Significance
Color pattern mimicry in Heliconius butterflies provides a key example of trait evolution via selection on large effect genes. To understand how individual genes drive the evolution of new, complex traits, we functionally characterized 5 enhancers of the red color pattern gene in Heliconius. We discovered these enhancers are all necessary for color patterning and influence multiple elements of wing patterning rather than driving distinct wing pattern modules. Enhancer variants are shared among similarly colored disjunct populations via hybridization. These enhancers also appear to be evolving in parallel in distantly related co-mimic species, suggesting their ancient role in Heliconius wing pattern diversity. We propose a model for how interacting enhancers and their upstream modifiers can generate wing pattern diversity.
Keywords: mimicry, adaptation, butterfly, enhancer, evolution
Abstract
Color pattern mimicry in Heliconius butterflies is a classic case study of complex trait adaptation via selection on a few large effect genes. Association studies have linked color pattern variation to a handful of noncoding regions, yet the presumptive cis-regulatory elements (CREs) that control color patterning remain unknown. Here we combine chromatin assays, DNA sequence associations, and genome editing to functionally characterize 5 cis-regulatory elements of the color pattern gene optix. We were surprised to find that the cis-regulatory architecture of optix is characterized by pleiotropy and regulatory fragility, where deletion of individual cis-regulatory elements has broad effects on both color pattern and wing vein development. Remarkably, we found orthologous cis-regulatory elements associate with wing pattern convergence of distantly related comimics, suggesting that parallel coevolution of ancestral elements facilitated pattern mimicry. Our results support a model of color pattern evolution in Heliconius where changes to ancient, multifunctional cis-regulatory elements underlie adaptive radiation.
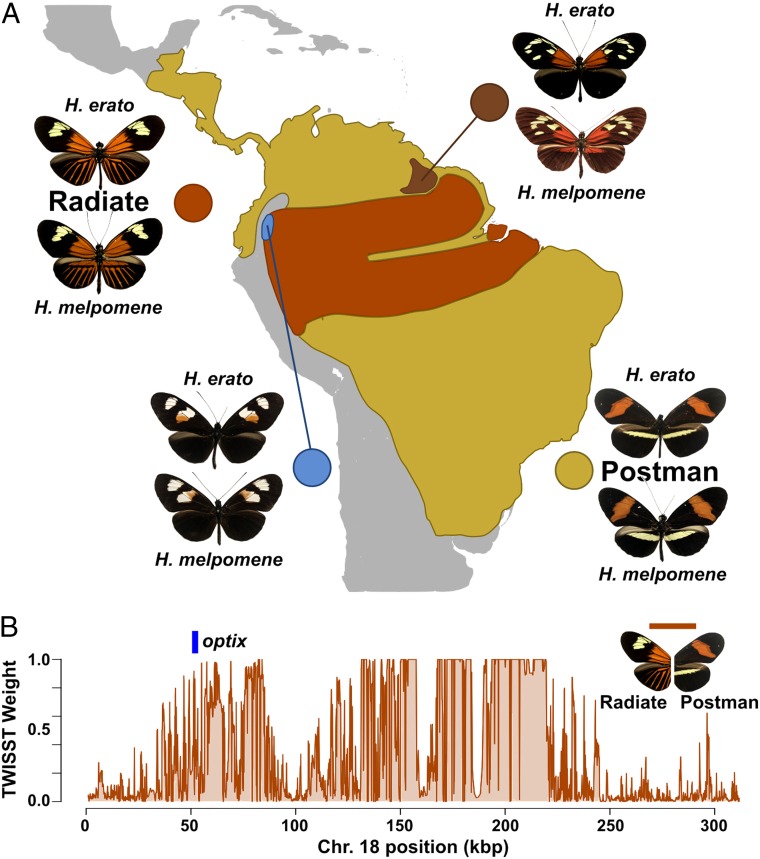
Neotropical Heliconius butterflies are a classic example of an adaptive radiation driven by few genes of large effect (1–3). These butterflies are one of the defining examples of Müllerian mimicry, where local populations have evolved warning coloration to mimic other local toxic butterfly and moth species (4, 5). Approximately 10 to 15 MYA, Heliconius spread across Central and South America, evolving red, black, and yellow aposematic wing patterns to warn predators of their toxicity (6). Incredibly, red color pattern diversity in the genus Heliconius is largely controlled by divergent expression of a single gene, optix, making Heliconius an important case study of how variation at a single major effect locus can drive adaptation of a complex trait both within and between species (Fig. 1A) (7). Population genetic scans and comparative gene expression studies suggest that cis-regulatory differences underlie adaptive variation of red color patterns, and recent work has sought to characterize the specific noncoding regions around optix responsible for this (Fig. 1B) (1, 5, 8, 9). A general prediction of evolutionary genetics is that gene regulation should evolve largely through changes in independent cis-regulatory “modules” that segregate developmental roles while minimizing deleterious pleiotropic side effects of mutation (10, 11). Following this expectation, it has been proposed that discrete cis-regulatory elements (CREs) adjacent to optix activate independent red color pattern subelements, such as rays or bands, potentially explaining how this single gene can drive such complex pattern evolution (5, 9, 12). This model has not been tested experimentally, however, and may even be contradicted by some lines of genetic evidence (SI Appendix, Appendix A).
Fig. 1.
Red color pattern variation in Heliconius maps to the optix locus. (A) Map showing the geographical distribution of major red color pattern types of H. erato, along with the comimic H. melpomene. Two major phenotypes, radiate and postman, comprise the majority of red color patterns in the 2 species. (B) Phylogenetic weighting plot between radiate and postman H. erato populations shows loci separating these 2 morphs around the gene optix.
Here we functionally characterize 5 optix CREs that underlie Heliconius color pattern variation. Contrary to prior predictions, we discovered that all 5 elements are pleiotropic and functionally interdependent, and at least 4 of the elements are necessary for development of multiple red color pattern elements and even wing venation. We found that introgression likely propagated cis-regulatory alleles throughout the Amazon, and that orthologous regulatory elements are implicated in parallel coevolution between comimetic species. Our findings provide a case study of how parallel evolution of ancient, multifunctional regulatory elements can facilitate the rapid diversification of complex phenotypes and lead us to propose a revised model of Heliconius color pattern architecture that is characterized by epistatic interactions and regulatory pleiotropy.
Results and Discussion
Candidate CREs Occur in Color Pattern-Associated Regions.
To characterize genomic regions that control color pattern-related optix expression in Heliconius erato we used a combination of assay for transposase-accessible chromatin sequencing (ATAC-seq) (13, 14), chromatin immunoprecipitation and sequencing (ChIP-seq) (15), and a type of chromosome conformation capture (Hi-C) (16) data to identify cis-regulatory elements (CREs) acting on optix (SI Appendix, Table S1). We hypothesized that optix CREs should be active early to midway through pupal development, in association with the initiation of optix expression in color patterns, and would likely differ in accessibility between early and late developmental stages and between wings with different color pattern elements (1, 17). Thus, we generated chromatin accessibility (ATAC-seq) data, which indicate cis-regulatory activity (14), for midpupal (3 d postpupation) and late-pupal (ommochrome stage, ∼7 d postpupation) stages of both forewing and hindwing tissues for 3 color pattern morphs of the H. erato clade (H. erato lativitta, Heliconius himera, and H. erato petiverana), including samples of the common “radiate” (red triangle at the base of the forewing, rayed pattern on the hindwing) and “postman” (red band on the forewing) phenotypes (Fig. 2A and SI Appendix, Fig. S1) (5, 18). Consistent with previous studies (13, 19), our initial results showed little detectable chromatin accessibility variation between forewings and hindwings with different color patterns, suggesting that differences in chromatin accessibility between color pattern elements is not driving optix expression variation between wings of a single individual.
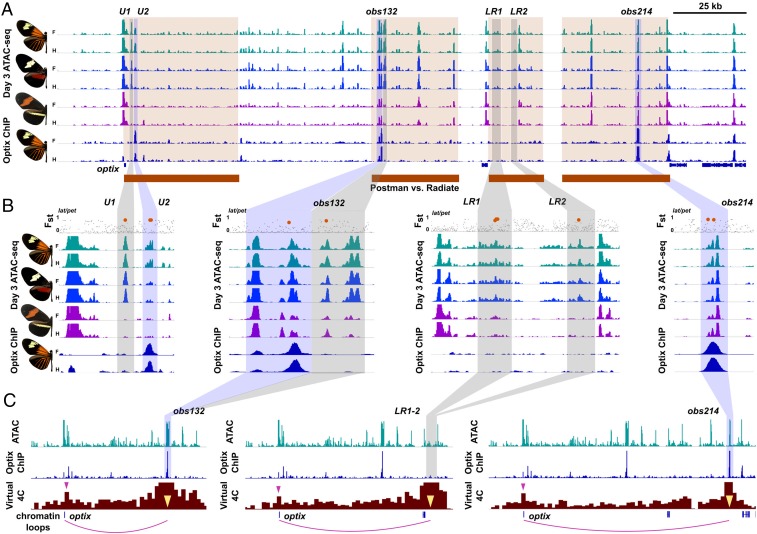
Fig. 2.
Accessibility, occupancy, and promoter contacts identify color pattern-associated optix CREs. (A) Day 3 ATAC-seq data for 3 H. erato populations with 6 candidate optix CREs identified in shaded boxes. Gray boxes mark elements binding unknown factors; blue boxes identify CREs with optix binding sites identified with ChiP-seq; blue bars show gene annotations with optix identified. No variation was observed between forewing and hindwing color patterns. Red bars highlight loci separating radiate from postman morphs in Fig. 1B. (B) U1, U2, obs132, LR1, and LR2 show variation in accessibility associated with radiate and postman morphs. U2, obs132, and obs214 show autoregulatory optix binding in ChIP-seq assays. Fst for all SNPs between 2 radiate and postman populations is shown above the ATAC-seq tracks, with high-Fst SNPs inside of ATAC-seq peaks highlighted in orange. (C) Virtual 4C Hi-C tracks for obs132, LR1-2, and obs214 show looping of distal elements to the optix promoter.
We next intersected our ATAC-seq data with the large number of whole-genome DNA resequencing datasets for H. erato to identify changes in chromatin accessibility correlated with geographic color pattern variation (Fig. 2A). Phylogenetic weighting (topology weighting by iterative sampling of sub-trees [TWISST]), which determines the frequency of phylogenies matching a specified topology, using the radiate reference genome identified 4 intervals that strongly associate with separation of radiate and postman color morphs. Repeating this TWISST analysis including additional races and species with additional less common nonradiate red forewing and hindwing patterns (see ref. 5) also revealed a smaller set of loci simply associated with the presence/absence of red hindwing and forewing phenotypes across species (SI Appendix, Fig. S2). We then identified 5 CREs that varied in accessibility between radiate and postman phenotypes and also fell within regions of hybrid zone population divergence (5, 8).
Two candidate elements were within the 3′ UTR of the optix coding region (Fig. 2B, U1 and U2); 1 cluster of accessible peaks was located 80 kb downstream of optix (Fig. 2B, obs132), while the final 2 elements fell within a distal region ∼125 kb 3′ of optix that associated specifically with separation of red hindwing phenotypes in a larger clade analysis, including morphs with less common red wing patterns (Fig. 2B, LR1 and LR2, SI Appendix, Fig. S2) (5). While there is distinct variation in chromatin accessibility in comparisons of radiate vs. postman phenotypes, which involve differences in the location of forewing red coloration and the presence of hindwing rays (Fig. 1), analysis of the presence of a red hindwing bar instead of the radiate phenotype (H. himera vs. H. e. lativitta) showed little difference in chromatin accessibility. We also found no evidence of accessible site variation positively correlated with the red forewing band between any morphs, even when ATAC-seq data were aligned to a postman (Heliconius erato demophoon) reference genome (5); however, there was 1 strong nonvariable candidate CRE in a very narrow interval associated with species-level variation in red forewing band phenotypes (SI Appendix, Fig. S3) that we will assess in a future study. We focus subsequent analyses on radiate versus postman phenotypes, in part due to the strength of chromatin accessibility associations.
The persistent expression pattern of optix over wing development, which initiates ∼2 to 3 d postpupation and remains stable in red wing pattern elements throughout most of the pupal period (17), led us to predict that optix could have an autoregulatory function to maintain precise long-term expression. To test this, we performed ChIP-seq to characterize optix protein binding in midpupal forewing and hindwing tissue from a radiate morph butterfly. optix ChIP-seq highlighted one additional CRE of interest (Fig. 2B, obs214), as well as the previous candidates U2 and obs132. The optix binding site obs214 falls within the radiate–postman association interval and also maps to a narrow locus associated with variation in multiple red forewing patterns between species, but shows little variation in chromatin accessibility between populations despite strong signatures of nucleotide association (see below). All candidate CREs showed correlated accessibility profiles at all pupal timepoints we sampled, tentatively suggesting that pattern formation and autoregulatory functions are integrated, and not stage specific. Neither the degree of chromatin accessibility at a locus, nor the conservation of accessibility between races, was a universal predictor of optix binding at autoregulatory elements.
Candidate CREs Interact Directly with optix Promoter.
To determine whether obs132, LR1-2, and obs214 are direct regulators of the optix promoter, we produced Hi-C data from midpupal forewing and hindwing tissue to generate virtual 4C plots (U1 and U2 are too proximal to the promoter for this approach). Our Hi-C data, which assay physical contacts between regulatory loci such as enhancers and promoters (16), showed broad, substantial enrichment in contacts between the optix promoter and the 3′ distal gene desert as would be expected for a locus with multiple CREs acting on a single promoter (SI Appendix, Fig. S4). To verify the regulatory role of the distal elements, we generated virtual 4C plots centered on these loci. As expected, all 3 showed substantial enrichment of physical contacts with the optix promoter (Fig. 2C and SI Appendix, Fig. S5), though Hi-C lacks the resolution to distinguish between promoter contacts with LR1 and LR2 individually. Ultimately, our Hi-C data confirm that optix is regulated by multiple CREs in a complex network that includes both autoregulatory and nonautoregulatory elements.
optix Color Pattern CREs Are Pleiotropic and Interdependent.
We next sought to experimentally define the role of optix CREs in wing pattern development and to test the model that modular regulatory sequences activate individual color pattern subelements in a population with a radiate color pattern (SI Appendix, Appendix A). To do this, we used CRISPR/Cas9-mediated targeted embryonic deletions in a radiate morph (H. e. lativitta) to produce somatic knockout mosaics for 5 of the CREs identified above: U1, obs132, LR1, LR2, and obs214 (SI Appendix, Figs. S6–S8). All reported CRE phenotypes were supported by multiple (3 to 7) mutant individuals (SI Appendix, Table S4). While some variation in red pattern elements is expected between individuals, patterns within an individual are highly symmetrical, allowing us to use left/right wing comparisons to test for knockout phenotypes in mosaic mutant individuals. Deletions of each element displayed multiple phenotypic effects. U1 had the most limited range of phenotypes in our experiments: loss of hindwing veins and related reduction and merging of ray color patterns (Fig. 3A). Deletions at all 4 of the other elements showed broader ranges of pleiotropic effects (SI Appendix, Table S4), including vein abnormalities as with U1 (e.g., Fig. 3B), but also black clones in both forewing and hindwing red pattern elements (Fig. 3C). This red/black color switch phenotype precisely phenocopies loss-of-function optix mutations (20), and thus leads us to infer that these elements function in an enhancer-like manner, i.e., as positive regulators of optix in red pattern elements.
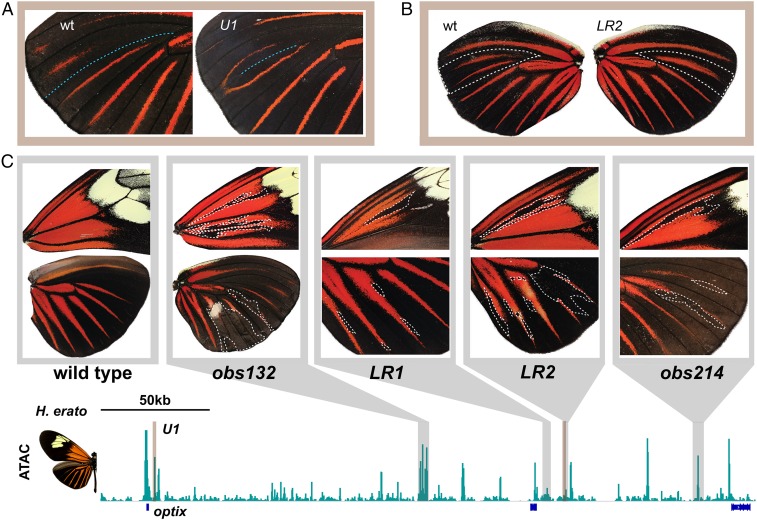
Fig. 3.
optix CREs are pleiotropic and interdependent. (A) A U1 mosaic deletion that shows partial loss of the hindwing M1 vein (blue dotted line), with associated merging of ray color patterns. (B) An LR2 mosaic deletion caused complete loss of the hindwing M1 vein, with associated loss of a ray pattern. The wings shown represent a left/right comparison from the same mosaic individual. (C) Mosaic deletions of obs132, LR1, LR2, and obs214 demonstrate that all 4 regulatory elements are required for development of both forewing and hindwing radiate color patterns. White dotted lines annotate deletion clones as inferred from asymmetrical differences between opposing wings (SI Appendix, Fig. S5).
Two important conclusions follow from these results. First, the optix CREs we characterized are all pleiotropic, with mutants for all 5 elements exhibiting multiple phenotypes, most of which affect all major forewing and hindwing red pattern elements as well as wing venation (Fig. 3C). Second, these CREs appear to be functionally interdependent. Surprisingly, our results show that deletion of any single distal regulatory element can cause loss of any red pattern element in F0 mutant clones, indicating that all distal regulatory loci we surveyed are necessary for normal development of all major red color pattern elements and provide another example of loci that require multiple regulatory loci for gene activation (21). While future studies may identify additional wing pattern CREs, our current data provide no support for previous modularity models where single enhancers independently activate discrete, individual pattern subelements (5, 10, 11). Instead, our evidence supports a data-driven model of highly pleiotropic regulatory landscape composed of multifunctional, interdependent CREs of large effect.
Genomic Differentiation of optix CREs Varies Between Hybrid Zones.
To better understand patterns of selection and divergence at the optix locus within the H. erato clade, we tested to what extent specific optix CREs show population differentiation and structure at color pattern hybrid zones. Due to strong natural selection on pure phenotypes, the entire gene desert between optix and the 3′ gene cluster often shows elevated population-associated genomic differentiation (Fst) between red morphs (5, 8). Despite this, individual hybrid zones frequently display some degree of variation in Fst at different CREs and the optix coding region (SI Appendix, Fig. S9). To accurately determine population divergence at our CRE loci while controlling for linked selection, we calculated the mean Fst for all SNPs within each CRE locus, then subtracted the mean Fst of the local genomic background (shown with a black bar, SI Appendix, Fig. S9) to generate a measure of average population differentiation for each CRE above the mean effect of linked selection.
As would be predicted, comparison of parapatric or hybridizing populations with the same red color patterns showed little evidence of elevated population differentiation. In contrast, in hybrid zones where the red color patterns of the parental populations differed, all optix CREs showed a substantial increase in Fst over the local background in one or more comparisons (0.14 to 0.42, actual Fst values ranged from 0.46 to 0.74 in these populations, Fig. 4A). Our results showed little evidence of consistent differentiation patterns for specific CREs, or by geography, across the 5 hybrid zones where parental patterns differed. Similarly, structural variation associated with red pattern variation was found in all 4 distal regulatory loci (SI Appendix, Figs. S10–S13). These results suggest that population divergence at optix is not predictably partitioned by individual CREs, but instead by regionally variable sets of CREs. While this pattern could indicate the presence of additional regulatory loci, our analysis provides little evidence for a model where individual, recombining CREs are sufficient to determine individual pattern subelements and is instead more consistent with functional interdependence where sets of multiple CREs are required to generate patterns.
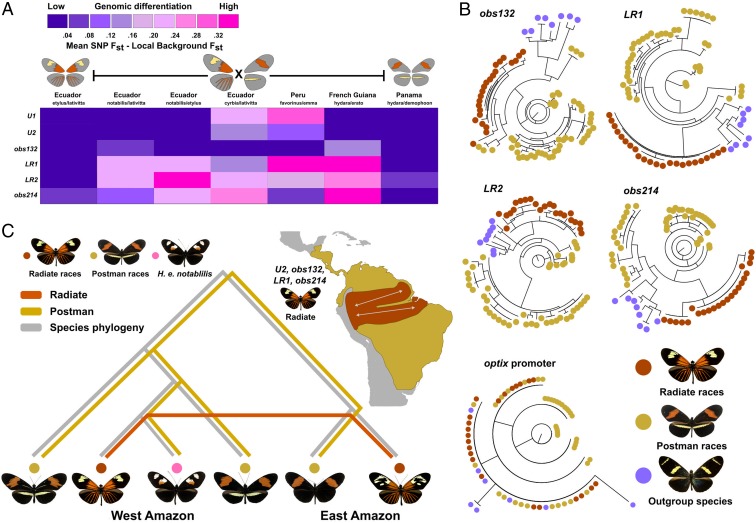
Fig. 4.
Derived optix CREs spread through introgression and mark loci of population differentiation at hybrid zones. (A) All annotated optix CREs show evidence of strong population differentiation in one or more hybrid zones. To reduce the effect of linked selection on evidence of population differentiation, the heatmap presents the mean SNP Fst of the entire optix locus (black bar) subtracted from the mean SNP Fst within a CRE. (B) Maximum likelihood phylogenies of distal optix CREs obs132, LR1, LR2, and obs214 form distinct clades for radiate populations (orange samples). (C) Collapsed phylogeny for Amazonian clade butterflies gives a summary of coalescent models for U2, obs132, LR1, and obs214, indicating that the radiate phenotype is derived from the basal postman morph and spread via introgression through the Amazonian populations.
CRE Phylogenies Reflect Origin, History, and Introgression of Color Patterns.
A previous study found that phylogenies of the optix coding sequence recapitulate color pattern groups instead of actual species phylogenies for both H. erato and the comimic Heliconius melpomene, and proposed a single evolutionary origin for radiate color patterns in each species (18). Consistent with this, our focused phylogenetic analyses of obs132, LR1-2, and obs214 illustrate strong relationships between color pattern and allele phylogenies within H. erato (Fig. 4B). In each tree, CRE alleles separate radiate morphs into a distinct clade that differs substantially from both the species phylogeny (5) (Fig. 4C) and the relationship between populations at the optix promoter, suggesting nonneutral evolution via either incomplete lineage sorting of an ancestral polymorphism or introgression of regulatory alleles across the Amazon basin (SI Appendix, Figs. S14–S21). Phylogenies including additional morphs and species support the same conclusion (SI Appendix, Figs. S14–S21), while postman populations consistently showed a non-monophyletic structure, consistent with this phenotype being the ancestral form within the species. The proximal U1 and U2 elements displayed similar phylogenetic signals, separating all radiate from most postman morphs, but included the French Guiana postman population (SI Appendix, Fig. S14). This suggests that this region—in which 3 different morphs hybridize—may have a more complex evolutionary history. This view is supported by the less consistent differentiation of U1 and U2 at hybrid zones.
To distinguish between incomplete lineage sorting and introgression between the radiate morphs, we used coalescence analysis to estimate the branch lengths of genomic sequences at each CRE. Coalescent trees consistently grouped individuals with radiate color patterns as seen in our phylogenies, and no population structure was apparent between geographically separated radiate populations. Despite the large effective population size and fast generation time of H. erato (22), coalescence time for radiate individuals at optix CREs was low, where radiate pattern haplotypes at most CRE loci coalesced some time in the past 20,000 to 30,000 y (SI Appendix, Figs. S22–S28). Thus, evidence supports the hypothesis that radiate CREs arose as a derived trait within the Amazonian clade of H. erato and spread throughout the Amazon region via introgression, with a subsequent free flow of the radiate alleles between populations (Fig. 4C). Consistent with the postman-like Heliconius hermathena being the sister species to H. erato, the postman phenotype individuals coalesce much later, again indicating that the postman phenotype was the ancestral state.
Parallel Evolution of optix Enhancers Between Comimic Species.
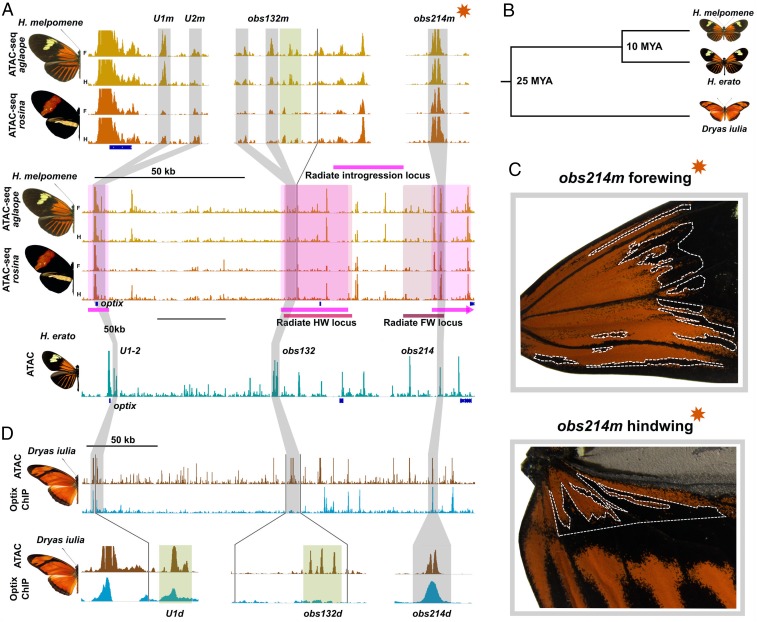
The question of coevolution between comimic butterfly species H. erato and H. melpomene has been a major focus of Heliconius biology (23–25). Prior studies have proposed mostly nonoverlapping genomic regions in H. melpomene (versus H. erato) associated with the convergent evolution of radiate color patterns (5, 9). Nonetheless, it is still unresolved to what extent orthologous regulatory elements may contribute to mimicry between species. To address this question, we used synteny analysis to align the radiate H. erato reference assembly to the H. m. melpomene assembly and then performed ATAC-seq on midpupal forewings and hindwings for radiate (H. m. aglaope) and postman (H. m. rosina) individuals of H. melpomene to test whether there was a shared, ancestral accessibility profile for the H. erato optix CREs. Our data indicate that the U1-2, obs132, and obs214 elements (Fig. 5A) are conserved and likely functional regulatory elements in H. melpomene despite ∼10 MY of divergence (Fig. 5B) (6). Accessibility profiles for these 4 elements also displayed similar patterns of variation between postman and radiate morphs as observed in H. erato.
Fig. 5.
Parallel coevolution of optix CREs between comimics H. erato and H. melpomene. (A) ATAC-seq comparison between radiate and postman morphs of H. melpomene show that U1, U2, obs132, and obs214 are conserved between H. erato and H. melpomene. Conserved optix CREs mark loci of SNP association with radiate patterns in the H. melpomene clade (red bars) and fall within loci associated with the introgression of radiate morphology from H. melpomene into H. timareta (pink bars). Gray boxes denote alignable DNA sequence conservation; green boxes shows presumptive functional conservation based on conserved flanking DNA sequences. (B) Trimmed phylogeny showing evolutionary relationships between H. erato, H. melpomene, and outgroup species D. iulia. (C) Mutant individual for obs214m shows clonal loss of the forewing and hindwing red subelements similar to H. erato mutants for obs214. (D) Deep conservation of U1, U2, obs132, and obs214 CREs provides additional evidence for parallel evolution between H. erato and H. melpomene via remodeling of ancestral regulatory elements.
To test whether the conserved CREs associated with color pattern evolution in H. melpomene, we compared our candidate CREs to loci previously linked to red wing pattern phenotypes in the H. melpomene clade. Supporting a functional role for these elements, the conserved CREs mark 3 distinct regions of introgression between radiate morphs in the H. melpomene species group (from H. melpomene into Heliconius elevatus) previously associated with the interspecific transfer of the radiate phenotype (Fig. 5A, pink shading) (26). We then compared our results to loci associated with red pattern divergence at the H. melpomene clade (H. melpomene and Heliconius timareta) hybrid zones (9). We found that the H. melpomene ortholog of obs132 (obs132m) maps within the proposed hindwing radiate phenotype SNP association interval in the H. melpomene clade and contains a series of 3 accessible sites showing varying degrees of conservation with H. erato (Fig. 5A, light red shading). Similarly, obs214 is conserved in H. melpomene (obs214m), and maps within a region previously associated with the red radiate forewing pattern (Fig. 5A, dark red shading). obs214m precisely marks the intersection of SNP associations for the red radiate forewing phenotype and the most distal locus associated with interspecific transfer of the radiate phenotype. As with H. erato, phylogenetic analysis of H. melpomene clade individuals shows that both obs132m and obs214m form monophyletic lineages by color pattern, including sorting the postman and radiate morphs of multiple H. melpomene clade species appropriately (SI Appendix, Figs. S29–S33).
Because phylogenetic and population analyses strongly implicate several orthologous CREs in mimicry between H. erato and H. melpomene, we aimed to experimentally validate the wing patterning role of 1 orthologous element in H. melpomene. To do this we used CRISPR/Cas9 mutagenesis to produce mosaic deletions of obs214m in a radiate H. melpomene comimic (H. m. aglaope) of H. erato. Our screen produced 3 obs214m mutants showing that this CRE is indeed necessary for red wing pattern formation in H. melpomene. These mutants displayed disruption of the red forewing radiate phenotype and the proximal portion of the radiate hindwing phenotype (Fig. 5C), very similar phenotypes to those seen in obs214 deletions in H. erato. Although other CREs remain to be tested, this result confirms that at least some orthologous CREs perform similar roles in generating convergent wing patterns between comimics.
Deep Ancestry of Color Pattern CREs.
To determine the degree to which optix CREs are ancestral in the heliconiines, we assayed chromatin accessibility and optix binding in pupal wings of the basal heliconiine, Dryas iulia, which diverged from the Heliconius ancestor ∼25 MYA (Fig. 5B) (6). Clear ATAC-seq peaks were observed in the conserved positions of U1, U2, and obs132, although the genomic sequences at the accessible sites were unalignable, suggesting substantial sequence evolution due to drift or selection (Fig. 5D). Furthermore, optix binding was absent at obs132, suggesting that this autoregulatory function was either derived in Heliconius or secondarily lost in D. iulia. obs214 was strongly conserved in both accessibility and DNA sequence, suggesting a deeply conserved ancestral function for this element. Taken together, our cross-species comparative results indicate some degree of parallel coevolution of a set of shared, deeply ancestral regulatory loci contributed to the evolution of red warning color mimicry between H. erato and H. melpomene.
Enhancer Shuttering: An Epistatic Model of Wing Pattern Evolution in Heliconius.
Finally, we sought to reconcile our H. erato findings with the modular enhancer hypothesis previously inferred from haplotype associations. This previous model proposes that individual enhancers are sufficient to activate color pattern subelements, and that red wing pattern variation is produced through differential recombination, or “shuffling,” of these enhancers (5). As described above, however, we were surprised when many of our results were not consistent with predictions of this model. Here we propose an alternative model inspired by Gilbert’s “window/shutter” concept (27), where epistatic interactions between early wing patterning genes and optix can readily explain both our current results, as well as results from previous studies (SI Appendix, Appendix A).
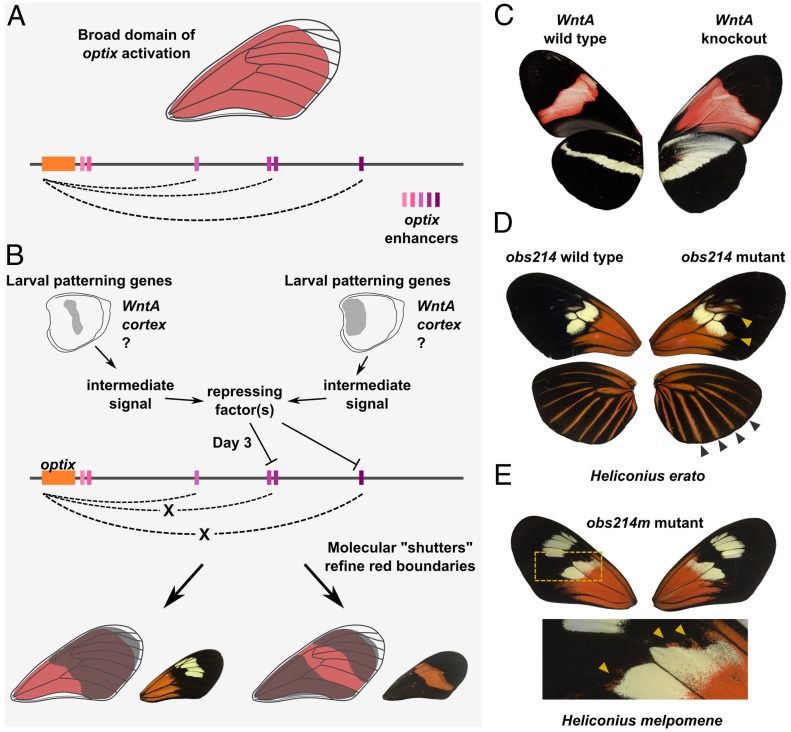
Specifically, our epistasis hypothesis of optix CRE function posits that a set of pleiotropic and functionally interdependent enhancers work together to generate a broad domain of potential optix expression, a “window” from Gilbert’s metaphor (Fig. 6A). This domain of potential optix expression is then refined by the epistatic action of repressing “shutter” factors that inhibit optix expression in some regions of the window (Fig. 6B). Under this model, different red color pattern morphs would evolve through gain and loss of shutter-responsive silencer elements (i.e., repressor binding sites) linked to optix enhancers. Thus, instead of color pattern evolution occurring through gain and loss of enhancers that individually activate pattern subelements, pattern evolution occurs via 2 processes: 1) gain or loss of repressor binding sites (i.e., silencers) that refine the final red patterns from a larger domain of potential optix expression, and 2) differential expression of the upstream repressors themselves (i.e., the shutters). These 2 mechanisms together form a molecular basis for Gilbert’s concept. In recognition of Gilbert’s original idea, we refer to this molecular model as “enhancer shuttering.”
Fig. 6.
Epistatic model of red color pattern determination in H. erato. (A) Representation of the domain of potential optix expression, or window of expression, driven by optix enhancers in the epistatic model. (B) Visual representation of epistatic early patterning genes (shutters) repressing optix enhancer activity to refine red wing patterns. Broken dashed lines show the loss of CRE activity on optix, leading to a lack of red patterning. (C) Wild-type and WntA knockout butterflies (reprinted from ref. 20) exemplify the epistatic role of WntA on red scale patterning in H. e. demophoon. (D) Wild-type and obs214 mutant forewings and hindwings from the same mosaic H. erato individual show pleiotropic expansion of red distally from basal forewing and expansion outward of the hindwing rays. (E) H. melpomene mutant for obs214m with distal expansion of basal forewing red. Asymmetric expansion of red patterning is highlighted in panel below.
Current Evidence for Enhancer Shuttering.
While further work is necessary to rigorously test an enhancer shuttering model, there is evidence that favors an epistatic model over recombination of modular enhancers as a mechanism of intraspecific Heliconius color pattern evolution. We provide a more thorough discussion in SI Appendix, Appendix A, but highlight 2 primary arguments here: First, and most importantly, knockout of the WntA black color pattern gene in Heliconius is sufficient to cause novel red color patterns to appear (Fig. 6C), as would be expected of a repressive shutter gene. WntA knockouts show that epistasis alone can generate phenotypes that resemble a hypothesized modular pattern subelement, even in the absence of novel optix enhancer alleles.
A second critical line of evidence for enhancer shuttering comes from closer examination of CRE deletion phenotypes. A prediction of enhancer shuttering is that some enhancers should have tightly linked or overlapping silencer elements—i.e., shutter factor binding sites that repress enhancer activity so that red patterns are inhibited where the shutter gene is expressed. In support of this, we generated a mutant H. erato where deletion of a CRE (obs214) resulted in expansion of red forewing patterns and widening of hindwing rays (Fig. 6D). This suggests that individual optix CREs can have both enhancer- and silencer-like functions. One H. melpomene mutant for the orthologous CRE (obs214m) displayed a similar expansion of red patterns (Fig. 6E), providing additional validation and suggesting that enhancer shuttering is potentially generalizable to both comimetic species.
This enhancer shuttering model of optix function is mechanistically very different from the modular enhancer model, yet would still produce similar phylogenetic and association profiles between morphs. Previously proposed “modular enhancers” would represent loci of local adaptation that allow phenotypes to modulate between neighboring color pattern forms, rather than literal enhancer elements individually sufficient for a gain of phenotype in any population. In hybrid zones where the local genetic background (i.e., upstream wing patterning loci) becomes uniform due to frequent admixture between adjacent morphs, these loci effectively act as local switches that can toggle between regional morphs via presence and absence of specific shutter-responsive silencer sites. One consequence of this hypothesis would be the necessity of coevolution of the underlying epistatic network during the Heliconius radiation, suggesting that the color pattern loci driving isolation between morphs should be more interconnected than previously suspected.
Conclusion
The work described here provides a significantly revised understanding of the cis-regulatory architecture of color pattern mimicry in Heliconius and proposes an updated model for the genetic basis of red color pattern evolution. Our experimental results provide little support for the current modular enhancer shuffling model of wing pattern development. Instead, we found CREs that are highly pleiotropic and interdependent. The lack of additivity or robustness in the cis-regulatory architecture of optix is surprising because these are common features of many regulatory systems, including recently characterized pleiotropic enhancer case studies in Drosophila (28–30). Expansion of some red color patterns in response to CRE deletion indicates that some optix CREs have additional silencer-like activity, similar to the behavior of hunchback enhancers during Drosophila embryogenesis (31). This is consistent with a growing body of association studies and gene knockouts that lead us to speculate that epistasis and enhancer shuttering is an important mechanism of red color pattern determination in Heliconius.
From an evolutionary perspective our findings highlight several surprises that could be of general importance for understanding the genetic architecture of adaptation. First, our results indicate that genomic mechanisms maintaining adaptive population divergence may be more genetically complex than often expected. Indeed, future studies with realistic population models will likely be critical to fully understand the adaptive benefit of the developmental mechanisms described here. Second, our work joins a growing list of recent studies demonstrating that enhancers, including those driving evolutionary change, can be highly pleiotropic (28, 32, 33). This contrasts with a traditional prediction of evolutionary developmental biology that enhancers driving phenotypic evolution should have largely segregated, modular phenotypic effects. Finally, our data suggesting adaptation via selection on mostly preexisting regulatory loci support a CRE “tinkering” model, with important implications for future studies searching for the genetic loci of adaptation. Modification of preexisting CREs by gaining and losing various enhancer- and silencer-like functions may be a more efficient route to adaptation than evolving CREs de novo.
Methods
Preparation of ATAC-Seq, ChIP-Seq, and Hi-C Libraries.
ATAC-seq libraries, which assays accessible regulatory elements in sufficiently frequent cell types, were prepared for H. melpomene and D. iulia as previously described with minor modifications to the original protocol (13, 14). In brief, day 3 (H. melpomene) and day 5 (D. iulia) wings were dissected in cold PBS and homogenized. Cells were then incubated with Tn5 following the original protocol, amplified for 10 to 11 cycles, and libraries were purified with 1.8× Ampure beads.
ChIP-seq libraries were prepared using a sonication-based protocol modified from Lewis et al. (15). Wing tissue was dissected from day 3 (H. erato) and day 5 (D. iulia, when optix expression in the wing increases) and fixed for 6 min while rocking at room temperature in 1% formaldehyde. Tissue samples were dounced briefly and cells lysed with ATAC-seq lysis buffer. Chromatin was sheared with a Bioruptor, after which 20 to 50 μg sheared chromatin was used for each immunoprecipitation assay. optix antibody was added to each immunoprecipitation (IP), after which the IP assay was performed following the previous protocol. Library preparation of immunoprecipitated fragments and an input DNA control was performed using the NEBNext DNA Ultra II kit following manufacturer recommendations. No size selection was performed.
Hi-C libraries were made using the protocol of Rao et al. (16) with some modifications: Nuclei were permeabilized using 0.1% SDS for 10 min at 62 °C, and DpnII was used for a 3- to 4-h restriction digest at 37 °C. Following purification of proximity ligated DNA, an additional step was added to remove any unligated biotin as described in ref. 34. Samples were sheared using a Covaris S2 sonicator and size selection of sheared fragments was performed using 1.2× Ampure beads. Libraries were prepared with the NEB DNA Ultra II library prep kit.
Data Analysis for ATAC-Seq, ChIP-Seq, and Hi-C Samples.
ATAC-seq and ChIP-seq data were processed as previously described (13). Peaks and variable loci between populations for ATAC-seq and ChIP-seq samples were manually curated around the optix locus. Orthology of ATAC-seq peaks between species was determined using Satsuma2 (35) for synteny analysis. Peaks were classified as wholly conserved if synteny aligned peaks in both species, and were considered functionally conserved (DNA sequence is not alignable, but peak presence appears conserved) if a peak in H. melpomene or D. iulia was present in a genomic interval with flanking conserved sites between H. erato and the other species.
Hi-C libraries were aligned using the Juicer pipeline as described in ref. 36. Virtual 4C plots were generated using a custom python script for a window around optix with reads placed into 3-kb bins (37). Interactions between distal enhancer elements and the optix promoter were manually curated in a genome browser.
CRISPR Mutagenesis and Genotyping.
CRISPR/Cas9 mutagenesis assays were performed based off of a previously described protocol (38). Injected larvae were reared at 28 °C in incubators and fed Passiflora biflora. Select mutants for each regulatory element screened were then genotyped for positive CRISPR mutagenesis using PCR. The TIDE software package (39) was used to confirm mutagenesis efficiency and to verify mutation around expected cut sites compared with wild-type (WT) individuals.
Phylogenetic and Coalescent Analyses.
Whole-genome 100-bp paired-end Illumina resequencing data of 109 H. erato individuals from 16 populations with distinct color patterns were obtained from Van Belleghem et al. (5, 40) and aligned to the H. e. lativitta reference. Genotypes were extracted for CREs and phylogenetic relationships among individuals were inferred for these intervals using maximum likelihood (ML) trees. The best likelihood tree was chosen from 100 trees generated from a distinct starting tree using a GTR model with CAT approximation of rate heterogeneity and the support values of this tree were inferred with 100 bootstrap replicates. Phylogentic weighting analysis used to refine the predicted regulatory intervals for optix was performed as previously described (5) using alignments against the H. e. lativitta genome assembly.
To determine coalescence time at CREs, ARGweaver was run on the same 109 genomes, masking out regions of the genome with repeats. ARGweaver was run for 2,000 iterations using a mutation rate of 2.9e-9/bp/generation, a recombination rate of 5.5e-8/bp/generation, and a diploid population size of 2e6. The discrete time points of the ARGweaver model were distributed on a logarithmic time scale, with a maximum coalescence time of 2e8 generations. The trees produced in this paper were taken from the final iteration of the ARGweaver anaysis and colored by population.
See SI Appendix, Supplemental Methods for complete methods.
Supplementary Material
Acknowledgments
We thank Arnaud Martin for his early assistance and insights on this project, and Joe Hanly for comments on the manuscript. We thank Carolina Concha and Arnaud Martin for providing WntA mutant images and James Mallet for permission to reprint natural hybrids of H. melpomene and H. numata. This work was supported by National Science Foundation (NSF) grants DEB-1354318, DEB-1546049, and IOS-1656514 to R.D.R.; IOS-1656389 (to R.P.); and OIA-1736026 (to R.P. and B.A.C.). M.J.H. was supported by NSF Graduate Research Fellowship Program grant DGE-1650441.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. H.M.H. is a guest editor invited by the Editorial Board.
Data deposition: ATAC-seq, ChIP-seq, and Hi-C datasets are available at NCBI-GEO: GSE123700, GSE123701, GSE123703, and GSE109889; and at the Dryad Digital Repository, https://doi.org/10.5061/dryad.h9w0vt4db.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907068116/-/DCSupplemental.
References
- 1.Reed R. D., et al. , optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333, 1137–1141 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Martin A., et al. , Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. U.S.A. 109, 12632–12637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nadeau N. J., et al. , The gene cortex controls mimicry and crypsis in butterflies and moths. Nature 534, 106–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mallet J., Gilbert L. E., Why are there so many mimicry rings? Correlations between habitat, behaviour and mimicry in Heliconius butterflies. Biol. J. Linn. Soc. Lond. 55, 159–180 (1995). [Google Scholar]
- 5.Van Belleghem S. M., et al. , Complex modular architecture around a simple toolkit of wing pattern genes. Nat. Ecol. Evol. 1, 0052 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak K. M., et al. , Multilocus species trees show the recent adaptive radiation of the mimetic Heliconius butterflies. Syst. Biol. 64, 505–524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin A., et al. , Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. EvoDevo 5, 7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Supple M. A., Papa R., Hines H. M., McMillan W. O., Counterman B. A., Divergence with gene flow across a speciation continuum of Heliconius butterflies. BMC Evol. Biol. 15, 204 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallbank R. W. R., et al. , Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 14, e1002353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prud’homme B., Gompel N., Carroll S. B., Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. U.S.A. 104 (suppl. 1), 8605–8612 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll S. B., Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell 134, 25–36 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Merrill R. M., et al. , Genetic dissection of assortative mating behavior. PLoS Biol. 17, e2005902 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis J. J., Reed R. D., Genome-wide regulatory adaptation shapes population-level genomic landscapes in Heliconius. Mol. Biol. Evol. 36, 159–173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis J. J., van der Burg K. R. L., Mazo-Vargas A., Reed R. D., ChIP-seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in Lepidoptera. Cell Rep. 16, 2855–2863 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Rao S. S. P., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hines H. M., et al. , Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics 13, 288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hines H. M., et al. , Wing patterning gene redefines the mimetic history of Heliconius butterflies. Proc. Natl. Acad. Sci. U.S.A. 108, 19666–19671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Burg K. R. L., et al. , Contrasting roles of transcription factors Spineless and EcR in the highly dynamic chromatin landscape of butterfly wing metamorphosis. Cell Rep. 27, 1027–1038.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Zhang L., Mazo-Vargas A., Reed R. D., Single master regulatory gene coordinates the evolution and development of butterfly color and iridescence. Proc. Natl. Acad. Sci. U.S.A. 114, 10707–10712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montavon T., et al. , A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Flanagan N. S., et al. , Historical demography of Mullerian mimicry in the neotropical Heliconius butterflies. Proc. Natl. Acad. Sci. U.S.A. 101, 9704–9709 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoyal Cuthill J. F., Charleston M., Wing patterning genes and coevolution of Müllerian mimicry in Heliconius butterflies: Support from phylogeography, cophylogeny, and divergence times. Evolution 69, 3082–3096 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Cuthill J. H., Charleston M., Phylogenetic codivergence supports coevolution of mimetic Heliconius butterflies. PLoS One 7, e36464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kronforst M. R., Kapan D. D., Gilbert L. E., Parallel genetic architecture of parallel adaptive radiations in mimetic Heliconius butterflies. Genetics 174, 535–539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heliconius Genome Consortium , Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487, 94–98 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert L. E., “Adaptive novelty through introgression in Heliconius wing patterns: Evidence for a shared genetic ‘tool box’ from synthetic hybrid zones and a theory of diversification” in Ecology and Evolution Taking Flight: Butterflies as Model Systems, Boggs C. L., Watt W. B., Ehrlich P. R., Eds. (University of Chicago Press, 2003), pp. 281–318. [Google Scholar]
- 28.Preger-Ben Noon E., et al. , Comprehensive analysis of a cis-regulatory region reveals pleiotropy in enhancer function. Cell Rep. 22, 3021–3031 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Cannavò E., et al. , Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 26, 38–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osterwalder M., et al. , Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry M. W., Boettiger A. N., Levine M., Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc. Natl. Acad. Sci. U.S.A. 108, 13570–13575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy O., et al. , Correlated evolution of two copulatory organs via a single cis-regulatory nucleotide change. Curr. Biol. 28, 3450–3457.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabarís G., Laiker I., Preger-Ben Noon E., Frankel N., Actors with multiple roles: Pleiotropic enhancers and the paradigm of enhancer modularity. Trends Genet. 35, 423–433 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Belaghzal H., Dekker J., Gibcus J. H., Hi-C 2.0: An optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grabherr M. G., et al. , Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics 26, 1145–1151 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durand N. C., et al. , Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray J., et al. Chromatin conformation remains stable upon extensive transcriptional changes driven by heat shock. Proc. Natl. Acad. Sci. 116, 19431–19439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L., Reed R. D., Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat. Commun. 7, 11769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brinkman E. K., Chen T., Amendola M., van Steensel B., Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Belleghem S. M., et al. , Patterns of Z chromosome divergence among Heliconius species highlight the importance of historical demography. Mol. Ecol. 27, 3852–3872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.