Significance
Direct in vivo identification and characterization of extracellular vesicles (EVs) in the authentic tumor microenvironment is essential for understanding cancer progression and developing new clinical biomarkers. Here, we introduce an optical-signature-based approach for visualizing, characterizing, and tracking EVs in unperturbed living systems by profiling their intrinsic metabolic and structural contrasts. Imaging of living tumor-bearing animals and fresh excised human breast tissue revealed abundance of NAD(P)H-rich EVs within the tumor, near the tumor boundary, and around vessel structures. In addition, the percentage of NAD(P)H-rich EVs is highly correlated with the human breast cancer diagnosis, which emphasizes the important role of metabolic imaging for EV characterization as well as for clinical applications.
Keywords: extracellular vesicles, NAD(P)H, in situ imaging, human breast cancer, nonlinear microscopy
Abstract
Despite extensive interest, extracellular vesicle (EV) research remains technically challenging. One of the unexplored gaps in EV research has been the inability to characterize the spatially and functionally heterogeneous populations of EVs based on their metabolic profile. In this paper, we utilize the intrinsic optical metabolic and structural contrast of EVs and demonstrate in vivo/in situ characterization of EVs in a variety of unprocessed (pre)clinical samples. With a pixel-level segmentation mask provided by the deep neural network, individual EVs can be analyzed in terms of their optical signature in the context of their spatial distribution. Quantitative analysis of living tumor-bearing animals and fresh excised human breast tissue revealed abundance of NAD(P)H-rich EVs within the tumor, near the tumor boundary, and around vessel structures. Furthermore, the percentage of NAD(P)H-rich EVs is highly correlated with human breast cancer diagnosis, which emphasizes the important role of metabolic imaging for EV characterization as well as its potential for clinical applications. In addition to the characterization of EV properties, we also demonstrate label-free monitoring of EV dynamics (uptake, release, and movement) in live cells and animals. The in situ metabolic profiling capacity of the proposed method together with the finding of increasing NAD(P)H-rich EV subpopulations in breast cancer have the potential for empowering applications in basic science and enhancing our understanding of the active metabolic roles that EVs play in cancer progression.
Extracellular vesicles (EVs) are small membrane-enclosed packages that are secreted by a variety of cell types (1–4). Over the past decade, EVs have been actively studied and shown to be associated with cancer growth and metastasis, including recruiting stromal cells, suppressing the immune response, and determining organotrophic metastasis with distinct biophysical and regional differences (5–12). Direct visualization of EVs within the context of the authentic tumor microenvironment is essential for understanding the distribution and dynamics of EVs, which are highly heterogeneous, and carry profound biological and clinical implications. The state of the art for EV visualization has been expanding mainly due to the rapid development of EV markers and labels (12–17). However, as with other imaging applications, marker-based methods are fundamentally limited by the complex tissue distribution of the markers, unknown disturbance of physiological functions, and unavoidable artifacts of nonspecific false-positive binding (18, 19).
In this paper, we visualize and characterize EVs in living cells and tissues with label-free optical imaging and investigate this utility in the scope of breast cancer research. The challenge of low contrast and specificity (due to small size, little cargo, and complex tissue context) of these nanometer-scale EVs was overcome by using efficient and simultaneous excitation of autofluorescence and multiharmonic signals (15, 20–22). Using a tailored optical fiber source with unique wavelengths (1080–1140 nm), pulse shaping, and repetition rate (10 MHz), the metabolic signatures of the EVs via 2-photon fluorescence (2PF) of FAD, 3-photon fluorescence (3PF) of NAD(P)H (20–24), and the structural properties (lipid–water interface) of the EVs via THG (optical heterogeneity) (25) were captured with precise temporal and spatial coregistration (see more in SI Appendix, Methods). By fully utilizing the intrinsic metabolic and structural properties of EVs, we achieve label-free visualization and characterization of EVs in validated fractioned EV samples, living cells, living animals, and fresh human tissues and found a consistent increase in NAD(P)H-rich EV populations in breast cancer. Lastly, we explored the potential for analyzing the dynamic behaviors (uptake, release, and movement) of EVs in live cells and tissues.
Results
Characterization of Fractioned EVs.
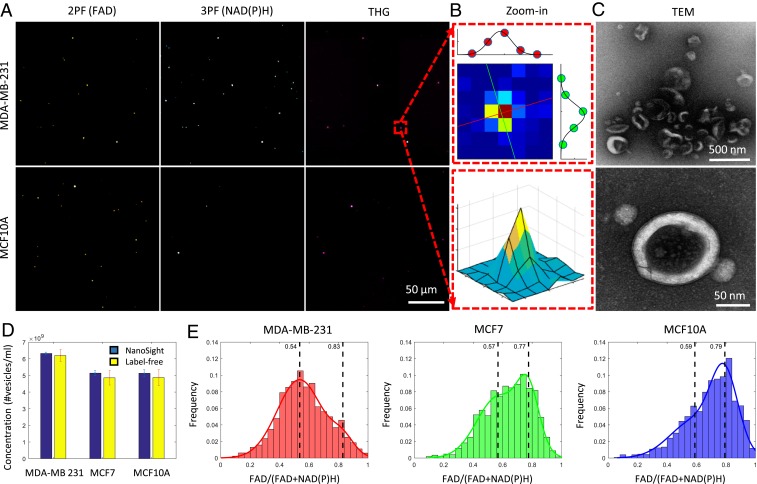
To evaluate the efficiency for detecting EVs from mammary cells of varying breast cancer status, EVs were isolated from the conditioned media of 2 human breast cancer cell lines (MDA-MB-231 and MCF-7) and a nontumorigenic human breast cell line (MCF10A) using differential ultracentrifugation (Fig. 1) (7, 26, 27). Most EVs exhibited circular/oval morphology with bilayer membranes visualized by TEM (Fig. 1C). Based on nanoparticle tracking analysis (NTA) (SI Appendix, Fig. S1), EVs (retrieved from 10-K pellets) showed an average diameter of 227.4 ± 1.2 nm. As the majority of EVs are smaller than the optical resolution (600–700 nm) of multiphoton microscopy (20), they appear as individual diffraction-limited bright punctate pixels in multiphoton microscopy images (Fig. 1 A and B). With no exogenous markers or labels, the contrast of the multiphoton images originates from the intrinsic molecular properties of individual EVs with the intensity of 2PF and 3PF representing the cargo concentrations of FAD and NAD(P)H within the EVs (17–19), respectively, and the THG being generated from the lipid layers/structures of each EV (optical heterogeneity at the aqueous–lipid interface) (25). As expected, the multiphoton image of a negative control (just media centrifuged and collected) is dark in all of the channels (SI Appendix, Fig. S1), in sharp contrast to the distinct punctate points of autofluorescence and harmonic-generated light in the fractioned EV samples. To examine the concordance of these intrinsic optical signatures for identifying EVs, fractionated EVs were labeled with membrane-specific fluorescent markers PKH26 (MilliporeSigma) and imaged using the multiphoton system. Signals generated by PKH26 (excitation 551 nm and emission 567 nm) can be detected in the original 2PF channel, and intrinsic signals from THG and 3PF (NADH) can be detected using the original setting (THG emission 370 nm and 3PF emission 450 nm). We observed spatial coregistration between the fluorescent markers and the intrinsic optical signatures (SI Appendix, Fig. S1), demonstrating the high sensitivity (91%) and specificity (99%) of the proposed label-free detection method. To further quantify the efficiency of this label-free EV detection scheme, we compared the number of EVs detected by NTA and multiphoton microscopy using EV samples of varying densities from 3 commonly used mammary cell lines. Measurements from multiphoton microscopy were largely consistent with NTA (<10% difference), regardless of the cancer status of the mammary cells, suggesting the potential of label-free multiphoton microscopy as a reliable imaging approach for detecting individual EVs (Fig. 1D). The capability of multiphoton microscopy for reliably detecting individual EVs in the context of varying densities and cell origins without the use of exogenous markers demonstrates the strong potential for direct imaging of EVs in complex biological systems for investigative studies.
Fig. 1.
Imaging and characterization of isolated EVs by multiphoton microscopy, transmission electron microscopy (TEM), and NTA. (A) Multiphoton imaging of isolated EVs from MDA-MB-231 cells and MCF10A cells. (B) Zoomed-in image of 1 representative EV with 2D (Upper) and 3D (Lower) visualization maps. The Jet color map was used to highlight the spatial contrast of the diffraction-limited punctate signal. Each pixel is 500 nm in size. (C) TEM images of the same batch of EVs with different magnification. Lower shows the bilayer membrane of an EV. (D) Comparison of EV concentration measured by NTA and multiphoton microscopy (mean ± SD; n = 3 experiments). (E) Histograms of the FAD/[FAD+NAD(P)H] values of EVs isolated from different cell lines. The histograms of EV optical signatures are modeled via a bimodal Gaussian distribution with 2 vertical dashed lines indicating the means of the 2 modes.
Unique Optical Signature for EVs from Breast Cancer Cells.
While unbiased detection of EVs from varying cell origins is desirable, correlation of optical signatures of the EVs and cancer status will enable unprecedented opportunities for the discovery of new biomarkers as well as for a more accurate understanding of the roles that EVs play in cancer. There have been reports of EVs carrying or even synthesizing various metabolic products (28–30). With the capability to simultaneously measure the metabolic [FAD by 2PF and NAD(P)H by 3PF, SI Appendix, Figs. S2 and S3] and structural (THG) properties of each EV, we examined whether multiphoton microscopy could differentiate between EVs isolated from human breast cancer cells and EVs from healthy human breast epithelial cells based on their individual label-free optical signatures. The intensities of each optical channel were analyzed for individual EVs. It was observed that EVs from breast cancer cells have significantly higher 3PF intensity [NAD(P)H concentration] compared to nontumorigenic cells (Fig. 1 A and E). A normalized ratio between FAD and NAD(P)H was used for quantitative analysis (31, 32) (Fig. 1E). As shown in Fig. 1E, EVs from cancer cells (isolated from MDA-MB-231 cells) displayed a significantly lower ratio [approximately higher NAD(P)H concentration, P < 0.01 by the Kolmogorov–Smirnov test] compared to EVs from nontumorigenic cells (isolated from MCF10A cells), while the EVs from noninvasive cancer cells (isolated from MCF-7 cells) have a mixed distribution.
In Vivo Optical Signature Reveals Abundance of NAD(P)H-Rich EVs in the Tumor Microenvironment.
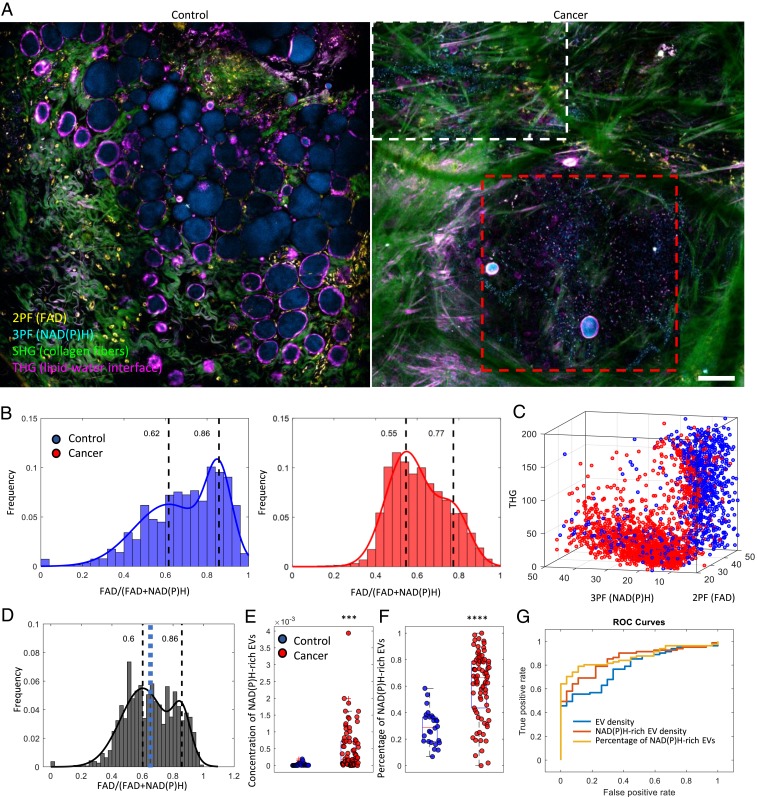
To investigate the correlation of the optical signature of EVs and the cancer status in the authentic living mammary cancer microenvironment, N-Nitroso-N-methylurea was injected into the peritoneal cavity of rats to induce the growth of mammary tumors (33, 34). Surgically exposed tumors were subsequently imaged in vivo using label-free multiphoton microscopy. The large field-of-view and 3D visualization (Fig. 2A, SI Appendix, Figs. S4 and S5, and Movie S1) showed a 500-μm-sized tumor densely packed with tumor cells, vesicles, and tumor-infiltrating immune cells. While most of the THG-high EVs were randomly scattered throughout the tumor mass as well as in the stroma, a significant portion of the NAD(P)H-high EVs were organized in a linear streamlike manner. In addition, the EVs in proximity to the endothelial cells were mostly NAD(P)H enriched, which is likely the result of an antioxidant defense mechanism stimulated by the increased oxidative stress in the tumor endothelial EVs (35).
Fig. 2.
In vivo visualization and characterization of EVs from cancer tissue in rat mammary tumors. (A) In vivo imaging of control rats and tumor-bearing rats by label-free multiphoton microscopy. (B) FAD/[FAD+NAD(P)H] values of EVs from control (n = 5 animals) and tumor-bearing rats (n = 5 animals). (C) Scatter plot of individual EVs with intensities from the 3 channels. (D) Combined histogram of vesicle optical signatures from cancer and normal tissue. The blue dashed line indicates the computed threshold value (0.65) for identifying the subpopulation of NAD(P)H-rich EVs by using Ostu’s method. (E) Concentration and (F) percentage of NAD(P)H-rich EVs in relation to their cancer status (2-tailed Student’s t test, ***P < 0.001, and ****P < 0.0001; 26 imaging sites from the control group and 82 imaging sites from the cancer group). (G) Receiver operating characteristic (ROC) curve of cancer prediction by logistic regression using different features. The feature of EV density yields an area under the ROC curve (AUC) of 0.786, the NAD(P)H-rich EV density yields an AUC of 0.849, and the percentage of NAD(P)H-rich EVs yields an AUC of 0.868 (Scale bar: 100 µm.).
Consistent with EVs isolated from breast cancer cell cultures (Fig. 1), analysis of individual EV content (via optical signatures) reveals significantly (P < 0.01 by the Kolmogorov–Smirnov test) higher concentrated NAD(P)H in EVs from mammary cancer tissue (Fig. 2B). In addition, EVs from tumor-bearing animals and EVs from control animals appear to form 2 distinct clusters in the 3-channel intensity space with minimal overlap (Fig. 2C), implicating the existence of 2 distinct subpopulations of EVs carrying different levels of metabolic products. In addition, the cluster of NAD(P)H-rich EVs is strongly associated with EVs from cancerous tissue. Based on the histograms of the optical (metabolic) signatures from both cancer and healthy EVs (Fig. 2D), a threshold value was computed using Otsu’s method to separate the 2 subpopulations of EVs. For simplicity, any EV that had a signature lower than the threshold value was defined as an NAD(P)H-rich EV in this paper. Thus, the concentration and the percentage of NAD(P)H-rich EVs can be readily calculated for each site (see more in Materials and Methods). As observed, the level of NAD(P)H-rich EVs was highly correlated with mammary cancer status (Fig. 2 E–G) and yielded far better cross-validation AUC (area under curve) values for the corresponding classification task than EV concentration alone.
This high concentration of NAD(P)H has been observed in cancer cells (31, 36) previously as a result of the Warburg effect (37) (increased glycolysis) as well as an antioxidant defense mechanism (38). To study the correlation between the NAD(P)H levels of EVs and that of the potential parental cancer cells, we compared cancer cells with tumor-bordering EVs as well as EVs outside the visible tumor area (SI Appendix, Fig. S6). The relative NAD(P)H concentration was estimated based on the known linear dependence of 3PF signals on the NAD(P)H concentration (39). Intensity analysis reveals that the 3PF intensity [correlated with NAD(P)H concentration] from the EVs was, on average, 5-fold higher than that of the neighboring cancer cells (SI Appendix, Fig. S6). In addition, EVs that were not in the proximity of any NAD(P)H-rich cells (SI Appendix, Figs. S6 and S7 and Movie S2) were observed to have an even higher 3PF intensity (7-fold higher than cancer cells). The difference in NAD(P)H levels between cancer cells and corresponding EVs was also observed in breast cancer cell cultures (SI Appendix, Fig. S8). This is an intriguing observation that highlights the possibly unique metabolic functions and signatures of EVs in breast cancer (28–30), but future studies are needed to elucidate exactly what mechanism results in this unexpected higher NAD(P)H concentration from certain cancer-associated EVs. Together, these results demonstrate the importance of directly mapping the metabolically and spatially heterogeneous EVs in the complex highly interactive and continuously evolving tumor microenvironment.
Abundance of NAD(P)H-Rich EVs Highly Correlated with Diagnosis of Human Breast Cancer.
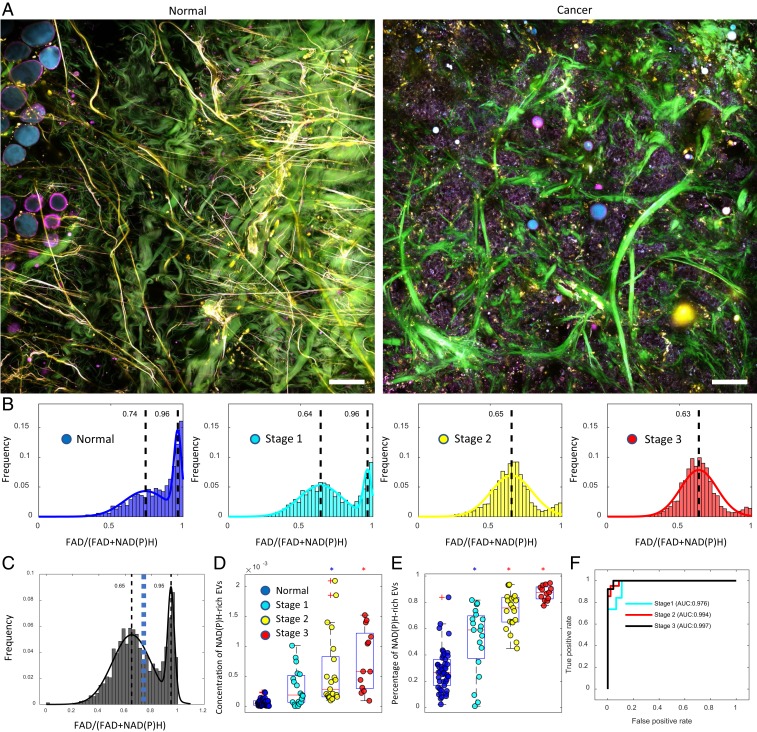
To explore the clinical potential of the finding of NAD(P)H-enriched EVs in cancer, we imaged fresh human breast tissue excised from female subjects with invasive ductal carcinoma as well as from female subjects undergoing breast reduction surgery with no history of cancer (Fig. 3A). Consistent with the preclinical study, the EVs from breast cancer patients have significantly higher NAD(P)H concentration than those from cancer-free subjects (Fig. 3 A and B). This observation remains true for both early- and late-stage cancer and for tissue that was both close to (within the tumor or adjacent to the tumor boundary) and far from (>5 cm away from tumor boundary) the tumor margin, which supports the hypothesis and findings that EVs have both an early and physically far-reaching influence in carcinogenesis (10, 11, 40). Similar to the EVs from rat mammary tumors, the histogram generated by combining EVs from human breast cancer and cancer-free subjects appeared bimodal, and a threshold value was determined by Ostu’s method to separate out the subpopulation of NAD(P)H-rich EVs (Fig. 3C). Statistical analysis of this cohort revealed that the level of NAD(P)H-rich EVs distinguished subjects with early-stage breast cancer from healthy subjects as well as subjects with late-stage breast cancer (Fig. 3 D and E). When comparing patients with Stages 1–3 breast cancer to healthy subjects, ROC curves show that NAD(P)H-rich EVs exhibit high accuracy for discriminating each stage of breast cancer (Fig. 3F, AUC Stage 1: 97.6%; Stage 2: 99.4%; and Stage 3: 99.7%) from healthy tissue, supporting the utility of these optical signatures also as a biomarker for the various stages and progression of human breast cancer.
Fig. 3.
High levels of NAD(P)H-rich EVs are correlated with early- and late-stage human breast cancer. (A) Label-free multiphoton imaging of fresh normal tissue and cancer tissue. (B) FAD/[FAD+NAD(P)H] values of EVs from subjects with different cancer statuses (19 subjects in total, Normal [n = 7], Stage 1 [n = 5], Stage 2 [n = 3], and Stage 3 [n = 4]). (C) Combined histogram of vesicles optical signature from cancer and normal tissue. The blue dashed line indicates a computed threshold value (0.73) for identifying the subpopulation of NAD(P)H-rich EVs by using Ostu’s method. (D) Concentration and (E) percentage of NAD(P)H-rich EVs in relation to their cancer status (ANOVA, post hoc Tukey–Kramer test; 105 imaging sites in total, Healthy [n = 52], Stage 1 [n = 19], Stage 2 [n = 21], Stage 3 [n = 13]). The colored circles correspond to the legend in D. The blue asterisks indicate statistical significance compared to healthy subjects, while the red asterisks indicate statistical significance compared to both healthy subjects and Stage 1 breast cancer subjects. (F) ROC curve of cancer prediction by logistic regression for different cancer stages (Stage 1 AUC: 0.978, Stage 2 AUC: 0.994, Stage 3 AUC: 0.997) (Scale bar: 100 µm.).
EV Dynamics in Living Cancer Cells and Animals.
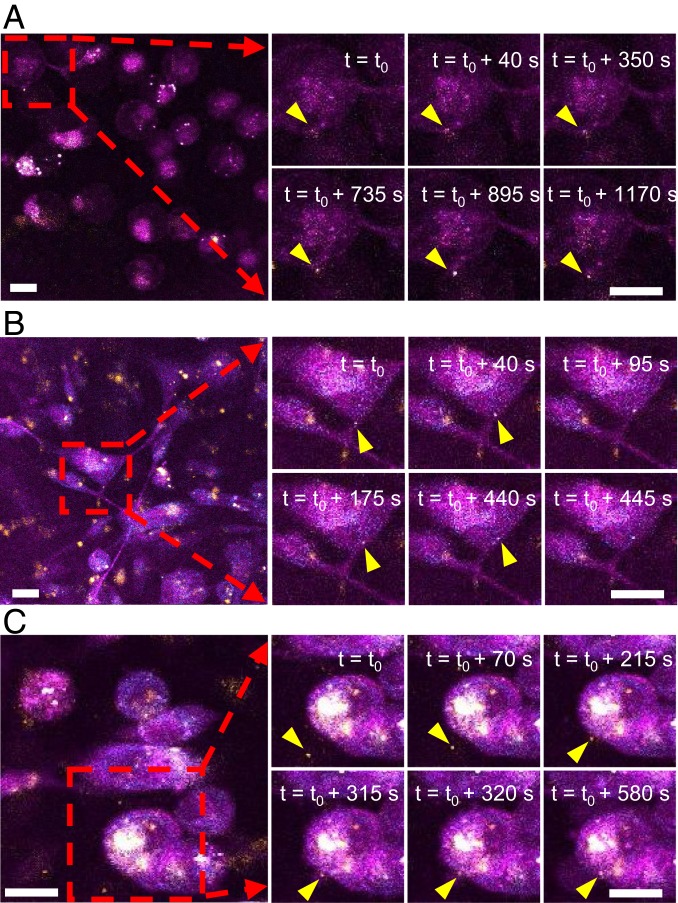
In addition to characterizing the metabolic properties of individual EVs, label-free multiphoton microscopy promises to monitor the dynamic behavior of EVs in live cells and tissues without perturbation. We performed live cell imaging targeting the cellular release and uptake of EVs in living MDA-MB-231 cancer cells without the aid of exogenous markers. As expected, we observed vesicles of varying sizes inside and outside the cells with most vesicles appearing to actively and randomly move within the cells. Vesicles were also observed being secreted or uptaken (Fig. 4 A–C and Movies S3–S5). Fig. 4A shows the dynamic process of vesicle release (Movie S3). Fig. 4B shows the migration of a vesicle from the end of a protrusion from the cell body through a tube-like channel (Movie S4). Fig. 4C shows the process of vesicle uptake—a vesicle moving freely in the media, approaching a target cell, appearing to be tethered to the cell, and 2 min later, entering the cell (Movie S5).
Fig. 4.
EV dynamics in MDA-MB-231 cells in vitro. (A) Release of a vesicle (Movie S3). (B) Migration of a vesicle through a cell protrusion (Movie S4). (C) Uptake of a vesicle (Movie S5) (Scale bar: 20 µm.).
EV visualization is further motivated by the potential to perform EV tracking in living animal models to enable more comprehensive and authentic observations of EV dynamics and vesicle-cell-stromal interactions. EV locomotion in the tumor microenvironment was tracked with time-lapse imaging at multiple intratumor sites and tumor-neighboring sites. In contrast to the active release, uptake, and migration of EVs observed in cell cultures (Fig. 4 A–C and Movies S3–S5), the majority of EVs in the living tumor microenvironment appear immobilized and firmly attached to the extracellular matrix and vessel walls (SI Appendix, Fig. S9 and Movies S6–S9). This observation is not consistent with previous reports that EVs systemically disseminate to phenocopy metastatic behavior or to condition secondary sites (6, 16, 17). This discrepancy might be attributed to the heterogeneity of dynamic functions in EVs as well as the possibility of other vesicle migration mechanisms. More in-depth studies are needed to investigate the mechanisms behind these motility patterns.
Discussion
Complementary to the widely used isolation-based EV assays, direct visualization and characterization of EVs in intact living cells and tissues promises to add more comprehensive and informative insight into their heterogeneous properties and distributions in unprocessed tissue, as well as their spatial–temporal incorporation and role in the highly interactive and ever-evolving tumor microenvironment. We expect that these methods and results will open doors for the exploration of the dynamic roles of cancer-associated vesicles in both basic research and clinical applications. The in situ metabolic profiling capacity of the proposed method together with the finding of the increasing NAD(P)H-rich EV subpopulation in mammary cancer can empower applications in the basic sciences as well as enhance our understanding of the active metabolic roles that EVs play in cancer progression. For clinical applications, the potential of this method is demonstrated via its sample-preparation-free nature and its strong correlation with clinical diagnosis. However, more studies are needed to prove that this method and this finding can be effectively used for improving clinical diagnosis, screening, and monitoring. The system instrumentation, imaging, and analysis will need to be standardized and pipelined to be demonstrated as a reliable and user-friendly clinical instrument. Investigations of liquid biopsy samples will be conducted to fully explore the clinical utility of this method for improving breast cancer screening and diagnosis. It is also possible that the abundance of NAD(P)H-rich EVs is not only specific to breast cancer, but also applicable to other types of cancers. However, we should be careful about generalizing these breast cancer based findings to other cancer types without extensive targeted studies as different cancer types are known to have different metabolic alterations and profiles (41, 42). Dedicated investigations will be performed in the future to systematically identify the metabolic signatures of EVs for other cancer types.
Materials and Methods
All experiments involving human breast tissue were conducted in accordance with a protocol approved by the Institutional Review Boards at the University of Illinois at Urbana–Champaign and Carle Foundation Hospital, Urbana, IL. All human tissue samples were obtained from subjects who preoperatively provided informed consent permitting the investigational use of their tissues. All experiments involving animals were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee at the University of Illinois at Urbana–Champaign. Detailed methods are available in the SI Appendix.
Data Availability.
The data and codes to perform the analyses have been deposited in OSF (https://osf.io/r5kdt/?view_only=ac9b10f32f31475abf8c78d1a682fa25) (43).
Supplementary Material
Acknowledgments
We thank Crislyn D’Souza-Schorey, James W. Clancy, and Edita Aksamitiene for providing expertise and insight for the EV isolation procedures. We thank Darold Spillman for his technical support. We thank Anna Higham, Kimberly Cradock, Natasha Luckey, and Z. George Liu from Carle Foundation Hospital for assisting in the acquisition and histological assessment of the human tissue specimens in this study. This work was supported, in part, by grants from the National Institutes of Health (R01 CA166309, R01 CA213149, and R01 EB023232 to S.A.B.). Additional information can be found at http://biophotonics.illinois.edu.
Footnotes
Competing interest statement: S.A.B., H.T., and S.Y. are named inventors on patents filed by the University of Illinois at Urbana–Champaign related to the laser source technology and the imaging and quantification of extracellular vesicles. S.A.B. and H.T. are co-founders and hold equity interest in LiveBx, LLC, Champaign, IL, which is commercializing the laser source technology used in this study. Requests for materials should be addressed to S.A.B. All other authors declare that they have no competing financial interests.
This article is a PNAS Direct Submission. R.W. is a guest editor invited by the Editorial Board.
Data deposition: The data and codes to perform the analyses have been deposited in OSF (https://osf.io/r5kdt/?view_only=ac9b10f32f31475abf8c78d1a682fa25).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909243116/-/DCSupplemental.
References
- 1.Zaborowski M. P., Balaj L., Breakefield X. O., Lai C. P., Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience 65, 783–797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lötvall J., et al. , Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 3, 26913 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raposo G., Stoorvogel W., Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crescitelli R., et al. , Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles 2, 20677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker A., et al. , Extracellular vesicles in cancer: Cell-to-cell mediators of metastasis. Cancer Cell 30, 836–848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoshino A., et al. , Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muralidharan-Chari V., Clancy J. W., Sedgwick A., D’Souza-Schorey C., Microvesicles: Mediators of extracellular communication during cancer progression. J. Cell Sci. 123, 1603–1611 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbins P. D., Morelli A. E., Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195–208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tkach M., Théry C., Communication by extracellular vesicles: Where we are and where we need to go. Cell 164, 1226–1232 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Willms E., Cabañas C., Mäger I., Wood M. J. A., Vader P., Extracellular vesicle heterogeneity: Subpopulations, isolation techniques, and diverse functions in cancer progression. Front. Immunol. 9, 738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa-Silva B., et al. , Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pucci F., et al. , SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science 352, 242–246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai C. P., et al. , Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 6, 7029 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao H., et al. , New technologies for analysis of extracellular vesicles. Chem. Rev. 118, 1917–1950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y., et al. , Intraoperative visualization of the tumor microenvironment and quantification of extracellular vesicles by label-free nonlinear imaging. Sci. Adv. 4, eaau5603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zomer A., et al. , In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 161, 1046–1057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Vos K. E., et al. , Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol. 18, 58–69 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitamura T., Pollard J. W., Vendrell M., Optical windows for imaging the metastatic tumour microenvironment in vivo. Trends Biotechnol. 35, 5–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takov K., Yellon D. M., Davidson S. M., Confounding factors in vesicle uptake studies using fluorescent lipophilic membrane dyes. J. Extracell. Vesicles 6, 1388731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.You S., et al. , Intravital imaging by simultaneous label-free autofluorescence-multiharmonic microscopy. Nat. Commun. 9, 2125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You S., et al. , Slide-free virtual histochemistry (Part I): Development via nonlinear optics. Biomed. Opt. Express 9, 5240–5252 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You S., et al. , Slide-free virtual histochemistry (Part II): Detection of field cancerization. Biomed. Opt. Express 9, 5253–5268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tu H., et al. , Stain-free histopathology by programmable supercontinuum pulses. Nat. Photonics 10, 534–540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu H., et al. , Concurrence of extracellular vesicle enrichment and metabolic switch visualized label-free in the tumor microenvironment. Sci. Adv. 3, e1600675 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weigelin B., Bakker G.-J., Friedl P., Third harmonic generation microscopy of cells and tissue organization. J. Cell Sci. 129, 245–255 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Théry C., Amigorena S., Raposo G., Clayton A., Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter 3, 22 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Clancy J. W., et al. , Regulated delivery of molecular cargo to invasive tumour-derived microvesicles. Nat. Commun. 6, 6919 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H., et al. , Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife 5, e10250 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodega G., et al. , Young and especially senescent endothelial microvesicles produce NADPH: The fuel for their antioxidant machinery. Oxid. Med. Cell. Longev. 2018, 3183794 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iraci N., et al. , Extracellular vesicles are independent metabolic units with asparaginase activity. Nat. Chem. Biol. 13, 951–955 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skala M. C., et al. , In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. U.S.A. 104, 19494–19499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., et al. , Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast. Sci. Adv. 4, eaap9302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gullino P. M., Pettigrew H. M., Grantham F. H., N-nitrosomethylurea as mammary gland carcinogen in rats. J. Natl. Cancer Inst. 54, 401–414 (1975). [PubMed] [Google Scholar]
- 34.Cha R. S., Thilly W. G., Zarbl H., N-nitroso-N-methylurea-induced rat mammary tumors arise from cells with preexisting oncogenic Hras1 gene mutations. Proc. Natl. Acad. Sci. U.S.A. 91, 3749–3753 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larson M. C., Hillery C. A., Hogg N., Circulating membrane-derived microvesicles in redox biology. Free Radic. Biol. Med. 73, 214–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgakoudi I., Quinn K. P., Optical imaging using endogenous contrast to assess metabolic state. Annu. Rev. Biomed. Eng. 14, 351–367 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Vander Heiden M. G., Cantley L. C., Thompson C. B., Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panieri E., Santoro M. M., ROS homeostasis and metabolism: A dangerous liason in cancer cells. Cell Death Dis. 7, e2253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng L.-C., Horton N. G., Wang K., Chen S.-J., Xu C., Measurements of multiphoton action cross sections for multiphoton microscopy. Biomed. Opt. Express 5, 3427–3433 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Loghlen A., Role for extracellular vesicles in the tumour microenvironment. Philos. Trans. R. Soc. B Biol. Sci. 373, 20160488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Budhu A., et al. , Metabolic profiles are principally different between cancers of the liver, pancreas and breast. Int. J. Biol. Sci. 10, 966–972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halama A., et al. , Metabolic signatures differentiate ovarian from colon cancer cell lines. J. Transl. Med. 13, 223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.You S., Multiphoton imaging of extracellular vesicles. Open Science Framework. https://osf.io/r5kdt/?view_only=ac9b10f32f31475abf8c78d1a682fa25. Deposited 19 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and codes to perform the analyses have been deposited in OSF (https://osf.io/r5kdt/?view_only=ac9b10f32f31475abf8c78d1a682fa25) (43).