Significance
Triplication of human chromosome 21, or trisomy 21 (T21), causes the condition known as Down syndrome (DS). People with DS show a markedly different disease spectrum relative to typical people, being highly predisposed to conditions such as Alzheimer’s disease, while being protected from other conditions, such as most solid malignancies. Interestingly, people with DS are affected by high rates of autoimmune disorders, whereby the immune system mistakenly attacks healthy tissues. This manuscript reports an exhaustive characterization of the T cells of people with DS, demonstrating many alterations in this key immune cell type that could explain their high risk of autoimmunity. These results reveal opportunities for therapeutic intervention to modulate T cell function and improve health outcomes in DS.
Keywords: trisomy 21, T cells, autoimmunity, type I interferon, inflammation
Abstract
Trisomy 21 (T21) causes Down syndrome (DS), a condition characterized by high prevalence of autoimmune disorders. However, the molecular and cellular mechanisms driving this phenotype remain unclear. Building upon our previous finding that T cells from people with DS show increased expression of interferon (IFN)-stimulated genes, we have completed a comprehensive characterization of the peripheral T cell compartment in adults with DS with and without autoimmune conditions. CD8+ T cells from adults with DS are depleted of naïve subsets and enriched for differentiated subsets, express higher levels of markers of activation and senescence (e.g., IFN-γ, Granzyme B, PD-1, KLRG1), and overproduce cytokines tied to autoimmunity (e.g., TNF-α). Conventional CD4+ T cells display increased differentiation, polarization toward the Th1 and Th1/17 states, and overproduction of the autoimmunity-related cytokines IL-17A and IL-22. Plasma cytokine analysis confirms elevation of multiple autoimmunity-related cytokines (e.g., TNF-α, IL17A–D, IL-22) in people with DS, independent of diagnosis of autoimmunity. Although Tregs are more abundant in DS, functional assays show that CD8+ and CD4+ effector T cells with T21 are resistant to Treg-mediated suppression, regardless of Treg karyotype. Transcriptome analysis of white blood cells and T cells reveals strong signatures of T cell differentiation and activation that correlate positively with IFN hyperactivity. Finally, mass cytometry analysis of 8 IFN-inducible phosphoepitopes demonstrates that T cell subsets with T21 show elevated levels of basal IFN signaling and hypersensitivity to IFN-α stimulation. Therefore, these results point to T cell dysregulation associated with IFN hyperactivity as a contributor to autoimmunity in DS.
Down syndrome (DS) is caused by trisomy 21 (T21), the most common chromosomal abnormality in the human population, affecting ∼1 in 700 newborns (1). Besides developmental delays and highly variable cognitive deficits, T21 causes an altered disease spectrum, wherein people with DS are protected from certain diseases, such as most solid malignancies (2), but highly predisposed to others, such as Alzheimer’s disease (AD) (3). Remarkably, people with T21 are highly predisposed to develop a specific spectrum of autoimmune conditions, including autoimmune thyroid disease, celiac disease, type I diabetes, and a range of autoimmune and autoinflammatory skin conditions (4–6). With the exception of AD, the high prevalence of which is due to the presence of the amyloid precursor protein gene (APP) on chromosome 21 (chr21), the molecular and cellular bases of this different disease spectrum are unclear.
Several genes involved in immune control are encoded on chr21, including 4 of the 6 interferon (IFN) receptor subunits (IFNRs): the 2 type I IFNRs (IFNAR1, IFNAR2), one type II IFNR (IFNGR2), and IL10RB, which encodes a subunit of the receptors for type III IFN ligands, IL-10, IL-22, and IL-26 (7). The role of IFN signaling in development of autoimmunity is well documented, with many genome-wide association studies revealing strong genetic associations between polymorphisms in components of the IFN pathway and autoimmune disorders (reviewed in ref. 8). Recently, transcriptome analysis of 4 different cell types from people with and without DS, including T cells, demonstrated that T21 causes a consistent activation of the IFN transcriptional response (9). Additionally, plasma proteomics analyses demonstrated that people with DS have signs of chronic autoinflammation, including elevated levels of proinflammatory cytokines, such as IL-6, TNF-α, MCP-1, and IL-22 (10). Thus, it is clear that people with DS display signs of chronic inflammation, even in the absence of diagnosed autoimmune disorders.
Less is known about alterations in cellular immunity that could contribute to the high prevalence of autoimmune disorders and the chronic inflammatory state in DS. Histopathologic studies of thymic tissue from people with DS showed hypoplasia and compromised architecture, even in newborns (11, 12), which could explain the lower thymic output of mature αβ+ thymocytes observed in DS (13). This decreased thymic output is consistent with decreased naïve T cell percentages; however, so is the reported increase in central memory and terminally differentiated T lymphocytes (13–17). Although it has been shown that individuals with T21 exhibit increased CD8/CD4 ratios and higher frequencies of regulatory CD4+ T cells (Tregs) in peripheral blood (17–20), characterization of CD8+ and CD4+ T cell subsets has been limited to the expression of a few phenotypic markers in the pediatric population (14, 17, 19, 21).
In this study, we used multiparametric, high-dimensional flow cytometry to comprehensively characterize the peripheral T cell compartment of individuals with DS by measuring the expression of a large panel of phenotypic markers. We also performed functional assays, cytokine measurements, and transcriptome analyses. We found that CD8+ T cells from individuals with T21 express markers of activated and senescent states and respond to stimulation more potently than their euploid counterparts, overproducing autoimmunity-related cytokines. Conventional CD4+ T cells (Tconvs) show a polarized state toward increased production of IL-17A, and plasma levels of IL-17A, IL-17B, IL-17C, and IL-17D are elevated in DS. Interestingly, we found that Tregs from individuals with T21 show neither significant phenotypic differences nor impaired functional capacity relative to control Tregs. Indeed, our results from allogeneic “criss-cross” experiments indicate that the functional deficiency previously reported for Tregs with T21 is not cell-autonomous (18). Instead, we find that T21 CD8+ T and CD4+ Tconv cells are resistant to Treg-mediated suppression, regardless of Treg karyotype. Transcriptome analysis of bulk white blood cells (WBCs) and T cells revealed gene expression signatures indicative of T cell differentiation and activation which correlated positively with IFN activity. Finally, mass cytometry experiments revealed that T cells of people with DS display both elevated basal IFN signaling and hyperactivation upon IFN-α stimulation. Altogether, these results reveal that multiple alterations observed in the T cell compartment of adults with T21 mimic those observed in autoimmune conditions, even in the absence of clinical manifestation of autoimmunity, concurrent with IFN hyperactivity.
Results
Trisomy 21 Reshapes the Peripheral T Cell Compartment away from the Naïve State.
In order to investigate the impact of T21 on the peripheral T cell compartment, we used flow cytometry to define alterations in the numbers and/or frequencies of different functional subsets of T cells present in blood from adult individuals, including total T cells (CD3+), CD8+, CD4+ Tconvs, and Tregs, as well as a plethora of phenotypic markers (Dataset S1) (22). First, we analyzed the numbers and frequencies of major T cell subtypes in adults with T21 and age- and sex-matched euploid controls (D21). We recruited adults to avoid observations that could be affected by differences in early immune development. Our research participants presented a range of comorbidities more common in DS (6), but none of them presented signs of infection in the weeks prior to blood draw, and all tested negative for cytomegalovirus (CMV) infection (Dataset S2).
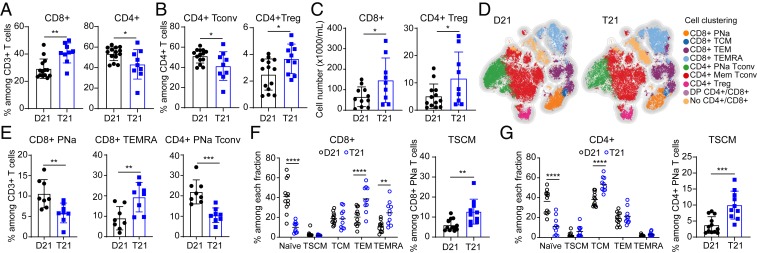
Although T21 does not significantly impact the frequency of CD3+ T cells among all viable peripheral blood mononuclear cells (PBMCs) or the total number of T cells, individuals with T21 have a higher percentage of CD8+ T cells and a decreased frequency of CD4+ T cells within the CD3+ population, leading to an increased CD8/CD4 ratio (Fig. 1A and SI Appendix, Fig. S1 A–C). When we classified CD4+ T cells into Tconvs and Tregs, we observed a decrease in the percentage of Tconvs and an increase in the percentage of Tregs (Fig. 1B and SI Appendix, Fig. S1A). Then, we evaluated the number of cells per milliliter, which revealed that people with DS have a higher number of CD8+ T and Tregs (Fig. 1C), with no significant differences in the number of total CD4+ T and CD4+ Tconvs (SI Appendix, Fig. S1D). We also observed a positive correlation between the numbers of CD8+ T cells and Tregs in both groups (SI Appendix, Fig. S1E). Interestingly, concurrent increases in CD8+ T cells and Tregs have been observed in autoinflammatory conditions (23, 24).
Fig. 1.
Adults with trisomy 21 show dysregulated T cell homeostasis. (A) Frequencies of CD8+ and CD4+ T cells within the CD3+ T cells in euploid controls (D21) versus people with trisomy 21 (T21) (n = 14 D21; n = 9 T21). (B) Percentage of CD4+ Tconv (CD4+ CD25low CD127high) and CD4+ Treg (CD4+ CD25high CD127low FOXP3+) cells among CD4+ T cells. (C) Numbers of CD8+ and CD4+ Treg cells in peripheral blood in people with and without T21. (D) A t-SNE map showing a random subsample of circulating T cells from individuals with and without T21, analyzed with FlowSOM clustering. (E) Frequencies of CD8+ PNa, CD8+ TEMRA, and CD4+ PNa Tconv cells within the CD3+ T cells obtained by FlowSOM clustering. (F and G) (Left) Frequencies of (F) CD8+ and (G) CD4+ T cells subsets based on expression of CCR7, CD45RA, CD45RO, CD27, and CD95 as shown in SI Appendix, Fig. S2C. (Right) Proportion of TSCMs among potentially naïve CD8+ and CD4+ T cells (n = 12 D21; n = 10 T21). Data in A–C, E, F, Right, and G, Right are shown as mean ± SEM with significance determined by unpaired t test; data in Left F and G are shown as mean ± SEM with significance determined by 2-way ANOVA with Sidak’s posttest. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
T cells are classified as naïve or memory, depending on their location, functional status, cytokine expression, and their history of antigen-induced activation. To obtain an overview of the T cell subsets in people with DS, we reduced the flow cytometry data to 2 dimensions by applying the t-distributed stochastic neighbor embedding (t-SNE) algorithm, where we considered the differential expression of 12 parameters, including surface markers, transcription factors, and signaling and activation molecules (SI Appendix, Fig. S2 A and B). These analyses were performed in conjunction with Flow Self-Organizing Maps (FlowSOM) clustering (25), which resolved 9 T cell subpopulations: potentially naïve (PNa, CD45RA+ CCR7+ cells, which include true naïve and stem cell memory-like T cells or TSCMs), central memory (TCM), effector memory (TEM), and terminally differentiated effector (TEMRA) CD8+ T cells; PNa and memory CD4+ Tconvs; Tregs; double-positive CD4+/CD8+ T cells; and non-CD4+/CD8+ T cells (Fig. 1D and SI Appendix, Fig. S2B). The most prominent change among CD3+ T cells in people with T21 was a reduction in the proportion of PNa cells among CD8+ and CD4+ T cells, concurrent with an increase in the frequency of TEMRA CD8+ T cells (Fig. 1E). We next analyzed the CD8+ and CD4+ T cell subsets in greater detail by manual gating using surface markers to discriminate the following 5 subpopulations (SI Appendix, Fig. S2C): truly naïve, TSCM, TCM, TEM, and TEMRA (26). This exercise confirmed that, among total CD8+ T cells, people with DS have lower levels of naïve CD8+ T cells and higher levels of TEM and TEMRA populations (Fig. 1F). When analyzed in terms of frequency among all CD8+ T cells, TSCMs were not found to be different; however, when calculated as a fraction of all CD45RA+ CCR7+ cells [commonly identified as naïve T cells in the literature (27)], there is an increase in TSCMs in people with DS (Fig. 1F). This discrepancy can be explained by the fact that the increase in TSCMs occurs in the background of decreased CD45RA+ CCR7+ cells (SI Appendix, Fig. S2D). Among CD4+ T cells, there was a similar depletion of naïve cells and an increase in the TCM population along with an increase in TSCMs among PNa CD4+ T cells (Fig. 1G). Next, we investigated alterations in Tregs by measuring expression of FOXP3, a key transcription factor in their development and function (28, 29). Since FOXP3+ T cells may not be functionally homogeneous, due to the transient expression of FOXP3 during activation of Tconvs, we used the expression of CD45RA and FOXP3 to distinguish 3 phenotypically distinct subpopulations (30): FOXP3+ non-Tregs (CD45RA-/FOXP3low), PNa Tregs (CD45RA+/FOXP3low), and effector Tregs (CD45RA-/FOXP3high) (SI Appendix, Fig. S2E). Although there is no redistribution among the PNa and effector Tregs, effector Tregs express more FOXP3 in people with DS (SI Appendix, Fig. S2F). As expected, effector Tregs express the most FOXP3 relative to the other populations (SI Appendix, Fig. S2F), showing the classical CD25high CD127low phenotype and producing the lowest levels of IFN-γ (SI Appendix, Fig. S2G).
Altogether, these results indicate that T21 disrupts peripheral T cell homeostasis, affecting the frequencies and total numbers of key T cell subtypes, thus justifying a deeper characterization of these populations.
Trisomy 21 Drives CD8+ T Cell Differentiation toward a Hyperactivated State.
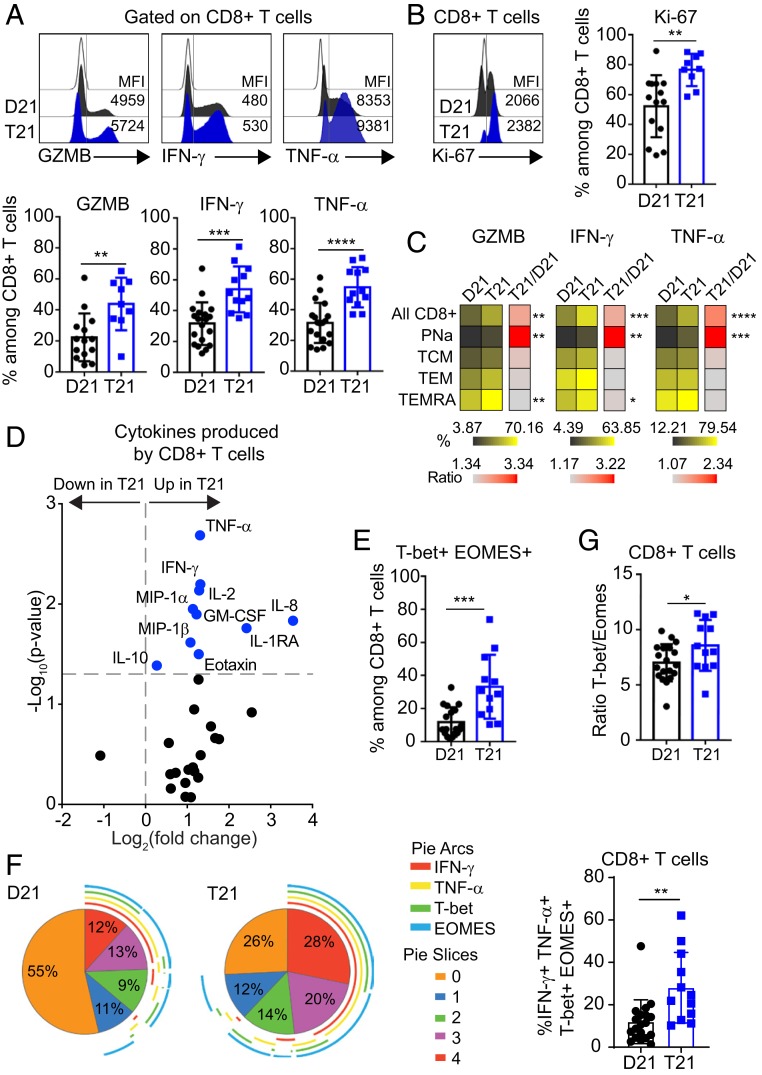
Having observed an increase in both frequency and number of total CD8+ T cells in people with DS, along with a redistribution away from the naïve state, we investigated the expression of markers of activation, proliferation, and senescence in this T cell subset (31). First, we evaluated the levels of granzyme B (GZMB), IFN-γ, and TNF-α after stimulating CD8+ T cells with phorbol 12-myristate 13-acetate (PMA)/Ionomycin and found that all 3 effector markers were elevated in cells from people with T21 compared to D21 (Fig. 2A). We also found increased frequency of CD8+ T cells that simultaneously produced 2 of these effectors, GZMB/IFN-γ or IFN-γ/TNF-α (SI Appendix, Fig. S3A), illustrating the polyfunctionality of these cells. CD8+ T cells from people with DS also expressed more Ki-67, consistent with increased cell proliferation upon activation (Fig. 2B). We also found increased levels of effector molecules and Ki-67 in some of the phenotypically matched subsets defined by CD45RA and CCR7 marker expression (PNa, TCM, TEM, and TEMRA) (Fig. 2C and SI Appendix, Fig. S3B). Whereas the increases in the CD45RA+ CCR7+ subset could be attributed, in part, to the increased fraction in TSCMs, even TEMRAs showed increases in GZMB, IFN-γ, and Ki-67 (Fig. 2C and SI Appendix, Fig. S3C).
Fig. 2.
CD8+ T cells from adults with trisomy 21 show signs of hyperactivation. Fresh PBMCs from individuals with and without DS were stimulated ex vivo with PMA/Ionomycin before flow cytometry analysis. (A) (Top) Representative histograms showing the expression levels (mean fluorescence intensity [MFI]) of GZMB, IFN-γ, and TNF-α on total CD8+ T cells from individuals with T21 (blue) and typical controls (D21, black) compared to fluorescence minus one (FMO) on CD8+ T cells (white). (Bottom) Scatter plots display frequencies of GZMB (n = 14 D21; n = 9 T21), IFN-γ, and TNF-α (n = 19 D21; n = 12 T21) positive events among CD8+ T cells. (B) (Left) Representative histogram and (Right) frequencies of Ki-67 expression among CD8+ T cells (n = 14 D21; n = 9 T21). (C) Heatmap showing the frequencies of GZMB, IFN-γ, and TNF-α positive events within the CD8+ T cell subsets, as defined in SI Appendix, Fig. S2C, in individuals with and without T21. (D) Volcano plot showing fold changes (log2 T21 over D21) and P values (log10) for cytokine levels produced by CD8+ T cells after being stimulated with anti-CD3/CD28, detected by MSD. Vertical dashed line represents the no-change midline. Horizontal dashed line represents P value of 0.05 as calculated by Student t test. (E) Scatter plot showing the coexpression of T-bet and EOMES among total CD8+ T cells (n = 19 D21; n = 12 T21). (F) (Left) Pie charts illustrating the expression pattern of IFN-γ, TNF-α, T-bet, and EOMES within the CD8+ T cell population. Pie-section colors correspond to the number of markers expressed on a cell. Arcs around the pie represent which marker(s) are expressed. (Right) Coexpression of IFN-γ, TNF-α, T-bet, and EOMES among CD8+ T cells. (G) Ratio of T-bet+ to EOMES+ among CD8+ T cells in people with and without T21 (n = 19 D21; n = 12 T21). Data in A–G are shown as mean ± SEM with significance determined by unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Next, we examined cytokines produced by CD8+ T cells in vitro upon stimulation with anti-CD3/CD28. Remarkably, 28 of the 29 cytokines detected were more abundant in the supernatant of T21 CD8+ T cells, 10 of them significantly (Fig. 2D, SI Appendix, Fig. S3D, and Dataset S3). We observed elevated levels of cytokines known to be produced by CD8+ T cells, including TNF-α, IFN-γ, IL-2, and MIP-1α (32–35). Notably, overproduction of GM-CSF by CD8+ T cells has been observed in autoimmune conditions (36), and overproduction of antiinflammatory IL-10 is indicative of negative feedback within this subset (37). Others and we have observed elevation of many of the cytokines overproduced by CD8+ T cells in the plasma of people with DS, including TNF-α, IL-2, MIP-1α, IL-10, Eotaxin, and IL-8 (SI Appendix, Fig. S3E) (10, 38). However, extended analysis of cytokine profiles of people with DS revealed no differences between those with diagnoses of autoimmune conditions and those without (Dataset S2 and SI Appendix, Fig. S3F).
Lastly, we measured the expression of the transcription factors T-bet (T-box 21, TBX21) and Eomesodermin (EOMES), which play well-recognized roles in CD8+ T cell differentiation, senescence, and exhaustion (39). In CD8+ T cells, T-bet is up-regulated upon activation and is associated with induction of effector functions, including cytotoxicity, whereas EOMES expression is higher in long-term, stable, self-renewing memory CD8+ T cells, and has also been associated with a dysfunctional phenotype (39). We observed that the frequency of T-bet+ EOMES+ CD8+ T cells is increased in people with DS (Fig. 2E). We also examined how the expression of T-bet and EOMES associated with effector markers of CD8+ T cell function and found an increase in IFN-γ+ TNF-α+ T-bet+ EOMES+ CD8+ T cells in people with DS (Fig. 2F). Moreover, the T-bet/EOMES ratio is higher in DS, consistent with an enhanced effector phenotype (Fig. 2G).
In sum, these findings indicate that CD8+ T cells from people with DS have a markedly altered program of differentiation toward hyperactivation, cytokine overproduction, and increased functionality.
CD8+ T Cells from People with DS Show Signs of Increased Senescence.
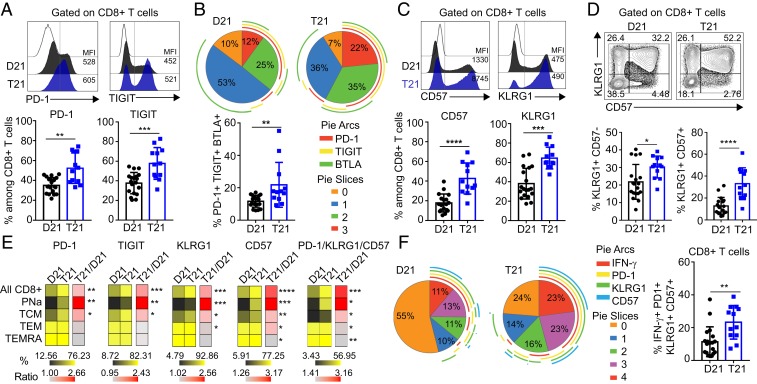
The fact that people with DS showed an increase in differentiated CD8+ T cell subsets prompted us to determine whether these cells had acquired a senescent phenotype, as is observed in chronic inflammatory settings (40). To test this, we measured expression of the inhibitory receptors PD-1, TIGIT, and BTLA, and the senescence markers CD57 and KLRG1. Samples with T21 showed an increased frequency of CD8+ T cells expressing PD-1 and TIGIT (Fig. 3A), but not BTLA (SI Appendix, Fig. S4A), which is expressed by most naïve CD8+ T cells (41). Next, we investigated coexpression of these receptors and found a greater proportion of CD8+ T cells coexpressing all 3 inhibitory receptors in T21 samples (Fig. 3B). Individuals with DS also have a higher percentage of CD8+ T cells positive for CD57 and KLRG1 (Fig. 3C), as well as higher numbers of cells expressing both markers (Fig. 3D). Of note is that individuals with T21 also exhibit a higher frequency of KLRG1+ CD57− CD8+ T cells, again indicative of an activated state (42) (Fig. 3D). Importantly, many of these increases in senescence markers were also observed when comparing phenotypically matched subsets of CD8+ T cells (Fig. 3E and SI Appendix, Fig. S4 B and C).
Fig. 3.
CD8+ T cells from adults with trisomy 21 show increased expression of inhibitory receptors and senescence markers. Fresh PBMCs from individuals with and without DS were stimulated ex vivo with PMA/Ionomycin before flow cytometry analysis. (A) (Top) Representative histograms showing the expression levels (MFI) of PD-1 and TIGIT on total CD8+ T cells from individuals with T21 (blue) and typical controls (D21, black) compared to FMO on CD8+ T cells (white). (Bottom) Display frequencies of PD-1 and TIGIT positive events among CD8+ T cells (n = 19 D21; n = 12 T21). (B) (Top) Pie charts illustrating the mean percentage of inhibitory receptor coexpression on total CD8+ T cells from people with and without T21 (n = 19 D21; n = 12 T21). Pie chart colors correspond to the number of inhibitory receptors expressed on a cell. Arcs around the pie represent which inhibitory receptor(s) are expressed. (Bottom) Coexpression of PD-1, TIGIT, and BTLA among CD8+ T cells. (C) Representative (Top) histograms and (Bottom) frequencies of CD57 and KLRG1 expression among CD8+ T cells (n = 19 D21; n = 12 T21). (D) Representative (Top) flow cytometric data and (Bottom) frequencies for coexpression of the senescence markers KLRG1 and CD57 among CD8+ T cells. (E) Heatmap showing the frequencies of PD-1, TIGIT, KLRG1, CD57, and PD1/KLRG1/CD57 positive events within the different subsets of CD8+ T cells, as defined in SI Appendix, Fig. S2C, in individuals with and without T21. (F) (Left) Pie charts as in B showing the coexpression pattern of the effector molecule IFN-γ and the indicated inhibitory receptors and senescence markers on the CD8+ T cell population (n = 19 D21; n = 12 T21). (Right) Coexpression of IFN-γ with PD-1, KLRG1, and CD57 among CD8+ T cells. Data in A–F are shown as mean ± SEM with significance determined by unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
In the CD8+ T cell response, the effector state precedes the memory state and may divert to a senescent phenotype upon chronic stimulation. Accordingly, cells can exist in an intermediate state where they express both activation and inhibitory/senescent markers. When we measured coexpression of these markers, samples with T21 showed an elevated frequency of cells that coexpress IFN-γ or TNF-α with PD-1, KLRG1, and CD57 (Fig. 3F and SI Appendix, Fig. S4D). Finally, we measured the expression of T-bet and EOMES in CD57+ and KLRG1+ CD8+ T cells, which revealed that adults with DS exhibit increased T-bet (but not EOMES) in KLRG1+ CD57+ and KLRG1+ CD57− CD8+ T cells (SI Appendix, Fig. S4E). This intermediate phenotype has been associated with senescence of CD8+ T cells in aged individuals (43).
Overall, these findings indicate that CD8+ T cells from people with DS show signs of differentiation toward activated and senescent states. However, upon ex vivo stimulation, they remain capable of proliferating and producing effector cytokines to a greater extent than their euploid counterparts.
Conventional T Cells from People with Trisomy 21 Are Polarized toward the Th1 and Th1/17 States.
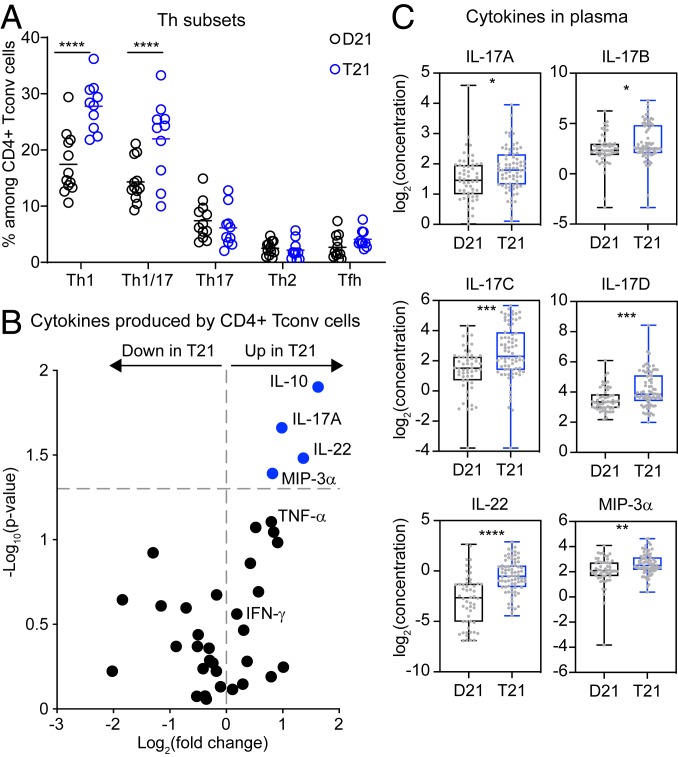
Given our findings that individuals with T21 show anomalies in the distribution of naïve/memory CD4+ T cells (Fig. 1G), we next investigated their polarization into biologically distinct subsets by evaluating the expression of chemokine receptors (SI Appendix, Fig. S5A). Th1 and Th1/17 subsets were enriched among CD4+ Tconvs from people with T21, with no differences in Th17, Th2, and Tfh subsets (Fig. 4A). Evaluation of cytokines produced by CD4+ Tconvs stimulated in vitro with anti-CD3/CD28 revealed elevated levels of IL-10, IL-17A, IL-22, and MIP-3α in T21 samples (Fig. 4B, SI Appendix, Fig. S5B, and Dataset S3). Other IL-17 isoforms were below detection limits in these assays (Dataset S3). The increased levels of IL-10 are consistent with self-regulation within this subset (44), and the elevated levels of IL-17A and IL-22 are consistent with the observed increase in the Th1/17 subset (45, 46). The increase in MIP-3α (CCL20, the ligand for CCR6) is indicative of increased CCR6-dependent signaling, a hallmark of the Th1/17 and Th17 subsets (45, 47). Differences in Th1 cytokines, such as TNF-α and IFN-γ, did not reach statistical significance (Fig. 4B). Of note is that the Tfh cytokine IL-21 was not detected in these assays (Dataset S3). We next investigated plasma levels of several IL-17 isoforms, IL-22, MIP-3α, and IL-21, which revealed increases in IL-17A, IL-17B, IL-17C, IL-17D, IL-22, and MIP-3α, but not in IL-17E, IL-17F, and IL-21 in people with DS (Fig. 4C and Dataset S3). We found no differences in IL-17 isoforms or IL-22 between people with DS with or without a confirmed diagnosis of an autoimmune condition (SI Appendix, Fig. S5C). Thus, dysregulation of the CD4+ compartment could also contribute to the autoimmunity-related cytokine profile of DS.
Fig. 4.
Conventional CD4+ T cells from adults with trisomy 21 are polarized toward the Th1 and Th1-like Th17 states. Fresh PBMCs from individuals with and without DS were stained and analyzed by flow cytometry. (A) Scatter plots showing the frequencies of cells expressing the indicated chemokine receptor combinations among CD4+ Tconv cells (n = 12 D21; n = 10 T21). (B) Volcano plot showing fold changes (log2 T21 over D21) and P values (log10) for cytokines produced by CD4+ Tconv cells after being stimulated with anti-CD3/CD28 detected by MSD. Vertical dashed line represents the no-change midline. Horizontal dashed line represents P value of 0.05 as calculated by Student t test. (C) Box and whisker plots showing the levels (log2 concentration) of IL-17A, IL-17B, IL-17C, IL-17D, IL-22, and MIP-3α in plasma of individuals with and without T21 (n = 54 D21; n = 74 T21) measured by MSD. Data in A are shown as mean ± SEM with significance determined by 2-way ANOVA with Sidak’s posttest; data in B and C are shown as mean ± SEM with significance determined by unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Effector T Cells with Trisomy 21 Are Resistant to Treg-Mediated Suppression.
The enhanced activation phenotype observed in CD8+ and CD4+ T cells of people with DS, which is accompanied by increased numbers of FOXP3+ Tregs (Fig. 1C), prompted us to test for differences in Treg functionality. We analyzed 14 phenotypic markers to study the proliferation, activation, inhibitory potential, functional specialization, and effector properties of Tregs, which revealed minor effects of karyotype (SI Appendix, Fig. S5 D and E). PNa Tregs expressed more of both Ki-67 and the activation marker Helios, but less PD-1 (SI Appendix, Fig. S5 D and E). In contrast, effector Tregs, which express more FOXP3 in people with DS (SI Appendix, Fig. S2F), also express more of the costimulatory molecule GITR (SI Appendix, Fig. S5 D and E).
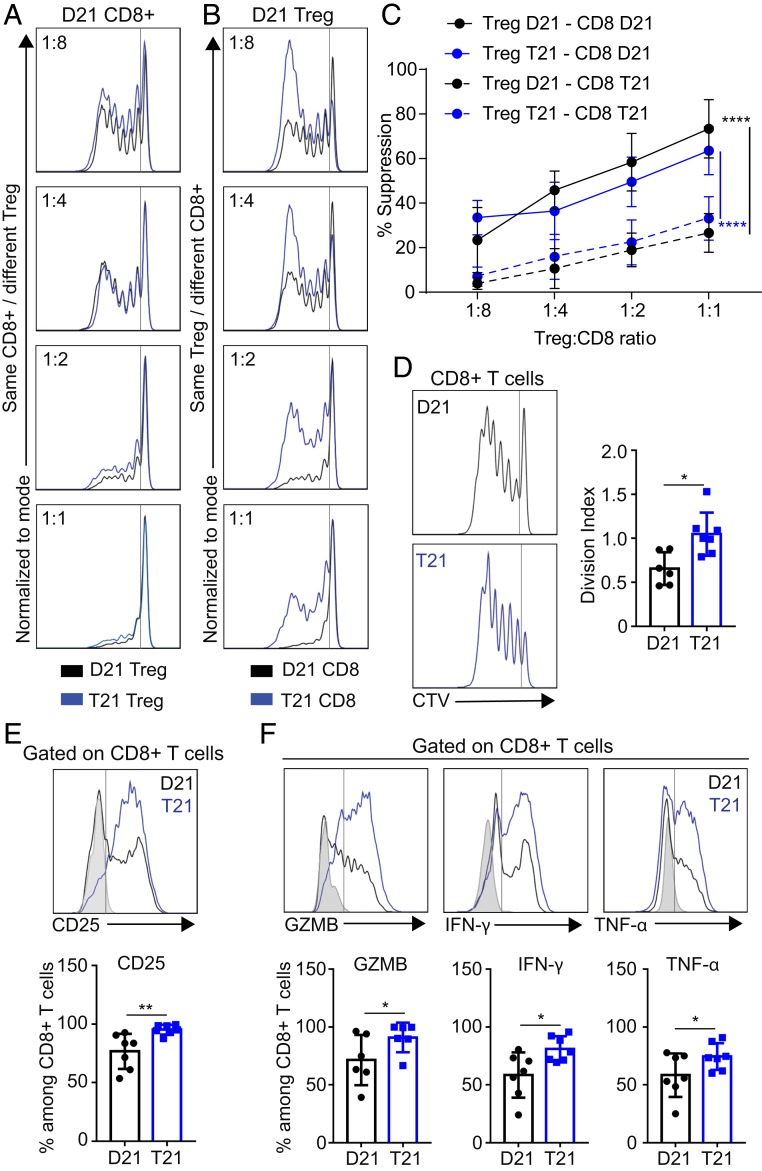
To further test Treg functionality, we performed allogeneic “criss-cross” suppression assays (24, 48–50), in which Tregs from people with and without DS were isolated and titrated into responder cells with or without T21. In this way, we could independently define the impact of karyotype on responder cells (either CD8+ T or CD4+ Tconv cells) versus impacts on Treg suppressive capacity. We observed that Tregs of either karyotype showed a similar ability to suppress responder CD8+ T and CD4+ Tconv cells from D21 individuals (Fig. 5A and SI Appendix, Fig. S6A). In contrast, both CD8+ T and CD4+ Tconv cells with T21 were more resistant to Treg-mediated suppression, regardless of the Treg karyotype (Fig. 5 B and C and SI Appendix, Fig. S6 B and C). Further analysis of effector cells upon stimulation with anti-CD3/CD28 in the absence of Tregs revealed that T21 CD8+ T cells display a higher rate of proliferation (Fig. 5D), an increased expression of the activation marker CD25 (IL-2 receptor alpha) (Fig. 5E), and increased expression of the effector molecules GZMB, IFN-γ, and TNF-α (Fig. 5F). For CD4+ Tconvs, we observed increased proliferation as well as increased expression of CD25 and IFN-γ (SI Appendix, Fig. S6 D–F).
Fig. 5.
Effector T cells from people with trisomy 21 are resistant to Treg suppression. CD4+ Treg and CD8+ T cells from individuals with and without DS were titrated in criss-cross combinations in anti-CD3/CD28-stimulated cultures, and proliferation of CD8+ T cells was subsequently measured based on CellTrace Violet (CTV) dilution. (A and B) Representative examples comparing (A) CD4+ Treg from D21 and T21 samples titrated against the same violet-labeled D21 CD8+ T cells and (B) same CD4+ Treg from D21 titrated against violet-labeled T21 or D21 CD8+ T cells. (C) Percentage of suppression in the criss-cross combinations cultured for 5 d in the presence of anti-CD3/CD28 at the indicated ratios of Treg:CD8+ T cells. (D) Representative histogram showing (Left) CTV dilution and (Right) division indices of CD8+ T cells from individuals with and without DS, after 5 d of culture in presence of anti-CD3/CD28. (E and F) Representative (Top) flow cytometric analysis and (Bottom) frequency of (E) CD25 and (F) GZMB, IFN-γ, and TNF-α among gated CD8+ T cells, after 5 d of culture in presence of anti-CD3/CD28 and stimulated for the last 4 h with PMA/Ionomycin. Data in C are shown as mean ± SEM with significance determined by 2-way ANOVA with Tukey’s posttest; data in D–F are shown as mean ± SEM with significance determined by unpaired t test (n = 7 D21 and 7 T21). *P < 0.05; **P < 0.01; ****P < 0.0001.
Altogether, these data show that effector T cells from people with DS are less sensitive to suppression by Tregs, which could further contribute to the inflammatory state in those with T21.
Transcriptome Analysis of Trisomy 21 Cells Reveals Signatures Indicative of T Cell Activation and Differentiation Associated with IFN Hyperactivity.
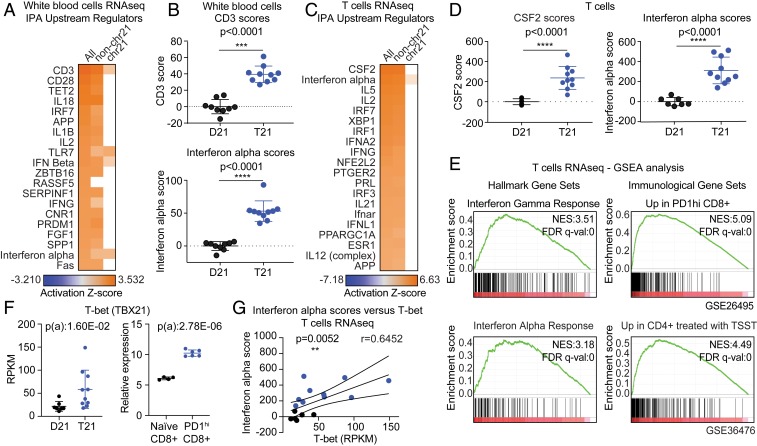
In order to gain mechanistic insights into the signaling pathways dysregulated by T21 that could contribute to the observed changes in the T cell compartment, we analyzed transcriptome data from WBCs from 19 individuals, 10 of them with T21 (Datasets S2 and S4) (51). DESeq2 analysis of differentially expressed genes (DEGs) revealed that T21 causes a distinct gene expression signature consisting of 497 down-regulated messenger RNAs (mRNAs) and 406 up-regulated mRNAs (false discovery rate [FDR] < 10%, SI Appendix, Fig. S7A). Of note is that 96 of the significantly up-regulated genes, but none of the significantly down-regulated genes, are encoded on chr21, consistent with the expected effects of increased gene dosage (SI Appendix, Fig. S7A). However, ∼90% of DEGs are encoded elsewhere in the genome, indicating the existence of signaling cascades dysregulated by the trisomy. In order to identify these signaling pathways, we employed the Upstream Regulator analysis tool of the Ingenuity Pathway Analysis (IPA) software (52), a bioinformatics tool that uses thousands of available gene expression datasets and myriad other experimental evidence to predict the “upstream regulators” (i.e., driving factors) responsible for the gene expression changes observed in a given experimental condition (52). IPA revealed that the top 2 upstream regulators predicted to be activated in WBCs with T21 are CD3 and CD28, the coreceptors mediating T cell activation (Fig. 6A). Therefore, although the samples were processed immediately after blood draw without any stimulation, samples with T21 resemble samples known to have activated T cells. Importantly, these predictions are identical when we employed only DEGs not encoded on chr21, indicating that these predictions are driven by the genome-wide signature (non-chr21, Fig. 6A). Although variable among individuals with T21, the CD3 scores (calculated from a gene signature composed of 38 mRNAs; Dataset S5) are ∼40-fold higher in people with DS (Fig. 6B), indicating that, while the total number of CD3+ cells is not elevated in DS, when looking at the entire WBC population, the most prominent gene expression changes are consistent with a more activated T cell compartment. Examples of genes in the CD3 signature elevated in samples with T21 are shown in SI Appendix, Fig. S7B. Other upstream regulators predicted to be activated in people with DS include cytokines (IL-18, IL-1β, IL-2) and components of the IFN signaling pathway (IRF7, TLR7, IFN-beta, IFNG, IFN alpha) (see Fig. 6A and B for IFN alpha scores). Although CD3 signatures and IFN alpha signatures are clearly different (Dataset S5), there was a strong positive correlation between CD3 and IFN scores in WBCs (SI Appendix, Fig. S7C).
Fig. 6.
Transcriptome analysis reveals gene expression signatures indicative of T cell activation in adults with trisomy 21. RNAseq analysis was performed using bulk WBCs (n = 9 D21; n = 10 T21). (A) Heatmap displaying the results of examining the RNAseq data using the Upstream Regulator tool of the IPA software. Factors predicted to drive the gene expression changes observed are labeled to the left of the heatmap, with CD3 on top. From left to right, the analysis was performed using all DEGs, DEGs not encoded on chr21 (non-chr21), and DEGs encoded on chr21. (B) Dot and whisker plots showing (Top) CD3 activation scores and (Bottom) IFN alpha scores derived from WBC samples with and without T21. (C) Upstream regulator analysis as in A using RNAseq data derived from bulk T cells (n = 7 D21; n = 10 T21). (D) Dot and whisker plots of (Left) CSF2 scores and (Right) IFN alpha scores derived from T cells with and without T21. (E) Enrichment plots from GSEA of T cell RNAseq data, using (Left) the Hallmarks gene sets or (Right) Immunological Signature gene sets. Normalized Enrichment Score (NES) and FDR-corrected q values are shown. (F) Dot and whisker plots for T-bet mRNA levels from (Left) T cell RNAseq data and (Right) the GSE26495 dataset. Benjamini−Hochberg adjusted P values were calculated using DESeq2 and GEO2R (ebayes), respectively. (G) Pearson correlation analysis for IFN alpha scores versus T-bet mRNA expression in T cells. Data in B, D, and F are shown as mean ± SD with significance determined by unpaired t test; data in G are shown as correlation with significance determined by Pearson’s correlation. **P < 0.01; ***P < 0.001 ****P < 0.0001.
To further investigate this phenomenon, we employed the same analysis on RNA sequencing (RNAseq) data previously generated from bulk T cells (CD45+CD3+CD14−CD19−CD56−) (9), which revealed that 5 of the top 10 upstream regulators predicted to be activated in T21 samples belong to the IFN pathway (IFN alpha, IRF7, IRF1, IFNA2, IFNG) (Fig. 6C). Interestingly, the top upstream regulator is CSF2/GM-CSF (Fig. 6 C and D), a cytokine overproduced by CD8+ T cells with T21 (Fig. 2D and SI Appendix, Fig. S3D). Once again, these signatures are driven mostly by DEGs not encoded on chr21, and enriched for canonical IFN-stimulated genes, leading to highly elevated IFN scores in T cells of people with DS (Fig. 6D).
Next, we performed Gene Set Enrichment Analysis (GSEA) to further explore to what degree the gene expression changes observed are consistent with differences in cellular composition and/or specific signaling events. We completed this analysis for both WBCs and T cells, which produced very similar results (Dataset S6). When we analyzed the T cell transcriptome data with the GSEA Hallmark gene sets, the most prominently enriched gene sets were IFN gamma, mTORC1, and IFN alpha (Fig. 6E, SI Appendix, Fig. S7D, and Dataset S6). Of note is that mTOR has been shown to be chronically activated in DS (53) and to be required for types I and II IFN responses (54, 55). Next, we interrogated the T cell transcriptome with the GSEA Immunological Signatures gene sets, which revealed signs of T cell activation and differentiation in samples with T21 (Fig. 6E and Dataset S6). For example, among the top signatures associated with T21 were genes elevated in PD-1 high CD8+ T cells over naïve CD8+ T cells (e.g., T-bet/TBX21, EOMES) and genes up-regulated during exposure of CD4+ T cells to the superantigen toxic shock syndrome toxin (e.g., Ki-67/MKI67 and other cell cycle genes) (Fig. 6F, SI Appendix, Fig. S7 E and F, and Dataset S6). Of note is that these signatures are mostly nonoverlapping (SI Appendix, Fig. S7E), indicating that different fractions of the T21-associated signature can be attributed to shifts in the CD8+ and CD4+ subsets toward activated effector states. Lastly, the T cell RNAseq not only confirmed transcriptional induction of T-bet/TBX21 and EOMES in T cells of people with DS but also revealed that expression of both factors correlated positively with the IFN alpha scores (Fig. 6 F and G and SI Appendix, Fig. S7 F and G).
Altogether, these results indicate that the clear signs of T cell activation and differentiation in people with DS correlate positively with increased IFN signaling.
T Cell Subsets with Trisomy 21 Are Hypersensitive to IFN Stimulation.
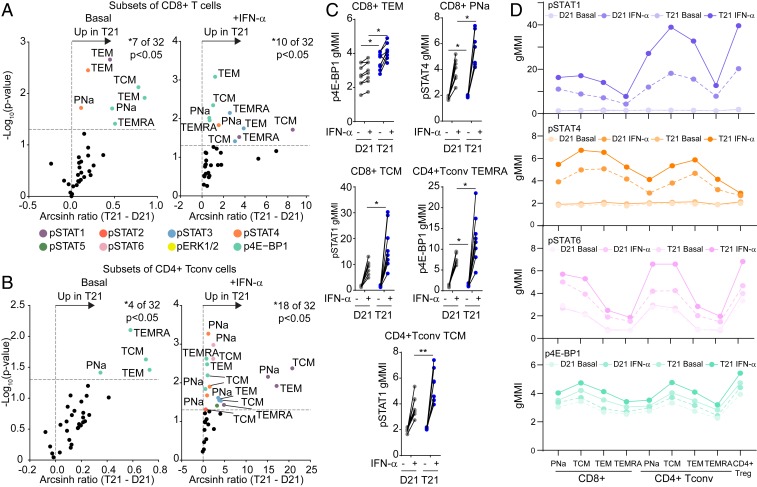
We next hypothesized that the IFN signature observed in T cells of people with DS could be driven by the IFNRs encoded on chr21, which we previously demonstrated to be overexpressed in bulk T cells at the mRNA level (9). We also showed previously that fibroblasts with T21 respond to lower doses of types I and II IFN ligands relative to D21 counterparts (9). In order to investigate whether T cell subsets with T21 are also hypersensitive to IFN stimulation, we employed mass cytometry to measure IFN-α−inducible intracellular signaling cascades using phosphospecific antibodies against the transcription factors STAT1-6, the kinases ERK1-2, and the translation factor 4E-BP1, all of which are known targets of IFN-inducible phosphorylation. Our antibody panel also enabled us to discriminate 9 different T cell subsets by manual gating, including 4 subsets of CD8+ T and CD4+ Tconv cells (PNa, TEM, TCM, and TEMRA), and Tregs (Datasets S1B and S2 and SI Appendix, Fig. S8A) (56). First, we monitored the effect of IFN stimulation on D21 samples, which confirmed that most phosphoepitopes are indeed IFN-inducible in T cell subsets (SI Appendix, Fig. S8B). Evaluation of the impact of T21 on basal IFN signaling revealed that most epitopes are elevated in T cell subsets from people with DS prior to any IFN stimulation (Fig. 7 A and B and SI Appendix, Fig. S8C). In CD8+ T cells, the most basally elevated epitopes were p4E-BP1 in all 4 subsets, pSTAT1 in TEMs, and pSTAT4 in PNa and TEM subsets (Fig. 7 A, C, and D). Among CD4+ Tconvs, p4E-BP1 was the most elevated epitope at baseline in all 4 subtypes (Fig. 7 B–D). In Tregs, the most elevated epitope at baseline was pSTAT6 (Fig. 7D and SI Appendix, Fig. S8 C and D).
Fig. 7.
T cells of people with trisomy 21 are hypersensitive to IFN-α stimulation. Whole blood was incubated directly ex vivo for 30 min with (+IFN-α) or without (basal) IFNα-2A, before being processed for mass cytometry analysis of 8 different phosphoepitopes in 9 different T cell subsets (see SI Appendix, Fig. S8A for gating strategy). (A and B) Volcano plots showing fold changes by karyotype (T21 versus D21) either (Left) under basal conditions or (Right) after stimulation with IFNα-2A on subsets of (A) CD8+ T cells and (B) CD4+ Tconv cells. Fold change was calculated as the arcsinh ratio of samples from people with T21 minus that of samples of typical people (D21). All data represent one Helios run containing a total of n = 8 biologically independent replicates per group. Vertical dashed line represents the no-change midline. Horizontal dashed line represents P value of 0.05 as calculated by Student t test. (C) Dot plots displaying geometric mean metal intensity (gMMI) for the indicated epitopes and cell types, with lines connecting the basal and IFN-α−stimulated values for each sample. (D) Scatter plots showing the gMMI of pSTAT1, pSTAT4, pSTAT6, and p4E-BP1 among the indicated T cell subsets in people with and without T21 before and after stimulation with IFNα-2A. Data in C are shown as mean ± SEM with significance determined by 2-way ANOVA with Sidak’s posttest. *P < 0.05; **P < 0.01.
The greatest impacts of T21 were observed upon IFN-α stimulation, with T cell subsets from people with DS showing hyperactivation of most epitopes tested (compare x axis scales in Basal versus +IFN-α volcano plots in Fig. 7 A and B and SI Appendix, Fig. S8C); however, IFN-inducible phosphorylation events were different across distinct T cell subpopulations, with some epitopes being more abundant in some cell types than others (Fig. 7D). To visualize this phenomenon more clearly, we employed FlowSOM clustering to identify diverse communities of T cells (SI Appendix, Fig. S8E) and then overlaid the information from the phosphoepitopes. This exercise revealed a heterogenous distribution of signaling events across T cell subsets, with clear differences among phosphoepitopes. For example, while pSTAT1 has maximum signals in the CD4+ TCM subset, pSTAT4 peaks in CD8+ subsets (Fig. 7D and SI Appendix, Fig. S8F).
Altogether, these results reveal that T cells from individuals with T21 show higher levels of basal IFN signaling and are also hyperresponsive to type I IFN stimulation.
Discussion
Despite substantial research efforts, the mechanisms by which T21 causes the immune anomalies reported in people with DS remain unclear. The situation is even more complex when investigating the adult immune system, as it becomes harder to dissect the direct effects of the extra chromosome versus environmental and lifestyle variables working across the lifespan of people with DS. Most of what is known regarding the peripheral immune system in people with DS comes from studies in children. These reports agree on an immune profile that resembles one of older typical people: a skewed distribution of the naïve/effector memory subsets, an increased CD8/CD4 ratio of unknown etiology, and a dysfunctional immune response (13, 57, 58). However, little is known about the alterations in the T cell compartment of fully developed adults with DS. Given the remarkable increase in life expectancy for individuals with DS (1), and the interesting observation that they display a different disease spectrum relative to the typical adult population (6), a deep study of their T cell compartment is amply justified. Here, we report that the peripheral T cell compartment of adults with DS is highly dysregulated, with multiple events associated with the development of autoimmunity in the typical population.
Our results demonstrate that adults with DS have a higher number of CD8+ T cells, leading to a persistently high CD8/CD4 ratio, which correlates positively with increased frequency and number of CD4+ Tregs. We found that CD8+ T cells from T21 individuals are functional and more proliferative than their D21 counterparts after polyclonal stimulation, and overproduce cytokines tied to autoimmunity (e.g., TNF-α). They also show signs of polyfunctionality, defined by higher expression of several effector molecules and more than one inhibitory receptor on their surface. These results suggest that the CD8+ T cell compartment of people with DS shares features observed in autoimmune and autoinflammatory conditions (23, 59, 60). Interestingly, the phenotype observed in the CD8+ T cells from adults with DS is consistent with chronic type I IFN hyperactivity, as type I IFN signaling cannot only lead to CD8+ T cell activation but also to expression of negative immune regulators during prolonged viral infections, such as PD-1/PD-L1 (61). Importantly, cytokine analysis showed that inflammatory markers overproduced by CD8+ T cells with T21 are also elevated in plasma of people with DS. Future studies will be required to define whether CD8+ T cells with T21 are more capable of killing target cells, which could reveal why this population is protected from diverse solid malignancies (2).
Previous efforts to characterize the polarization of CD4+ Tconvs in adults with T21 have led to contradictory conclusions (19, 21). We show here that CD4+ Tconvs from people with DS are polarized toward the Th1 and Th1/17 states, overproducing IL-17A and IL-22, concurrent with higher levels of circulating IL-17A-D and IL-22 in plasma compared to controls. The role of IL-17−expressing cells in the pathogenesis of inflammatory and autoimmune disease is well known, particularly in autoimmune diseases characterized by high levels of type I IFN signaling, such as in systemic lupus erythematosus (SLE) and dermatomyositis (62). Furthermore, it has been hypothesized that type I IFN and IL-17 act in concert to sustain and amplify autoimmune and inflammatory responses (62), making them a dangerous combination involved in the pathogenesis of autoimmune diseases, which could explain the increased incidence of autoimmune conditions observed in DS. Interestingly, the increase in IL-10 could be indicative of dampening mechanisms preventing the widespread development of autoimmunity in DS (44).
Our findings demonstrate that Tregs show increased numbers and higher FOXP3 expression in people with T21, yet our analysis did not detect major phenotypic differences. Furthermore, when allogenic criss-cross suppression assays were performed, T21 Tregs were perfectly capable of suppressing D21 CD8+ T cells and CD4+ Tconv cells. However, both effector CD8+ T and CD4+ Tconv cells from people with DS were resistant to Treg-mediated suppression, independent of Treg karyotype. Resistance to suppression has previously been reported in different autoimmune diseases such as type I diabetes (63) and SLE (50). Although the mechanisms leading to effector T cell resistance in DS are unknown at this point, continued exposure to cytokines that are elevated in people with DS, such as TNF-α (10), have been shown to induce T cell resistance to suppression (64). Moreover, both CD8+ and CD4+ Tconv cells from people with DS show increased proliferation rates upon stimulation compared to controls while producing higher levels of effector molecules, which could potentially explain their resistance to Treg-mediated suppression.
Repeatedly, our work points to hyperactive IFN signaling as a potential driver of the observed dysregulation of T cell homeostasis in DS. RNAseq analysis revealed a positive correlation between IFN transcriptional scores, CD3/CD28 activation scores, and expression of T-bet and EOMES (39, 43). Furthermore, GSEA analysis revealed that T cells from adults with DS have transcriptome signatures indicative of both increased differentiation and hyperactivation. The previously observed overexpression of IFNRs in T cells and other cell types with T21 (9) and the clear hyperreactivity that diverse T cell subsets show upon IFN-α stimulation could have profound impacts in shaping the T cell compartment.
Finally, our results point to inhibition of IFN signaling as a therapeutic strategy in DS, particularly for the T cell-driven autoimmune conditions that are more prevalent in this population. IFN signaling can be blocked by a number of strategies, including inhibitors of JAK kinases which are approved for the treatment of autoimmune conditions such as rheumatoid arthritis (65). In fact, we recently reported 2 cases of JAK inhibition in individuals with DS, with remarkable therapeutic benefit for alopecia areata, a T cell-driven autoimmune disorder more prevalent in DS (66). We believe our results justify a thorough investigation of the potential therapeutic benefits of anti-IFN strategies in people with DS.
Methods
Study Approval.
All donors were enrolled under a study protocol approved by the Colorado Multiple Institutional Review Board, known as the Crnic Institute’s Human Trisome Project (COMIRB #15-2170, www.trisome.org). Written informed consent was obtained from parents or guardians of each participant, and assent was obtained from participants over the age of 7 y who were cognitively able to assent. Cohort information can be found in Dataset S2.
Flow Cytometry.
Blood samples were processed and stained according to standard procedures. For more details on antibodies, flow cytometry, and software, see SI Appendix, Materials and Methods.
In Vitro Assays.
In vitro suppression assays were performed according to a protocol modified from ref. 67. For more details, see SI Appendix, Materials and Methods.
RNAseq.
For RNAseq of WBCs, blood samples were immediately processed using a previously described protocol (10). For more details, see SI Appendix, Materials and Methods.
Mass Cytometry Antibody Staining and Data Acquisition.
The procedure for mass cytometry analyses and the antibody−metal conjugates used are detailed in SI Appendix, Materials and Methods.
Meso Scale Discovery Assays (MSD).
Cytokines were measured from plasma samples of 128 individuals (Datasets S2E and S3C) and from supernatant of in vitro stimulated CD8+ and CD4+ Tconv cells of 20 individuals with anti-CD3/CD28 (Datasets S2B and S3 A and B) using V-PLEX immunoassays and a Meso QuickPlex SQ120 from Meso Scale Diagnostics per manufacturer’s instructions.
Statistical Analysis.
All statistical analyses are detailed in SI Appendix, Materials and Methods.
Data Availability.
RNAseq datasets have been deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE128622 for WBCs (68) and GSE84531 for T cells (51). All other datasets and code have been deposited in the Open Science Framework database and can be accessed at https://osf.io/rzpyu/ (22) and https://osf.io/p5kfm/ (56).
Supplementary Material
Acknowledgments
We thank staff members of the Linda Crnic Institute for Down Syndrome, the Sie Center for Down Syndrome, the Global Down Syndrome Foundation, the Clinical and Translational Research Center, the University of Colorado Cancer Center Flow Cytometry Shared Resource, and the Human Immune Monitoring Shared Resource for their support in various aspects of the work. We thank Burkhard Becher for discussion and technical assistance. This work was supported by NIH Grants R01AI150305, R01AI145988, R01AI141662, T32CA190216, UL1TR002535, and P30CA046934. Additional funding was provided by NSF Grant MCB1817582, the Linda Crnic Institute for Down Syndrome, the Global Down Syndrome Foundation, the Anna and John J. Sie Foundation, the Human Immunology and Immunotherapy Initiative, the GI & Liver Innate Immune Program, Proyectos de Investigación Científica y Tecnológica Subsidio 2016-2414, Secretaria de Ciencia y Tecnología de la Universidad Nacional de Córdoba, CONICET, and the Boettcher Foundation. N.G.N. received a fellowship from the University Research Priority Program.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (accession nos. GSE84531 and GSE128622) and the Open Science Framework (https://osf.io/rzpyu/ and https://osf.io/p5kfm/).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908129116/-/DCSupplemental.
References
- 1.de Graaf G., Buckley F., Skotko B. G., Estimation of the number of people with Down syndrome in the United States. Genet. Med. 19, 439–447 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hasle H., Friedman J. M., Olsen J. H., Rasmussen S.A., Low risk of solid tumors in persons with Down syndrome. Genet. Med. 18, 1151–1157 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Hartley D., et al. , Down syndrome and Alzheimer’s disease: Common pathways, common goals. Alzheimers Dement. 11, 700–709 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Madan V., Williams J., Lear J. T., Dermatological manifestations of Down’s syndrome. Clin. Exp. Dermatol. 31, 623–629 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Mårild K., et al. , Down syndrome is associated with elevated risk of celiac disease: A nationwide case-control study. J. Pediatr. 163, 237–242 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Alexander M., et al. , Morbidity and medication in a large population of individuals with Down syndrome compared to the general population. Dev. Med. Child Neurol. 58, 246–254 (2016). [DOI] [PubMed] [Google Scholar]
- 7.de Weerd N. A., Nguyen T., The interferons and their receptors–Distribution and regulation. Immunol. Cell Biol. 90, 483–491 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen K., Liu J., Cao X., Regulation of type I interferon signaling in immunity and inflammation: A comprehensive review. J. Autoimmun. 83, 1–11 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Sullivan K. D., et al. , Trisomy 21 consistently activates the interferon response. eLife 5, e16220 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan K. D., et al. , Trisomy 21 causes changes in the circulating proteome indicative of chronic autoinflammation. Sci. Rep. 7, 14818 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin S., et al. , Thymic deficiency in Down’s syndrome. Pediatrics 63, 80–87 (1979). [PubMed] [Google Scholar]
- 12.Larocca L. M., et al. , Morphological and immunohistochemical study of Down syndrome thymus. Am. J. Med. Genet. Suppl. 7, 225–230 (1990). [DOI] [PubMed] [Google Scholar]
- 13.Bloemers B. L., et al. , Decreased thymic output accounts for decreased naive T cell numbers in children with Down syndrome. J. Immunol. 186, 4500–4507 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Roat E., et al. , Homeostatic cytokines and expansion of regulatory T cells accompany thymic impairment in children with Down syndrome. Rejuvenation Res. 11, 573–583 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Murphy M., Epstein L. B., Down syndrome (DS) peripheral blood contains phenotypically mature CD3+TCR alpha, beta+ cells but abnormal proportions of TCR alpha, beta+, TCR gamma, delta+, and CD4+ CD45RA+ cells: Evidence for an inefficient release of mature T cells by the DS thymus. Clin. Immunol. Immunopathol. 62, 245–251 (1992). [DOI] [PubMed] [Google Scholar]
- 16.Barrena M. J., Echaniz P., Garcia-Serrano C., Cuadrado E., Imbalance of the CD4+ subpopulations expressing CD45RA and CD29 antigens in the peripheral blood of adults and children with Down syndrome. Scand. J. Immunol. 38, 323–326 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Guazzarotti L., et al. , T lymphocyte maturation is impaired in healthy young individuals carrying trisomy 21 (Down syndrome). Am. J. Intellect. Dev. Disabil. 114, 100–109 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Pellegrini F. P., et al. , Down syndrome, autoimmunity and T regulatory cells. Clin. Exp. Immunol. 169, 238–243 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoch J., et al. , Quantitative, phenotypical, and functional characterization of cellular immunity in children and adolescents with Down syndrome. J. Infect. Dis. 215, 1619–1628 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Cossarizza A., et al. , Precocious aging of the immune system in Down syndrome: Alteration of B lymphocytes, T-lymphocyte subsets, and cells with natural killer markers. Am. J. Med. Genet. Suppl. 7, 213–218 (1990). [DOI] [PubMed] [Google Scholar]
- 21.Jakubiuk-Tomaszuk A., et al. , Decrease of interleukin (IL)17A gene expression in leucocytes and in the amount of IL-17A protein in CD4+ T cells in children with Down Syndrome. Pharmacol. Rep. 67, 1130–1134 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Araya P., Characterization of circulating T cells in people with and without Down syndrome. Open Science Framework. https://osf.io/rzpyu/. Deposited 17 October 2019.
- 23.Lili Y., et al. , Global activation of CD8+ cytotoxic T lymphocytes correlates with an impairment in regulatory T cells in patients with generalized vitiligo. PLoS One 7, e37513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wehrens E. J., et al. , Functional human regulatory T cells fail to control autoimmune inflammation due to PKB/c-akt hyperactivation in effector cells. Blood 118, 3538–3548 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Van Gassen S., et al. , FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Mahnke Y. D., Brodie T. M., Sallusto F., Roederer M., Lugli E., The who’s who of T-cell differentiation: Human memory T-cell subsets. Eur. J. Immunol. 43, 2797–2809 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Thome J. J., et al. , Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159, 814–828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontenot J. D., Gavin M. A., Rudensky A. Y., Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Hori S., Nomura T., Sakaguchi S., Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Miyara M., et al. , Functional delineation and differentiation dynamics of human CD4+ T cells expressing the FoxP3 transcription factor. Immunity 30, 899–911 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Halle S., Halle O., Förster R., Mechanisms and dynamics of T cell-mediated cytotoxicity in vivo. Trends Immunol. 38, 432–443 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Sa Q., Woodward J., Suzuki Y., IL-2 produced by CD8+ immune T cells can augment their IFN-γ production independently from their proliferation in the secondary response to an intracellular pathogen. J. Immunol. 190, 2199–2207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saunders K. O., et al. , Secretion of MIP-1β and MIP-1α by CD8+ T-lymphocytes correlates with HIV-1 inhibition independent of coreceptor usage. Cell. Immunol. 266, 154–164 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prévost-Blondel A., Roth E., Rosenthal F. M., Pircher H., Crucial role of TNF-alpha in CD8 T cell-mediated elimination of 3LL-A9 Lewis lung carcinoma cells in vivo. J. Immunol. 164, 3645–3651 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Takeda K., et al. , IFN-γ is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat. Commun. 8, 14607 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiomi A., Usui T., Pivotal roles of GM-CSF in autoimmunity and inflammation. Mediators Inflamm. 2015, 568543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith L. K., et al. , Interleukin-10 directly inhibits CD8+ T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., et al. , Aberrations in circulating inflammatory cytokine levels in patients with Down syndrome: A meta-analysis. Oncotarget 8, 84489–84496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLane L. M., et al. , Differential localization of T-bet and Eomes in CD8 T cell memory populations. J. Immunol. 190, 3207–3215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krieg C., Han P., Stone R., Goularte O. D., Kaye J., Functional analysis of B and T lymphocyte attenuator engagement on CD4+ and CD8+ T cells. J. Immunol. 175, 6420–6427 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Ibegbu C. C., et al. , Expression of killer cell lectin-like receptor G1 on antigen-specific human CD8+ T lymphocytes during active, latent, and resolved infection and its relation with CD57. J. Immunol. 174, 6088–6094 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Dolfi D. V., et al. , Increased T-bet is associated with senescence of influenza virus-specific CD8 T cells in aged humans. J. Leukoc. Biol. 93, 825–836 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jankovic D., Kugler D. G., Sher A., IL-10 production by CD4+ effector T cells: A mechanism for self-regulation. Mucosal Immunol. 3, 239–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duhen T., Campbell D., Human CCR6+CXCR3+ “Th1/17” cells are polyfunctional and can develop from Th17 cells under the influence of IL-1β and IL-12 (P4122) J. Immunol. 190 (suppl. 1.), 191.7 (2013). [Google Scholar]
- 46.Duhen T., Campbell D. J., IL-1β promotes the differentiation of polyfunctional human CCR6+CXCR3+ Th1/17 cells that are specific for pathogenic and commensal microbes. J. Immunol. 193, 120–129 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh S. P., Zhang H. H., Foley J. F., Hedrick M. N., Farber J. M., Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J. Immunol. 180, 214–221 (2008). [DOI] [PubMed] [Google Scholar]
- 48.D’Alise A. M., et al. , The defect in T-cell regulation in NOD mice is an effect on the T-cell effectors. Proc. Natl. Acad. Sci. U.S.A. 105, 19857–19862 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider A., et al. , The effector T cells of diabetic subjects are resistant to regulation via CD4+ FOXP3+ regulatory T cells. J. Immunol. 181, 7350–7355 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venigalla R. K., et al. , Reduced CD4+,CD25- T cell sensitivity to the suppressive function of CD4+,CD25high,CD127 -/low regulatory T cells in patients with active systemic lupus erythematosus. Arthritis Rheum. 58, 2120–2130 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Sullivan K. D., Pandey A., Smith K. P., Espinosa J. M., RNAseq from disomic and trisomic T cells and monocytes. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84531. Deposited 18 July 2016.
- 52.Krämer A., Green J., Pollard J. Jr, Tugendreich S., Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Domenico F., et al. , mTOR in Down syndrome: Role in Aß and tau neuropathology and transition to Alzheimer disease-like dementia. Free Radic. Biol. Med. 114, 94–101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaur S., et al. , Critical roles for Rictor/Sin1 complexes in interferon-dependent gene transcription and generation of antiproliferative responses. J. Biol. Chem. 289, 6581–6591 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kroczynska B., et al. , Interferon γ (IFNγ) signaling via mechanistic target of rapamycin complex 2 (mTORC2) and regulatory effects in the generation of type II interferon biological responses. J. Biol. Chem. 291, 2389–2396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waugh K. A., Whole blood from adults with Down syndrome stimulated with IFNa-2a. Open Science Framework. https://osf.io/p5kfm/. Deposited 17 October 2019. [Google Scholar]
- 57.Joshi A. Y., Abraham R. S., Snyder M. R., Boyce T. G., Immune evaluation and vaccine responses in Down syndrome: Evidence of immunodeficiency? Vaccine 29, 5040–5046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bloemers B. L., van Bleek G. M., Kimpen J. L., Bont L., Distinct abnormalities in the innate immune system of children with Down syndrome. J. Pediatr. 156, 804–809.e5 (2010). [DOI] [PubMed] [Google Scholar]
- 59.Xing L., et al. , Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat. Med. 20, 1043–1049 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blanco P., et al. , Increase in activated CD8+ T lymphocytes expressing perforin and granzyme B correlates with disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 52, 201–211 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Snell L. M., Brooks D. G., New insights into type I interferon and the immunopathogenesis of persistent viral infections. Curr. Opin. Immunol. 34, 91–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ambrosi A., Espinosa A., Wahren-Herlenius M., IL-17: A new actor in IFN-driven systemic autoimmune diseases. Eur. J. Immunol. 42, 2274–2284 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Ihantola E. L., et al. , Effector T cell resistance to suppression and STAT3 signaling during the development of human type 1 diabetes. J. Immunol. 201, 1144–1153 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Mercadante E. R., Lorenz U. M., Breaking free of control: How conventional T cells overcome regulatory T cell suppression. Front. Immunol. 7, 193 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwartz D. M., et al. , JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat. Rev. Drug Discov. 17, 78 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rachubinski A. L., et al. , Janus kinase inhibition in Down syndrome: 2 cases of therapeutic benefit for alopecia areata. JAAD Case Rep. 5, 365–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McMurchy A. N., Levings M. K., Suppression assays with human T regulatory cells: A technical guide. Eur. J. Immunol. 42, 27–34 (2012). [DOI] [PubMed] [Google Scholar]
- 68.Sullivan K. D., et al. , RNAseq transcriptome analysis of White Blood Cells (WBCs) from individuals with and without trisomy 21. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128622. Deposited 20 March 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq datasets have been deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE128622 for WBCs (68) and GSE84531 for T cells (51). All other datasets and code have been deposited in the Open Science Framework database and can be accessed at https://osf.io/rzpyu/ (22) and https://osf.io/p5kfm/ (56).