, Kholodenko BN, Cascante M. Bistability from double phosphorylation in signal transduction: kinetic and structural requirements. FEBS J. 2006;273(17):3915–3926. doi: 10.1111/j.1742-4658.2006.05394.x. [DOI] [PubMed] [Google Scholar]
- 41.Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, et al. Digital signaling and hysteresis characterize Ras activation in lymphoid cells. Cell. 2009;136(2):337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams WW, Loustaunau P. An Introduction to Gröbner Bases. Graduate Studies in Mathematics, vol. 3: American MATHEMATICAl Society. American Mathematical Society; 2000.
- 43.Winkler F. The method of Gröebner bases. In Polynomial Algorithms in Computer Algebra, Texts and Monographs in Symbolic Computation, chapter 8. Wien: Springer; 1996. [Google Scholar]
- 44.Khalil HK. Nonlinear systems: Prentice Hall. Pearson Higher Education; 2014.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this manuscript. Given the data, the Gröbner bases were calculated by calling the GroebnerBasis routine available in MATHEMATICA or the gbasis routine available in the Symbolic Math Toolbox of MATLAB. No special software is needed except to enter the data reported in the manuscript in MATLAB or MATHEMATICA.