Significance
CLL is the most common human leukemia. We identified tsRNA signatures in aggressive and indolent CLL. We further analyzed the expression of mature tRFs and found that mature tRFs are drastically dysregulated in CLL. Thus, we conclude that tsRNAs and mature tRFs, 2 classes of small noncoding RNAs, may be associated with the development of CLL.
Keywords: tsRNAs, tRNA fragments, tRFs
Abstract
Chronic lymphocytic leukemia (CLL) is the most common human leukemia, and dysregulation of tRNA-derived short noncoding RNA (tsRNA) (tRF-1) expression is an accompanying event in the development of this disease. tsRNAs are fragments originating from the 3′ end of tRNA precursors and do not contain mature tRNA sequences. In contrast to tsRNAs, mature tRFs (tRF-3s, tRF-5s, and internal tRFs) are produced from mature tRNA sequences and are redundant fragments. We investigated tsRNA expression in CLL and determined tsRNA signatures in indolent CLL and aggressive CLL vs. normal B cells. We noticed that both ts-43 and ts-44 are derived from distinct genes of pre-tRNAHis, and are down-regulated in CLL 3- to 5-fold vs. normal B cells. Thus, we investigated expression levels of tRF-5 fragments from tRNAHis in CLL samples and healthy controls, and determined that such fragments are down-regulated by 5-fold in CLLs vs. normal controls. Given these results, we investigated the expression of all mature tRFs in CLLs vs. normal controls. We found a drastic dysregulation of the expression of mature tRFs in CLL. In aggressive CLL, for the top 15 up-regulated fragments, linear fold change varied from 2,053- to 622-fold. For the top 15 down-regulated fragments in CLL, linear fold change varied from 314- to 52-fold. In addition, 964 mature tRFs were up-regulated at least 2-fold in CLL, while 701 fragments were down-regulated at least 2-fold. Similar results were obtained for indolent CLL. Our results suggest that mature tRFs may have oncogenic and/or tumor suppressor function in CLL.
Chronic lymphocytic leukemia (CLL) is the most common human adult leukemia. CLL is a heterogeneous disease characterized by the expansion of CD19+/CD5+ B cells (1). Generally, aggressive CLLs express high levels of zeta-chain–associated protein kinase 70 (ZAP-70) and unmutated Ig heavy-chain variable-region genes, while indolent CLLs display opposite characteristics (2). High expression of the TCL1 oncogene is a critical element in the development of aggressive CLL, and high TCL1 expression correlates with the aggressive clinical course of the disease (3). These data are supported by the development of transgenic mice expressing TCL1 in B cells that develop the aggressive form of CLL (4, 5). While investigating possible microRNAs (miRNAs) targeting TCL1 expression, we found that miR-3676, located in 17p13 in proximity to TP53 and codeleted with the TP53 gene in CLL, targets TCL1 expression (6). We also found that miR-3676 binds Ago1 and Ago2 and can function as a microRNA, albeit being produced as the result of cleavage in the 3′ region of a tRNA precursor by endonuclease Z (7). We later renamed this noncoding RNA ts-53 and determined that it is a member of a small family (∼100 members) of tRNA-derived short noncoding RNAs (tsRNAs), produced from the 3′ region of tRNA precursors (7). tRNA genes are located in multiple copies throughout the entire human genome. Indeed, more than 500 human tRNA genes are encoded by the human genome overall and contain 49 different anticodons. In addition, there are more than 300 tRNA pseudogenes in humans (8). tRNA genes are transcribed by RNA polymerase III. Mature tRNAs are then processed by several endonucleases from transcribed pre-tRNA molecules. This process produces additional small RNA molecules. The fragments produced from the 3′ trailer of tRNA precursors by endonuclease RNase Z represent unique sequences, tsRNAs, also called tRF-1s. tsRNAs do not contain mature tRNA sequences (9, 10). In contrast to tsRNAs, mature tRFs (tRF-3s and tRF-5s, representing 3′ and 5′ tRFs, respectively, and internal tRFs) are produced from mature tRNA sequences and are redundant fragments (7).
Despite recent advances in the biology of tRNA fragments, their molecular function is currently not entirely clear. Recently, Zhang et al. (11) reported that in monocytes a tRF-5 (tRF-5030c) interacts with PIWI proteins and causes histone H3K9 methylation in the CD1A promoter region by recruiting histone methyltransferases in a sequence-specific manner. In addition, Maute et al. (12) have demonstrated that a tRF-3 (named CU1276 or tRF-3027) is underexpressed in human B cell lymphomas and represses the expression levels of the tumor suppressor RPA1 in an miRNA-like manner. Our previous studies determined tsRNA signatures in several types of cancer, and identified ts-43, ts-44, and ts-46 as potential tumor suppressors (6, 7). Goodarzi et al. (13) demonstrated that specific tRFs bind to the oncogenic RNA-binding protein YBX1, displacing prooncogenic transcripts and resulting in their destabilization. In addition, Lee et al. (14) showed that the expression of ts-36 (also termed tRF-1001) accelerates cell-cycle transition in prostate cancer cells. In recent years, the advent of next-generation sequencing technology has provided the opportunity to collect a substantially greater amount of data at a higher level of precision than previously, resulting in more reliable information and insight, especially regarding noncoding RNA (15, 16). In this study, we analyzed the expression of tsRNAs and mature tRFs in aggressive CLL, indolent CLL, and normal controls by small RNA sequencing (sRNA-seq) and found a massive dysregulation of tRNA-derived fragments (tsRNAs and mature tRFs) in CLL.
Results
tsRNA Signatures in Aggressive and Indolent CLLs.
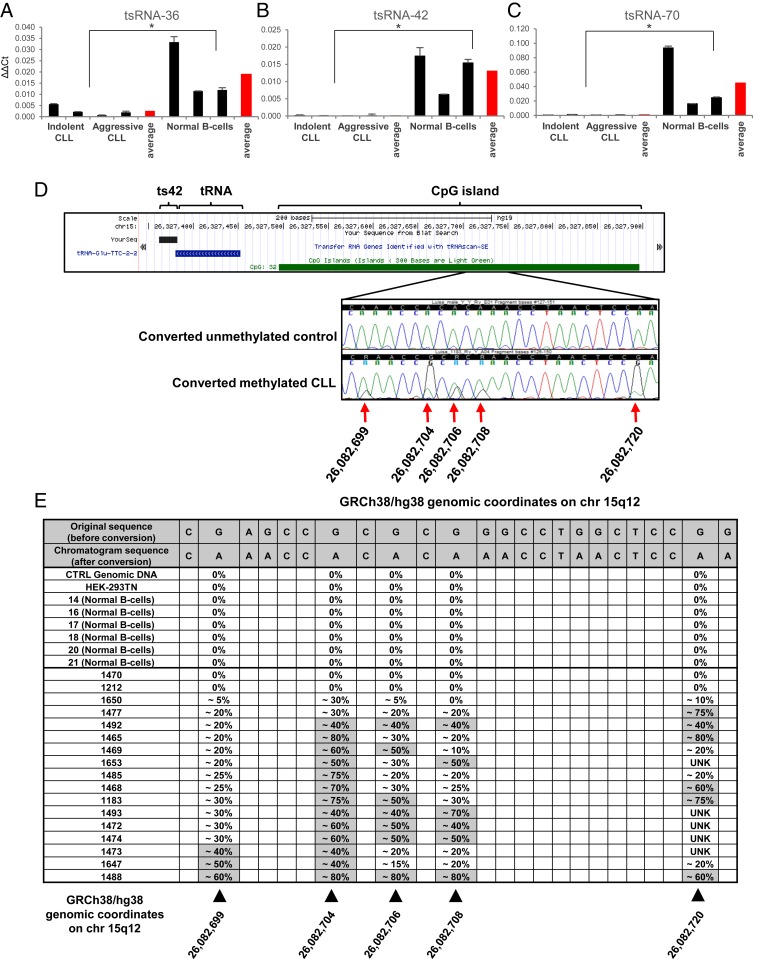
We previously studied tsRNA expression in CLL using custom microarray technology (7, 17). However, data obtained using such technology do not provide precise counts for each molecule. Thus, to obtain tsRNA signatures in CLL more accurately, we selected 28 RNA samples from 10 indolent and 10 aggressive CLL patients and 8 normal controls extracted from CD19+ B cells of healthy donors (SI Appendix, Table S1) and carried out small RNA-seq expression analysis. Fastq files generated via the sRNA-seq protocols were preprocessed by first trimming adapter sequences via the tool Cutadapt (18), and then quality filtering trimmed reads via the tool ConDeTri (19). Preprocessed samples were analyzed by leveraging a modified version of the detection tool tDRmapper (20), allowing for the exact mapping of reads on a custom-made annotation of the pre-tRNA genomic space (50 nt before and after known mature tRNA sequences, human genome version GRCh37), accounting for both reference and single occurrences of common SNP (single-nucleotide polymorphism) versions of the known set of 113 tsRNA sequences (7). Raw counts were extracted and collapsed through in-house scripts. Furthermore, raw counts for all tsRNA molecules which were detected having SNPs were all combined, along with their reference sequence, into a single collapsed read count per sample. A minimum expression filtering was applied on raw counts before normalization, with tsRNA molecules retained if they reported at least 5 raw reads in each of at least 8 samples. As recommended (21), filtered molecules were normalized by applying the TMM method (trimmed mean of M values) and statistical analysis of the data was carried out within the R studio environment (version 1.1.423), employing the software package edgeR (22), and by applying the Mann–Whitney–Wilcoxon nonparametric test in order to assess significant differential expression for each molecule between 2 groups. Table 1 shows differentially expressed tsRNAs in aggressive CLL vs. normal controls with a P value < 0.05 and a linear fold-change cutoff of 1.5-fold, along with their corresponding fold change in the indolent CLL vs. normal controls comparison. The sequences of all dysregulated fragments can be found in ref. 7. We found that in aggressive CLL vs. normal CD19+ B cells, 11 tsRNAs were up-regulated, while 12 tsRNAs were down-regulated. Similarly, in indolent CLL vs. normal CD19+ B cells, 8 tsRNAs were up-regulated, while 14 tsRNAs were down-regulated (SI Appendix, Table S2). Only 4 tsRNAs were differentially expressed in aggressive CLL vs. indolent CLL (SI Appendix, Table S3). To confirm our results, we proceeded to further verify the expression of ts-42, ts-70 (the 2 most down-regulated tsRNAs), and ts-36 [a tsRNA potentially involved in cell-cycle progression (14)]. Fig. 1 A–C shows the expression of ts-36, ts-42, and ts-70 in representative CLL and normal control samples by custom real-time RT-PCR. In CLL, ts-42 expression was down-regulated ∼245-fold, ts-70 was down-regulated ∼61-fold, while ts-36 was down-regulated ∼7-fold, confirming the sRNA-seq data. Additionally, their expression (as well as that of tRFs) in normal B cells was found to be heterogeneous, most likely due to the fact that B cells from normal individuals are polyclonal and numerous, while most CLL cells are monoclonal and/or consist of very few clones.
Table 1.
sRNA-seq differential expression results (P < 0.05) for aggressive CLL vs. normal CD19+ B cells, including corresponding indolent CLL vs. normal CD19+ B cell fold change
| tsRNA ID | Linear fold change | P value | ||
| Aggressive vs. normal | Indolent vs. normal | Aggressive vs. normal | Indolent vs. normal | |
| ts81_Glu-CTC-1-1* | 11.37 | 11.15 | 0.00005 | 0.00137 |
| ts32_tRNA4-ValTAC | 8.34 | 9.81 | 0.00018 | 0.00009 |
| ts20_tRNA135-ThrAGT | 4.47 | 3.96 | 0.00087 | 0.00005 |
| ts37_tRNA17-ValTAC* | 3.75 | 3.48 | 0.00306 | 0.00055 |
| ts55_tRNA15-CysGCA | 3.58 | 4.37 | 0.00032 | 0.00206 |
| ts66_tRNA154-IleAAT | 3.11 | 2.49 | 0.02052 | 0.16403 |
| ts67_tRNA59-IleAAT | 2.53 | 2.19 | 0.01554 | 0.00032 |
| ts58_tRNA13-AlaCGC* | 2.53 | 2.77 | 0.00439 | 0.08314 |
| ts109_tRNA1-ArgCCG | 2.00 | 1.94 | 0.02665 | 0.06760 |
| ts34_tRNA6-ValTAC | 1.71 | 1.09 | 0.02052 | 0.89675 |
| ts53_tRNA8-ThrAGT | 1.65 | 1.94 | 0.00306 | 0.12199 |
| ts36_tRNA2-SerTGA* | −1.64 | −1.66 | 0.00306 | 0.01554 |
| ts101_tRNA7-SerGCT | −1.75 | −2.15 | 0.00018 | 0.00306 |
| ts51_tRNA21-ArgCCT | −1.83 | −1.77 | 0.02665 | 0.00622 |
| ts27_His-GTG-1-3 | −2.04 | −2.48 | 0.00306 | 0.00855 |
| ts80_tRNA14-ThrCGT | −2.32 | −4.82 | 0.02340 | 0.00115 |
| ts94_tRNA7-ArgACG* | −2.36 | −2.40 | 0.00206 | 0.00306 |
| ts56_tRNA26-CysGCA | −2.58 | −3.20 | 0.00032 | 0.00032 |
| ts6_tRNA56-ThrTGT | −2.93 | −2.43 | 0.00018 | 0.00005 |
| ts44_tRNA8-HisGTG | −3.39 | −6.41 | 0.00005 | 0.00005 |
| ts43_tRNA9-HisGTG* | −4.61 | −5.01 | 0.00009 | 0.00055 |
| ts70_tRNA96-PheGAA | −7.26 | −20.46 | 0.00032 | 0.00005 |
| ts42_tRNA11-GluTTC* | −107.54 | −92.50 | 0.00378 | 0.00468 |
tsRNAs with collapsed SNP reads.
Fig. 1.
Down-regulation of tsRNA expression in CLL. (A–C) Real-time RT-PCR analysis shows a significant decrease of ts-36, ts-42, and ts-70 expression in patient samples (indolent CLL, IDs 1473 and 1212; aggressive CLL, IDs 1493 and 1653) compared with normal B cells. One-tailed Wilcoxon rank-sum test was applied for statistical analysis to all 3 experiments. *P < 0.05 was considered statistically significant (P = 0.02857 for each of the 3 tsRNAs considered). Controls are derived from 3 distinct healthy individuals. (D) Graphic representation of the methylation status of the CpG islands inside the ts-42 promoter region (human build GRCh38/hg38). Methylated cytosine residues in the representative chromatograms are indicated by arrows. (E) The table shows the methylation rate for selected genomic sites inside the ts-42 promoter region. We used commercial genomic DNA (Clontech), the HEK-293TN cell line, and normal B cells as controls. Error bars indicate the SD for the 3 replicates.
ts-42 was the most down-regulated tsRNA in CLL. It is located immediately downstream of tRNA11GluTTC at 15q12 (Fig. 1D). Bioinformatics analysis revealed the presence of a CpG island immediately upstream of tRNA11GluTTC (Fig. 1D). Since DNA methylation of promoters is one of the most common mechanisms of gene silencing, we investigated the methylation status of the ts-42 promoter in 17 CLL samples (out of 20 samples employed for sRNA-seq) by sequencing of bisulfite-converted DNA. We found that 14 of 17 CLL samples showed at least 40% methylation of the ts-42 promoter in at least 1 of 4 methylated CpGs (we considered 100% methylation to be when both alleles were completely methylated) (Fig. 1 D and E). No methylation was observed in 6 normal controls (Fig. 1E). We concluded that ts-42 is inactivated in CLL mostly by promoter methylation.
tRF-5 of tRNAHis Is Down-Regulated in CLL.
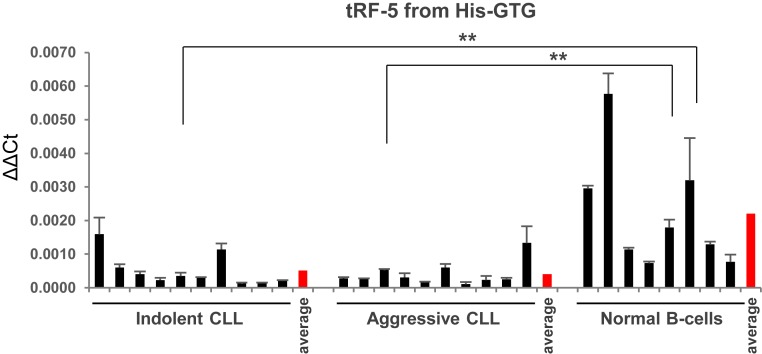
As mentioned above, sRNAs are fragments originating from the 3′ end of tRNA precursors and do not contain mature tRNA sequences. On the other hand, mature tRFs (tRF-3s, tRF-5s, and internal tRFs) are produced from mature tRNA sequences and are redundant fragments. Very recently, Guzzi et al. (23) studied the modification by pseudouridylation of the tRF-5 derived from tRNAAla/Cys/Val (a mature tRNA fragment derived from 5′ tRNAAla/Cys/Val). They determined that this tRF-5 represses translation in stem cells, plays a significant role in early embryogenesis, and impacts hematopoietic commitment, with these effects being dependent on the pseudouridylation levels of this tRF-5 (23). We observed that ts-43 and ts-44 are derived from distinct transcripts of pre-tRNAHis, and are down-regulated in CLL with an average of 3- to 5-fold (Table 1). Previously, we determined that ts-46, which derives from another transcript of pre-tRNAHis, is also down-regulated in CLL (7). We thus speculated that a mature tRNA fragment(s) derived from mature tRNAHis might also be differentially expressed in CLL. Because of the important biological role of the tRF-5 originating from tRNAAla/Cys/Val, we investigated the expression levels of tRF-5 from tRNAHis in CLLs and normal controls. Fig. 2 shows that tRF-5 from tRNAHis is down-regulated ∼5-fold in CLLs vs. normal CD19+ controls.
Fig. 2.
Down-regulation of tRF-5 expression from tRNAHis in CLL. One-tailed Wilcoxon rank-sum test was applied for statistical analysis. P < 0.05 was considered statistically significant (with a P value of 0.001651 for indolent CLL vs. normal B cells, and a P value of 0.0006553 for aggressive CLL vs. normal B cells). Annotations for **P < 0.01 are provided accordingly. Controls are derived from 8 distinct healthy individuals. Error bars indicate the SD for the 3 replicates.
Dysregulation of Mature tRNA Fragments in CLL.
tsRNAs are produced from the 3′ ends of pre-tRNAs by endonuclease RNase Z, and represent unique sequences starting precisely at the 3′ ends of tRNAs and ending at the sequence of 4 consecutive T nucleotides (7). On the other hand, mature tRFs (tRF-3s, tRF-5s, and internal tRFs) are produced from mature tRNA sequences and are not unique sequences (24). We believed mature tRFs less likely to be tightly regulated, considering that each such molecule is expressed from multiple tRNA genes. However, our results showed that a mature tRNA fragment, namely tRF-5 from tRNAHis, is down-regulated in CLLs vs. normal CD19+ controls (Fig. 2). Given these results, we decided to investigate the expression of all mature tRFs in CLLs vs. normal CD19+ controls.
We analyzed the same 28 small RNA-seq files as above for the expression of all mature tRFs. Preprocessed fastq files were analyzed by leveraging the recently published tool MINTmap (25), applied with standard parameters (default genome assembly GRCh37), in order to mine for and quantify mature tRFs (tRF-3s, tRF-5s, and internal tRFs). The tool allows profiling all possible subsequences with lengths between 16 and 50 nt that overlap the 640 human mature tRNA sequences, taking into account the posttranscriptional addition of a single nucleotide to the 5′ end, as well as the nontemplated CCA (26). Raw counts were extracted and collapsed through in-house scripts. A minimum expression filtering was applied on raw counts before normalization, with tRF molecules retained if they reported at least 5 raw reads in each of at least 8 samples, as per tsRNA molecules. As recommended (21), filtered molecules were normalized by applying the TMM method and statistical analysis of the data was carried out within the R studio environment (version 1.1.423), employing the software package edgeR (22), and by applying the Mann–Whitney–Wilcoxon nonparametric test in order to assess significant differential expression for each molecule between 2 groups.
Table 2 shows drastic up- and down-regulation of mature tRFs in aggressive CLL vs. normal controls. For the top 15 up-regulated fragments, the linear fold change varied from 2,053- to 622-fold. For the top 15 down-regulated tRFs in CLL, the linear fold change varied from 314- to 52-fold. Full annotation and additional results are reported in Dataset S1. Similarly, for the indolent CLL vs. normal controls comparison, for the top 15 up-regulated fragments, the linear fold change varied from 2,329- to 618-fold (SI Appendix, Table S4; see Dataset S2 for full annotation and additional results information), while for the top 15 down-regulated, the linear fold change varied from 273- to 94-fold (SI Appendix, Table S4). In aggressive CLL vs. normal controls comparison, a total of 964 mature tRFs were up-regulated at least 2-fold in CLL, while 701 fragments were down-regulated at least 2-fold (Dataset S1). Similarly, for the indolent CLL vs. normal controls comparison, a total of 999 mature tRFs were up-regulated at least 2-fold in CLL, while 914 fragments were down-regulated at least 2-fold (Dataset S2). In the aggressive CLL vs. indolent CLL comparison, we found 107 mature tRFs to be up-regulated in aggressive CLL at least 2-fold, while 62 fragments were down-regulated. The top linear fold changes were 51 and 21, respectively (Dataset S3).
Table 2.
sRNA-seq differential expression results for the top 15 significantly deregulated (P < 0.05) mature tRFs in aggressive CLL vs. normal controls
| tRF MINTmap ID | Fragment sequence, 5′-3′ | Length, nt | tRF type | Linear fold change | P value | ||
| Aggressive vs. normal | Indolent vs. normal | Aggressive vs. normal | Indolent vs. normal | ||||
| tRF-34-5YXENDBP1IUU1J | GATTGTGAATCTGACAACAGAGGCTTACGACCCC | 34 | i-tRF | 2,053.45 | 1,499.29 | 0.00030 | 0.00114 |
| tRF-20-V25Z2IUI | TAGGAGATTTCAACTTAACT | 20 | i-tRF | 1,927.74 | 1,409.13 | 0.00030 | 0.00114 |
| tRF-18-5YXENDR | GATTGTGAATCTGACAAC | 18 | i-tRF | 1,786.08 | 2,329.87 | 0.00030 | 0.00030 |
| tRF-33-5YXENDBP1IUUDJ | GATTGTGAATCTGACAACAGAGGCTTACGACCC | 33 | i-tRF | 1,339.26 | 954.66 | 0.00030 | 0.00114 |
| tRF-27-08F4BDNZ8O4 | ACCGGAGATGAAAACCTTTTTCCAAGG | 27 | i-tRF | 1,129.54 | 822.20 | 0.00909 | 0.00209 |
| tRF-19-5YXEND0U | GATTGTGAATCTGACAACA | 19 | i-tRF | 1,110.67 | 1,401.93 | 0.00030 | 0.00030 |
| tRF-16-5YXENDB | GATTGTGAATCTGACA | 16 | i-tRF | 893.64 | 757.54 | 0.00042 | 0.00042 |
| tRF-25-HMI8W47W1R | ATATCATTGGTCGTGGTTGTAGTCC | 25 | i-tRF | 837.55 | 788.87 | 0.00035 | 0.00035 |
| tRF-20-2IUIX1Q7 | CAACTTAACTTGACCGCTCT | 20 | i-tRF | 837.47 | 618.80 | 0.00039 | 0.00039 |
| tRF-25-M0IBB7Z92K | CGGAGATGAAAACCTTTTTCCAAGG | 25 | i-tRF | 835.19 | 661.14 | 0.00030 | 0.00379 |
| tRF-17-FN7BWU2 | AGATTGTGAATCTGACA | 17 | i-tRF | 808.16 | 870.72 | 0.00030 | 0.00030 |
| tRF-16-B2Y0FDD | AAAGATTAAGAGAACC | 16 | i-tRF | 764.07 | 1,142.20 | 0.00030 | 0.00030 |
| tRF-26-325UB1ZZK00 | CCGGAGATGAAAACCTTTTTCCAAGG | 26 | i-tRF | 687.57 | 556.40 | 0.00030 | 0.00379 |
| tRF-19-H6Z3NPIJ | ATCCATTGGTCTTAGGCCC | 19 | i-tRF | 676.43 | 876.26 | 0.00030 | 0.00379 |
| tRF-21-DNB2Y0FDD | AAGTTAAAGATTAAGAGAACC | 21 | i-tRF | 622.10 | 579.68 | 0.00030 | 0.00030 |
| tRF-26-KVNMEH623K0 | CCCGCCTGTCACGCGGGAGACCGGGG | 26 | i-tRF | −52.97 | −48.61 | 0.00024 | 0.00018 |
| tRF-16-RK9P4PE | GGGGGTGTAGCTCAGT | 16 | 5′-tRF | −55.60 | −133.42 | 0.00055 | 0.00044 |
| tRF-18-OUR83004 | GACGAGGTGGCCGAGTGG | 18 | 5′-tRF | −59.07 | −20.03 | 0.00005 | 0.00137 |
| tRF-16-YU4RRW0 | TTCGACTCCCGGTGTG | 16 | i-tRF | −69.74 | −20.86 | 0.00039 | 0.00005 |
| tRF-19-RKVP4PJZ | GGGGGTATAGCTCAGTGGT | 19 | 5′-tRF | −74.65 | −44.26 | 0.00035 | 0.00035 |
| tRF-20-RK9P4P9L | GGGGGTGTAGCTCAGTGGTA | 20 | 5′-tRF | −76.64 | −258.08 | 0.00042 | 0.00024 |
| tRF-17-YU4RRQ4 | TTCGACTCCCGGTATGG | 17 | i-tRF | −84.37 | −12.90 | 0.00034 | 0.00213 |
| tRF-23-RK9P4P9LDS | GGGGGTGTAGCTCAGTGGTAGAG | 23 | 5′-tRF | −84.39 | −225.24 | 0.00035 | 0.00024 |
| tRF-22-RKVP4P9LL | GGGGGTATAGCTCAGTGGTAGA | 22 | 5′-tRF | −89.79 | −273.41 | 0.00042 | 0.00030 |
| tRF-22-PZK0JNOR4 | GCCTTCCAAGCAGTTGACCCGG | 22 | i-tRF | −141.65 | −33.28 | 0.00018 | 0.00024 |
| tRF-17-31QJ3KQ | CCGCCGCGGCCCGGGTT | 17 | i-tRF | −152.75 | −40.41 | 0.00018 | 0.00024 |
| tRF-38-RPFQRDWXHR13M80E | GGTAGAGCATGGGACTCTTAATCCCAGGGTCGTGGGTT | 38 | i-tRF | −161.18 | −201.05 | 0.00012 | 0.00018 |
| tRF-21-RKVP4P9L0 | GGGGGTATAGCTCAGTGGTAG | 21 | 5′-tRF | −188.88 | −126.57 | 0.00030 | 0.00030 |
| tRF-16-RKVP4P0 | GGGGGTATAGCTCAGG | 16 | 5′-tRF | −242.56 | −222.28 | 0.00042 | 0.00039 |
| tRF-18-7SERM404 | GTTCGACTCCCGGTGTGG | 18 | i-tRF | −314.03 | −54.18 | 0.00018 | 0.00024 |
Corresponding indolent CLL vs. normal control fold changes are reported. i-TRF, wholly internal tRF.
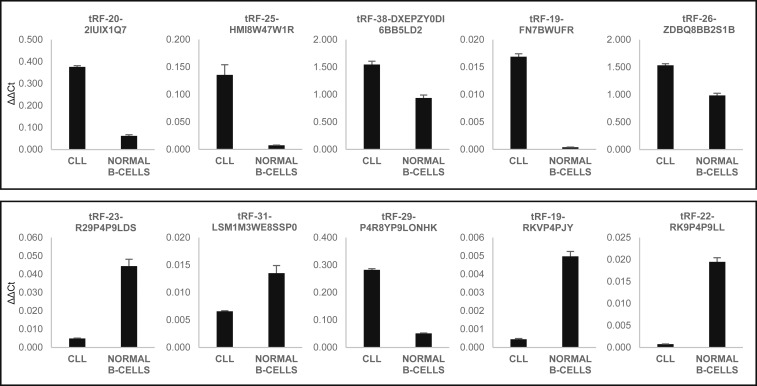
To confirm our results, we selected to further assess the expression of 5 up-regulated and 5 down-regulated tRFs in CLL via custom real-time RT-PCR (Dataset S1, in red and green). First, we used 1 indolent CLL sample and 1 normal B cell sample. CLL sample 1477 showed the highest expression levels for mature tRFs overexpressed in CLL, and close to average expression levels for mature tRFs underexpressed in CLL. Normal B cell sample 6139 showed close to average expression levels of all mature tRFs. The results are shown in Fig. 3. We were able to confirm the overexpression of all 5 tRFs up-regulated in CLL (Fig. 3, Top), and of 4 out of 5 tRFs down-regulated in CLL (Fig. 3, Bottom). We then selected 6 out of these 10 fragments to carry out custom real-time RT-PCR using all 28 samples (Dataset S1, in green). These experiments confirmed all 3 tRFs underexpressed in CLL (P < 0.05) (SI Appendix, Fig. S1 D–F). For 2 out of 3 tRFs overexpressed in CLL, we observed no statistically significant trends (SI Appendix, Fig. S1 B and C), and overexpression of the sixth fragment was not confirmed (SI Appendix, Fig. S1A). We speculated that the discrepancy between sRNA-seq and custom real-time RT-PCR data is due to the presence of very similar but not differentially expressed mature tRFs which can be separately detected by sequencing but not by real-time RT-PCR. To assess the expression profile of tRFs which included the fragments we considered, we performed a sequence alignment analysis leveraging the National Center for Biotechnology Information Megablast algorithm, specifying very stringent alignment and scoring parameters, with no sequence masking nor filters, in order to identify those tRFs which passed the minimum expression threshold and whose sequence significantly overlaps with that of those tRFs we considered for real-time RT-PCR. We identified mostly longer, nondifferentially expressed tRFs containing within their sequence 100% of the differentially expressed fragments in question (SI Appendix, Fig. S2). Specifically, SI Appendix, Fig. S2 A and B shows highly but nondifferentially expressed similar tRFs containing tRF-20-2IU and tRF-19-FN7. In both these cases, real-time RT-PCR results were not statistically significant (SI Appendix, Fig. S1). On the contrary, SI Appendix, Fig. S2 C and D shows homologous tRFs containing tRF-23-RKV and tRF-22-RK9. In these instances, the counts for these additional tRFs did not interfere significantly with the counts of tRF-23-RKV and tRF-22-RK9. And, in fact, real-time RT-PCR validation for these 2 tRFs was statistically significant (SI Appendix, Fig. S1).
Fig. 3.
Dysregulation of selected mature tRFs in CLL by real-time RT-PCR. We selected 10 tRFs dysregulated in CLL for custom real-time RT-PCR. (Top) Expression of 5 mature tRFs up-regulated in CLL. (Bottom) Expression of 5 mature tRFs down-regulated in CLL as well. The single CLL sample belongs to the group of indolent CLLs, and its ID is 1477. The normal B cell sample is 6139. Error bars indicate the SD for the 3 replicates.
Discussion
In this report, we first determined tsRNA signatures in CLL. We previously studied tsRNA expression in CLL employing custom microarray technology (7, 17). However, data obtained using such technology do not provide precise counts for each molecule. The data shown in this study are more precise and accurate. tsRNAs are unique sequences which are produced from the 3′ ends of pre-tRNAs by endonuclease RNase Z, beginning exactly at the 3′ end of tRNAs and ending at the sequence of 4 consecutive T nucleotides (7, 17). Other small tRFs (tRF-3s, tRF-5s, and internal tRFs) are derived from mature tRNA sequences, encoded by multiple genes, and are not unique fragments (24). We previously considered mature tRFs as less likely to be up- or down-regulated, as each of these fragments is expressed from several tRNA genes located in multiple distinct loci of the human genome. Interestingly, during our investigation of tsRNA expression in CLL, we determined that a mature tRNA fragment, specifically a tRF-5 originating from tRNAHis, is down-regulated in CLLs compared with normal controls. This result prompted us to investigate the expression of all mature tRFs in CLL. Here we determined that the expression of almost 1,000 mature tRFs is drastically altered in CLL. Thus, tsRNAs and mature tRFs could be exploited as diagnostic, prognostic, and predictive biomarkers in CLL.
The molecular function of these fragments is currently not clear. A recent report showed that in monocytes, a tRF-5 (tRF-5030c) interacts with PIWI proteins and recruits histone methyltransferases to the CD1A promoter, increasing histone H3K9 methylation (11). Another recent study reported that another tRF-5 deriving from tRNAAla/Cys/Val represses translation in stem cells, impacts hematopoietic commitment, and plays an essential role in early embryogenesis (23). Interestingly, the pseudouridylation of this tRF-5 plays a regulatory role in these processes (23). Another report demonstrated that specific tRFs bind to the oncogenic RNA-binding protein YBX1, displacing prooncogenic transcripts and resulting in their destabilization (13). In addition, Maute et al. (12) showed that a tRF-3 (CU1276 or tRF-3027) is down-regulated in human B cell lymphomas and functions in an miRNA-like manner, by targeting a tumor suppressor, RPA1.
For any up- or down-regulated coding or noncoding RNAs, the next and very important question is whether these changes are original events causing these malignancies, or whether they are consequences of dysregulation of many genes during malignant transformation. Even in the case that dysregulation of different classes of tRFs in CLL is a downstream event, these fragments may represent valuable markers of CLL progression, drug resistance, and/or prognosis. The fact that almost 2,000 mature tRFs are dysregulated in comparison with usually a few tens of microRNAs suggests that statistically mature tRFs will have a much better chance of serving as CLL biomarkers. Additional studies are necessary in order to determine if dysregulated tRFs are critical tumor suppressors/oncogenes in CLL or if alterations of their expression are downstream events of disease progression.
Methods
Patient and Healthy Control Sample Selection.
All human samples (both from CLL patients and healthy donors) utilized in this study were collected and analyzed under the protocol approved by the Institutional Review Board of The Ohio State University. The CLL Research Consortium provided us with a total of 20 samples collected from CLL patients enrolled upon written informed consent. According to IgVH mutational gene status, ZAP-70 expression, karyotype analysis, and clinical features (SI Appendix, Table S1), 10 of these samples were classified as clinically indolent CLL and 10 as clinically aggressive CLL. CLL samples were mostly (>95%) composed of CD5+/CD19+ B cells. Normal CD19+ B cell samples used for real-time RT-PCR and RNA sequencing came from frozen peripheral blood mononuclear cells of 8 healthy donors described in ref. 7. RNA was extracted using TRIzol Reagent (Thermo Fisher Scientific) following the standard protocol. The quality of the RNAs of each sample was analyzed on the Agilent 2100 Bioanalyzer. For ts-42 promoter-methylation analysis, normal CD19+ B cell samples were collected from the whole blood of 6 healthy donors using RosetteSep Human B Cell Enrichment Mixture (STEMCELL Technologies) according to the manufacturer’s instructions. DNA was extracted with the DNeasy Blood & Tissue Kit (Qiagen).
Reverse Transcription and Real-Time RT-PCR.
The expression of tRNA-derived fragments and tsRNAs was analyzed by real-time RT-PCR after the design of custom TaqMan Small RNA Assays (Thermo Fisher Scientific) (SI Appendix, Table S5), according to the standard protocol. The data were normalized using RNU44 (assay ID 001094; 4427975; Thermo Fisher Scientific). We applied the 1-tailed Wilcoxon rank-sum test to all real-time PCR experiments. The unit of measurement employed for dispersion is the SD. P < 0.05 was considered statistically significant.
Methylation Analysis.
DNA (1.5 µg) previously extracted with the DNeasy Blood & Tissue Kit (Qiagen) was subjected to bisulfite conversion and subsequent purification using the EpiTect Fast DNA Bisulfite Kit (Qiagen), according to the manufacturer’s protocol. After bisulfite conversion, the ts-42 promoter region was analyzed by PCR amplification and sequencing. PCR was performed using 50× Advantage 2 Polymerase Mix (Clontech) and the following primer sequences: ts42metY_F: 5′-GGTAAAATGGGGTTTAATGG-3′, ts42metY_R: 5′-CTACTAACACTACCCAATTC-3′; ts42metB_F: 5′-AGAAGGAGTATATTTTTATTAATAAG-3′, ts42metB_R: 5′-CTTCAAATCTCCACACATCC-3′. After purification with ExoSAP-IT PCR Product Cleanup Reagent (Thermo Fisher Scientific), PCR products were subjected to Sanger sequencing and analyzed with Sequencer 5.4.6 software to evaluate the C-to-T conversion rate.
Next-Generation Sequencing of tRNA-Derived Fragments.
Total RNA (2 µg) for each sample was analyzed by next-generation sequencing (NGS) after quality control on the Agilent 2100 Bioanalyzer. Universal cDNA synthesis, cDNA cleanup, library amplification, and library cleanup were performed using the QIAseq miRNA Library Kit (Qiagen) according to the manufacturer’s protocol. Small RNA-seq libraries were then sequenced using the Illumina NGS HiSeq 2500 System.
Preprocessing of Raw Sequencing Files.
Fastq files generated via the QIAseq Small RNA Library and sequencing protocols were first merged (to attain sufficient sequencing depth per sample), preprocessed by trimming the QIAseq 3′ adapter (5′-AACTGTAGGCACCATCAAT-3′) as well as the 3′ universal Illumina adapter (5′-AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC-3′) via the tool Cutadapt (18) (parameters: min length=12, max length=50, min_quality=25, -n 1 -O 5 ‐‐match-read-wildcards), and finally quality-filtered via the tool ConDeTri (19) (parameters: -pb = fq -lq = 20 -hq = 30 -minlen = 15 -sc = 33).
tsRNA Mapping and Quantification.
Preprocessed fastq files were then analyzed by leveraging a modified version of the tRNA fragment detection tool tDRmapper (20), allowing exclusively for the exact mapping of filtered sequencing reads on a customized version of the tRNA genomic space. Such space comprises the original mature tRNA sequences provided with tDRmapper, together with 616 pre-tRNA sequences downloaded from the University of California, Santa Cruz Table Browser (https://genome.ucsc.edu/cgi-bin/hgTables?) for the GRCh37/hg19 assembly (group: gene and gene predictions; track: tRNA genes; table: tRNAs) and characterized by 50-nt sequence extensions flanking both sides of known mature tRNA sequences. Finally, the custom tRNA space was also augmented in order to account for single occurrences of common SNP (as reported by dbSNP v150) (27) occurring on any of the tsRNA sequences, with an additional total of 114 common SNP-including pre-tRNA sequences.
After mapping, in-house scripts were employed to parse tDRmapper output files (.mapped files) to collect and collapse raw counts which mapped to locations within pre-tRNA molecules contained in the provided tRNA space and which were annotated to be tsRNA loci, by leveraging a custom annotation file associating tsRNA IDs and their location and length with pre-tRNA IDs as reported in the custom tRNA space. Raw counts for all tsRNA molecules which were detected having SNPs were all combined, along with their reference tsRNA sequence count when detected, into a single collapsed read count per sample (marked with an asterisk at the end of their tsRNA ID). All raw counts per sample were collected into a single table which was employed as input for the differential expression analysis.
tRF Mapping and Quantification.
Preprocessed fastq files were analyzed by leveraging the recently published tool MINTmap (25), applied with standard parameters (default genome assembly GRCh37), in order to mine for and quantify internal tRNA fragments (25). Raw counts were extracted and collapsed through in-house scripts.
Differential Expression Analysis.
A minimum expression filtering was applied on raw counts prior to normalization, with detected molecules retained if they reported at least 5 raw reads in each of at least 8 samples. As recommended (21), filtered molecules were normalized by applying the TMM method and statistical analysis of the data was carried out within the R studio environment (version 1.1.423), employing the software package edgeR (22), and by applying an unpaired, 2-tailed Mann–Whitney–Wilcoxon nonparametric test with continuity correction in order to assess significant differential expression for each molecule between any 2 groups among aggressive, indolent, and normal samples. Molecules differentially expressed with P < 0.05 were considered for further investigation.
BLASTn Analysis to Assess Homology between tRF Sequences.
Sequence alignment analysis between 4 selected tRF molecules (tRF-20-2IUIX1Q7, tRF-19-FN7BWUFR, tRF-23-R29P4P9LDS, and tRF-22-RK9P4P9LL) and the 3,079 tRF molecules which survived the minimum expression threshold prior to normalization was performed leveraging the Blastn suite (https://blast.ncbi.nlm.nih.gov/Blast.cgi) by specifying the optimization for highly similar sequences (Megablast) and automatically adjusting word size and other parameters to improve results for short queries, with an expected number of 50 chance matches in a random model, 16 as the length of the seed that initiates an alignment, 0 matches to a query range, a match score of 4, a mismatch score of −5, gap existence cost of 12 and extension cost of 8, and no additional parameters specified.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute Grants P01-CA81534 (to T.J.K. and C.M.C.) and R35-CA197706 (to C.M.C.). We thank Dr. John Byrd for providing CD19+ B cell controls.
Footnotes
Competing interest statement: N.C. and T.J.K. are coauthors on a 2018 guidelines paper. N.C. and C.M.C. are coinvestigators on a grant application but have not begun a collaboration yet.
Data deposition: The data reported in this paper have been deposited in NCBI’s Gene Expression Omnibus (accession no. GSE138889).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913695116/-/DCSupplemental.
References
- 1.Döhner H., et al. , Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 343, 1910–1916 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Rassenti L. Z., et al. , ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N. Engl. J. Med. 351, 893–901 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Herling M., et al. , TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia 20, 280–285 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Bichi R., et al. , Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc. Natl. Acad. Sci. U.S.A. 99, 6955–6960 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyer K. K., et al. , Dysregulated TCL1 promotes multiple classes of mature B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 99, 14392–14397 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balatti V., et al. , TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 112, 2169–2174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pekarsky Y., et al. , Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 5071–5076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parisien M., Wang X., Pan T., Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 10, 1853–1867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haussecker D., et al. , Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martens-Uzunova E. S., Olvedy M., Jenster G., Beyond microRNA—Novel RNAs derived from small non-coding RNA and their implication in cancer. Cancer Lett. 340, 201–211 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Zhang X., et al. , IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J. Immunol. 196, 1591–1603 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Maute R. L., et al. , tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 110, 1404–1409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodarzi H., et al. , Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y. S., Shibata Y., Malhotra A., Dutta A., A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veneziano D., Nigita G., Ferro A., Computational approaches for the analysis of ncRNA through deep sequencing techniques. Front. Bioeng. Biotechnol. 3, 77 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veneziano D., et al. , Noncoding RNA: Current deep sequencing data analysis approaches and challenges. Hum. Mutat. 37, 1283–1298 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Balatti V., et al. , tsRNA signatures in cancer. Proc. Natl. Acad. Sci. U.S.A. 114, 8071–8076 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 1–3 (2011). [Google Scholar]
- 19.Smeds L., Künstner A., ConDeTri—A content dependent read trimmer for Illumina data. PLoS One 6, e26314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selitsky S. R., Sethupathy P., tDRmapper: Challenges and solutions to mapping, naming, and quantifying tRNA-derived RNAs from human small RNA-sequencing data. BMC Bioinformatics 16, 354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzzi N., et al. , Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173, 1204–1216.e26 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Venkatesh T., Suresh P. S., Tsutsumi R., tRFs: miRNAs in disguise. Gene 579, 133–138 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Loher P., Telonis A. G., Rigoutsos I., MINTmap: Fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 7, 41184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loher P., Telonis A. G., Rigoutsos I., Accurate profiling and quantification of tRNA fragments from RNA-seq data: A vade mecum for MINTmap. Methods Mol. Biol. 1680, 237–255 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Sherry S. T., et al. , dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.