Significance
Host immunity exerts strong selection pressure on viruses, often leading to the evolution of immune antagonists in viral genomes. In insects, this is exemplified by antiviral RNA interference (RNAi), a major antiviral defense system to which many viruses have evolved suppressor proteins. Yet, the physiological functions of viral suppressors of RNAi remain incompletely understood, as experimental systems to manipulate the viral genome of interest are often lacking. Here we generate an RNAi suppressor-defective mutant of a complex DNA virus, invertebrate iridescent virus-6. Using this mutant virus, we found that RNAi suppressor proteins can effectively and completely block the antiviral immune system of the host.
Keywords: RNAi, viral suppressor of RNAi, insect immunity, insect virus, antiviral defense
Abstract
Coevolution of viruses and their hosts may lead to viral strategies to avoid, evade, or suppress antiviral immunity. An example is antiviral RNA interference (RNAi) in insects: the host RNAi machinery processes viral double-stranded RNA into small interfering RNAs (siRNAs) to suppress viral replication, whereas insect viruses encode suppressors of RNAi, many of which inhibit viral small interfering RNA (vsiRNA) production. Yet, many studies have analyzed viral RNAi suppressors in heterologous systems, due to the lack of experimental systems to manipulate the viral genome of interest, raising questions about in vivo functions of RNAi suppressors. To address this caveat, we generated an RNAi suppressor-defective mutant of invertebrate iridescent virus 6 (IIV6), a large DNA virus in which we previously identified the 340R protein as a suppressor of RNAi. Loss of 340R did not affect vsiRNA production, indicating that 340R binds siRNA duplexes to prevent RNA-induced silencing complex assembly. Indeed, vsiRNAs were not efficiently loaded into Argonaute 2 during wild-type IIV6 infection. Moreover, IIV6 induced a limited set of mature microRNAs in a 340R-dependent manner, most notably miR-305–3p, which we attribute to stabilization of the miR-305–5p:3p duplex by 340R. The IIV6 340R deletion mutant did not have a replication defect in cells, but was strongly attenuated in adult Drosophila. This in vivo replication defect was completely rescued in RNAi mutant flies, indicating that 340R is a bona fide RNAi suppressor, the absence of which uncovers a potent antiviral immune response that suppresses virus accumulation ∼100-fold. Together, our work indicates that viral RNAi suppressors may completely mask antiviral immunity.
Viruses often evade, avoid, or suppress antiviral immunity to ensure efficient replication and transmission. For example, such interactions have led to viral strategies that inhibit antigen presentation, prevent interferon production, and suppress antiviral effector functions in mammals (1). Likewise, viruses of plants and insects may encode suppressors of the antiviral RNA interference (RNAi) response (2, 3). In antiviral RNAi in insects, viral double-stranded RNA (dsRNA) is processed into viral small interfering RNAs (vsiRNAs) by the nuclease Dicer-2. These small interfering RNAs (siRNAs) are incorporated into Argonaute 2, the catalytic component of the RNA-induced silencing complex (RISC), where they guide the recognition of complementary viral RNA, resulting in target RNA cleavage by the nuclease activity of Argonaute-2 (2, 3).
Most, if not all, viruses produce dsRNA during their life cycle (4, 5), and it is thus likely that RNAi has antiviral activity across a broad range of viruses. Indeed, vsiRNAs are produced during infections with RNA viruses with single-stranded genomes (both of positive and negative polarity), as well as viruses with dsRNA genomes (2). In these cases, intermediates of replication or the viral genomic RNA are the substrates for vsiRNA production. Hence, Drosophila and mosquito mutants with defects in RNAi genes, such as Dicer-2 (Dcr-2) and Argonaute 2 (AGO2), are hypersensitive to positive-sense and negative-sense RNA viruses, and dsRNA viruses, with higher virus-induced mortality rates in RNAi mutants than in wild-type (WT) controls (6–11). As a consequence, many viruses evolved antagonists of antiviral RNAi, which seems to be a convergent trait that emerged independently in multiple viral taxa (2, 3).
Although DNA viruses do not replicate through a dsRNA intermediate, dsRNA is produced due to the relative compactness of their genomes producing overlapping, converging transcripts or to the production of (noncoding) RNAs that fold into duplex hairpin RNA structures (4, 12). In agreement, vsiRNAs have been detected during infections with insect DNA viruses from several families including iridoviruses, baculoviruses, and nudiviruses (10, 13–18), and, for those cases analyzed, mortality rates are higher in RNAi mutants than in control flies (10, 13, 19). Yet, these observations should be interpreted with care because of the reduced stress resistance and shorter life span of RNAi mutant flies (20), especially given the relatively long time required for significant virus-induced mortality to occur during these infections. Moreover, increased mortality in RNAi mutant flies was associated with only minor (10, 13) or transient (19) effects on viral DNA accumulation. This is in striking contrast to RNA virus infections, during which differences in mortality rates are often associated with strong increases in viral titers in RNAi mutant flies (6–9). These observations may be explained by a relatively modest antiviral efficacy of RNAi against DNA viruses, vsiRNA-mediated silencing of specific viral genes that modulate pathogenicity but not replication per se, or potent viral antagonism of the effector stage of RNAi downstream of vsiRNA production.
Viral suppressors of RNAi have indeed been identified in DNA viruses of insects. For example, p35 of the baculovirus Autographa californica multiple nucleopolyhedrovirus was proposed as an inhibitor of both apoptosis and RNAi (18). In addition, we previously showed that the 340R protein of invertebrate iridescence virus-6 (IIV6; genus Iridovirus, family Iridoviridae) has potent RNAi suppressive activity in vitro and in cellular RNAi reporter assays (21). 340R contains a canonical dsRNA-binding domain (dsRBD, IPR014720) and, using biochemical assays, we found that the protein suppresses RNAi by binding dsRNA and siRNA, preventing their cleavage and loading into Argonaute 2, respectively.
In this study, we generated an RNAi suppressor-defective mutant of IIV6 (Δ340R), allowing us to study the in vivo importance of a virus-encoded RNAi suppressor in the context of a DNA virus infection in Drosophila. We found that IIV6 Δ340R does not have major replication defects in cells, but that replication is strongly attenuated in flies, which is rescued in an RNAi mutant background. Moreover, small RNA analyses indicate that 340R does not affect vsiRNA production but sequesters siRNA duplexes. In addition, we found that an asymmetrically loaded microRNA (miRNA) (miR-305) is stabilized as a miRNA 5p:3p duplex, which occurred in a miRNA sequence-dependent manner. Our results indicate that an RNAi suppressor protein can completely block the antiviral activity of RNAi immunity, shielding the phenotype of loss-of-function mutations in WT virus infections. Furthermore, our results indicate that RNAi suppressor proteins can sequester cellular miRNAs in a sequence-specific manner at the posttranscriptional level.
Results
An IIV6 Mutant Lacking Its RNAi Suppressor Protein Does Not Have a Replication Defect In Vitro.
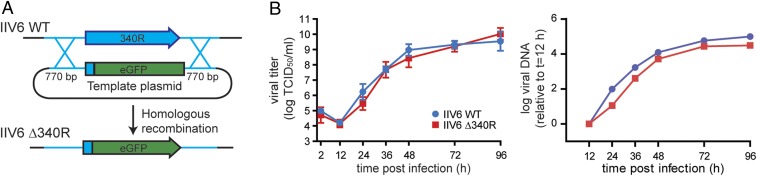
We used homologous recombination to generate the mutant IIV6 in which the 340R-coding sequence was replaced by green fluorescent protein (GFP) (IIV6 Δ340R; Fig. 1A and SI Appendix, Supplementary Text and Fig. S1). We assessed replication kinetics of WT and Δ340R IIV6 in Drosophila S2 cells by qPCR to assess intracellular viral DNA accumulation and by end-point dilution to assess production of infectious virus in the supernatant. We found that both IIV6 WT and Δ340R replicated efficiently in S2 cells, showing similar kinetics and producing similar titers, although intracellular DNA levels were slightly lower for IIV6 Δ340R over the time course of the experiment (Fig. 1B). These observations indicate that, although vsiRNAs are produced in S2 cells (10), the IIV6 mutant lacking its proposed RNAi suppressor 340R does not have a major replication defect in cell culture.
Fig. 1.
No replication defect in vitro of IIV6 lacking its RNAi suppressor protein 340R. (A) Strategy to generate an RNAi suppressor-defective IIV6 mutant (IIV6 Δ340R). The recombination template plasmid contained the GFP transgene fused to 14 N-terminal amino acids of 340R, flanked by homology arms at both the 5′ and 3′ ends. (B) Replication kinetics of IIV6 WT and Δ340R in Drosophila S2 cells. Titers (Left) are presented as means ± SD of 3 biological replicates. qPCR data (Right) were normalized to housekeeping gene rp49 and presented relative to the 12 hpi time point (log-transformed data). qPCR data are means of 2 technical replicates.
340R Does Not Affect the Accumulation of vsiRNAs.
Our previous study indicates that 340R can bind both long dsRNA and siRNAs to prevent dsRNA processing into siRNAs and to prevent siRNA loading into Argonaute-2, respectively (21). To establish the relative importance of both activities in the context of IIV6 infection, we analyzed small RNA profiles in IIV6 WT and Δ340R-infected S2 cells (22). If long dsRNA binding and inhibition of Dicer-2 function is the predominant mechanism, increased vsiRNA production is anticipated in IIV6 Δ340R infection. If siRNA binding and inhibition of RISC loading is the predominant mechanism, no major difference in vsiRNA levels is expected.
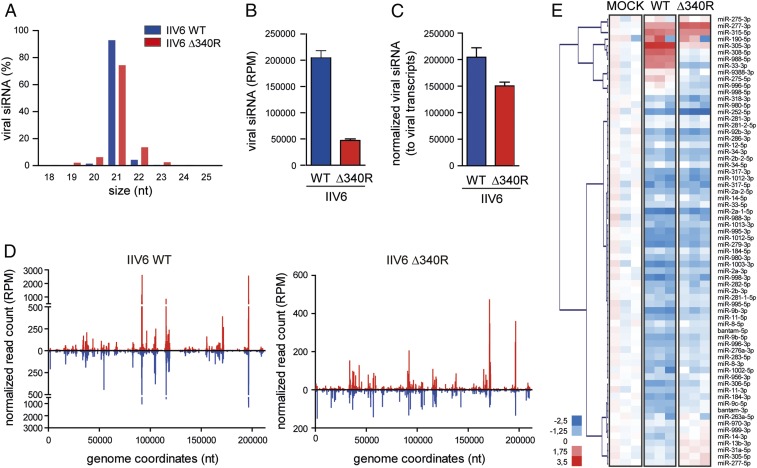
As observed before (13), virus-derived small RNAs were predominantly 21 nucleotides (nt) in length, the typical size of Dicer-2 products, in both WT and Δ340R IIV6 infection, although the sizes seem to be less precise with more 20- and 22-nt reads in mutant IIV6 infection (Fig. 2A). The 21-nt vsiRNAs were 4.3-fold less abundant in IIV6 Δ340R than in IIV6 WT infection (Fig. 2B), which reflects the slightly reduced intracellular viral DNA levels in IIV6 Δ340R infection (Fig. 1B). We thus normalized vsiRNA levels to viral transcripts (average of 4 transcripts, individually presented in SI Appendix, Fig. S2A) and found only a modest reduction in vsiRNA levels in IIV6 Δ340R infection (Fig. 2C). Similar results were obtained when normalizing total vsiRNAs or gene-specific vsiRNAs to individual transcripts (SI Appendix, Fig. S2B). Moreover, the distribution of vsiRNA across the viral genome was similar in WT and mutant IIV6 infection (Fig. 2D). Together, these data suggest that 340R does not suppress the production of vsiRNAs in the context of an infection.
Fig. 2.
Small RNA profiles in WT and Δ340R IIV6 in Drosophila S2 cells. (A) Size profile of virus-derived small RNAs from IIV6 WT and Δ340R-infected S2 cells at 72 hpi. Viral small RNAs were mapped to the IIV6 genome, allowing 1 mismatch, and their size distribution as the percentage of total viral small RNAs is presented. Bars are mean ± SD of 3 independent libraries. (B) Number of viral 21-nt siRNAs presented as mean ± SD per million total reads (RPM). (C) Numbers of viral siRNAs normalized to library size and to relative viral transcript levels in the samples used for sequencing (averaged over 4 viral genes) (mean ± SD of 3 independent libraries). (D) Distribution of 21-nt vsiRNAs across the IIV6 genome, with vsiRNAs mapping to the R and L strands of the genome in red and blue, respectively. The average counts (3 experiments) of 5′ ends of small RNA reads at each nucleotide position are indicated. (E) Heatmap of the changes in miRNA abundance in IIV6 WT and 340R-infected S2 cells. Three libraries are presented separately. Hierarchical clustering is based on the averages of the 3 libraries. Color coding indicates log2-transformed fold-changes relative to mock infection.
IIV6 Induced a Small Set of miRNAs in an 340R-Dependent Manner.
In plants, viral suppressors of RNAi affect cellular miRNAs, which may be explained by convergence of antiviral RNAi and miRNA pathways on AGO1 (23). In contrast, the miRNA and siRNA pathways in Drosophila rely predominantly on dedicated Dicer and Argonaute proteins, Dicer-1 and AGO1 for the miRNA pathway and Dicer-2 and AGO2 for the siRNA pathway (24, 25). We thus analyzed whether IIV6 infection affects cellular miRNAs and the role of 340R therein. For the majority of cellular miRNAs, we observed a reduction in their numbers in both IIV6 WT and Δ340R infection (Fig. 2E). In contrast, 2 smaller subsets of miRNAs were induced upon IIV6 infection. The first group, represented by miR-277–3p and miR-315–5p, were induced (>1.5-fold) by both IIV6 WT and Δ340R infection. The other, most intriguing group, consisting of miR-33–3p, miR-988–5p, miR-308–5p, and miR-305–3p, was induced by IIV6 WT infection (range: 3.1- to 8.2-fold), but not by IIV6 Δ340R infection, suggesting 340R-dependent miRNA induction. Intriguingly, the mature miRNA derived from the other arm of these pre-miRNAs was not enhanced, but even reduced, upon IIV6 infection (range: 0.3- to 0.7-fold). Among this group, miR-305–3p was most abundant and showed the strongest induction (8.2-fold). Using an independent, publicly available dataset from IIV6-infected S2 cells from another laboratory (10), we confirmed that miR-305–3p, but not miR-305–5p, was induced by IIV6 infection (SI Appendix, Fig. S3). Together, our small RNA profiles indicate that 340R does not affect vsiRNA production, but strongly induces the levels of a select set of mature miRNAs, most notably, miR-305–3p.
Accumulation of miR-305–3p in IIV6-Infected Cells.
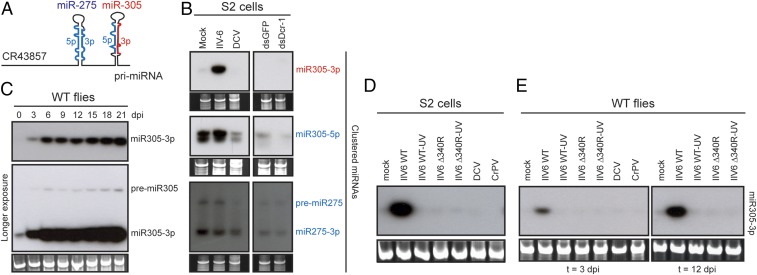
miR-305 seemed to be expressed along with miR-275 from a single, spliced primary miRNA (CR43857, Fig. 3A and SI Appendix, Fig. S4) (26, 27). To confirm our observations from small RNA sequencing, we analyzed expression of miR-305–3p, along with other miRNAs expressed from the same cluster (miR-305–5p, miR-275) as well as a series of additional miRNAs by Northern blot analyses in IIV6-infected S2 cells. IIV6 infection induced strong accumulation of miR-305–3p, whereas the 5p arm seems to be only slightly more abundant than in mock infected cells (Fig. 3B). In contrast, miR-275–3p and pre–miR-275 were expressed at lower levels in IIV6-infected cells. As these miRNAs are expressed from the same primary transcript, these data suggest that IIV6 affects miR-305–3p expression at the posttranscriptional level. This does not seem to be due to a generalized effect on miRNAs, as expression of 7 other miRNAs was reduced upon IIV6 infection (SI Appendix, Fig. S5 A and B). In cells infected with Drosophila C virus, used as a control in these experiments, all miRNAs including miR-305–3p were less abundant. This is likely due to cytopathic effects and RNA degradation, as can be appreciated from the ribosomal RNA (rRNA) images.
Fig. 3.
Increased expression of a mature miRNA in IIV6-infected cells. (A) Schematic representation of the primary miRNA transcript encoding miR-275 and miR-305. See SI Appendix, Fig. S6E, for miRNA sequence and predicted structure. (B) Northern blot analyses of the clustered miR-275 and miR-305 in Drosophila S2 cells infected with IIV6 at 3 dpi. As controls, noninfected cells treated with dsRNA targeting Dicer-1 (dsDcr-1) or GFP (dsGFP, as a control) were run in parallel. rRNA was used as a loading control. (C) Northern blot analyses of miR-305–3p over the course of IIV6 infection in WT flies (genotype, w1118). (Lower) A longer exposure of the same blot, allowing visualization of the pre-miRNA. (D) miR-305–3p levels in S2 cells at 3 dpi and (E) adult female flies (w1118) at 3 and 12 dpi with the indicated viruses.
We next validated these results in adult flies infected with IIV6 by intrathoracic injection and found a strong induction of miR-305–3p over a time course of 21 d post infection (dpi) (Fig. 3C). In contrast to findings in S2 cells, miR-305–3p induction was accompanied by a slight increase in pre–miR-305 expression, which seemed insufficient to explain the strongly increased miR-305–3p levels. Moreover, induction of miR-305–3p was also induced in flies with defects in the RNAi genes AGO2 and Dicer-2 (SI Appendix, Fig. S5C) and required active replication (Fig. 3 D and E). As observed in small RNA sequence data, miR-305–3p was not induced by the IIV6 Δ340R mutant (Fig. 3 D and E). Unexpectedly, 340R alone seems to be required and sufficient for enhanced miR-305–3p levels as assessed after transfection of an expression plasmid in S2 cells (SI Appendix, Fig. S5D). In contrast, 340R mutants with defects in dsRNA binding [K86A, K89A (21)] did not induce miR-305–3p, nor did other insect virus RNAi suppressors with demonstrated dsRNA-binding activity (Drosophila C virus 1A, Drosophila X virus VP3, Culex Y virus VP3, mosinovirus B2, and Flock House virus B2) (6, 28–30) (SI Appendix, Fig. S5D).
To validate these observations using an independent method, we set up a stemloop RT-qPCR assay to quantify expression of both 5p and 3p arms of the clustered miR-305 and miR-275, as well as miR-2b-1–5p, miR-995–5p, and bantam-3p as controls (SI Appendix, Fig. S6 A and B). In line with our previous observations, we found that miR-305–3p was strongly enhanced by IIV6 WT infection (128.5-fold at 12 dpi in WT flies), but not by IIV6 Δ340R infection (1.1-fold at 12 dpi). In contrast, no or low induction was observed for the other miRNAs tested (range: 1.1- to 6.4-fold) (SI Appendix, Fig. S6 C and D). Together, these data indicate that IIV6 infection results in strongly enhanced expression of miR-305–3p and slightly enhanced levels of miR-305–5p. Surprisingly, viral RNAi suppressor 340R is required and sufficient for these effects.
340R Efficiently Stabilizes miR-305 5p:3p Duplexes.
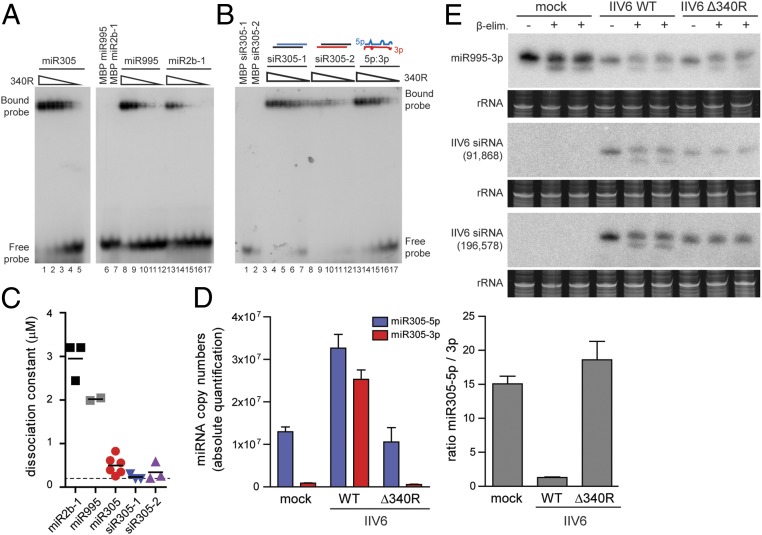
The 340R is a dsRNA-binding protein (21), and our data indicate that residues involved in dsRNA binding are required to alter miR-305–3p levels. We thus hypothesized that 340R may bind miRNA 5p:3p duplexes in a sequence-specific manner. To test this hypothesis, we compared 340R binding to miR-305 and 2 control miRNAs, miR-995 and miR-2b-1 (sequences in SI Appendix, Fig. S6E), by electrophoretic mobility shift assays (EMSAs) using recombinant 340R fused to maltose-binding protein (MBP). We found that 340R binds all tested miRNA duplexes, but that the affinity for miR-305 duplexes is 5.9- and 8.6-fold higher than for miR-2b-1 and miR-995 (P < 0.001 and P = 0.002, respectively, Student’s t test) Fig. 4 A and C). Residues known to interact with dsRNA, K86 and K89, were required for miRNA duplex binding (SI Appendix, Fig. S7A, lanes 8 to 12), and no binding was observed to the mature, single-stranded RNAs miR-305–5p and miR-305–3p (SI Appendix, Fig. S7B, lanes 8 to 12 and 13 to 17), suggesting that 340R uses its canonical dsRBD for miRNA duplex binding.
Fig. 4.
340R efficiently binds miR-305 and siRNA duplexes. (A) EMSA of 5p:3p duplexes of miR-305, miR-995, and miR-2b-1 with recombinant 340R. (B) EMSA of the miR-305 5p:3p duplex (lanes 13 to 17) and miR-305–derived siRNAs with 340R. The siRNAs were designed by annealing miR-305–5p or miR-305–3p with perfectly complementary RNA to generate siRNAs with 2-nt 3′ overhangs (siR305-1 and siR305-2, respectively; lanes 3 to 7 and lanes 8 to 12). Two-fold dilutions of recombinant protein were tested, starting from a concentration of 3.2 μM. Incubations in buffer only (−) and MBP were included as controls, as indicated. (C) Dissociation constants of 340R for the indicated miRNA and siRNA duplexes. Each symbol represents an independent experiment. Horizontal lines indicate means. The dashed line is the lowest concentration tested. (D) Absolute quantification of miR-305–5p and miR-305–3p by stemloop-qPCR in mock infected cells and in IIV6 WT and Δ340R-infected S2 cells at 72 hpi. For each miRNA, a standard curve of synthetic RNA was used for quantification. Expression of the miRNAs was normalized to expression of the U6 small nuclear RNA. (E) Northern blot analysis of miR-995–3p and 2 IIV6-derived siRNAs (positions in the genome indicated) in total RNA of Drosophila S2 cells infected with IIV6 WT or Δ340R at 3 dpi. Where indicated, the samples were subjected to β-elimination (+) or mock treated (−), with 2 technical replicates on the same RNA for the β-elimination reaction. Ethidium-bromide–stained rRNA was used as a loading control.
We previously observed that 340R binds both dsRNA and siRNAs in EMSAs (21). We thus compared 340R binding affinity for the miR-305 duplex to its affinity for 2 siRNAs, designed as either miR-305–5p or miR-305–3p annealed to a fully complementary RNA with 2-nt overhangs at each 3′ end (siR305-1 and siR305-2, respectively). This experiment indicated that, although 340R efficiently binds miR-305 duplexes, affinity for duplex RNA without bulges is higher (Fig. 4 B and C, average dissociation constants of 0.495 μM for miR-305 5p:3p and <0.236 μM and <0.342 μM for siR305-1 and siR305-2, respectively).
These data indicate that 340R has high specificity for miR-305, which led us to hypothesize that it binds siRNAs as well as miR-305 duplexes and stabilizes them in vivo to prevent their incorporation in RISC. The increased levels of miR-305–3p in IIV6 WT, but not IIV6 Δ340R, infection, would then be due to stabilization of an otherwise asymmetrically loaded miRNA duplex. To test this hypothesis, we quantified absolute levels of each miRNA strand in mock and IIV6 (WT and Δ340R) infection. In line with our earlier findings (SI Appendix, Fig. S6), strongly enhanced levels of miR-305–3p and slightly enhanced levels of miR-305–5p were observed in IIV6 WT infection (29.4- and 2.5-fold over mock infection, respectively), which reverted to mock levels in IIV6 Δ340R infection (Fig. 4 D, Left). Importantly, the miR-305 5p-to-3p ratios were highly skewed in mock and IIV6 Δ340R infection (ratios of 15.1 and 18.6, respectively), but neared 1 in IIV6 WT infection (ratio 1.3, Fig. 4 D, Right) consistent with our hypothesis of miRNA duplex stabilization.
Our model that miR-305 duplexes are sequestered before AGO1 loading predicts that miR-305 function is suppressed during IIV6 infection. To analyze this, we generated reporters with target sites for miR-305–5p and miR-305–3p in the 3′ UTR of firefly luciferase and found that the miR-305–5p target site resulted in a modest, but consistent, reduction in luciferase activity (mean: 66% of scrambled control) (SI Appendix, Fig. S8 A and B). In IIV6 WT infection, however, miR-305–5p silencing was reduced compared to IIV6 Δ340R infection, confirming that IIV6 interferes with miR305 function in a 340R-dependent manner (SI Appendix, Fig. S8C). In contrast to the 5p reporter, the miR-305–3p target site did not induce silencing (SI Appendix, Fig. S8D), likely due to low expression of this miRNA in noninfected cells. Moreover, also in IIV6-infected cells, the miR305-3p reporter was not silenced compared to a scrambled control, consistent with the idea that this miRNA is induced in a nonfunctional form during infection.
To directly test whether 340R interferes with AGO2 loading, we analyzed sensitivity to β-elimination of viral siRNAs in IIV6 WT and Δ340R infection. In Drosophila, once incorporated in AGO2, siRNAs are 2′O-methylated at the 3′ terminal ribose by Hen1, which renders them resistant to β-elimination (31). We found that a large fraction of vsiRNAs in IIV6 WT infection are sensitive to β-elimination, as evident from the increased mobility on gel, indicating that they have not been loaded into AGO2. In contrast, vsiRNAs in IIV6 Δ340R infection are completely resistant to β-elimination, indicating that they are associated with AGO2 (Fig. 4E). miRNAs that are generally loaded into AGO1 lack this modification and are thus sensitive to β-elimination (31). Indeed, a control miRNA, miR-995–3p, was sensitive to β-elimination (Fig. 4E), albeit the reaction was incomplete, preventing us from drawing quantitative conclusions from this experiment. Together, these data are in line with our proposal that IIV6 340R binds siRNAs and selected miRNA duplexes to prevent their association with RISC, thus suppressing antiviral RNAi and interfering with the function of specific miRNAs.
340R Completely Masks the Antiviral Activity of RNAi.
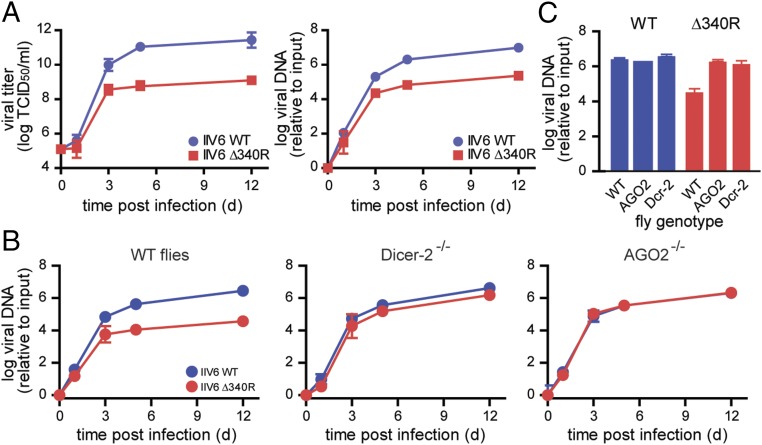
IIV6 Δ340R did not have a major replication defect in cells, indicating that 340R is not required for the basal replication machinery of the virus. To assess the importance of 340R in vivo, we injected IIV6 WT and Δ340R intrathoracically into adult Drosophila and monitored viral titers and DNA levels over time. A consistent and strong replication defect was observed, with 310.6- and 41.7-fold lower viral titers and DNA levels, respectively, at 12 dpi (Fig. 5A). Importantly, this replication defect was completely absent in flies defective in Dcr-2 and AGO2: WT and Δ340R replicated with similar kinetics and accumulated to similar levels in RNAi mutant flies (Fig. 5B). Again, in WT flies analyzed in parallel in the same experiment, IIV6 Δ340R accumulated to 74.2-fold lower levels than WT IIV6.
Fig. 5.
IIV6 Δ340R has a severe, RNAi-dependent replication defect in vivo. (A) Replication kinetics of IIV6 WT and Δ340R in WT (w1118) flies, as assessed by end-point dilution (Left) and intracellular DNA quantification (Right). qPCR data were normalized to the housekeeping gene rp49 and presented relative to a sample taken directly after inoculation (input). The data were log-transformed and presented as means and SD of 3 biological replicates of 5 flies. (B) Replication kinetics of IIV6 WT and Δ340R in WT (w1118), Dicer-2, and AGO2 mutant flies. The experiment was performed as described in A. (C) Data from 12 dpi from B were plotted and grouped by virus genotype (other time points in SI Appendix, Fig. S10).
To analyze which of the activities of 340R—sequestering miR-305 duplexes or preventing vsiRNA loading into AGO2—is dominant in vivo, we performed genetic rescue experiments in which we analyzed viral accumulation in flies lacking miR-305. We used 2 transheterozygous combinations of miR-275–305 alleles to generate flies with reduced miRNA expression (6-fold reduction for miR-305–5p and 6.7-fold reduction for miR-305–3p compared to control flies, SI Appendix, Fig. S9A) or flies completely lacking these miRNAs. In these flies, IIV6 Δ340R replicated to 1.9- and 5.9-fold higher levels at 3 dpi and 4.5- and 7.1-fold higher levels at 12 dpi, relative to control flies (SI Appendix, Fig. S9B). Together, these results suggest that 340R interferes with a putative antiviral activity of miR305, but that its major function is to suppress antiviral RNAi.
In a previous study, we observed that IIV6 WT accumulated to only slightly higher levels in RNAi mutant flies than in WT control flies (13), which argues against a strong antiviral effect of RNAi to this DNA virus. Grouping our current data separately for each virus genotype, we confirmed that, in the context of IIV6 WT infection, AGO2 and Dcr-2 mutant flies do not accumulate higher viral levels than WT flies (Fig. 5C and SI Appendix, Fig. S10). In sharp contrast, IIV6 Δ340R accumulated to 55.4- and 40.6-fold higher levels in RNAi-defective AGO2 and Dcr-2 mutant flies, respectively, than in WT flies. These data indicate that the viral RNAi suppressor can completely block the antiviral activity of RNAi and thus mask the virus hypersensitivity phenotype of RNAi mutant flies.
Discussion
Facilitated by their large coding capacity, coevolution of large (insect) DNA viruses with their hosts led to the evolution of virus-encoded antagonists or modulators of diverse immune pathways including RNAi, NF-κB pathways, and apoptosis (18, 19, 21, 32, 33). Here we use an RNAi suppressor-defective mutant of IIV6 to show that 340R sequesters vsiRNAs to prevent the effector mechanism of RNAi. This suppressor is sufficiently potent to completely block the antiviral function of RNAi, thus masking the virus sensitivity phenotype of RNAi-deficient flies. Furthermore, we find that the RNAi suppressor binds a highly selective set of miRNA duplexes in a sequence-specific manner and inhibits their function.
RNAi suppressor proteins have been identified in over 10 insect-specific viruses, most of which are RNA viruses. Due to the lack of an experimental system to manipulate the viral genomes of interest, RNAi suppressor proteins are often characterized in isolation upon transgenic expression, using recombinant proteins, or in heterologous systems (8, 21, 28, 34, 35). By generating an RNAi suppressor-defective virus, our study indicates that the in vivo function of RNAi suppressor proteins may differ from that deduced from such studies. Specifically, whereas our previous results indicated that 340R binds both long dsRNA and vsiRNAs to counteract antiviral RNAi (21), our current data indicate that the in vivo function of 340R is to sequester vsiRNAs to prevent their incorporation into RISC.
The use of an RNAi suppressor-defective mutant virus allowed us to establish the antiviral potential of RNAi against a DNA virus. Whereas in the context of WT IIV6, our previous data suggested that RNAi is not strongly antiviral against DNA viruses (13), our current data indicate that RNAi is in fact an extremely potent antiviral mechanism against DNA viruses. An important implication of our work, therefore, is that a virus-encoded immune antagonist can veil the phenotype of a bona fide antiviral pathway. Thus, the antiviral potency against IIV6 can only be truly appreciated when using an RNAi suppressor-defective virus mutant. This is different from observations with insect RNA viruses, in which hypersensitivity phenotypes of RNAi mutants are apparent also when using WT viruses (e.g., refs. 6–9). This notion may extend to other immune pathways in which putative virus-encoded immune antagonists may mask transcriptional responses or survival and viral load phenotypes of immune-deficient mutants. For example, absence of a viral load phenotype in Toll pathway mutants upon infection with Kallithea virus, a DNA virus from the Nudivirus family, may be explained by its virus-encoded suppressor of the Toll pathway (19).
An unexpected observation from our study is that IIV6 modifies expression of a select set of cellular miRNAs in an RNAi suppressor-dependent manner. Viral manipulation of the cellular miRNA repertoire has been reported before for DNA viruses, such as herpesviruses that encode their own miRNAs to modulate the host (reviewed in ref. 36). In addition, several herpesviruses encode a viral (noncoding) RNA that induces the degradation of a specific cellular miRNA, miR-27 (37–40). Additionally, poxviruses manipulate the host-cell miRNA repertoire in both insects and mammals, through a mechanism in which the viral poly(A) polymerase adds nontemplated adenosines (tailing) to miRNAs to induce their degradation (41). Our data provide an additional mechanism by which DNA viruses manipulate cellular miRNAs at the posttranscriptional level. The 340R binds 5p:3p duplex miRNAs in a sequence-specific manner, thereby sequestering both mature arms of miR-305 to prevent their functions. A window of opportunity to do so is provided by the (on average) ∼1-h delay between miRNA duplex production and RISC loading (42).
It is somewhat unexpected that 340R binds miRNA duplexes in a sequence-specific manner, as dsRBD interactions with dsRNA were initially proposed to be sequence nonspecific (43–45). Yet, the dsRBD-containing proteins Staufen in Drosophila and ADAR proteins (adenosine deaminase acting on RNA) in mice exert sequence specificity, which, in the case of ADAR, is based on the individual dsRBDs (46–48). In particular, dsRNA structure distortions, such as bulges observed in miRNA duplexes, widen the major groove of dsRNA, offering the opportunity to interact in a sequence-specific manner (49). Moreover, the dsRBD in 340R is flanked by 30 and 73 amino acids at its N and C terminus, respectively, and it is possible that these extensions contribute to or mediate miRNA specificity.
Although the main function of 340R is to bind vsiRNA to inhibit antiviral RNAi, our data suggest that miR-305 sequestration does impact virus accumulation. In this respect, it is intriguing that miR-305 is evolutionarily conserved among insects. It seems to play a role in immunity in the mosquito species Anopheles gambiae (50) and has been reported to be dynamically regulated upon dengue and chikungunya virus infection of Aedes mosquitoes (51, 52). Moreover, the miRNA-275–305 cluster has been proposed to be under posttranscriptional control in noninfected flies, suggesting that expression of these miRNAs requires precise control (53). Which miR-305 target genes necessitate a dedicated mechanism for miRNA sequestration remains to be defined. Of interest in this regard are 2 well-characterized targets of miR-305–5p, PI3K (phosphatidyl-inositol-3-kinase) and p53 (26, 54), which are often exploited and modulated by viruses for effective replication (55, 56).
Materials and Methods
Please refer to SI Appendix, Materials and Methods for a complete description of the materials and methods used in this study. Oligonucleotide sequences are provided in SI Appendix, Table S1. Homologous recombination was used to produce IIV6 Δ340R. Fifty nanoliters of undiluted virus stock (IIV6 WT: titer 6.34 × 108 TCID50/mL) or an equivalent of the 340R mutant virus was injected into the thorax of adult flies (57). Small RNA sequencing libraries were prepared from Drosophila S2 cells (Invitrogen) infected with IIV6 WT or Δ340R mutant virus at a multiplicity of infection of 1. RNA was isolated at 72 h post infection (hpi) using RNA-Solv reagent (Omega Biotek).
Data Availability.
Small RNA data have been submitted to the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA574259.
Supplementary Material
Acknowledgments
We thank current and past members of the laboratory for fruitful discussions, especially Koen van Cleef for advice on gene deletion in DNA viruses. We thank Carla Saleh (Pasteur Institute) for hosting Drosophila injections in her laboratory, and Kathryn Rozen-Gagnon (Rockefeller University) for discussions. This work was financially supported by a fellowship from the Radboud Institute for Molecular Life Sciences (Radboud University Medical Center) and by a Consolidator Grant from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (ERC CoG 615680). Sequencing was performed by the GenomEast platform, a member of the “France Génomique” consortium (ANR-10-INBS-0009).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA574259).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909183116/-/DCSupplemental.
References
- 1.Randall R. E., Goodbourn S., Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89, 1–47 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Bronkhorst A. W., van Rij R. P., The long and short of antiviral defense: Small RNA-based immunity in insects. Curr. Opin. Virol. 7, 19–28 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Guo Z., Li Y., Ding S. W., Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 19, 31–44 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Weber F., Wagner V., Rasmussen S. B., Hartmann R., Paludan S. R., Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80, 5059–5064 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Son K. N., Liang Z., Lipton H. L., Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA viruses. J. Virol. 89, 9383–9392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Rij R. P., et al. , The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 20, 2985–2995 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dostert C., et al. , The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila. Nat. Immunol. 6, 946–953 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Wang X. H., et al. , RNA interference directs innate immunity against viruses in adult Drosophila. Science 312, 452–454 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller S., et al. , RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 19390–19395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemp C., et al. , Broad RNA interference-mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 190, 650–658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuel G. H., Wiley M. R., Badawi A., Adelman Z. N., Myles K. M., Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc. Natl. Acad. Sci. U.S.A. 113, 13863–13868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathews M. B., Shenk T., Adenovirus virus-associated RNA and translation control. J. Virol. 65, 5657–5662 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bronkhorst A. W., et al. , The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. U.S.A. 109, E3604–E3613 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabin L. R., et al. , Dicer-2 processes diverse viral RNA species. PLoS One 8, e55458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma M., et al. , Discovery of DNA viruses in wild-caught mosquitoes using small RNA high throughput sequencing. PLoS One 6, e24758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster C. L., et al. , The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 13, e1002210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayachandran B., Hussain M., Asgari S., RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 86, 13729–13734 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mehrabadi M., Hussain M., Matindoost L., Asgari S., The baculovirus antiapoptotic p35 protein functions as an inhibitor of the host RNA interference antiviral response. J. Virol. 89, 8182–8192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer W. H., et al. , Induction and suppression of NF-κB signalling by a DNA virus of Drosophila. J. Virol. 93, e01443-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim D. H., et al. , The endogenous siRNA pathway in Drosophila impacts stress resistance and lifespan by regulating metabolic homeostasis. FEBS Lett. 585, 3079–3085 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Bronkhorst A. W., van Cleef K. W., Venselaar H., van Rij R. P., A dsRNA-binding protein of a complex invertebrate DNA virus suppresses the Drosophila RNAi response. Nucleic Acids Res. 42, 12237–12248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bronkhorst A. W., et al. , Small RNA sequencing of Invertebrate iridescent virus 6 WT and Δ340R in Drosophila melanogaster cells. National Center for Biotechnology Information Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA574259. Deposited 26 September 2019.
- 23.Csorba T., Kontra L., Burgyán J., Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 479–480, 85–103 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Okamura K., Ishizuka A., Siomi H., Siomi M. C., Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 18, 1655–1666 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee Y. S., et al. , Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Foronda D., Weng R., Verma P., Chen Y. W., Cohen S. M., Coordination of insulin and Notch pathway activities by microRNA miR-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes Dev. 28, 2421–2431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berezikov E., et al. , Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 21, 203–215 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Cleef K. W., et al. , Mosquito and Drosophila entomobirnaviruses suppress dsRNA- and siRNA-induced RNAi. Nucleic Acids Res. 42, 8732–8744 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster S., et al. , A unique nodavirus with novel features: Mosinovirus expresses two subgenomic RNAs, a capsid gene of unknown origin, and a suppressor of the antiviral RNA interference pathway. J. Virol. 88, 13447–13459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Li W. X., Ding S. W., Induction and suppression of RNA silencing by an animal virus. Science 296, 1319–1321 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Horwich M. D., et al. , The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Clem R. J., Viral IAPs, then and now. Semin. Cell Dev. Biol. 39, 72–79 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Lamiable O., et al. , Cytokine Diedel and a viral homologue suppress the IMD pathway in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 113, 698–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayak A., et al. , Cricket paralysis virus antagonizes Argonaute 2 to modulate antiviral defense in Drosophila. Nat. Struct. Mol. Biol. 17, 547–554 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Mierlo J. T., et al. , Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog. 8, e1002872 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kincaid R. P., Sullivan C. S., Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathog. 8, e1003018 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Libri V., et al. , Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. U.S.A. 109, 279–284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cazalla D., Yario T., Steitz J. A., Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 328, 1563–1566 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buck A. H., et al. , Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 16, 307–315 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcinowski L., et al. , Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 8, e1002510 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backes S., et al. , Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe 12, 200–210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichholf B., et al. , Time-resolved small RNA sequencing unravels the molecular principles of microRNA homeostasis. Mol. Cell 75, 756–768.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryter J. M., Schultz S. C., Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17, 7505–7513 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blaszczyk J., et al. , Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure 12, 457–466 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Bevilacqua P. C., Cech T. R., Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35, 9983–9994 (1996). [DOI] [PubMed] [Google Scholar]
- 46.Higuchi M., et al. , Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Ferrandon D., Elphick L., Nüsslein-Volhard C., St Johnston D., Staufen protein associates with the 3'UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221–1232 (1994). [DOI] [PubMed] [Google Scholar]
- 48.Liu Y., Lei M., Samuel C. E., Chimeric double-stranded RNA-specific adenosine deaminase ADAR1 proteins reveal functional selectivity of double-stranded RNA-binding domains from ADAR1 and protein kinase PKR. Proc. Natl. Acad. Sci. U.S.A. 97, 12541–12546 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian B., Bevilacqua P. C., Diegelman-Parente A., Mathews M. B., The double-stranded-RNA-binding motif: Interference and much more. Nat. Rev. Mol. Cell Biol. 5, 1013–1023 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Dennison N. J., BenMarzouk-Hidalgo O. J., Dimopoulos G., MicroRNA-regulation of Anopheles gambiae immunity to Plasmodium falciparum infection and midgut microbiota. Dev. Comp. Immunol. 49, 170–178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Campbell C. L., Harrison T., Hess A. M., Ebel G. D., MicroRNA levels are modulated in Aedes aegypti after exposure to Dengue-2. Insect Mol. Biol. 23, 132–139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shrinet J., Jain S., Jain J., Bhatnagar R. K., Sunil S., Next generation sequencing reveals regulation of distinct Aedes microRNAs during chikungunya virus development. PLoS Negl. Trop. Dis. 8, e2616 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryazansky S. S., Gvozdev V. A., Berezikov E., Evidence for post-transcriptional regulation of clustered microRNAs in Drosophila. BMC Genomics 12, 371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrio L., Dekanty A., Milán M., MicroRNA-mediated regulation of Dp53 in the Drosophila fat body contributes to metabolic adaptation to nutrient deprivation. Cell Rep. 8, 528–541 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Diehl N., Schaal H., Make yourself at home: Viral hijacking of the PI3K/Akt signaling pathway. Viruses 5, 3192–3212 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sato Y., Tsurumi T., Genome guardian p53 and viral infections. Rev. Med. Virol. 23, 213–220 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Merkling S. H., van Rij R. P., Analysis of resistance and tolerance to virus infection in Drosophila. Nat. Protoc. 10, 1084–1097 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Small RNA data have been submitted to the National Center for Biotechnology Information Sequence Read Archive under accession number PRJNA574259.