Significance
Enzymes involved in the biosynthesis of peptide natural products may be used as platforms for regioselective and chemoselective modifications. These enzymes can catalyze the same chemical transformation on extremely diverse peptide substrates. Here, we show that recruitment of these modification enzymes to a given substrate is dictated by a minimal number of hydrophobic interactions. Installation of this motif can direct modification on unrelated peptide substrates, and establishes a framework for generating new peptidic compounds with drug-like properties.
Keywords: biochemistry, RiPPs, biosynthesis
Abstract
Enzymes that generate ribosomally synthesized and posttranslationally modified peptide (RiPP) natural products have garnered significant interest, given their ability to produce large libraries of chemically diverse scaffolds. Such RiPP biosynthetic enzymes are predicted to bind their corresponding peptide substrates through sequence-specific recognition of the leader sequence, which is removed after the installation of posttranslational modifications on the core sequence. The conservation of the leader sequence within a given RiPP class, in otherwise disparate precursor peptides, further supports the notion that strict sequence specificity is necessary for leader peptide engagement. Here, we demonstrate that leader binding by a biosynthetic enzyme in the lasso peptide class of RiPPs is directed by a minimal number of hydrophobic interactions. Biochemical and structural data illustrate how a single leader-binding domain can engage sequence-divergent leader peptides using a conserved motif that facilitates hydrophobic packing. The presence of this simple motif in noncognate peptides results in low micromolar affinity binding by binding domains from several different lasso biosynthetic systems. We also demonstrate that these observations likely extend to other RiPP biosynthetic classes. The portability of the binding motif opens avenues for the engineering of semisynthetic hybrid RiPP products.
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) constitute a large class of structurally diverse bioactive natural products in which chemical diversity is produced by extensive posttranslational modification on linear precursor peptides. Such precursors can often be demarcated into a highly conserved leader sequence that guides recognition by posttranslational modification enzymes and a highly variable core sequence where the modifications are actually installed (1). RiPP biosynthetic enzymes specifically recognize their corresponding leader sequence but are tolerant of variations in the core sequence. The importance of the leader in directing biosynthesis is underscored by the observation that several RiPP biosynthetic clusters, including those that produce lanthipeptides (2), cyanobactins (3), or lasso peptides (4), can encode multiple precursor peptides, which all share conserved leader sequences but contain distinct core sequences. Notably, each of these precursor peptides can be posttranslationally modified by the same set of enzymes within each biosynthetic cluster to yield structurally divergent products (1). Hence, the separation of the recognition element from the site of modification enables chemical diversity without sacrificing specificity during RiPP biosynthesis.
Leader sequences in RiPP precursor peptides are engaged by domains that may exist either as a stand-alone protein or as fusions with catalytic domains of the modification enzymes. Such leader-binding domains structurally fall into the PqqD superfamily (PF05402/IPR008792), so named for a stand-alone protein responsible for binding the peptide precursor for the bacterial redox cofactor pyrroloquinoline quinone (PQQ) (5–7). These binding domains have also been called RiPP precursor peptide recognition elements (RREs) when present in a RiPP biosynthetic cluster (7). Binding of the leader sequence is presumed to allow PqqD/RRE domains to present the core peptide to the catalytic domain/protein for modification (8, 9). The prevailing proposal for RiPP biosynthesis suggests that substrate peptides are recruited to the biosynthetic enzymes through sequence specific recognition of the leader peptide, enabled by the PqqD/RRE domain from the corresponding biosynthetic cluster (10, 11).
Lasso peptides are a class of RiPPs characterized by an isopeptide bond between the N-terminal α-amine and side chain carboxylate of an Asp or Glu residue located 6 to 8 amino acids downstream (12). The isopeptide linkage results in an internal lactam ring that encircles the remaining linear peptide to create a lasso-like shape. The minimal set of genes required for the biosynthesis of lasso peptides encodes the precursor peptide (A protein), a transglutaminase-like protease (B protein), and an asparagine synthase-like protein (C protein). In vitro reconstitution of the lasso peptides microcin J25 (13, 14) and fuscanodin/fusilassin (15, 16) provides a route for understanding lasso peptides biosynthesis. First, the B-protein protease binds to and cleaves the leader peptide from the A-protein precursor peptide to yield the core peptide. Next, the side chain of a Glu/Asp C-terminal to the cleavage site (located 7 residues downstream in the case of microcin J25) is activated with adenosine 5′-triphosphate (ATP) by the C protein to form an adenylyl intermediate. Nucleophilic attack by the newly exposed α-amine of the core peptide onto the Glu/Asp side chain adenylate generates the macrolactam ring. This ring encircles the C-terminal linear portion of the core peptide to yield the characteristic lasso structure. An efflux pump (the D protein) extrudes the mature product from the producing organism.
In some lasso peptide biosynthetic clusters, the B protein is a single polypeptide, but, in others, such as those that produce lassomycin (17), lariatin (18), and fuscanodin/fusilassin (15, 16), this gene is split into 2 open reading frames (ORFs), B1 and B2. These 2 genes are similar to the N and C termini, respectively, of full-length B. Bioinformatics analyses of B-protein sequences identifies that the B1 protein (and the corresponding N-terminal region of fused B proteins) contains hallmarks of an RRE (7). Binding studies of proteins from different split lasso peptide biosynthetic clusters demonstrate that the B1 proteins from each cluster bind to the cognate leader peptides with submicromolar affinity (7, 19, 20). Coincubation studies with heterologously purified lasso peptide biosynthetic proteins from the paeninodin and fuscanodin/fusilassin clusters demonstrates that proteolysis of the precursor peptide by the B2 protein only occurs in the presence of the corresponding B1 protein, indicating that leader recognition precedes and dictates peptide processing (15, 16, 20).
The bifurcation of the recognition site from the site of enzymatic modification in the precursor peptide substrate has focused research interest in the engineering of designer, or even hybrid, RiPPs. However, such efforts have been hindered by a paucity of data regarding the exact determinants for leader recognition. Here, we identify a plausible lasso peptide biosynthetic cluster from Thermobacculum terrenum American Type Culture Collection (ATCC) BAA-798 (Tbi) that encodes 2 putative precursor peptides with leaders that are sequence-divergent. We demonstrate that the single leader recognition domain from the Tbi cluster can engage both precursor peptides as competent substrates for subsequent processing. Crystal structures and binding studies identify a simple motif in the leader peptide that directs leader recognition by primarily hydrophobic interactions and steric complementarity. Implantation of this motif into a noncognate peptide results in leader recognition and processing by the Tbi protease machinery. Notably, we show that recognition proteins from divergent lasso peptide pathways can engage the unrelated TbiAα leader peptide, and we carry out analysis suggesting that hydrophobic packing is the guiding force for leader peptide recognition. These studies open avenues for further design of hybrid leader sequences that facilitate precursor recognition and modification by orthogonal classes of RiPP biosynthetic enzymes.
Results
Identification and Verification of Therbactin Biosynthetic Cluster.
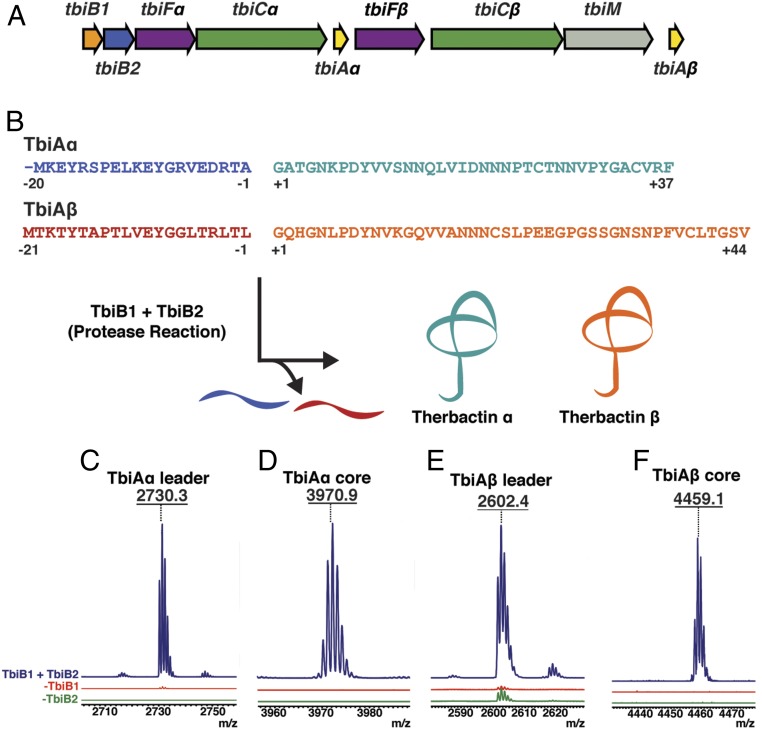
In order to identify systems amenable for biochemical studies, we manually curated putative lasso peptide biosynthetic clusters from thermophilic organisms. Of particular interest was a putative gene cluster from the thermophilic bacteria T. terrenum ATCC BAA-798, which we denote here as tbi (Fig. 1A and SI Appendix, Table S1). The gene cluster includes the B1 protein (tbiB1) that engages the leader sequence in the precursor peptide, the peptidase (tbiB2) that excises the leader from the precursor peptide, and an efflux pump (tbiD) for export of the mature lasso product. The cluster contains 2 different genes that resemble the requisite isopeptide-forming lasso cyclase (tbiCα and tbiCβ). Other genes include plausible tailoring enzymes encoding a methyltransferase and 2 kinases (tbiM, and tbiFa/β, respectively) (21). Unexpectedly, the cluster also encodes 2 putative precursor peptides (tbiAα and tbiAβ) that possess sequence-divergent leader and core sequences, sharing only 50% sequence similarity (Fig. 1 A and B and SI Appendix, Fig. S1). This contrasts with observations from other RiPP clusters that contain multiple precursor peptides, in which the core sequences may be divergent but the leader sequences are highly conserved (10, 22). For example, the 30 different precursor peptides encoded in the prochlorosin biosynthetic cluster have a sequence similarity of >90% among the leader sequences, but no discernable conservation between core peptides (2). The gene organization of the tbi cluster suggests that the single split B protein can process both precursor peptides, despite the divergence in their leader sequences. Further modification of the 2 processed precursors may be catalyzed by the 2 different C proteins to yield the corresponding lasso products. While these final products were not isolated, we have named the theoretical mature lasso peptides therbactin α and therbactin β (Fig. 1B).
Fig. 1.
(A) The therbactin gene cluster of T. terrenum. (B) Proposed biosynthetic route for therbactin α and therbactin β. (C−F) MALDI-TOF-MS analysis of the TbiB1- and TbiB2-mediated protease reaction observing formation of (C) TbiAα leader: 2730.3 m/z; (D) TbiAα core: 3970.9 m/z; (E) TbiAβ leader: 2602.4 m/z; and (F) TbiAβ core: 4459.1 m/z.
To decipher the strategy underlying the biosynthesis of therbactins α and β, we carried out reconstitution of the split B-protein activity using both precursor peptides as substrates. Given the low conservation between the 2 putative leader sequences, we speculated that TbiB1 would bind to one of the 2 precursor peptides via the leader sequence and present the core sequence to TbiB2 for proteolysis. Using matrix-assisted laser desorption ionization, matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) analysis, we demonstrate that the TbiAα precursor is indeed a precursor and that the combination of both TbiB1 and TbiB2 is necessary for processing of this peptide (Fig. 1 C and D and SI Appendix, Fig. S2). No activity was observed upon omission of either of the 2 proteins. Surprisingly, TbiB1+TbiB2 could also process the TbiAβ precursor (Fig. 1 E and F and SI Appendix, Fig. S3), despite the lack of extensive sequence conservation among the leader sequences of the TbiAα and TbiAβ precursor peptides. Lastly, although ATP is a required cosubstrate for processing by the fused B proteins (14), proteolysis by the therbactin system is consistent with other split B proteins, as it does not require ATP (15, 16, 20).
The above data suggest that a single leader-binding protein can engage multiple sequence-divergent leader peptides, each of which can be processed by a single leader peptidase. In order to provide direct evidence of leader binding, we used fluorescence polarization assays to quantify the affinities of TbiB1 for both TbiAα and TbiAβ. Binding measurements to an N-terminally fluorescein isothiocyanate (FITC)-labeled TbiAα leader peptide yields a Kd of 266 nM (SI Appendix, Table S2 and Fig. S4). Competition assays with unlabeled TbiAα gave a Ki of 359 nM, suggesting that the FITC tag did not significantly alter the peptide binding affinities of TbiB1 (SI Appendix, Fig. S5A). The protein bound to the TbiAβ leader somewhat more tightly with a Ki of 227 nM (SI Appendix, Fig. S5B). Hence, even though the leader sequences of TbiAα and TbiAβ are divergent, TbiB1 binds both peptides with comparable affinity.
Molecular Basis for Promiscuity in Leader Peptide Engagement.
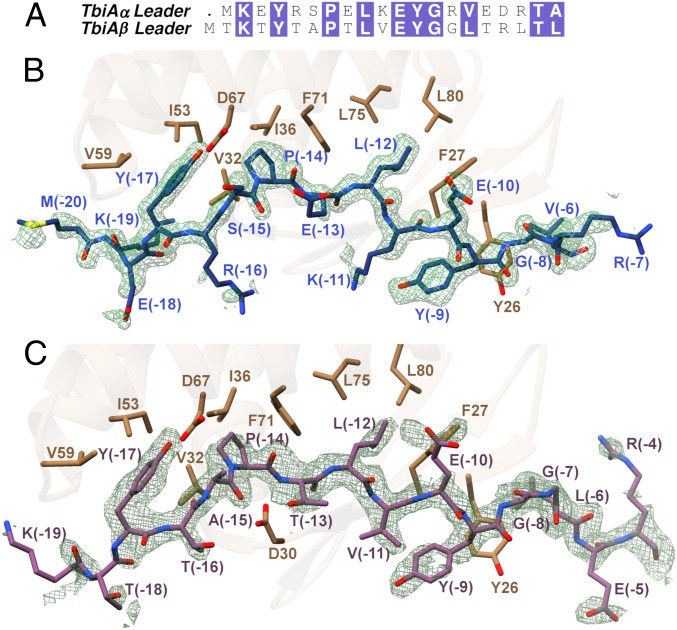
Prior characterization of RiPP biosynthesis has fostered the notion that the biosynthetic enzymes are specific for the cognate leader peptide. However, studies also suggest that the interactions between the leader peptide and the RRE can be tolerant to mutations (23, 24) and thus be amenable to engineering efforts (25). Our studies of the processing of substrates with divergent leader sequences by the Tbi system (Fig. 2A) support this notion of leader peptide permissiveness. To elucidate the basis for the leader peptide sequence tolerance of TbiB1, we determined cocrystal structures of TbiB1 in complex with the leader sequences of TbiAα (PDB ID code 5V1V, 1.35-Å resolution) and of TbiAβ (PDB ID code 5V1U, 2.05-Å resolution) (SI Appendix, Fig. S6). It should be noted that the mutant TbiAβ T(-5)E leader peptide was used to facilitate crystallization (see SI Appendix for details). The structure of TbiB1 consists of a winged helix-turn-helix motif as observed in the leader-binding domains of the cyanobactin heterocyclases TruD (26) and LynD (27), the microcin C7 synthase MccB (28), the lanthipeptide dehydratase NisB (29), the radical S-adenosyl-L-methionine enzymes involved in the biosynthesis of streptide (SuiB) (30), and sactipeptide (CteB) (31), as well as the PqqDs from Xanthomonas campestris (32) and Methylobacterium extorquens (33).
Fig. 2.
(A) Sequence alignment of TbiAα and TbiAβ leader peptides. The (B) TbiAα (blue) and (C) TbiAβ T(-5)E (purple) leader peptides interact through steric complementarity through insertion of hydrophobic residues into corresponding pockets found in TbiB1. Simulated annealing omit map (contoured at 2σ above background) calculated with Fourier coefficients (Fobs − Fcalc) with phases from the final models minus the coordinates of the leader peptides omitted prior to calculations.
Binding of the leader is mediated largely through hydrophobic residues wherein select residues of TbiA fit into pockets located along the periphery of TbiB1 (Fig. 2 B and C). Residues Lys(-11) through Arg(-7) of TbiAα (corresponding to Val(-11) through Gly(-7) of TbiAβ) form an antiparallel β-sheet with Thr25 through Arg30 of TbiB1 (Fig. 2 B and C). The amino acids of the leader peptide were numbered relative to the excision site, starting with (-1) as the C terminus of the leader. In each of the cocrystal structures, 3 residues from the leader sequence are positioned in the following manner: Tyr(-17) and Pro(-14) are encapsulated in a pocket formed by Val32, Ile36, Ile53, Val59, and Phe71, and Leu(-12) is settled into a second pocket formed by Phe27, Phe71, Leu75, and Leu80 (Fig. 2 B and C). In addition to these contacts, hydrogen bonding interactions are observed with the side chains of Tyr(-17), and Glu(-10) of either peptide.
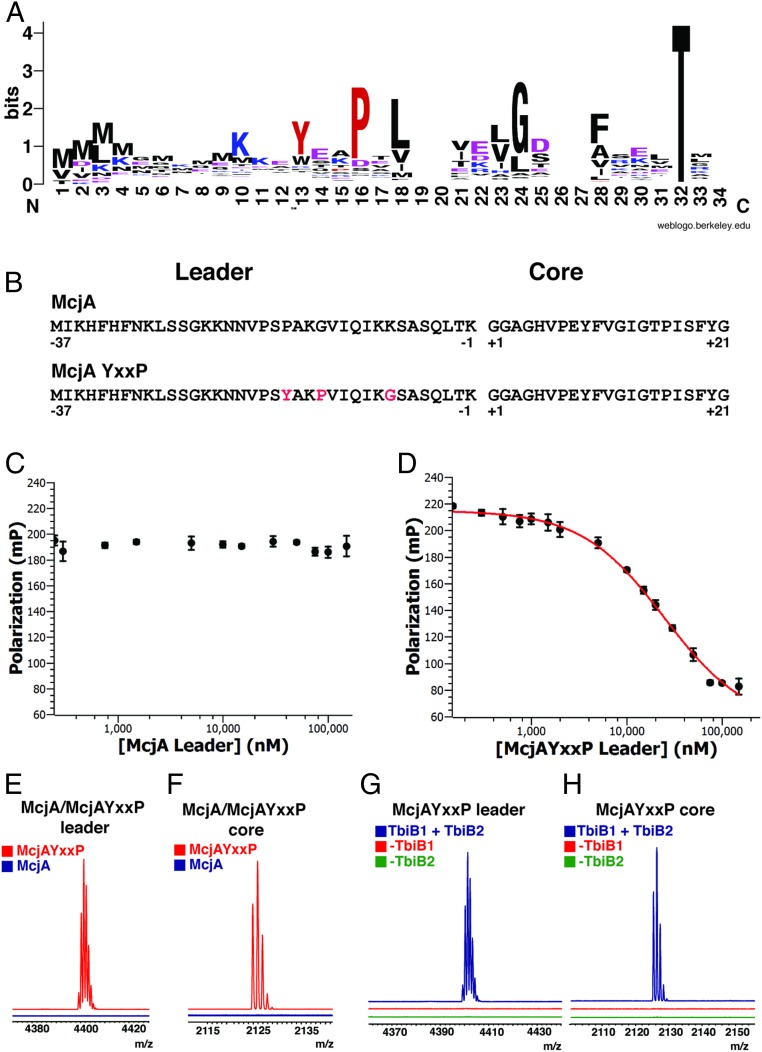
Previous bioinformatics studies on precursors from >1,000 split B-protein gene clusters identified a prevalent Y-(X)2-P-X-L-(X)3-G motif (34) (Fig. 3A), but fused B-protein clusters lack this motif (SI Appendix, Fig. S7). Our structural data reconciles Tyr, Pro, and Leu as the “knobs” that fit into the hydrophobic “holes” in the leader-binding TbiB1 protein (Fig. 2 B and C). The conserved Gly of this motif (Gly(-8) in TbiAα) enables the leader sequence to form a kink around Tyr26 of TbiB1 (Fig. 2B). The above data demonstrate that TbiB1 can engage leader peptides of seemingly disparate sequence through steric complementarity by employing multiple hydrophobic pockets to engage appropriately spaced hydrophobic residues in the leader (Fig. 2A).
Fig. 3.
(A) Weblogo analysis (45) of 400 aligned leader sequences demonstrates the conserved motif. (B) Sequences of the McjA peptide and the engineered McjAYxxP peptides. The 3 mutated residues in McjAYxxP are indicated in red. (C and D) Competitive binding assays for the (C) truncated McjA leader peptide and (D) truncated McjAYxxP leader peptide indicate that the orthogonal McjA leader peptide cannot bind, and insertion of the conserved binding motif is sufficient to binding to TbiB1. (E and F) MALDI-TOF-MS analysis of protease action observing (E) McjA and McjAYxxP leader peptide and (F) McjA and McjAYxxP core peptide. (G and H) MALDI-TOF-MS analysis of TbiB1- and TbiB2-dependent processing of (G) McjAYxxP leader peptide and (H) McjAYxxP core peptide.
In order to verify the importance of the “knobs-into-holes” packing criterion for leader engagement, we carried out binding assays with TbiAα leader derivatives against wild-type TbiB1 and variants. The binding affinity of TbiB1 for the Tyr(-17)Ala and Pro(-14)Ala variants of the TbiAα leader peptide was too weak to be accurately determined, indicating the importance of these 2 residues in this conserved motif (SI Appendix, Table S2 and Fig. S4). Of the 7 mutations of TbiB1 tested, alteration to residues in the hydrophobic pockets had the greatest impact on the binding affinity for the TbiAα leader peptide (SI Appendix, Table S2 and Fig. S4). A sequence alignment of 25 other putative B1 proteins illustrates that the residues significant for leader binding (Asp67, Phe71, Leu75, and Leu80) are largely conserved (SI Appendix, Fig. S8). Asp67 interacts with Tyr(-17) of the precursor peptide and represents the only hydrogen bonding interaction with a side chain of the leader peptide. Mapping the sequence alignment of these 26 B1 proteins onto the TbiB1 structure reveals that the areas that interact with the peptide have the greatest sequence conservation, suggesting a common mode of peptide binding across split B-protein gene clusters (SI Appendix, Fig. S9).
Universality and Portability of Sterically Guided Leader Peptide Engagement.
Our biochemical and structural data suggest that the precursor peptide substrate is recognized by the TbiB1 leader-binding domain through primarily hydrophobic packing interactions, and is largely independent of sequence context. To probe whether the observed motif is indeed sufficient to instill leader binding, we engineered the Y-(X)2-P-X-L-(X)3-G motif into the leader sequence of McjA, which is the precursor peptide for the unrelated lasso peptide microcin J25 (Fig. 3B and SI Appendix, Fig. S10). McjA is produced from a fused B-protein microcin J25 lasso peptide cluster (35), and neither can bind to TbiB1 (Fig. 3C and SI Appendix, Table S1) nor is a substrate for the TbiB1+TbiB2 protease reaction (Fig. 3 E and F). Insertion of this motif into the McjA leader required only 3 mutations, Pro(-10)Tyr, Lys(-14)Pro, and Lys(-8)Gly, and yielded the variant that we termed McjAYxxP (Fig. 3B). While wild-type McjA leader peptide failed to demonstrate any detectable binding to TbiB1, the McjAYxxP triple-mutant variant bound to TbiB1 with low micromolar affinity (Ki = 13.3 μM) (Fig. 3D and SI Appendix, Table S2).
We next sought to investigate whether installation of the binding motif into this noncognate precursor could also facilitate processing by the TbiB2 protease. Incubation of the McjAYxxP variant with both TbiB1+TbiB2 resulted in proteolytic processing, whereas the wild-type precursor peptide could not be processed (Fig. 3 E–H). Importantly, this reaction required the presence of both TbiB1 and TbiB2, indicating the reaction was contingent on binding of the leader sequence and was not the result of spurious background hydrolysis by TbiB2. These studies demonstrate that 3 mutations in an otherwise inert peptide resulted in both binding of the leader sequence and subsequent processing, demonstrating that RiPP leader binding is portable in the context of this lasso biosynthetic system.
Based on the above data, we speculated that steric complementarity was a general feature for leader peptide engagement across all split B-protein lasso peptide biosynthetic clusters. To this end, we tested the binding of the FITC-labeled TbiAα leader peptide to the B1 proteins from 2 other lasso peptide biosynthetic clusters found in the readily available strains Thermobifida cellulosilytica TB100 (TcB1) and Streptomyces natalensis ATCC 27448 (SnB1). The B1 proteins from these clusters are divergent, and show low sequence identity (less than 40%) with each other and with TbiB1 (SI Appendix, Fig. S11). More importantly, the corresponding precursor peptides show little similarity within their respective leader sequences, outside of the Y-(X)2-P-X-L-(X)3-G motif (SI Appendix, Fig. S12A). Surprisingly, each of the 2 orthologous B1 proteins bound the FITC TbiAα peptide with affinities in the low micromolar range, Kd = 11.1 ± 1.1 μM for TcB1 and Kd = 13.8 ± 0.7 μM for SnB1 (SI Appendix, Fig. S12 B and C). The tight binding further demonstrates that the Y-(X)2-P-X-L-(X)3-G motif is not only necessary but also sufficient for peptide binding in TbiB1 but is present across diverse B1-protein family members. Notably, these values are similar to the binding affinity observed between TbiB1 and the engineered McjAYxxP peptide, suggesting that the basal dissociation constant between B1 proteins and the Y-(X)2-P-X-L-(X)3-G motif is ∼10 μM.
Efforts in Maturing a Chimeric Lasso Peptide.
We have demonstrated, for lasso peptides, that leader peptide binding is directed primarily by hydrophobic interactions, as insertion of the Y-(X)2-P-X-L-(X)3-G motif in noncognate peptides has resulted in binding to lasso peptide RREs. Therefore, we sought to generate a fully processed lasso peptide using a chimeric biosynthetic approach. We defined a chimeric system using the following criteria: 1) The native precursor peptide does not possess a Y-(X)2-P-X-L-(X)3-G motif; 2) the core peptide belongs to the fused lasso system; and 3) the engineered precursor peptide must be processed by the B1/B2 proteins from the split lasso pathway, and by the C protein from a fused B-protein lasso peptide pathway. Using this criteria, we attempted to generate microcin J25 (14) and klebsidin (36) using an engineered system. Specifically, chimeric precursor peptides were generated, which contained the TbiAα leader sequence and either the microcin J25 or klebsidin core sequences. Attempts were made to process these precursors to lasso peptides using TbiB1, TbiB2, and either the microcin J25 or klebsidin cyclase. However, efforts to mature these chimeras failed in vitro and in vivo for both of these systems (SI Appendix, Figs. S13–S16). We can exclude the possibility that the C protein is incapable of catalyzing the isopeptide forming reaction, as the core peptide that is processed in these experiments was native to the C-protein cyclase. Likewise, our experiments also show that the B1/B2 system is capable of both binding to and processing these chimeric peptides (SI Appendix, Figs. S13 and S15). We conclude that, as the cleaved core no longer contains a leader peptide, direct protein−protein interactions between B2 and C proteins may be required to facilitate substrate transfer. This hypothesis is supported by work that suggests that B and C proteins can form complexes (14) and soluble C-protein production can require coexpression of the B2 protein (16).
Binding Motif Presence across PqqD Protein Family.
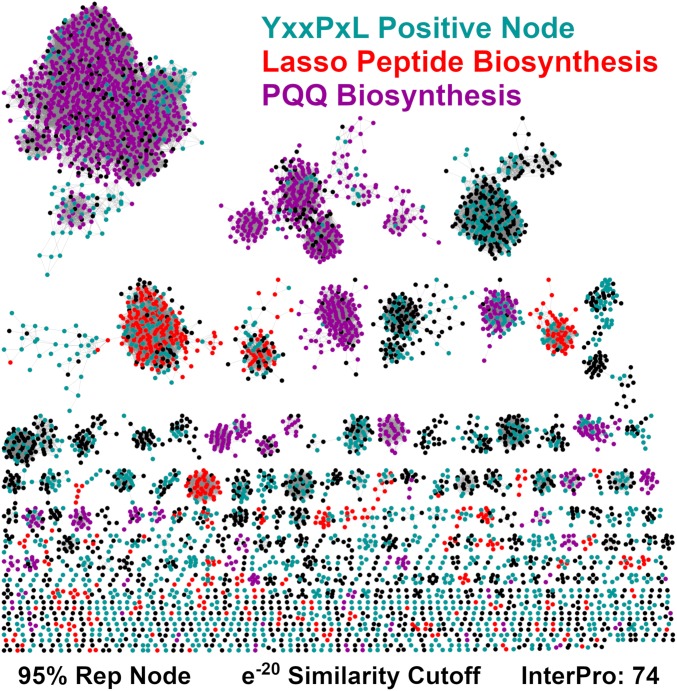
While our structural and biochemical analyses have demonstrated a clear motif in several split B-protein lasso peptide systems, we wanted to explore the prevalence of the motif across all lasso peptide clusters. To generate a list of putative lasso peptides to perform this analysis, we constructed a Sequence Similarity Network (SSN) using the entire PqqD protein family (Fig. 4) (37). Nodes that colocalized with a B2 and C protein were selected, as they possessed the minimal biosynthetic machinery to afford a lasso peptide. These nodes were then queried using RODEO (Rapid ORF Description & Evaluation Online) to identify putative precursor peptides (34), followed by manual curation to produce a final list of ∼1,800 putative precursor peptides. Of these, 57% of the sequences contained the Y-(X)2-P-X-L/V/I/F/A motif, while 24% contained a variant in the first position of the motif, W-(X)2-P-X-L/V/I/F/A (Dataset S1). When this analysis was correlated to our entire set of putative lasso peptide B1 proteins, we found that 43% of all split B-protein lasso peptides utilize the Y-(X)2-P-X-L/V/I/F/A motif, while 20% utilize the W-(X)2-P-X-L/V/I/F/A motif. These results suggest that the knobs-into-holes hydrophobic packing motif described for TbiB1 is generally conserved throughout split B-protein lasso peptide systems.
Fig. 4.
SSN of the entire PqqD protein family. SSN was constructed at a 95% representative node (Rep Node) cutoff, using the InterPro 74.0 database. Cyan nodes are colocalized near short ORFs that contain a Y/W-(X)2-P-X-L/V/I/A/F/Y motif. Red and purple nodes are predicted to be part of lasso peptide and PQQ biosynthesis, respectively. These nodes also contain the YxxPxL motif. Remaining nodes are colored black.
Even though the motif is conserved in split B-protein lasso peptides, these systems only represent 22% of the entire PqqD protein family. Therefore, we sought to evaluate the prevalence of this motif across the entire PqqD Pfam, which represents different classes of potential RiPPs. We employed the same strategy as detailed above to analyze the remainder of the PqqD Pfam SSN for peptides which contain the Y/W-(X)2-P-X-L/V/I/A/F/Y motif, henceforth abbreviated as YxxPxL. Nodes corresponding to PqqD homologs that are adjacent to putative peptides bearing this motif are highlighted in this SSN (Fig. 4). Because of amino acid variations at the final position in the motif, we included several bulky and hydrophobic residues, consistent with the knobs-into-holes model. The localization of the motif in nearly every node in a cluster suggests that it is contained within an authentic precursor peptide.
Examination of the SNN reveals that the YxxPxL motif is widespread throughout putative peptides near PqqD Pfam members. To quantify the abundance, we counted all clusters in which at least 75% of the nodes colocalized with a putative YxxPxL motif containing precursor peptide. Using this methodology, 247 of the 622 clusters appear to utilize the motif, representing 51% of the total nonredundant PqqD homologs. We also completed Genome Neighborhood Network analysis to determine whether a given cluster is a member of a known RiPP type (38). Classification of each SSN cluster reveals that PQQ biosynthetic clusters represent the single most abundant class of RiPPs in the Pfam, with 31% of nodes contained in PQQ SSN clusters. In many cases, the corresponding precursor peptide contains the YxxPxL binding motif. Using the same 75% node cutoff as before, 99.9% of PQQ nodes are contained within clusters that appear to utilize a variant of the YxxPxL motif. While NMR studies have shown that the location of the peptide binding site in PqqD overlaps with the one found in TbiB1 (33), molecular details of the residues guiding the interaction remain unclear. Our bioinformatic analysis indicates that a binding mode similar to the one observed in the therbactin lasso peptide system is likely operative in the interaction between PqqA and PqqD.
Leader Peptide Engagement in Other RiPP Biosynthetic Enzymes.
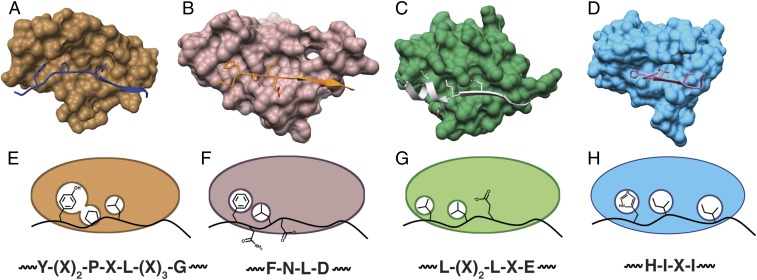
The mutational analysis of the TbiAα leader peptide and TbiB1 presented here demonstrates that a short hydrophobic packing motif found in a leader peptide guides precursor peptide binding by the B1 protein (Fig. 5 A and E). The natural variation in the tbi gene cluster further demonstrates that this short cassette is the feature driving the interaction and appears to be present throughout the Pfam. As the RRE is structurally conserved across multiple classes of RiPP biosynthetic clusters, we analyzed the structures from other RiPP systems and found similar steric complementarity packing strategies in their respective leader-binding domains. In the structure of lanthipeptide dehydratase NisB bound to NisA leader peptide (29), Leu(-16) and Phe(-18) from the conserved Phe-Asn-Leu-Asp motif occupy 2 hydrophobic cavities (Fig. 5 B and F). Likewise, the PatE′ leader peptide is engaged through the packing of Leu(-26) and Leu(-29) into hydrophobic pockets in the cyanobactin heterocyclase LynD (27) (Fig. 5 C and G). These Leu residues comprise a conserved Leu-(X)2-Leu-X-Glu motif abundant in cyanobactin leaders (39, 40). Lastly, in the case of CteA leader peptide, Ile(-15) and Ile(-17) fit into a hydrophobic cleft, and additional packing is achieved by the salt bridges between His(-16) and CteB (Fig. 5 D and H) (31).
Fig. 5.
(A−D) Surface models of (A) TbiB1 (copper) and TbiAα leader (blue), (B) NisB RRE (purple) and NisA leader (orange), (C) LynD RRE (green) and PatE’ (silver), and (D) CteB RRE (sky blue) and CteA (pink) indicate a conserved binding mode. (E−H) Cartoon depiction of the knob-into-hole binding of leader consensus motifs into the RREs of (E) TbiB1, (F) NisB, (G) LynD, and (H) CteB.
In order to determine whether a short motif was sufficient for leader peptide binding in one of these other RiPP classes, we carried out binding studies on TruD, the heterocyclase involved in the biosynthesis of the cyanobactin trunkamide (41). Prior biochemical studies establish that recognition sequence I (RSI) and recognition sequence II (RSII) of the trunakamide leader are necessary to direct heterocyclization (39), and the interaction of this region of the leader with the RRE domain is shown in the cocrystal structure of the aesturamide heterocyclase LynD (27, 42). We speculated that only a short segment of the full-length leader bearing an L-(X)2-L-(X)2-E motif will engage in both hydrophobic packing and hydrogen bonding to allow for leader peptide binding to the RRE domain. To confirm this hypothesis, we carried out fluorescence polarization measurements of an FITC-labeled 17-residue peptide containing a minimal region of RSI and RSII that harbors the above motif (SI Appendix, Fig. S17A). This peptide bound to TruD with a Kd = 5.61 ± 0.43 μM, indicating that the RRE of TruD can tightly bind to a short leader peptide region (SI Appendix, Fig. S17B).
While the basis for leader binding is not known for many RiPP classes, bioinformatics could potentially be used to identify putative RRE-engaging motifs. In NisB, LynD, TbiB1, and CteB, the residues of the leader that interact with the RRE are prominent in sequence alignments, consistent with their importance in binding (SI Appendix, Fig. S18). Therefore, we generated alignments of leader peptides to identify potential hydrophobic or charged regions that may interact with the RRE for several other classes of RiPPs (SI Appendix, Fig. S19). Further experiments will be required to validate that these short regions of conservation are the key drivers of a knobs-into-holes interaction.
Discussion
RiPP biosynthetic pathways provide highly flexible platforms for generating chemically diverse scaffolds in a high-throughout manner (43, 44). This diversity is possible because the posttranslational processing enzymes are guided to the precursor peptide substrate by the leader sequence and are permissive for alterations to the core peptide. For example, the lanthipeptide synthase ProcM was employed in a library of over one million precursor peptides to successfully screen for inhibitors of protein−protein interactions (43). Likewise, cyanobacterial biosynthetic enzymes can efficiently process fusion substrates in which the core sequence for a different cyanobacterial pathway is fused to the cognate leader sequence (39). Furthermore, RiPP tailoring enzyme promiscuity was recently exploited with precursor peptides that incorporated leader regions recognized by enzymes from divergent RiPP pathways, enabling production of chimeric nonnatural RiPP products (25). Efforts to understand the details of precursor−RRE interactions in other systems would further expand the prospects of producing new chimeric products.
Our results on lasso peptide leader binding indicate that leader recognition does not require extensive sequence specificity, but rather is guided by steric complementarity to facilitate hydrophobic packing. More significantly, the tenets for leader recognition are conserved across many orthologous split B-protein lasso clusters and may extend into other RiPP classes, such as PQQ biosynthesis or even novel pathways. We used these observations to engineer noncognate peptides as substrates for protease processing in these clusters by embedding a small, fairly nonintrusive Y-(X)2-P-X-L-(X)3-G motif. In contrast, efforts to make chimeric systems that can process a precursor peptide to a final lasso peptide product were not successful. Both our work and results from other systems suggests protein−protein interactions may be critical for proper processing. In principle, it may be possible to engineer an artificial leader sequence in which requisite recognition elements are embedded within each other, instead of concatenated one after another. This would produce shortened engineered leader sequences and may prove to be more effective and improve production of novel hybrid RiPP products that are not found in nature.
Materials and Methods
SI Appendix includes methods for expression and purification for TbiB1, TbiB2, substrate peptides, and other proteins used in this study. Methodologies for activity assays, crystallization, bioinformatics, binding studies, and attempts to create chimeric lasso peptides are also detailed therein.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM079038 to S.K.N.). We thank J. D. Hegemann for constructive discussions and assistance with instrumentation, and Keith Brister and colleagues at the Life Sciences Collaborative Access Team (LS-CAT) at the Advanced Proton Source (Argonne National Labs) for facilitating X-ray data collection.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The structure factors and coordinates have been deposited in the Protein Data Bank, www.wwpdb.org: TbiB1-TbiAalpha leader (PDB ID code 5V1V) and TbiB1-TbiAbeta leader (PDB ID code 5V1U). NCBI accession numbers for proteins discussed in this work are available in SI Appendix, Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908364116/-/DCSupplemental.
References
- 1.Arnison P. G., et al. , Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Q., Yang X., Wang H., van der Donk W. A., High divergence of the precursor peptides in combinatorial lanthipeptide biosynthesis. ACS Chem. Biol. 9, 2686–2694 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sivonen K., Leikoski N., Fewer D. P., Jokela J., Cyanobactins-ribosomal cyclic peptides produced by cyanobacteria. Appl. Microbiol. Biotechnol. 86, 1213–1225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmermann M., Hegemann J. D., Xie X., Marahiel M. A., Characterization of caulonodin lasso peptides revealed unprecedented N-terminal residues and a precursor motif essential for peptide maturation. Chem. Sci. (Camb.) 5, 4032–4043 (2014). [Google Scholar]
- 5.Latham J. A., Iavarone A. T., Barr I., Juthani P. V., Klinman J. P., PqqD is a novel peptide chaperone that forms a ternary complex with the radical S-adenosylmethionine protein PqqE in the pyrroloquinoline quinone biosynthetic pathway. J. Biol. Chem. 290, 12908–12918 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goosen N., Horsman H. P., Huinen R. G., van de Putte P., Acinetobacter calcoaceticus genes involved in biosynthesis of the coenzyme pyrrolo-quinoline-quinone: Nucleotide sequence and expression in Escherichia coli K-12. J. Bacteriol. 171, 447–455 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkhart B. J., Hudson G. A., Dunbar K. L., Mitchell D. A., A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 11, 564–570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barr I., et al. , Demonstration that the radical s-adenosylmethionine (SAM) enzyme PqqE catalyzes de novo carbon-carbon cross-linking within a peptide substrate PqqA in the presence of the peptide chaperone PqqD. J. Biol. Chem. 291, 8877–8884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wieckowski B. M., et al. , The PqqD homologous domain of the radical SAM enzyme ThnB is required for thioether bond formation during thurincin H maturation. FEBS Lett. 589, 1802–1806 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Li B., et al. , Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 10430–10435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donia M. S., et al. , Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2, 729–735 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Maksimov M. O., Pelczer I., Link A. J., Precursor-centric genome-mining approach for lasso peptide discovery. Proc. Natl. Acad. Sci. U.S.A. 109, 15223–15228 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duquesne S., et al. , Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem. Biol. 14, 793–803 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Yan K.-P., et al. , Dissecting the maturation steps of the lasso peptide microcin J25 in vitro. ChemBioChem 13, 1046–1052 (2012). [DOI] [PubMed] [Google Scholar]
- 15.DiCaprio A. J., Firouzbakht A., Hudson G. A., Mitchell D. A., Enzymatic reconstitution and biosynthetic investigation of the lasso peptide fusilassin. J. Am. Chem. Soc. 141, 290–297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koos J. D., Link A. J., Heterologous and in vitro reconstitution of fuscanodin, a lasso peptide from Thermobifida fusca. J. Am. Chem. Soc. 141, 928–935 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gavrish E., et al. , Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 21, 509–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inokoshi J., Matsuhama M., Miyake M., Ikeda H., Tomoda H., Molecular cloning of the gene cluster for lariatin biosynthesis of Rhodococcus jostii K01-B0171. Appl. Microbiol. Biotechnol. 95, 451–460 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Hegemann J. D., Schwalen C. J., Mitchell D. A., van der Donk W. A., Elucidation of the roles of conserved residues in the biosynthesis of the lasso peptide paeninodin. Chem. Commun. (Camb.) 54, 9007–9010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu S., et al. , The B1 protein guides the biosynthesis of a lasso peptide. Sci. Rep. 6, 35604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu S., et al. , Insights into the unique phosphorylation of the lasso peptide paeninodin. J. Biol. Chem. 291, 13662–13678 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegemann J. D., Zimmermann M., Xie X., Marahiel M. A., Caulosegnins I-III: A highly diverse group of lasso peptides derived from a single biosynthetic gene cluster. J. Am. Chem. Soc. 135, 210–222 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Khusainov R., Moll G. N., Kuipers O. P., Identification of distinct nisin leader peptide regions that determine interactions with the modification enzymes NisB and NisC. FEBS Open Bio 3, 237–242 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plat A., Kluskens L. D., Kuipers A., Rink R., Moll G. N., Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl. Environ. Microbiol. 77, 604–611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkhart B. J., Kakkar N., Hudson G. A., van der Donk W. A., Mitchell D. A., Chimeric leader peptides for the generation of non-natural hybrid RiPP products. ACS Cent. Sci. 3, 629–638 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koehnke J., et al. , The cyanobactin heterocyclase enzyme: A processive adenylase that operates with a defined order of reaction. Angew. Chem. Int. Ed. Engl. 52, 13991–13996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koehnke J., et al. , Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat. Chem. Biol. 11, 558–563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regni C. A., et al. , How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 28, 1953–1964 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ortega M. A., et al. , Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis K. M., et al. , Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition. Proc. Natl. Acad. Sci. U.S.A. 114, 10420–10425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grove T. L., et al. , Structural insights into thioether bond formation in the biosynthesis of sactipeptides. J. Am. Chem. Soc. 139, 11734–11744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai T. Y., Yang C. Y., Shih H. L., Wang A. H. J., Chou S. H., Xanthomonas campestris PqqD in the pyrroloquinoline quinone biosynthesis operon adopts a novel saddle-like fold that possibly serves as a PQQ carrier. Proteins 76, 1042–1048 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Evans R. L. 3rd, Latham J. A., Xia Y., Klinman J. P., Wilmot C. M., Nuclear magnetic resonance structure and binding studies of PqqD, a chaperone required in the biosynthesis of the bacterial dehydrogenase cofactor pyrroloquinoline quinone. Biochemistry 56, 2735–2746 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tietz J. I., et al. , A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 13, 470–478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solbiati J. O., et al. , Sequence analysis of the four plasmid genes required to produce the circular peptide antibiotic microcin J25. J. Bacteriol. 181, 2659–2662 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metelev M., et al. , Acinetodin and klebsidin, RNA polymerase targeting lasso peptides produced by human isolates of Acinetobacter gyllenbergii and Klebsiella pneumoniae. ACS Chem. Biol. 12, 814–824 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Gerlt J. A., et al. , Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zallot R., Oberg N. O., Gerlt J. A, ‘Democratized’ genomic enzymology web tools for functional assignment. Curr. Opin. Chem. Biol. 47, 77–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sardar D., Pierce E., McIntosh J. A., Schmidt E. W., Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth. Biol. 4, 167–176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Donia M. S., Schmidt E. W., Linking chemistry and genetics in the growing cyanobactin natural products family. Chem. Biol. 18, 508–519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donia M. S., Ravel J., Schmidt E. W., A global assembly line for cyanobactins. Nat. Chem. Biol. 4, 341–343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McIntosh J. A., Lin Z., Tianero M. D. B., Schmidt E. W., Aestuaramides, a natural library of cyanobactin cyclic peptides resulting from isoprene-derived Claisen rearrangements. ACS Chem. Biol. 8, 877–883 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., et al. , A lanthipeptide library used to identify a protein-protein interaction inhibitor. Nat. Chem. Biol. 14, 375–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt S., et al. , Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nat. Chem. Biol. 15, 437–443 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.