Abstract
WEE1 kinase is a key regulator of the G2/M transition. The WEE1 kinase inhibitor AZD1775 (WEE1i) induces origin firing in replicating cells. We show that WEE1i induces CDK1-dependent RIF1 phosphorylation and CDK2- and CDC7-dependent activation of the replicative helicase. WEE1 suppresses CDK1 and CDK2 kinase activities to regulate the G1/S transition after the origin licensing is complete. We identify a role for WEE1 in cell cycle regulation and important effects of AZD1775, which is in clinical trials.
Keywords: WEE1, cell cycle, DNA replication, CDK1, RIF1
The cell cycle is regulated by oscillating concentrations of cyclins that bind cyclin-dependent kinases (CDKs) to generate “activatable” complexes. Cyclin D–CDK4/CDK6 and cyclin E–CDK2 complexes are associated with G1 and the G1/S transition, respectively, cyclin A–CDK2 with S phase, and cyclin B–CDK1 with the G2/M transition. Since the concentration of CDK1 does not oscillate, and cyclin B levels increase through S and G2, additional levels of regulation are required to generate the timely and sharp activation of cyclin B–CDK1 that triggers mitosis.
WEE1 kinase and CDC25 phosphatase compete in a bistable system that activates cyclin B–CDK1 (1, 2). WEE1 and MYT1 kinases phosphorylate and inhibit CDK1 (3, 4). CDC25 reverses these phosphorylations and activates CDK1 (5). When the concentration of cyclin B exceeds a threshold, cyclin B–CDK1 inactivates WEE1 and MYT and activates CDC25 (1, 2, 6) and this shifts the equilibrium to activate all cyclin B–CDK1. Little is known about the role of CDK1 in G1 and S.
CDK1 activity in S phase is suppressed by ATR/CHK1 signaling and that this prevents excessive origin firing and premature S/G2 transition (7, 8). CDK2/CDC7 kinase activities, responsible for replication initiation in S phase, are opposed by PP1 phosphatase, and PP1 binding to the chromatin factor RIF1 limits origin firing (9). ATR and CHK1 kinase inhibitors (ATRi and CHK1i) induce CDK1 kinase-mediated phosphorylation on RIF1 Ser2205, disrupt RIF1–PP1 binding, and induce dormant origin firing in replicating cells (8). WEE1i AZD1775 (10) induces origin firing (11) with kinetics similar to ATRi and CHK1i (12–15), but the underlying mechanism has not been described.
Results and Discussion
WEE1 Limits Dormant Origin Firing in S Phase and Prevents Replication Initiation in G1.
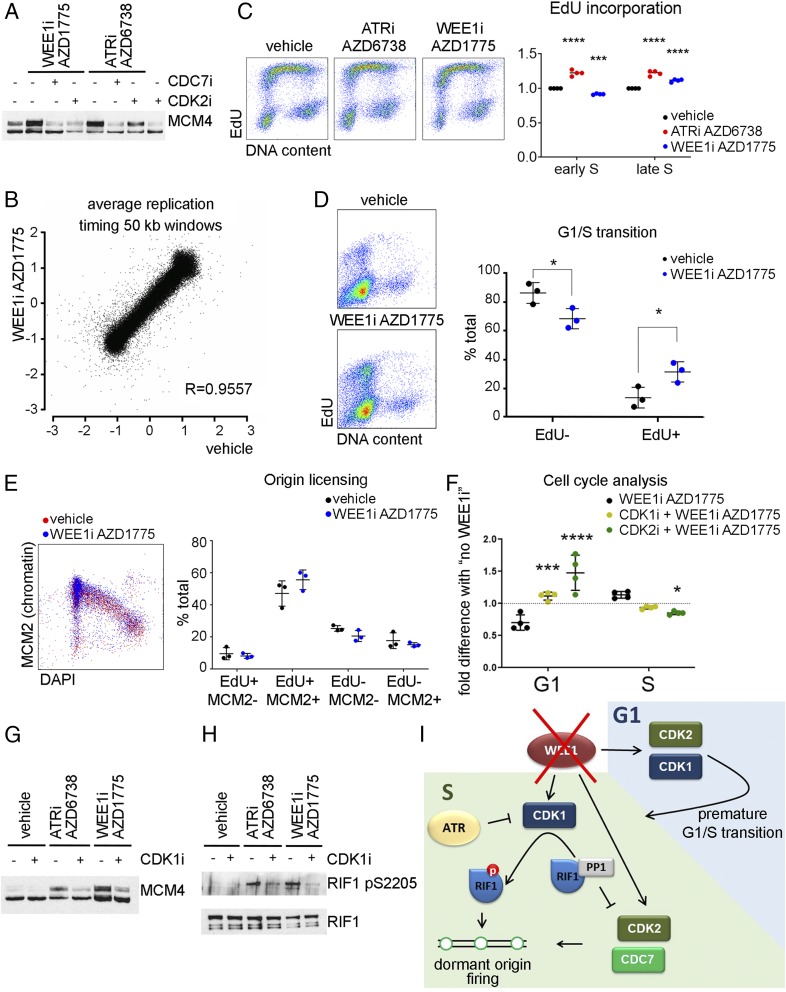
To determine whether WEE1i and ATRi (13, 16) have similar effects on replication in unperturbed cells, we treated 293T cells with WEE1i AZD1775 or ATRi AZD6738 after CDK2i CVT-313 or CDC7i PHA767491. WEE1i induced CDK2- and CDC7-dependent MCM4 hyperphosphorylation and ATRi induced CDC7-dependent MCM4 hyperphosphorylation (Fig. 1A). No statistically significant difference in the replication timing program was observed in cells treated with WEE1i (one-sided paired t test, P > 0.1), indicating that neither WEE1i nor ATRi cause late origin firing in early S (Fig. 1B) (8, 17). However, while ATRi increased DNA synthesis in early and late S equally (Fig. 1C) (8), WEE1i decreased EdU (5-ethynyl-2′-deoxyuridine) incorporation in early S (2N to 3N) and increased DNA synthesis in late S (3N to 4N) (Fig. 1C). This is attributed to WEE1i’s increasing the number of early S cells and decreasing the number of G1 cells (reduced 2N/EdU-negative and increased 2N/EdU-positive cells) (Fig. 1C). We synchronized U2OS cells and treated G1/early S-phase cells with WEE1i. This treatment induced DNA synthesis in G1 and a premature G1/S transition (Fig. 1D). DNA synthesis in cells that prematurely enter S (as well as untreated cells entering S after synchronization) is low and this decreases the total DNA synthesis observed in the early S (2N to 3N) fraction.
Fig. 1.
(A) The 293T cells were pretreated with vehicle (dimethyl sulfoxide), 10 µM CDK2i, or 10 µM CDC7i for 15 min before 5 µM ATRi or 500 nM WEE1i for 30 min. Immunoblot of the nuclease-insoluble fraction. (B) U2OS cells were treated with WEE1i for 60 min, and 5-bromodeoxyuridine was added for the last 30 min. Repli-Seq correlation between log ratio of early S to late S at LOESS smoothing step of 50-kb windows along the genome. R, Pearson correlation coefficient. (C) U2OS cells were treated with ATRi or WEE1i for 45 min, and 10 µM EdU was added for the last 15 min. Cells were fixed with ethanol and stained for EdU and DNA. (Left) Flow cytometry plots. (Right) Quantification of 4 independent experiments. (D). U2OS cells were synchronized by thymidine–nocodazole block. Three hours postrelease, WEE1i was added for 45 min, and EdU was added for the last 15 min. (Left) Flow cytometry plots. (Right) Quantification of 3 independent experiments. (E) U2OS cells were treated with WEE1i for 45 min, and EdU was added for the last 15 min. After extraction, cells were fixed and stained for chromatin-bound MCM2, EdU, and DNA content (DAPI). (Left) Flow cytometry plots. (Right) Quantification of 3 independent experiments. (F) U2OS cells were pretreated with 5 µM CDK1 or 5 µM CDK2 inhibitors for 15 min before the addition of WEE1i for 45 min, and EdU was added for the last 15 min. The effect of WEE1i on the percent of G1- or S-phase cells (normalized to samples with no WEE1i) is shown (4 independent experiments). (G) The 293T cells were pretreated with 5 µM CDK1i for 15 min before the addition of ATRi or WEE1i for 30 min. Immunoblot of the nuclease insoluble fraction. (H) The 293T cells were transfected with Rif1-GFP. Forty-eight hours after transfection, cells were pretreated with CDK1i for 15 min before the addition of ATRi or WEE1i for 1 h. RIF1 was immunoprecipitated using green fluorescent protein trap beads. (I) Cartoon depicting the roles of WEE1 in controlling replication initiation. *P < 0.05, ***P < 0.0005, ****P < 0.0001.
WEE1i Does Not Induce DNA Synthesis Prior to the Completion of Origin Licensing.
The MCM2-7 hexamer is loaded onto DNA in G1 to license origins (18). A licensing checkpoint ensures that MCM loading is complete before the G1/S transition (19). In order to check whether WEE1 induces DNA synthesis in G1 before licensing is complete, we quantified the association of MCM with chromatin. WEE1i did not induce DNA synthesis in underlicensed cells (Fig. 1E), indicating that WEE1 has no role in the licensing checkpoint.
WEE1 Suppresses Both CDK1 and CDK2 Activities to Extend G1 Phase after Origin Licensing.
CDK1 and CDK2 kinases are the main targets of WEE1. The G1/S transition is regulated by cyclin E–CDK2 (20). Since CDK1 kinase is actively suppressed in S (7, 8), we hypothesized that CDK1 may act with CDK2 to induce a premature G1/S transition in cells treated with WEE1i. We treated U2OS cells with CDK1i or CDK2i for 15 min and then added WEE1i for 45 min. DNA synthesis was labeled with EdU for the last 15 min of treatment. As expected, CDK2i delayed the G1/S transition (Fig. 1F). However, CDK1i also blocked the WEE1i-induced decrease in G1 cells and increase in S-phase cells (Fig. 1F), revealing a role for CDK1 in the premature G1/S transition. WEE1i- and ATRi-induced MCM4 hyperphosphorylation is CDK1-mediated (Fig. 1G). Moreover, WEE1i and ATRi induced RIF1 phosphorylation on Ser2205 (Fig. 1H). Therefore, WEE1i induces dormant origin firing through a mechanism similar to ATRi and CHK1i (8).
We have identified an intrinsic G1/S checkpoint enforced by WEE1 that is similar to the recently described intrinsic S/G2 checkpoint enforced by ATR (7). Our data and recent reports (7, 8) suggest that CDK1 drives the entire cell cycle with different mechanisms suppressing CDK1 activities in each stage: WEE1 in G1, WEE1/ATR in S, and ATR at the S/G2 transition. Accordingly, WEE1i and ATRi/CHK1i increase origin firing and this is associated with fork stalling and extensive regions of single-stranded DNA in cells that have yet to be treated with a DNA-damaging agent (13). These effects should be considered in the design and interpretation of clinical trials.
Materials and Methods
Antibodies.
Antibodies included MCM4 (3228; Cell Signaling), RIF1 (A300-568A; Bethyl), RIF1 pS2205 (8), and MCM2 (610700; BD Biosciences).
Chemicals.
Chemicals included AZD6738 and AZD1775 (AstraZeneca), Ro-3306 and PHA767491 (Selleckchem), and CVT-313 (Santa Cruz).
Cell culture, immunoblotting, Repli-Seq, and EdU fluorescence-activated cell sorting were as described (8, 13). Repli-Seq data have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE138998).
Acknowledgments
This work was supported by NIH grants R01 CA204173 (C.J.B.), R00 CA207871 (H.U.O.), and P30CA047904.
Footnotes
The authors declare no competing interest.
Data deposition: Repli-Seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE138998).
References
- 1.Sha W., et al. , Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc. Natl. Acad. Sci. U.S.A. 100, 975–980 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pomerening J. R., Sontag E. D., Ferrell J. E. Jr, Building a cell cycle oscillator: Hysteresis and bistability in the activation of Cdc2. Nat. Cell Biol. 5, 346–351 (2003). [DOI] [PubMed] [Google Scholar]
- 3.McGowan C. H., Russell P., Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 12, 75–85 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu F., Stanton J. J., Wu Z., Piwnica-Worms H., The human Myt1 kinase preferentially phosphorylates Cdc2 on threonine 14 and localizes to the endoplasmic reticulum and Golgi complex. Mol. Cell. Biol. 17, 571–583 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann I., Clarke P. R., Marcote M. J., Karsenti E., Draetta G., Phosphorylation and activation of human cdc25-C by cdc2–cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO J. 12, 53–63 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deibler R. W., Kirschner M. W., Quantitative reconstitution of mitotic CDK1 activation in somatic cell extracts. Mol. Cell 37, 753–767 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saldivar J. C., et al. , An intrinsic S/G2 checkpoint enforced by ATR. Science 361, 806–810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moiseeva T. N., et al. , An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication. Proc. Natl. Acad. Sci. U.S.A. 116, 13374–13383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamazaki S., et al. , Rif1 regulates the replication timing domains on the human genome. EMBO J. 31, 3667–3677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirai H., et al. , Small-molecule inhibition of Wee1 kinase by MK-1775 selectively sensitizes p53-deficient tumor cells to DNA-damaging agents. Mol. Cancer Ther. 8, 2992–3000 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Beck H., et al. , Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol. Cell. Biol. 32, 4226–4236 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couch F. B., et al. , ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27, 1610–1623 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moiseeva T., et al. , ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat. Commun. 8, 1392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petermann E., Woodcock M., Helleday T., Chk1 promotes replication fork progression by controlling replication initiation. Proc. Natl. Acad. Sci. U.S.A. 107, 16090–16095 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Syljuåsen R. G., et al. , Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25, 3553–3562 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiraga S. I., et al. , Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep. 18, 403–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moiseeva T. N., Qian C., Sugitani N., Osmanbeyoglu H. U., Bakkenist C. J., The effect of WEE1 inhibition on the replication timing program. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138998. Deposited 16 October 2019.
- 18.O’Donnell M., Langston L., Stillman B., Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 5, a010108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shreeram S., Sparks A., Lane D. P., Blow J. J., Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene 21, 6624–6632 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann I., Draetta G., Karsenti E., Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 13, 4302–4310 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]