Significance
Seasonal molts to winter-white pelage or plumage are key adaptations to seasonal snow in over 20 animal species. However, winter color polymorphism within species is crucial for adaptation to environmental heterogeneity and provides striking models to tackle the basis of repeated evolution. We show that the pigmentation gene Agouti, which underlies introgression-driven winter color variation in snowshoe hares, is also associated with winter-white/winter-grey polymorphism in mountain hares, possibly due to altered expression of the gene. The winter-grey variant introgressed into the mountain hare through hybridization with a noncolor changing species. These findings show that the same genomic region was repeatedly recruited to determine winter coat color in different hare species, highlighting the recurrent role of introgression in generating phenotypic variation.
Keywords: seasonal coat color change, agouti signaling protein gene, Lepus timidus, genotype-phenotype
Abstract
Changing from summer-brown to winter-white pelage or plumage is a crucial adaptation to seasonal snow in more than 20 mammal and bird species. Many of these species maintain nonwhite winter morphs, locally adapted to less snowy conditions, which may have evolved independently. Mountain hares (Lepus timidus) from Fennoscandia were introduced into the Faroe Islands in 1855. While they were initially winter-white, within ∼65 y all Faroese hares became winter-gray, a morph that occurs in the source population at low frequency. The documented population history makes this a valuable model for understanding the genetic basis and evolution of the seasonal trait polymorphism. Through whole-genome scans of differentiation and single-nucleotide polymorphism (SNP) genotyping, we associated winter coat color polymorphism to the genomic region of the pigmentation gene Agouti, previously linked to introgression-driven winter coat color variation in the snowshoe hare (Lepus americanus). Lower Agouti expression in the skin of winter-gray individuals during the autumn molt suggests that regulatory changes may underlie the color polymorphism. Variation in the associated genomic region shows signatures of a selective sweep in the Faroese population, suggesting that positive selection drove the fixation of the variant after the introduction. Whole-genome analyses of several hare species revealed that the winter-gray variant originated through introgression from a noncolor changing species, in keeping with the history of ancient hybridization between the species. Our findings show the recurrent role of introgression in generating winter coat color variation by repeatedly recruiting the regulatory region of Agouti to modulate seasonal coat color change.
Recurrent evolution has puzzled evolutionary biologists for decades (1, 2). While controlled experiments testing the repeated nature of evolution are possible in only a few models, comparative evolutionary studies of similar traits that may have evolved independently in closely related taxa provide powerful means to understand recurrent evolution at different levels, from processes to phenotypes and their molecular basis (3–5). Seasonal pelage or plumage color change is a key adaptive phenological trait of over 20 vertebrates, such as hares, weasels, and grouse, which inhabit environments with seasonal snow cover (6). Photoperiod-controlled pelage molts from summer-brown to winter-white coats allow the maintenance of camouflage year-round, reducing the fitness costs of increased predation due to coat-background color mismatch (7–9). However, genetically determined winter coat color polymorphism exists in many of these species, maintained by clinal selective pressures correlated with snow cover environmental variables (10). The occurrence of winter coat color variation across species provides valuable models to dissect pathways for recurrent phenotypic evolution. Also, such polymorphism may provide standing variation to enable rapid adaptation to snow cover reductions caused by climate change, which endangers winter-white populations (7, 10). Understanding the emergence of seasonal coat color polymorphisms is therefore crucial for quantifying the adaptive potential of seasonal coat color changing species facing climate change (10, 11).
Of the >30 species of hares (Lepus spp.), 6 undergo seasonal coat color molts, often maintaining within-species variation in winter pelage color (10). In snowshoe hares (Lepus americanus), winter-white/winter-brown polymorphism maps to the agouti signaling protein gene (ASIP; Agouti) and is driven by cis-regulatory changes influencing the expression of the gene (11). The winter-brown variant introgressed from the neighboring black-tailed jackrabbit (Lepus californicus), a winter-brown species, seeding adaptation to the warmer coastal habitat with less winter snow in the Pacific Northwest region in North America. Winter variation in coat color has also been studied in arctic foxes (Vulpes lagopus), where melanocortin 1 receptor gene (MC1R) mutations induce the expression of a blue-gray winter phenotype instead of the predominant winter-white (12). These results implicate the MC1R-Agouti melanin pigmentation system, often involved in permanent pelage color variation in mammals (13), in the evolution of seasonal changes in coloration. However, little is known about the evolution of seasonal coat color variation across species.
Here, we study the genetic basis and evolution of winter coat color polymorphism, using the winter-white/winter-gray coat color variation in mountain hares (Lepus timidus). In most of the species’ range, mountain hares molt to winter-white coats, but alternative winter color morphs occur in the species. For example, in Ireland mountain hares remain brown year-round, and in Fennoscandia a winter-gray morph exists (i.e., the heath-hare; also called blue-gray) in addition to the predominant winter-white morph (Fig. 1 A and B) (14, 15). Winter-gray mountain hares were common in southern Sweden but have become progressively rarer since the early 20th century, likely due to a combination of predation by the red fox (Vulpes vulpes), habitat degradation, and competition with the introduced European brown hare (Lepus europaeus) (16). In 1855, 4 mountain hare leverets were translocated from the Kragerø region in southern Norway to Tórshavn in the Faroe Islands, where no hares were present, and they multiplied rapidly (14, 17). Initially only winter-white hares were observed in the Faroe Islands, but years later winter-gray hares began to be seen, and by the 1870s hunting bags were composed by an equal number of winter-white and winter-gray hares. The percentage of winter-gray hares increased to 75% in 1882 and 95% in 1890, and the last winter-white hare was observed in 1916–1917 (18). Breeding experiments suggest that the winter-gray coat is inherited as a recessive trait (14), implying that the individuals that founded the Faroese hare population carried the causal allele. The rapid fixation of winter-gray morphs may have resulted from adaptation to the milder snow conditions of the Faroe Islands, influenced by the warm waters of the North Atlantic Current (17). Using whole-genome sequencing data, we map winter coat color polymorphism to a single genomic region and show that the winter-gray variant was introduced into the mountain hare through introgression.
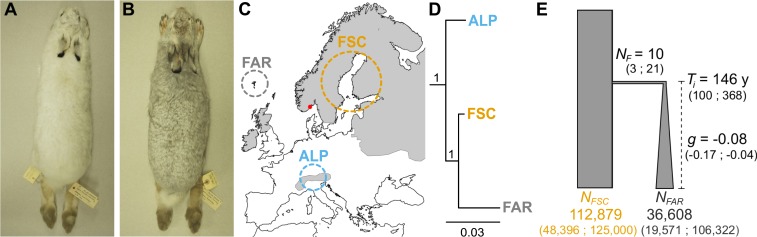
Fig. 1.
Mountain hare winter coat color morphs, sampling, and evolutionary relationships among populations. (A) Winter-white morph (specimen NRM588859, Swedish Museum of Natural History [NRM]). (B) Winter-gray morph (NRM588861, NRM). Pictures reproduced with permission of NRM. (C) Distribution of the mountain hare in Europe (gray area) and approximate sampling localities of individuals used in whole-genome analyses (SI Appendix, Table S1): ALP, the Alps (n = 20); FAR, the Faroe Islands (n = 20); FSC, Fennoscandia (n = 19). The red dot indicates the region of origin of the hares translocated to the Faroe Islands. (D) Population tree based on allele frequencies, with bootstrap supports. (E) Parameter estimates for the demographic history of Faroese hares: N, effective population sizes (number of diploid individuals) of the Fennoscandian (FSC), Faroese (FAR), and founder (F) populations; Ti, time of the introduction (years; assuming 2 y per generation); g, growth rate (negative value backward-in-time implies expansion forward-in-time) according to NF = NFAR egTi; mode of estimated parameters is shown and 95% HPD intervals are in parentheses (SI Appendix, Table S4).
Results and Discussion
Sequencing, Population Structure, and Evolutionary History.
Whole genomes of 59 mountain hare specimens were sequenced at low individual coverage from individually barcoded libraries. Three populations were sampled: hares from the Faroe Islands (winter-gray; n = 20 individuals; 16.8× population depth), Fennoscandia (the source population of the Faroese hares; winter-white; n = 19; 18.0×), and the Alps (winter-white; n = 20; 17.1×) (Fig. 1C and SI Appendix, Tables S1 and S2). One Faroese hare was sequenced at higher depth (6.9×; SI Appendix, Table S3). Reads were mapped to a hare pseudoreference genome (97% mapped reads on average per specimen; SI Appendix, Table S2), built through iterative mapping to the European rabbit (Oryctolagus cuniculus) genome (from ref. 19; ref. 20). Principal component analysis of genetic variation (123,995 single-nucleotide polymorphisms, SNPs) showed 3 groups, corresponding to the geographic origin of specimens (SI Appendix, Fig. S1A). Phylogenies based on population allele counts (136,834 sites) (Fig. 1D) and on individual pairwise differences (SI Appendix, Fig. S1B) showed that the Faroese mountain hares were closely related to Fennoscandian hares and formed a single clade, in agreement with the historical information about the introduction (14). Using Approximate Bayesian computation, we fitted a scenario compatible with the historical reports, but with wide priors to allow parameter inference (Fig. 1E). The historical records of the time of introduction and the size of the founding Faroese population are within the inferred 95% high posterior density intervals (Fig. 1E and SI Appendix, Figs. S2 and S3 and Table S4).
Association Region Includes Pigmentation Gene Agouti.
To identify genomic regions with exceptional differentiation in winter-gray Faroese hares, a genome scan of the population branch statistic (PBS) was performed, based on genotype likelihoods after filtering sites to represent a minimum of 6 individuals per population. Given 3 structured populations and the closer relationship of Faroese and Fennoscandian hares (Fig. 1D and SI Appendix, Fig. S1), this statistic localizes strong allele frequency changes specific to Faroese hares. The scan identified consecutive outlier 20-kb windows (top 0.01% of genome-wide values) in chromosome 4, and a few in chromosome 15 (Fig. 2A). These results were concordant with FST scans, Fisher exact tests of allele frequency differences (based on 14,102,734 SNPs, derived from pooled population data), and case-control association tests (8,129,498 SNPs; estimated from genotype likelihoods) (SI Appendix, Fig. S4). More than 700 variants with extreme allele frequency differences between Faroese and the other populations were found along a ∼360-kb region in chromosome 4. In these SNPs, the allele fixed in Faroese hares segregates at low frequencies in Fennoscandian hares and is generally absent in Alpine hares, as predicted by the geographic distribution of winter-gray morphs (14). This genomic region includes the pigmentation gene Agouti as well as EIF2S2, RALY, and AHCY genes, which have no known functions in pigmentation (Fig. 2B). An overlapping genomic interval has been implicated in winter coat color variation in the snowshoe hare (11). The second genomic region, with a few windows of increased differentiation, maps to chromosome 15 and is located ∼40 kb upstream of USP38. This gene has no known direct function in pigmentation but belongs to the ubiquitin proteasome system, which is involved in the regulation of skin pigmentation through endogenous degradation of tyrosinase (21).
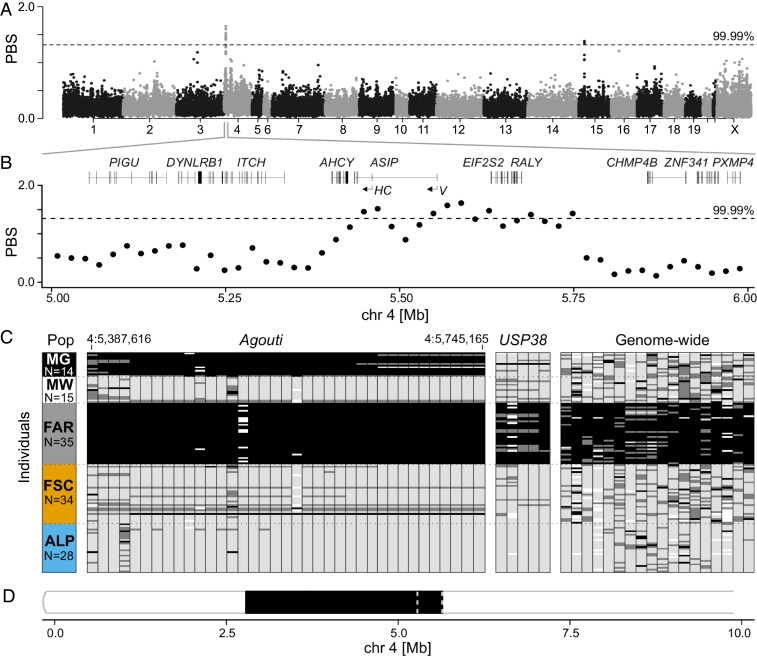
Fig. 2.
The candidate association region for winter-white/winter-gray coat color polymorphism in mountain hares. (A) Genome scan of PBS for Faroese hares in 20-kb nonoverlapping windows. (B) Zoomed-in view of PBS in the chromosome 4 candidate region and gene structure (noncoding exons of Agouti are marked as HC, hair-cycle specific isoform; V, ventral isoform). The dashed line represents the 0.01% genome-wide cutoff. (C) Genotypes at 60 loci spanning the Agouti (chromosome 4) and USP38 (chromosome 15) candidate regions, and other windows of high PBS along the genome (SI Appendix, Tables S6 and S7 and Fig. S5). Rows depict specimens and columns indicate genotyped loci (coordinates of first and last SNPs in the Agouti region are indicated). Black, homozygous for the Faroese variant; light gray, homozygous for the alternative variant; dark gray, heterozygous; white, missing data; ALP, Alps; FAR, Faroe Islands; FSC, Fennoscandia; MG, winter-gray museum specimens; MW, winter-white museum specimens. (D) Selective sweep detected by Pool-hmm in the Faroese hares on the first 10 Mb of chromosome 4; dashed lines delimit the association region (5.40–5.76 Mb).
To validate the association and examine allelic segregation patterns, we genotyped 59 SNPs showing large allele frequency differences between Faroese and other hare populations, located in windows of extreme PBS. These SNPs spanned the candidate regions in chromosomes 4 (36 SNPs; including Agouti and neighboring genes) and 15 (5 SNPs in the USP38 region), and some were scattered along the genome (18 SNPs). In addition, a ∼1.2-kb deletion, detected in the Agouti region of the Faroese hare genome using reads from the specimen sequenced at higher depth, was genotyped (SI Appendix, Fig. S5 and Tables S3, S5, and S6). Sampling was extended from 59 to 126 specimens (SI Appendix, Tables S6 and S7) and included 29 museum specimens sampled in Sweden and Russia (15 white and 14 gray; Swedish Natural History Museum; specimen codes in SI Appendix, Table S1) for which the winter coat color had been recorded (e.g., Fig. 1 A and B). The genome scans were performed in highly structured populations, which could result in the inference of spurious associations, so genotyping of white and gray museum specimens originating from overlapping localities (SI Appendix, Table S1) was carried out in order to provide an independent test of association, devoid of population structure. Thirty-six loci genotyped in the Agouti region were significantly associated with winter coat color in the museum specimens (recessive model, P < 0.05 and codominant model, P < 0.001). A ∼180-kb block (chromosome 4, 5,419,007–5,599,413), encompassing part of AHCY and the whole Agouti, showed the strongest association. In contrast, the remaining genotyped SNPs, including those from the USP38 region, showed no association with winter coat color in the museum specimens, although they did confirm the strong allele frequency differences between Faroese and other mountain hares (Fig. 2C and SI Appendix, Table S7). Two SNPs in the Agouti region (chromosome 4, positions 5,457,584 and 5,460,551) were perfectly associated with winter coat color in the museum specimens (Fig. 2C). In both, winter-gray hares were homozygous for the derived allele and the ancestral state was conserved at deep phylogenetic levels (SI Appendix, Table S8). This suggests that the winter-gray coat is inherited as a recessive trait, in agreement with breeding experiments (14). In the strongest association block, perfect association across genotyped loci was only disrupted in 1 specimen that was classified as winter-gray and heterozygous for most SNPs (Fig. 2C and SI Appendix, Table S6). While this suggests incomplete dominance of white over gray (14), it is also possible that the specimen was collected while in the process of molting to white.
Down-Regulation of Agouti Hair-Cycle Isoform in Faroese Hares.
Our results show an association between the Agouti genomic region and white/gray winter color variation. The agouti signaling protein antagonizes the melanocortin 1 receptor, shifting melanogenesis to the production of lighter phaeomelanin or inhibiting pigment production (22), and has been implicated in lighter permanent pigmentation in other organisms (13, 23). Moreover, cis-regulatory variation in Agouti has been associated with the determination of white or brown winter coats in snowshoe hares, where the winter-white allele is dominant and causes increased expression of Agouti relative to winter-brown during the autumn molt (11). In Faroese hares, the winter-gray coat is formed by a mixture of white hairs with short black tips and dark hairs with short white bands (SI Appendix, Fig. S6). No associated variants were found in the Agouti coding exons, and we hypothesized that the gray coat could be caused by an overall down-regulation of the Agouti hair cycle isoform (expressed on the dorsum; ref. 24) during the autumn molt.
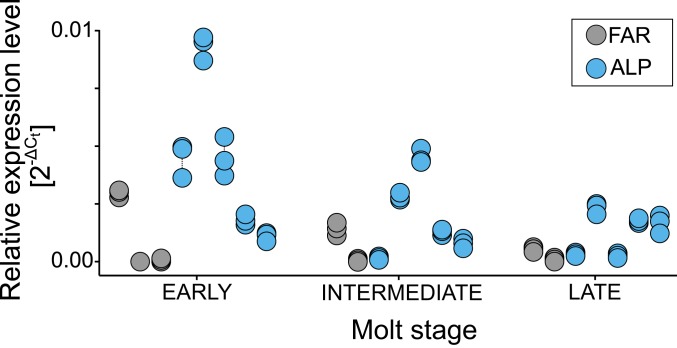
We quantified the expression of Agouti hair-cycle isoform in the skin of mountain hares undergoing the autumn molt, sampled in the Faroe Islands (n = 3 individuals) and the Alps (n = 5), normalized by the expression of reference genes (SI Appendix, Table S9). Three skin types were sampled in each specimen, representing 3 stages—early molt (brown coat patch), ongoing molt (intermediate), and late molt (white or gray coat patch)—to capture the expression changes during the molt (following 24). Notwithstanding the expected variance in quantitative measures of gene expression in a small number of animals sampled in the wild, and batch effects that could not be controlled for, our results suggest lower expression of Agouti in winter-gray hares (P = 0.028, Aligned Rank Transform) (Fig. 3 and SI Appendix, Fig. S7), most notably at the early stage of the molt when melanogenesis is active (25). These results indicate that, as in snowshoe hares (11), cis-regulatory changes down-regulating Agouti expression during the molt may cause nonwhite winter coats in mountain hares. These regulatory modifications should be confirmed with controlled essays, either by using a larger number of captive winter-gray and winter-white hares sampled at 1 time, or by estimating allele-specific gene expression in heterozygous individuals (as in ref. 11). Cis-regulatory evolution at single genes emerges as a mechanism responsible for repeated phenotype modifications (26, 27), which may result from fewer pleiotropic effects of such mutations (28).
Fig. 3.
Agouti hair-cycle isoform expression in mountain hares during the autumn molt (skin sampled at early, intermediate, and late molt per specimen, following 24). The expression level (2-ΔCt) is shown relative to reference gene ACTB (see additional analyses in SI Appendix, Fig. S7). Points represent relative measures and dashed lines connect technical replicates. ALP, winter-white Alpine mountain hares; FAR, winter-gray Faroese mountain hares.
Evidence of a Selective Sweep in Faroese Hares.
Our genotyping confirmed that the winter-gray variant segregates in Fennoscandian mountain hares (Fig. 2C), suggesting that the founder individuals of the Faroese population carried at least 1 copy of the causal allele, which became fixed after ∼65 y (14, 18). We looked for signals of selective sweeps along chromosome 4 in Faroese hares to understand whether fixation may have been driven by positive selection. Indeed, Agouti is at the edge of a long region (chromosome 4: 3.0–5.8 Mb) characterized by lower nucleotide diversity and Tajima’s D in Faroese hares than in Alpine and Fennoscandian hares and the rest of the Faroese hare genome (P < 0.001, 100,000 random samples; SI Appendix, Fig. S8 A and B). A selective sweep in Faroese hares but not in Fennoscandian or Alpine hares was confirmed by using a window-free Hidden Markov Model approach suitable for pooled data (Fig. 2D and SI Appendix, Figs. S8 C and D and S9 A and B) and a composite likelihood-ratio approach derived from genotype likelihoods (SI Appendix, Figs. S8E and S9 C and D). This signal remained when controlling for the demographic history of the Faroese hares using simulated data (SI Appendix, Fig. S8C). Given the stronger sweep signal at Agouti neighboring sites (SI Appendix, Fig. S8C), we cannot fully exclude other selection targets, but in nonequilibrium situations such as the introduction bottleneck (29) and with heterogeneous landscapes of recombination (30), the inferred sweeps may not be centered around the target of selection. Together with the PBS (Fig. 2 A and B), our results suggest that positive selection underlies the fixation of the winter-gray morph in the Faroese hares. In agreement with this hypothesis, forward demographic simulations suggested a low probability of fixation for an initially rare variant after 65 y of neutral evolution (P < 0.05; SI Appendix, Table S10). The translocation of mountain hares to an environment with less snow and with pressure for crypsis on the rocky screes imposed by intensive hunting by humans (31) may have led to the rapid adaptation from standing genetic variation determining winter color.
The Winter-Gray Variant Introgressed into the Mountain Hare.
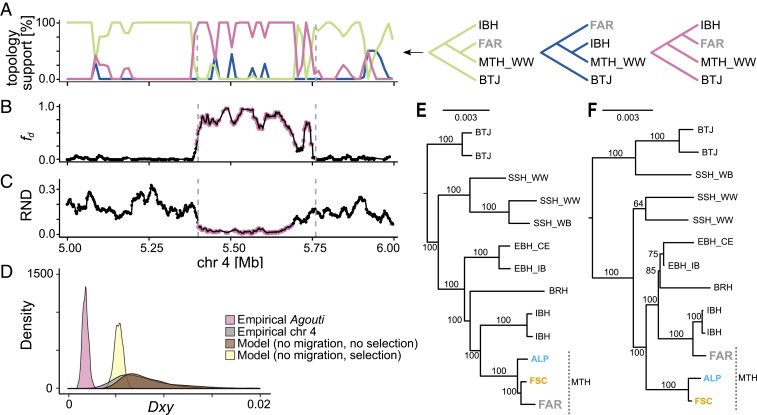
Our results suggest an overlapping genetic and functional basis of winter coat color polymorphism in mountain and snowshoe hares. We investigated the evolutionary origin of the concerned variants by combining higher individual coverage (6.1×–29.3×) whole-genome sequencing data from 6 hare species from western Europe (mountain hares, European brown hares, Iberian hares—Lepus granatensis—and the broom hare—Lepus castroviejoi) and North America (snowshoe hares and black-tailed jackrabbits) (from this work and refs. 11, 19, and 32; refs. 33–38) (SI Appendix, Table S3). In snowshoe hares, the winter-brown variant introgressed from the neighboring black-tailed jackrabbit, so the local Agouti phylogeny differs from the species tree (11). We detected a topological change at the Agouti association region (Fig. 4A and SI Appendix, Fig. S10), grouping the Faroese hares with the Iberian hares and not with other mountain hares. Maximum-likelihood phylogenies contrasting the whole chromosome 4 and the association region confirmed these results, and showed, as expected, grouping of the winter-brown snowshoe hares and black-tailed jackrabbits at Agouti (Fig. 4 E and F). The discordant local Agouti phylogeny suggests introgression from the Iberian hare. This hypothesis was further supported by patterns of variation at Agouti, namely (i) increased sharing of derived variants between Iberian hares and Faroese mountain hares (fd statistic; P < 0.001, 100,000 random samples) (Fig. 4B), and (ii) reduced scaled genetic distance (relative node depth [RND]; P < 0.001, 100,000 random samples) (Fig. 4C). Modeling the divergence between mountain and Iberian hares (SI Appendix, Table S11) and simulating the expected absolute sequence divergence (dXY) showed that reduced dXY in Agouti cannot be explained by incomplete lineage sorting alone, even if ancestral polymorphism had been reduced to a minimum by an extreme selective sweep before speciation (Fig. 4D). Collectively, these results show that the winter-gray variant introgressed from the Iberian hare into the mountain hare. Although the species are not currently in contact, genetic exchange during postglacial contact in southern Europe led to vast introgression into the Iberian hare genome (19). We found that part of the genomic block determining the winter-gray morph is also shared with the European brown hare from the Iberian Peninsula, but not with that from Central Europe (SI Appendix, Fig. S11). These results point to Iberia as the arena of the introgressive hybridization events, and to the Iberian hare as the source of the winter-gray allele. Still, we cannot fully exclude stepwise introgression pathways, for example with the brown hare, a species that is in contact with and hybridizes with both the Iberian hare (39) and the mountain hare (40), acting as a vehicle and transmitting the variant to the mountain hare (SI Appendix, Fig. S12).
Fig. 4.
The evolution of the winter-gray variant of the mountain hare: signatures of introgression from the Iberian hare. (A) Weightings for 3 tree topologies including the Faroese mountain hare (FAR), winter-white mountain hares (MTH_WW), the Iberian hare (IBH), and the black-tailed jackrabbit (BTJ), plotted with loess smoothing (span = 0.025). (B) Fraction of introgression (fd) between the Iberian hare and the Faroese mountain hare (20-kb windows with 2-kb steps). Pink dots indicate 1% highest values on chromosome 4. (C) RND between the Faroese mountain hare and the Iberian hare (20-kb windows with 2-kb steps). Pink dots indicate 1% lowest values in chromosome 4. (D) Distributions of dXY between the Faroese mountain hare and the Iberian hare on chromosome 4 (gray), at the Agouti region (pink), simulated under a model without migration (brown), and simulated with strong ancestral selection (yellow) (SI Appendix, Fig. S12A). (E) Maximum-likelihood tree with bootstrap supports for chromosome 4. BRH, broom hare; BTJ, black-tailed jackrabbit; EBH_IB and EBH_CE, European brown hares from the Iberian Peninsula and Central Europe, respectively; IBH, Iberian hare; MTH, mountain hares, from the Faroe Islands (FAR), Alps (ALP), and Fennoscandia (FSC); SSH_WW and SSH_WB, winter-white and winter-brown snowshoe hares, respectively. (F) Maximum-likelihood tree for the ∼360-kb Agouti association region.
Conclusions
Our work reveals that the evolution of nonwhite winter coats was remarkably parallel in 2 hare species with seasonally changing coat color, suggesting that evolution found similar solutions to the same problem (41). Nonwhite winter coats are determined by genetic variation at the region of the Agouti pigmentation gene both in snowshoe hares (11) and in mountain hares, most likely through similar regulatory changes that influence the expression of the gene during the autumn molt. These inferences confirm that the prominent role of Agouti in animal pigmentation (13, 42) extends to seasonal color polymorphism. In both hare species, the generation of winter coat color polymorphism resulted from a replicated process of introgressive hybridization with noncolor changing close relatives affecting the same genomic region. Independently of the nature of the winter-gray variant in the initially introgressed populations (neutral, advantageous, or possibly mildly deleterious), it persisted in mountain hare populations before reaching fixation in the Faroese hares. Given the slow emergence of novel adaptive mutations, standing introgressed variation has the potential to accelerate local adaptation (43, 44). In this context, the recurrent introgression of genetic variation, generating phenotypes upon which selection can act (11, 45, 46), may fuel replicated adaptive responses to rapidly changing environments.
Materials and Methods
Additional detailed information on materials and methods with associated references is provided in SI Appendix.
Samples and Sequencing.
Individually barcoded whole-genome sequence data at low individual coverage (Illumina) was generated for 59 mountain hares (L. timidus) collected from the Faroe Islands (n = 20 individuals; total population coverage 16.8×), Fennoscandia (n = 19; 21.7×) and the Alps (n = 20; 17.1×) (Fig. 1C). Samples were kindly donated by hunters during the regular permitted hunting season or were kindly provided by researchers; no animals were killed for the purpose of this research, and all samples were collected before the Nagoya Protocol came into force where applicable (procedures approved by the Organism Responsible for the Well-Being of Animals [ORBEA] of CIBIO). Higher individual coverage whole-genome sequence data from 1 Faroese hare and 6 Lepus species were also analyzed (6.1–29.3×; SI Appendix, Table S3). The reads were mapped to a hare pseudoreference genome built through iterative mapping (19).
Population Structure and Evolutionary Relationships.
Principal component analysis was performed using a single-read sampling approach. The evolutionary relationship among populations was determined inferring a neighbor-joining tree based on pairwise genetic distances between individuals and a population tree based on allele frequencies. The introduction of hares into the Faroe Islands was modeled using coalescent simulations and Approximate Bayesian computation.
Scans of Differentiation, SNP Genotyping, and Selection Analyses.
The PBS, FST, and Fisher exact tests were estimated in 20-kb nonoverlapping windows, and case-control association tests were performed, averaged across nonoverlapping windows of 100 SNPs. Fifty-nine SNPs (MassArray System) and 1 insertion-deletion (PCR) were genotyped in 127 mountain hares, including 29 museum specimens with recorded winter coat color (15 winter-gray and 14 winter-white; Swedish Museum of Natural History; specimen codes in SI Appendix, Table S1). Tajima’s D and nucleotide diversity were estimated in 200-kb windows. Scans for selective sweeps were performed along chromosome 4 using Hidden Markov Model and composite likelihood-ratio approaches, taking into account the inferred demographic model. Forward simulations under the demographic scenario were used to estimate the probability of fixation of a rare allele.
Gene Expression.
The expression of Agouti hair-cycle isoform was quantified in skin biopsies sampled in mountain hares from the Faroe Islands (n = 3 individuals) and the Alps (n = 5) during the autumn molt. Relative expression was measured using qPCR, normalized to reference genes ACTB and SDHA using a ΔCt approach, and estimating log2(abundance) using a Bayesian approach.
Phylogenies and Introgression Inferences.
Topology weighting analyses were performed along chromosome 4 using higher-coverage genome data from several Lepus species (SI Appendix, Table S3). Maximum-likelihood local Agouti and broad chromosome 4 phylogenies were inferred. Introgression was tested by estimating the fraction of introgression (fd) and the relative node depth (RND) in 20-kb overlapping windows of 2-kb steps along chromosome 4. The divergence between mountain hares and Iberian hares was modeled using higher-coverage whole genome data (n = 2 individuals per species), and the inferred parameters were used to perform coalescent simulations of the expected distribution of genetic distances.
Data Availability.
Previously generated sequencing data are available in the NCBI Sequence Read Archive (SAMN07526960, SAMN07526962, SAMN12618118, SAMN12618122, SAMN07526967, SAMN07526972, SAMN08146528 to SAMN08146531, and SAMN08146494). The hare pseudoreference genome is available in Dryad, https://doi.org/10.5061/dryad.x95x69pd8.
Supplementary Material
Acknowledgments
This work was supported by Portuguese national funds through Fundação para a Ciência e a Tecnologia (FCT) Project “CHANGE”, PTDC/BIA-EVF/1624/2014. Additional funding was provided by the European Social Fund (Programa Operacional Potencial Humano-Quadro de Referência Estratégica Nacional) and FCT IF/00033/2014/CP1256/CT0005 Investigador FCT Contract (to J.M.-F.) and Grant (to I.M.); FCT SFRH/BD/87126/2012 and SFRH/BD/115089/2016 PhD Grants (to F.A.S. and J.P.M., respectively); FCT PD/BD/108131/2015 BIODIV PhD Grant (to M.S.F.); and the European Regional Development Fund (ERDF) through COMPETE 2020 POCI-01-0145-FEDER-028124 and FCT PTDC/BIA-EVL/28124/2017 Research Contract (to J.P.). Access to specimens at the Swedish Museum of Natural History was supported by Synthesys (Access Grant SE-TAF-4695; EU FP7, Grant Agreement 226506). Additional support was obtained from Project NORTE-01-0145-FEDER-000007 (NORTE2020, PORTUGAL 2020, ERDF). The work benefited from the CIBIO NEW-GEN sequencing platform via European Union FP7 Research Potential Programme Grant 286431, and from the Montpellier Bioinformatics Biodiversity platform (LabEx CeMEB, an Agence Nationale de la Recherche “Investissements d’avenir” program, ANR-10-LABX-04-01). This research was conducted in the frame of the Laboratoire International Associé “Biodiversity and Evolution” supported by Institut Écologie et Environnement (Centre National de la Recherche Scientifique, France) and FCT (Portugal). We thank F. Suchentrunk, A. Angerbjörn, E. Randi, F. Ballesteros, and Z. Boratynski for kindly providing some of the samples used in this work; B. Muffat-Joly, P.-F. Galvin, and R. Gadient for their help in sampling molting hares; Daniela Kalthoff for access to the museum specimens; Marketa Zimova for help in sampling museum specimens; Nancy Jennings for revising the manuscript text; and Paulo C. Alves, Miguel Carneiro, Jeffrey M. Good, L. Scott Mills, Matthew R. Jones, and Pierre Boursot for valuable comments and discussions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All original high-throughput sequencing data reported in this article have been deposited in the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) under BioProjects PRJNA564335 (IDs SAMN12710256 to SAMN12710314) and PRJNA562432 (ID SAMN12640762). Genotyping data generated in this work are provided in SI Appendix, Tables S6 and S7.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910471116/-/DCSupplemental.
References
- 1.Gould S. J., Wonderful Life: The Burgess Shale and the Nature of History (W. W. Norton & Company, 1989). [Google Scholar]
- 2.Losos J., Improbable Destinies: How Predictable is Evolution? (Penguin Books, 2017). [Google Scholar]
- 3.Blount ZD, Lenski RE, Losos JB (2018) Contingency and determinism in evolution: Replaying life’s tape. Science 362, eaam5979. [DOI] [PubMed] [Google Scholar]
- 4.Hu Y., et al. , Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc. Natl. Acad. Sci. U.S.A. 114, 1081–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alves J. M., et al. , Parallel adaptation of rabbit populations to myxoma virus. Science 363, 1319–1326 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimova M., et al. , Function and underlying mechanisms of seasonal colour moulting in mammals and birds: What keeps them changing in a warming world? Biol. Rev. Camb. Philos. Soc. 93, 1478–1498 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Mills L. S., et al. , Camouflage mismatch in seasonal coat color due to decreased snow duration. Proc. Natl. Acad. Sci. U.S.A. 110, 7360–7365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimova M., Mills L. S., Nowak J. J., High fitness costs of climate change-induced camouflage mismatch. Ecol. Lett. 19, 299–307 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Atmeh K., Andruszkiewicz A., Zub K., Climate change is affecting mortality of weasels due to camouflage mismatch. Sci. Rep. 8, 7648 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills L. S., et al. , Winter color polymorphisms identify global hot spots for evolutionary rescue from climate change. Science 359, 1033–1036 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Jones M. R., et al. , Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science 360, 1355–1358 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Våge D. I., et al. , Two cysteine substitutions in the MC1R generate the blue variant of the Arctic fox (Alopex lagopus) and prevent expression of the white winter coat. Peptides 26, 1814–1817 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Hubbard J. K., Uy J. A., Hauber M. E., Hoekstra H. E., Safran R. J., Vertebrate pigmentation: From underlying genes to adaptive function. Trends Genet. 26, 231–239 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Bergengren A., On genetics, evolution and history of distribution of the heath-hare: A distinct population of the arctic hare, Lepus timidus Lin. Viltrevy (Stockh.) 6, 381–460 (1969). [Google Scholar]
- 15.Angerbjörn A., Flux J., Lepus timidus. Mamm. Species 495, 1–11 (1995). [Google Scholar]
- 16.Winiger A., “The apparent population crash in heath-hares Lepus timidus sylvaticus of southern Sweden–Do complex ecological processes leave detectable fingerprints in long-term hunting bag records?” MSc thesis, Swedish University of Agricultural Sciences, Umeå, Sweden (2014).
- 17.Degerbøl M., “Mammalia” in The Zoology of the Faroes, Jensen A. S., Lundbeck W., Mortensen T., Spärck R., Eds. (A. F. Høst & Søn, Copenhagen, 1940) Vol III, pp. LXV:1–133. [Google Scholar]
- 18.Reinert A., “Højere dyr på land” in Danmarks natur - Færøerne, Nørrevang A., Lundø J., Eds. (Poletikkens Forlag, Copenhagen, 1982), pp. 115–122. [Google Scholar]
- 19.Seixas F. A., Boursot P., Melo-Ferreira J., The genomic impact of historical hybridization with massive mitochondrial DNA introgression. Genome Biol. 19, 91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seixas F. A., Boursot P., Melo-Ferreira J., Data from “The genomic impact of historical hybridization with massive mitochondrial DNA introgression.” Dryad. 10.5061/dryad.x95x69pd8. Accessed 30 July 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ando H., Ichihashi M., Hearing V. J., Role of the ubiquitin proteasome system in regulating skin pigmentation. Int. J. Mol. Sci. 10, 4428–4434 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Pape E., et al. , Microarray analysis sheds light on the dedifferentiating role of agouti signal protein in murine melanocytes via the Mc1r. Proc. Natl. Acad. Sci. U.S.A. 106, 1802–1807 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linnen C. R., Kingsley E. P., Jensen J. D., Hoekstra H. E., On the origin and spread of an adaptive allele in deer mice. Science 325, 1095–1098 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontanesi L., et al. , Characterization of the rabbit agouti signaling protein (ASIP) gene: Transcripts and phylogenetic analyses and identification of the causative mutation of the nonagouti black coat colour. Genomics 95, 166–175 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Ferreira M. S., et al. , The transcriptional landscape of seasonal coat colour moult in the snowshoe hare. Mol. Ecol. 26, 4173–4185 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Kratochwil C. F., et al. , Agouti-related peptide 2 facilitates convergent evolution of stripe patterns across cichlid fish radiations. Science 362, 457–460 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Mazo-Vargas A., et al. , Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc. Natl. Acad. Sci. U.S.A. 114, 10701–10706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wray G. A., The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8, 206–216 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Pavlidis P., Jensen J. D., Stephan W., Searching for footprints of positive selection in whole-genome SNP data from nonequilibrium populations. Genetics 185, 907–922 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alachiotis N., Pavlidis P., RAiSD detects positive selection based on multiple signatures of a selective sweep and SNP vectors. Commun. Biol. 1, 79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patursson S., Haren paa Færøerne. Naturen. 28, 240–241 (1904). [Google Scholar]
- 32.Seixas F. A., “Genome admixture with massive mitochondrial DNA introgression in hares (Lepus spp.): The relative roles of demography and natural selection,” PhD thesis, University of Porto and University of Montpellier, Porto, Montpellier (2017).
- 33.Giska I., et al. , Lepus timidus WGS. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA564335. Deposited 6 September 2019.
- 34.Marques J. P., et al. , Genomic resources for L. castroviejoi, BioSample SAMN12640762. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/biosample/SAMN12640762. Deposited 27 August 2019.
- 35.Seixas F. A., Boursot P., Melo-Ferreira J., The genomic impact of historical hybridization with massive mitochondrial DNA introgression in the Iberian hare (Lepus granatensis). NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA399194. Accessed 1 August 2018.
- 36.Seixas F. A., et al. , Genomic resources for L. europaeus, BioSample SAMN12618118. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/biosample/SAMN12618118. Accessed 1 August 2018.
- 37.Seixas F. A., et al. , Genomic resources for L. europaeus, BioSample, SAMN12618122. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/biosample/SAMN12618122. Accessed 1 August 2018.
- 38.Jones M. R., et al. , Whole exome and whole genome sequencing of hares and jackrabbits (genus Lepus). NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA420081. Accessed 1 August 2018.
- 39.Melo-Ferreira J., et al. , Home-loving boreal hare mitochondria survived several invasions in Iberia: The relative roles of recurrent hybridisation and allele surfing. Heredity 112, 265–273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thulin C. G., Jaarola M., Tegelström H., The occurrence of mountain hare mitochondrial DNA in wild brown hares. Mol. Ecol. 6, 463–467 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Stern D. L., Orgogozo V., Is genetic evolution predictable? Science 323, 746–751 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoekstra H. E., Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97, 222–234 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Hedrick P. W., Adaptive introgression in animals: Examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22, 4606–4618 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Jagoda E., et al. , Disentangling immediate adaptive introgression from selection on standing introgressed variation in humans. Mol. Biol. Evol. 35, 623–630 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oziolor E. M., et al. , Adaptive introgression enables evolutionary rescue from extreme environmental pollution. Science 364, 455–457 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Feulner P. G. D., et al. , Introgression and the fate of domesticated genes in a wild mammal population. Mol. Ecol. 22, 4210–4221 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Previously generated sequencing data are available in the NCBI Sequence Read Archive (SAMN07526960, SAMN07526962, SAMN12618118, SAMN12618122, SAMN07526967, SAMN07526972, SAMN08146528 to SAMN08146531, and SAMN08146494). The hare pseudoreference genome is available in Dryad, https://doi.org/10.5061/dryad.x95x69pd8.