Significance
Neurons have an important role in human brain evolution. However, the contribution of other brain cell types to human brain evolution has been largely unexplored. In this study, we take advantage of recent advances in transcriptomic profiling techniques to characterize 2 distinct cell types (neurons and oligodendrocytes) from the prefrontal cortex of human, chimpanzee, and rhesus macaque brain tissue. Our data reveal that oligodendrocytes have undergone an increased acceleration in the human lineage compared with neurons. Moreover, we find that human-specific genes in oligodendrocytes are enriched for genes associated with neuropsychiatric disorders, underscoring the importance of oligodendrocytes in both human brain evolution and cognitive diseases.
Keywords: neurogenomics, comparative primate genomics, brain evolution, cell-type expression
Abstract
Recent discussions of human brain evolution have largely focused on increased neuron numbers and changes in their connectivity and expression. However, it is increasingly appreciated that oligodendrocytes play important roles in cognitive function and disease. Whether both cell types follow similar or distinctive evolutionary trajectories is not known. We examined the transcriptomes of neurons and oligodendrocytes in the frontal cortex of humans, chimpanzees, and rhesus macaques. We identified human-specific trajectories of gene expression in neurons and oligodendrocytes and show that both cell types exhibit human-specific up-regulation. Moreover, oligodendrocytes have undergone more pronounced accelerated gene expression evolution in the human lineage compared to neurons. We highlighted human-specific coexpression networks with specific functions. Our data suggest that oligodendrocyte human-specific networks are enriched for alternative splicing and transcriptional regulation. Oligodendrocyte networks are also enriched for variants associated with schizophrenia and other neuropsychiatric disorders. Such enrichments were not found in neuronal networks. These results offer a glimpse into the molecular mechanisms of oligodendrocytes during evolution and how such mechanisms are associated with neuropsychiatric disorders.
Increased brain size, accompanied by increased neuron numbers, has been a central theme in human brain evolutionary studies (1, 2). However, such changes alone are unlikely to entirely account for the evolved cognitive capabilities of humans (3). Changes in gene expression have been hypothesized as a key facet of human brain evolution (4, 5), and previous bulk transcriptome studies have shown that gene expression changes in neurons have been extensive (6–8). However, nonneuronal cell types, particularly oligodendrocytes, show altered functional and disease-related patterns in humans compared to other primates (9–11). For example, compared to nonhuman primates, human brains have greater than expected connectivity requiring myelination (9), myelination in human brains has a protracted developmental timing, and myelination and oligodendrocyte function has been implicated in neuropsychiatric diseases such as schizophrenia (SZ) (12). Moreover, ∼75% of nonneurons in the human cortex consist of oligodendrocytes (13, 14). Along with the growing appreciation of oligodendrocyte involvement in cognition (15, 16), this suggests that oligodendrocytes may have been important targets of change in human brain evolution.
Results
Cell-Type Evolutionary Trajectories Highlight Oligodendrocyte Acceleration in the Human Lineage.
To address the contribution of cell types to human brain evolution, we compared the cell-type-specific transcriptome profiling of sorted nuclei from humans to chimpanzees, our closest extant relative, using rhesus macaque as an outgroup. We analyzed genome-wide expression levels in adult human Brodmann area 46 (BA46, NeuN+: n = 27, OLIG2+: n = 22), and the homologous regions of chimpanzee (NeuN+: n = 11, OLIG2+: n = 10), and rhesus macaque (NeuN+: n = 15, OLIG2+: n = 10) (Dataset S1). Prefrontal area BA46 was selected due to its association with human-specific cognitive abilities and evolution as well as neuropsychiatric disorders (11, 17, 18). Cell-type-specific whole-transcriptome data were obtained using fluorescence-activated nuclei sorting (FANS) (19) with antibodies to either NeuN or OLIG2 to isolate neurons (NeuN+) or oligodendrocytes and their precursors (OLIG2+), respectively (SI Appendix, Fig. S1 A–E). Covariates such as age and sex and technical confounders such as RNA integrity number (RIN) explained only a small portion of the variance in both cell types (SI Appendix, Fig. S2A). While species contributed to a large proportion of the variance, the greatest amount of variance was contributed by unknown residual factors that could reflect many additive effects of small size or potentially other attributes of these postmortem samples for which we have incomplete information. Furthermore, we compared the distribution of the gene expression in each species for both expression of the 8372 NeuN+ genes and 7560 OLIG2+ genes, confirming that there are no apparent technical differences in ascertainment between NeuN+ and OLIG2+ cells (SI Appendix, Fig. S2B)
Comparisons of our data with single-cell transcriptome data from human brain (20) demonstrate that NeuN+ gene expression was representative of both inhibitory and excitatory neuronal expression signatures while OLIG2+ gene expression was primarily representative of oligodendrocyte expression signatures, supporting our FANS approach (SI Appendix, Fig. S2C).
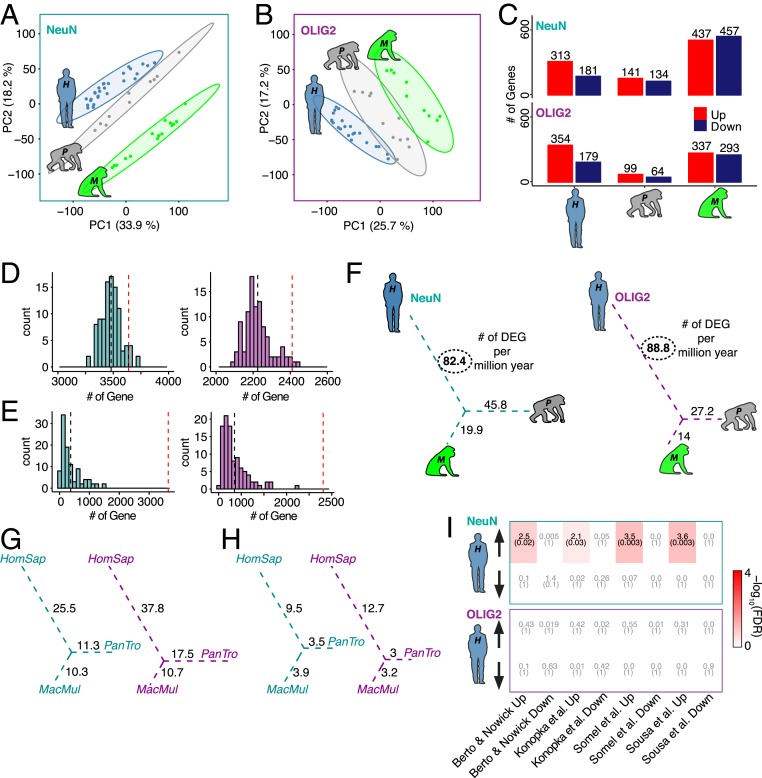
Using only high-confidence orthologous genes, we detected 8,759 protein-coding genes expressed in at least one species in NeuN+ cells and 7,362 protein-coding genes in OLIG2+ cells. Principal component analysis revealed that gene expression in each cell type separated by species (Fig. 1 A and B). Using a parsimony method, we detected species-specific differentially expressed genes (DEGs) (SI Appendix, Fig. S2D and Dataset S2 and Materials and Methods). The lineage connecting the rhesus macaque to the ancestor of humans and chimpanzees had the greatest number of DEGs, which is consistent with the idea that gene expression changes accumulate with divergence times (Fig. 1C) (6–8). Furthermore, a greater number of genes exhibited up-regulation compared to down-regulation in the human lineage for both NeuN+ and OLIG2+ samples in comparison with the nonhuman primates (χ2 test, P = 1 × 10−06 and P = 4 × 10−05, respectively) (Fig. 1C). While there were some genes that were altered in the same direction for both cell populations, the majority of species-specific cell-type genes were nonoverlapping (SI Appendix, Fig. S2E). We next compared the human-specific genes with single-cell transcriptome data from human brain (20). For neurons, the NeuN+ nuclei capture signals from both inhibitory and excitatory neurons to an equivalent extent among all 3 species. Interestingly, there appear to be more down-regulated genes among excitatory neuronal classes and more up-regulated genes among inhibitory classes (SI Appendix, Fig. S2F). On the other hand, OLIG2+ up- and down-regulated human-specific gene expression is explained by higher proportions of mature oligodendrocytes (SI Appendix, Fig. S2G). These results further confirm that NeuN+ and OLIG2+ signals of human-specific genes are not likely due to a bias in cell state or purity. In addition, a leave-one-out (LOO) and permutation approach were both applied to these data to ensure accurate interpretation of the comparisons (Fig. 1 D and E and Materials and Methods).
Fig. 1.
Cell-type-specific differential gene expression analysis of 3 primates. (A and B) Principal component analysis of NeuN+ (A) and OLIG2+ (B) nuclei. Blue, human (H is H. sapiens); gray, chimpanzee (P is P. troglodytes); green, rhesus macaque (M is M. Mulatta). (C) Bar plots representing species-specific DEGs divided by up- (red) and down-regulated (dark blue) for both NeuN and OLIG2. (D) LOO cross-validation based on 100 bootstraps for NeuN (cyan) and OLIG2 (magenta). Observed numbers of DEGs (red dashed line) were falling in the distribution of the LOO DEGs based on ANOVA. (E) Permutation analysis based on 100 permutation comparisons based on subject randomization for NeuN (cyan) and OLIG2 (magenta). Observed numbers of DEGs (red dashed line) were significantly different from the randomized DEGs based on ANOVA. (F) The number of DEGs per million years for the unrooted tree of the study species of NeuN (cyan) and OLIG2 (magenta). (G) Down-sampled DEGs per million years based on 100 permutations. Values are calculated based on the average of species-specific DEGs for NeuN (cyan) and OLIG2 (magenta). (H) Down-sampled DEGs per million years based on 100 permutations for NeuN (cyan) and OLIG2 (magenta). Values are calculated based on the species-specific observed DEGs supported by the downsampling P value in >90% of downsampled sets (HomSap, H. sapiens; Pan Tro, P. troglodytes; MacMul, M. mulatta). (I) Heat map showing FDR (parentheses) and OR of gene set enrichment for both NeuN (Top) and OLIG2 (Bottom). Enrichment is based on a Fisher’s exact test. The x axis shows the representative data included for this analysis (7, 8, 21, 22).
Interestingly, human-specific expression was more pronounced in the OLIG2+ samples compared to NeuN+ samples. Specifically, OLIG2+ cells exhibited greater effect sizes in pairwise comparisons [human versus chimpanzee, mean(log2(FC)): 0.26 NeuN+, 0.59 OLIG2+, P < 2 × 10−16, Kolmogorov–Smirnov (K-S) test; human versus rhesus macaque, mean(log2(FC)): 0.25 NeuN+, 0.58 OLIG2+, P < 2 × 10−16, K-S test; SI Appendix, Fig. S2H] as well as greater number of DEGs per million years (Fig. 1F) compared to NeuN+ cells. These differences in effect sizes between cell types are not likely driven by the fact that neurons express greater numbers of genes than oligodendrocytes as the distribution of species-relevant fold changes is similar between cell types (SI Appendix, Fig. S1I). In addition, the proportion of OLIG2+ human-specific DEGs is significantly greater than human NeuN+-specific DEGs (odds ratio [OR] = 1.21, P = 0.002). In contrast, OLIG2+-specific DEGs are not significantly more abundant than NeuN+-specific DEGs in chimpanzee and rhesus macaque lineages (PanTro [Pan troglodytes or chimpanzee]: OR = 0.65, P = 1; MacMul [Macaca mulatta or rhesus macaque]: OR = 0.71, P = 1). These results support the observation that human oligodendrocytes have undergone human-specific acceleration of gene expression evolution.
To ensure that these observations were not due to different numbers of samples in each species, we used the same numbers of samples between species, which effectively is a downsampling of the human and rhesus macaque samples (Fig. 1 G and H and Materials and Methods). Doing so reduced the number of DEGs per million years due to smaller sample size and heterogeneity within and between species; nevertheless, down-sampled DEGs show the same pattern of acceleration in OLIG2+ samples compared with NeuN+ samples, further supporting the result that oligodendrocytes have undergone an evolutionary acceleration on the human lineage. We next examined previous gene expression studies of frontal cortex evolution (7, 8, 21, 22) to assess how bulk tissue expression profiles may have been confounded by cell-type-specific trajectories. We found that human-specific up-regulated genes in the bulk tissue studies were enriched with human-specific NeuN+ up-regulated DEGs. In comparison, human-specific DEGs in OLIG2+ samples were not enriched in the previous studies (Fig. 1I), indicating that studies using bulk tissues may have been underpowered to detect oligodendrocyte-specific evolutionary trajectories. Carrying out deconvolution analysis, we found that these bulk RNA sequencing (RNA-seq) datasets were primarily comprised of neuronally derived gene-expression signatures (SI Appendix, Fig. S3 A–D). Thus, using a cell-type-specific approach, we detected a previously undiscovered signal of rapid acceleration of oligodendrocyte-gene expression compared with neurons in the human lineage.
Gene Coexpression Network Highlights Human-Specific Modules.
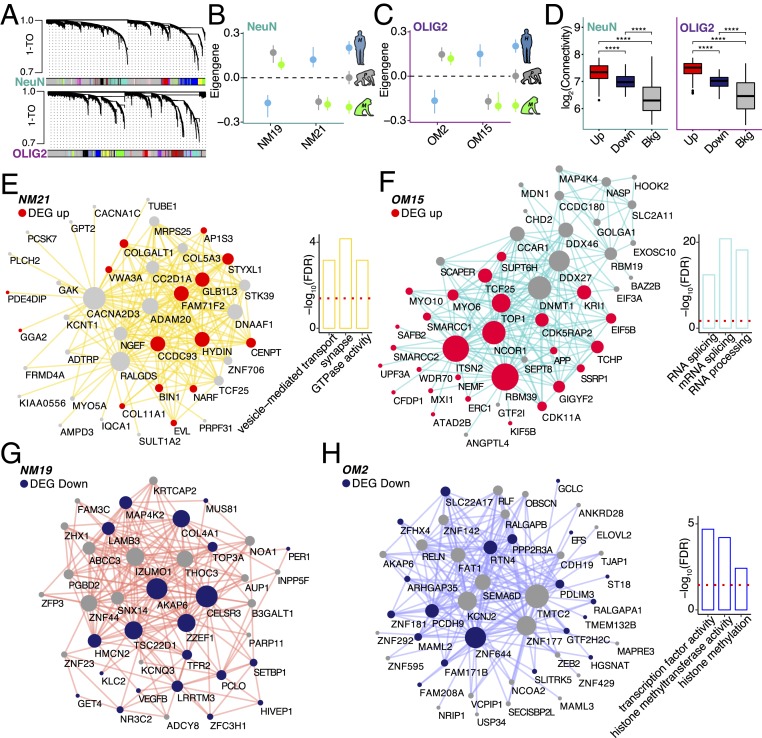
To place the human-specific changes within a systems-level context and identify the relevant biological processes associated with these changes, we next applied a permuted weighted gene coexpression analysis (23) to detect human-specific cell-type coexpression modules (Fig. 2A, Dataset S3, and Materials and Methods). Using the expression data that were adjusted for potential variation explained by covariates and surrogate variables, we defined 2 modules in NeuN+ samples and 2 modules in OLIG2+ samples that exhibited human-specific expression and showed a strong enrichment for human-specific DEGs (Fig. 2 B and C and SI Appendix, Fig. S4 A and B). These modules showed higher association with species than with other covariates (SI Appendix, Fig. S4 C and D). Human-specific DEGs in both NeuN+ and OLIG2+ samples exhibited significantly greater connectivity compared with other genes across all modules, indicating their pivotal roles in human frontal cortex transcriptional networks (Fig. 2D). NeuN+ human up-regulated module NM21 was enriched for genes involved in synaptic function and vesicular transport (Fig. 2E and Dataset S3). Interestingly, the OLIG2+ human up-regulated module OM15 was enriched for pathways implicated in RNA splicing, RNA metabolism, and chromatin remodeling (Fig. 2F and Dataset S3). Whereas down-regulated module NM19 was not enriched for any specific function (Fig. 2G), the OLIG2+ down-regulated module OM2 functions were related to transcriptional regulation, histone methylation, and modification (Fig. 2H and Dataset S3). Of note, both OLIG2+ modules that are associated with human-specific expression are significantly enriched for transcription factors and RNA binding proteins (SI Appendix, Fig. S4 E and F). These results suggest that alternative splicing and transcriptional regulation are biological functions linked with oligodendrocyte evolution in the human frontal cortex. However, it is interesting that while these modules contain genes with similar functional properties, the overall trend in expression is in opposite directions (up-regulated in OM15 versus down-regulated in OM2). These modules may represent convergent functions of up-regulated and down-regulated genes in human oligodendrocyte evolution. Indeed, the genes of these 2 modules might be subsets of 1 larger integrated module that has evolved but has been separated into 2 modules by expression direction due to using a “signed” coexpression network approach.
Fig. 2.
Coexpression analyses identify human-specific modules. (A) Representative network dendrograms for NeuN (Top) and OLIG2 (Bottom). (B and C) Dot plots with standard errors (SEs) demonstrate the association of the modules detected by parsimony with species for NeuN (B) and OLIG2 (C). SEs are calculated based on the eigengene across samples. Dots represent the mean eigengene for that module. (D) Box plots show the difference in connectivity between human-specific genes in NeuN (Left) and OLIG2 (Right) across the entire coexpression network compared with the background genes (****P < 0.001; Wilcoxon’s rank sum test). (E–H) Visualization of the top 200 connections ranked by weighted topological overlap values for NM21 (E), OM15 (F), NM19 (G), and OM2 (H). Node size corresponds to the number of edges (degree). Human-specific up-regulated genes are highlighted in red. Human-specific down-regulated genes are highlighted in blue. Side bar plots show the top 3 functions of the module based on −log10(FDR). Red dashed line corresponds to the FDR threshold of 0.05.
Oligodendrocyte Human-Specific Modules Are Enriched for Variants Associated with Neuropsychiatric Disorders.
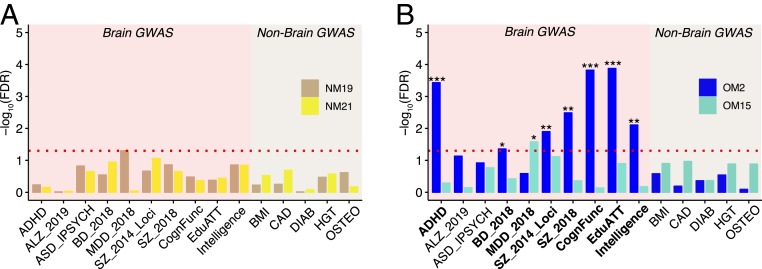
It is hypothesized that genes important for the evolution of human-specific cognitive abilities are linked with human-specific cognitive disorders (24, 25). To investigate this hypothesis, we assessed the enrichment of human-specific coexpression modules with genome-wide association study (GWAS) signals (Materials and Methods). While we did not find any enrichment for neuropsychiatric disorders GWAS signals in the human-specific neuronal modules NM19 and NM21 (Fig. 3A), the OLIG2+ human down-regulated module OM2 showed a strong enrichment for attention deficit hyperactivity disorder (ADHD), bipolar disorder (BD), and SZ loci as well as loci associated with cognitive traits, education attainment, and intelligence (Fig. 3B and Dataset S4). The up-regulated module OM15 showed enrichment for major depressive disorder (MDD) (Fig. 3B and Dataset S4). In comparison, neither NeuN+ nor OLIG2+ human-specific modules showed significant enrichment for GWAS signals associated with non-brain-related traits/disease. While very little is known about the role of oligodendrocytes in ADHD (26), BD, SZ, and MDD have been associated with alterations in white matter and differential regulation of oligodendrocyte-related genes (27–30). These results suggest that human-specific coexpressed genes that are under evolutionary expression trajectories in oligodendrocytes are at risk to be associated with cognitive disease-related variants.
Fig. 3.
Human-specific genes are enriched for cognitive disease risk variants. (A and B) Bar plots highlighting the enrichment for genetic variants [−log10(FDR)]. Bars correspond to (A) NeuN modules (NM19: tan, NM21: yellow) and (B) OLIG2 modules (OM2: blue, OM15: turquoise) species-specific modules (***FDR < 0.001, **FDR < 0.01, *FDR < 0.05; MAGMA statistics). Red dashed line corresponds to the FDR threshold of 0.05. The x axis shows the acronyms for the GWAS data utilized for this analysis. ALZ, Alzheimer's disease; ASD, autism spectrum disorder from IPSYCH (Integrative Psychiatric Research); CognFunc, cognitive functions; EduATT, educational attainment; BMI, body mass index; CAD, coronary artery disease; DIAB, diabetes; HGT, height; OSTEO: osteoporosis.
Human-Specific Modules Are Enriched for Neuropsychiatric DEGs.
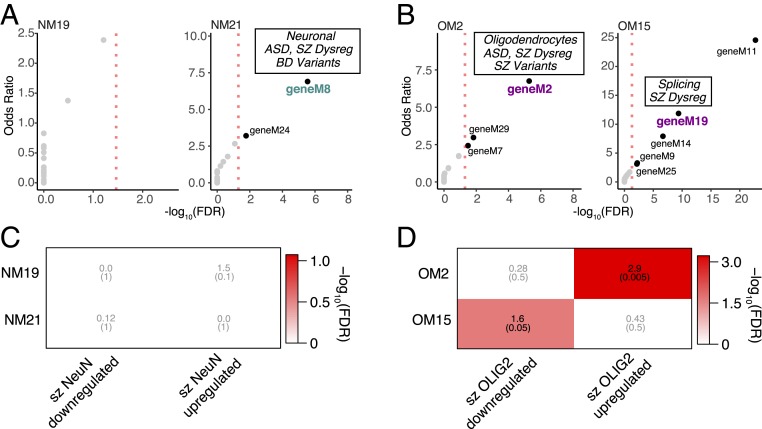
To further examine the potential relationship between dysregulation in neuropsychiatric disorders and human-specific changes using a large-scale gene expression dataset, we used recently published meta-analyses of cognitive disease brain gene expression from the PsychENCODE Consortium (31) (Materials and Methods). We found that the NeuN+ human-specific up-regulated module NM21 is overrepresented for genes in a neuronal module dysregulated in SZ and autism spectrum disorder (ASD) (geneM8; OR = 9.7, false discovery rate [FDR] = 1 × 10−08; Fig. 4A). In contrast, the OLIG2+ human-specific down-regulated module OM2 is enriched for genes in an oligodendrocyte module containing genes dysregulated in ASD, BD, and SZ (geneM2; OR = 9.5, FDR = 8 × 10−09; Fig. 4B). Interestingly, the OLIG2+ up-regulated module OM15 is enriched for genes in a module dysregulated in SZ and linked with splicing (geneM19; OR = 10.1, FDR = 1 × 10−09; Fig. 4B), reflecting the functional enrichment we described (Fig. 2F). We next assessed whether human-specific cell-type expression patterns are at risk in neuropsychiatric disorders using cell-type-specific disease-relevant gene expression data (Materials and Methods). We examined cell-type-specific whole-transcriptome data from BA46 from 23 patients with SZ, generated following identical experimental procedures (32). Using genes differentially expressed between SZ and healthy donors at the cell-type level (referred to as szDEGs; Dataset S5), we asked whether dysregulated genes in SZ were enriched for human-specific evolutionary changes of gene coexpression at the cell-type level. Whereas NeuN+ modules were not found enriched for cell-type SZ genes (Fig. 4C), we found that OLIG2+ szDEGs were enriched for human OLIG2+ modules (Fig. 4D). Specifically, the down-regulated module OM2 is enriched for SZ OLIG2+ up-regulated genes (OR = 2.9, FDR = 5 × 10−04) while the up-regulated module OM15 is moderately enriched for SZ OLIG2+ down-regulated genes (OR = 1.6, FDR = 0.05). Taken together, these observations highlight the link between oligodendrocyte evolution and neuropsychiatric disease etiologies.
Fig. 4.
Cell-type-specific expression in SZ is related to human-specific genes. (A) Bubble chart illustrates −log10(FDR) (x axis) and OR (y axis) of gene set enrichment for gene modules implicated in neuropsychiatric disorders (31) and NeuN human-specific modules. Marked the modules with functional conservation. Dysreg, dysregulated. (B) Bubble chart illustrates −log10(FDR) (x axis) and OR (y axis) of gene set enrichment for gene modules implicated in neuropsychiatric disorders (31) and OLIG2 human-specific modules. The modules with functional conservation with the PsychENCODE dataset are indicated. (C) Heat map illustrates FDR (parentheses) and OR of gene set enrichment (Fisher’s exact test). The x axis shows NeuN human-specific up-/down-regulated genes. The y axis shows SZ DEGs up-/down-regulated in NeuN. (D) Heat map illustrates FDR (parentheses) and OR of gene set enrichment. The x axis shows OLIG2 human-specific up-/down-regulated. The y axis shows SZ DEGs up-/down-regulated in OLIG2.
Discussion
These data provide insights into cell-type, species-specific expression patterns during primate brain evolution. Much of the recent focus on human brain evolution has highlighted changes in neuronal number and function in the human brain; however, the molecular characterization of the mechanisms driving such changes in neurons as well as other cell types is critical for understanding human brain evolution. Here, we show that NeuN+ human-specific DEGs encode genes important for synaptic function in line with previous data from bulk RNA-seq (6–8). Surprisingly though, we find that gene expression in oligodendrocytes has undergone a more dramatic acceleration in the human lineage compared with neurons. We also show that previous comparative primate gene expression studies were likely underpowered to detect these nonneuronal expression changes.
The human-specific oligodendrocyte genes are enriched for functional categories such as RNA metabolism and RNA processing. While such molecular functions are underexplored with respect to oligodendrocytes, there is increasing evidence that these functions are altered in cognitive diseases (33–35). Moreover, the brain GWAS and PsychENCODE enrichments for cell-type expression modules suggest that human-specific cell-type-specific evolutionary trajectories of gene expression are implicated in disease pathophysiology in multiple cognitive disorders. Using the only available disease cell-type expression dataset, we also observe SZ cell-type-specific down-regulation among human-specific oligodendrocyte genes. Together, these data suggest a role for human-specific oligodendrocyte genes in disorders including SZ, MDD, BD, and ADHD. Previous work has specifically singled out oligodendrocyte dysfunction in both SZ and MDD (36). In addition, human brains have undergone a volumetric expansion of white matter (9, 11), while these white matter volumes are significantly reduced in SZ (12, 37). While we have focused on the novelty of the human-specific oligodendrocyte genes, there are clearly evolutionarily relevant changes in neurons that are likely important for cognitive disorders such as SZ and ASD too. Since neuronal activity can direct oligodendrocyte development and myelination (38), the functional outcome of the interplay of gene expression changes in these 2 cell types may be important for multiple cognitive disorders. In addition to their well-studied role in myelination, oligodendrocytes are a critical component of brain metabolism. This function is intricately linked to maintenance of neuronal health and function, for example via transportation of lactate necessary for axonal activity (39). There is also strong evidence for oligodendrocytes playing a vital role in axonal mitochondrial function (40). Thus, while genetic and functional studies of cognitive disease have historically focused on the involvement of neurons in the pathology of these diseases, there is a clear disconnect in the literature from the studies demonstrating an essential role for nonneurons such as oligodendrocytes in brain development and function and the potential for these cell types to either facilitate or compensate for neuronal-related disease pathologies. While there is a burgeoning literature implicating microglia in cognitive diseases (41), here we implicate oligodendrocytes as having a potential role in such diseases, especially from the perspective of evolved gene expression patterns. As human neurons have increased relative size and more complex axonal morphologies compared to nonhuman primates or rodents (42, 43), it is perhaps not too surprising that myelinating oligodendrocytes might have to undergo similar genomic alterations to facilitate matching this evolutionary adaptation. Recent work has implicated that morphological changes in myelinated axons might be a factor in cortical folding that has evolved in the primate brain (44). Interestingly, several of the genes we have identified as changing specifically in human oligodendrocytes have been studied for their functional roles in mediating neuronal health and/or have been implicated in a number of cognitive functions or diseases. For example, SMARCA4 (i.e., BRG1), an important promoter of oligodendrocyte differentiation, is thought to alter addictive behavior by modulating neuronal plasticity (45). In line with this, imaging studies in humans have delineated a dynamic role for oligodendrocytes and myelin in task-related plasticity (46–49). FEZ1 is important for oligodendrocyte development and has been implicated in SZ (50). Mutations in SNX14, which is in OM2, causes a syndromic neurodevelopmental disorder characterized by intellectual disability with cerebellar atrophy and lysosome–autophagosome dysfunction (51). The expression of LRRK2 in oligodendrocytes is associated with neuronal loss in multiple system atrophy (52). Of note, one of the first human postmortem brain tissue microarray studies using tissue from SZ patients identified differential expression of myelin-related genes (53). At least one of those genes, gelsolin, GSN, is up-regulated in our study in human OLIG2+ samples and in OM15. Interestingly, recent work in macaques has shown that oligodendrocytes derive from the outer subventricular zone, or oSVZ (54). The production of oligodendrocytes by the oSVZ might be needed for myelinating the increased number of axons driving cortical gyrification in primates. How oligodendrocyte development from the oSVZ might have further evolved in the chimpanzee or human brain remains to be determined. One can imagine, though, that our results demonstrating a link between oligodendrocyte gene expression evolution and cognitive disorders such as SZ might also be relevant to disorders that affect gyrification such as lissencephaly and polymicrogyria. However, even though we have enriched for specific populations of brain-cell types using a FANS approach, we cannot rule out the potential contribution of cellular diversity or abundance at the species level (55). For example, OLIG2+ cells could contain oligodendrocyte precursors or oligodendrocytes undergoing different myelination states or NeuN+ cells can represent both excitatory and inhibitory neurons. A deconvolution analysis indicates that cell portions are similarly represented in all 3 species even among the DEGs (SI Appendix, Fig. S2 A, F, and G); however, this point warrants further analyses. Recent advances using single-cell approaches in all 3 species will be able to address this issue. In addition, future studies that connect changes in the functional properties of oligodendrocytes, for example at the level of RNA binding and/or processing, to either disease pathophysiology, white matter volume alterations, or response to neuronal activity will confirm the importance of the identified genes in human brain evolution. Our study highlights the importance of nonneuronal cell types in brain evolution and cognitive disorders.
Materials and Methods
Experimental Model and Subject Details.
Postmortem brain samples.
Human postmortem brain samples from BA46 were obtained from the National Institutes of Health NeuroBioBank (the Harvard Brain Tissue Resource Center, the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, and the University of Miami Brain Endowment Bank) and the University of Texas Neuropsychiatry Research Program (Dallas Brain Collection) (Dataset S1). Nonhuman primate tissue samples were obtained from Yerkes National Primate Research Center (Dataset S1).
Nuclei extraction, FANS, and RNA isolation.
Nuclei Isolation, FANS, and RNA isolation were performed as done previously (32).
RNA-seq.
RNA-seq was performed as described previously (32, 56) with some modifications. In order to determine the quality of the RNA from the nuclear samples, the RNA from the matched cytoplasmic fractions was extracted with the miRNeasy Mini kit (217004; Qiagen) according to the manufacturer’s instructions. Samples with a total cytoplasmic RNA average RIN value of 7.5 ± 0.16 were used for RNA-seq library preparation of the nuclear samples.
Computational Methods.
RNA-seq mapping, quality control, and expression quantification.
Reads from the 3 different primates were aligned to either the human hg19, chimpanzee PanTro4, or rhesus macaque RheMac8 reference genome using STAR 2.5.2b (57) with the following parameters: “–outFilterMultimapNmax 10–alignSJoverhangMin 10–alignSJDBoverhangMin 1–outFilterMismatchNmax 3–twopassMode Basic.” For each sample, a BAM file including mapped and unmapped reads that spanned splice junctions was produced. Secondary alignment and multimapped reads were further removed using in-house scripts. Only uniquely mapped reads were retained for further analyses. Quality control (QC) metrics were performed using RseqQC using the hg19 gene model provided. These steps include number of reads after multiple-step filtering, ribosomal RNA reads depletion, and defining reads mapped to exons, untranslated regions, and intronic regions. The Picard tool was implemented to refine the QC metrics (http://broadinstitute.github.io/picard/). CrossMap and liftOver were used to translate the nonhuman primate unique read coordinates into human coordinates based on hg19 (58, 59). Ensemble annotation for hg19 (version GRCh37.87) was used as reference alignment annotation and downstream quantification. Gene-level expression was calculated using HTseq version 0.9.1 using intersection-strict mode by Exons (60). Counts were calculated based on protein-coding genes from the Ensemble GRCh37.87 annotation file. Orthologous genes were downloaded from Ensemble Biomart portal (61). Orthologous genes were categorized using a high confidence score provided by ensemble and presence in known chromosomes in all 3 species analyzed. We removed sex chromosomes. A total of 14,212 genes were considered for downstream analysis.
Covariate adjustment and differential expression.
Counts were normalized using counts per million reads (cpm) with the edgeR package in R (62). Normalized data were log2-scaled with an offset of 1. Genes with no reads in human, chimpanzee, or rhesus macaque samples were removed. Normalized data were assessed for effects from known biological covariates (gender and age), technical variables related to sample processing (RIN), and technical variables related to surrogate variation (SVs). Other biological and technical covariates (e.g., hemisphere and postmortem interval [PMI]) were not considered for the analysis because these were confounded with species. Nonhuman primates’ ages were converted to human age referring to species life traits as maximal age reached, male sexual maturity, female sexual maturity, gestation, weaning, first reproduction, number of litters, teething deciduous first and last, and teething permanent first and last as proposed in ref. 22. Traits are stored in Dataset S1. A linear model was applied between species life traits. Human age was converted into nonhuman primates in R as
This method provided us with an accurate estimation of human age translated into nonhuman standard (e.g., a 25-y-old human corresponded to a 13.2-y-old chimpanzee and an 8.0-y-old rhesus macaque). Age was converted to categorical variables. Three groups were defined: less than 40 y old, between 40 and 60 y old, and more than 60 y old. SVs were calculated using SVA in R based on a “2-step” method with 100 iterations (63). The data were adjusted for technical covariates using a linear model:
Adjusted cpm values were used for coexpression analysis and visualization. Differential expression analysis was performed in R using linear modeling. To fit our parsimony approach, we performed pairwise analysis between the 3 species analyzed (e.g., human–chimpanzee, human–rhesus macaque, and chimpanzee–rhesus macaque). Additionally, we performed an ANOVA based on the 3 species (e.g., human–chimpanzee–rhesus macaque) as follows:
Fitting this model, we estimated log2 fold changes and P values. P values were adjusted for multiple comparisons using a Benjamini–Hochberg correction (FDR). This method was used to detect human-specific changes, chimpanzee-specific changes, and rhesus macaque-specific changes using a standard cutoff of |log2(fold-change)| > 0.3 and FDR < 0.05. For example, in human, we considered specific up-regulation where human showed log2(FC) > 0.3 and FDR < 0.05 in comparison with chimpanzee and rhesus macaque and where chimpanzee and rhesus macaque were not differentially expressed for FDR > 0.1. In addition, we considered in this paradigm the Bonferroni adjusted P value from ANOVA of <0.05. In contrast, for down-regulated genes we consider log2(FC) < −0.3 and FDR < 0.05 in comparison with chimpanzee and rhesus macaque and where chimpanzee and rhesus macaque were not differentially expressed for FDR > 0.1. For the up-regulated genes, we considered additional Bonferroni-adjusted P values from ANOVA of <0.05.
LOO and downsampling analysis.
To validate the robustness of our differential expression analysis, we applied a LOO cross-validation by subsampling our data with n = number of subjects per species − 1 with number of permutations = 100. −log10(observed ANOVA) strongly correlated with −log10(LOO ANOVA), underscoring that individual subjects are not driving differential expression detection. We additionally applied a permutation method by randomizing the subjects per species 200 times and recalculating the species-specific DEGs across subjects. The number of observed DEGs were significantly different for the randomized one for both cell types. A downsampling analysis was applied to confirm the more pronounced acceleration in OLIG2 compared with NeuN given the greater sample size for humans. Using the chimpanzee as the minimal number of subjects (NeuN = 11, OLIG2 = 10), we recalculated the species-specific DEGs with the number of permutations = 100. Due to the reduced sample size and high heterogeneity between and within species, the total number of species-specific DEGs was reduced. Nevertheless, this approach recapitulated the more pronounced acceleration in OLIG2, confirming the observed results based on the total number of samples.
Coexpression network analysis.
To identify modules of coexpressed genes in the RNA-seq data, we carried out weighted gene coexpression network analysis (WGCNA) (23). The same number of genes used for differential expression analysis were included for WGCNA (8,372 for NeuN+ cells and 7,560 for OLIG2+ cells; Dataset S4). Signed networks were used for both NeuN+ and OLIG2+ data. A soft-threshold power was automatically calculated for both NeuN+ and OLIG2+ samples to achieve approximate scale-free topology (R2 > 0.85). Networks were constructed with the blockwiseModules function with biweight midcorrelation (bicor). For NeuN data, we used corType = bicor, maxBlockSize = 10000, mergingThresh = 0.10, minCoreKME = 0.5, minKMEtoStay = 0.4, reassignThreshold = 1e-10, deepSplit = 4, detectCutHeight = 0.999, minModuleSize = 25, networkType = signed. For OLIG2 data, we used corType = bicor, maxBlockSize = 10000, mergingThresh = 0.10, minCoreKME = 0.5, minKMEtoStay = 0.4, reassignThreshold = 1e-10, deepSplit = 4, detectCutHeight = 0.999, minModuleSize = 35, networkType = signed.
The modules were then determined using the dynamic tree-cutting algorithm. To ensure robustness of the observed network, we used a permutation approach recalculating the networks 200 times with permuted gene expression. Observed connectivity per gene was compared with the randomized one. None of the randomized networks showed similar connectivity, providing robustness to the network inference. We refer to this approach as permWGCNA. Additional analysis using a bootstrapping approach was performed. Briefly, we recalculated networks resampling the initial set of samples 200 times and compared the observed connectivity per gene with the randomized one. As for the permutation, none of the randomized networks showed similar connectivity. This additional test was applied to further provide robustness of the network inference.
Module sizes (25/35 respectively) were chosen to detect small modules driven by potential noise on the adjusted data. Deep split of 4 was used to split more aggressively the data and create more specific modules. Spearman’s rank correlation was used to compute module eigengene–covariates associations. A parsimony approach was used to select the modules: Human-specific modules were significantly correlated with the 3 species but oppositely correlated between human and the nonhuman primates. Given the adjusted expression, covariates did not have effect on the variance explained by the gene of the detected modules. Modules were visualized based on the rank of the weight (weighted topological overlap value, WTO). The top 200 connections were selected for the visualizations. Node size was adjusted based on the degree (e.g., number of links). Visualization was rendered using Cytoscape (64). For measurements of differential connectivity, we used the number of expressed genes for each cell type as background (8,372 for NeuN+ cells and 7,560 for OLIG2+ cells).
Functional enrichment.
The functional annotation of differentially expressed and coexpressed genes was performed using ToppGene (65) as we have previously done (32). Analysis was replicated using GOstats in R (66). We used Gene Ontology and Kyoto Encyclopedia of Genes and Genomes databases. Pathways containing between 5 and 2,000 genes were retained. Orthologous genes (14,212) were used as custom background. A Benjamini–Hochberg FDR (P < 0.05) was applied as a multiple comparisons adjustment.
Evolutionary tree calculation and normalization.
Trees were calculated and visualized using ape in R (67). Branches length of the unrooted trees was calculated based on number of DEGs divided by divergence time (HomSap/PanTro = ∼6 My, MacMul = ∼45 My) (HomSap = Homo sapiens). For rhesus macaque, divergence time was calculated based on divergence of apes and Old World monkeys (26 My) (68) and divergence between common ancestor of apes and Old World monkeys and common ancestor of humans and chimpanzees (∼19/20 My) (69). Thus, we used the sum of 26 plus 19 My to estimate DEGs per million years on the branch leading to M. mulatta. This refined approach was used due to the faster evolutionary rates in Old World monkey lineages compared to apes (70, 71).
GWAS data and enrichment.
GWAS studies (72–83) were analyzed as we have previously done (32). In brief, summary statistics from the genetic data were downloaded from the Psychiatric Genomics Consortium (http://www.med.unc.edu/pgc/results-and-downloads) and GIANT (https://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files), and gene-level analyses were performed using MAGMA v1.04 (84). MAGMA statistics and –log10(FDR) values are reported in Dataset S4.
Primate data and enrichment.
Data were downloaded from respective National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) sources. Berto and Nowick (21) and Konopka et al. (8) provide species-specific DEGs within supplementary information. For the Somel et al. microarray dataset (22), raw data were downloaded and analyzed with Affy in R (85). Degradation and quality checks were performed to the data, detecting no significant differences between the 3 species analyzed. We next performed a computational mask procedure using the maskBAD in R (86). This method developed for microarray data removed probes with binding affinity differences between species. We considered only the probesets significantly detected in at least one individual (P < 0.05). A linear model used for our data was applied to the data detecting species-specific DEGs. For Sousa et al. (7) RNA-seq data, RPKM (reads per kilobase per million) data were provided by the first and corresponding authors of the study. To render the data comparable with the BA46 data in our study, we used human, chimpanzee, and rhesus macaque samples from dorsolateral prefrontal cortex. RPKM data were log2-scaled. Genes with RPKM = 0 in human, chimpanzee, or rhesus macaque samples were removed. A linear model was applied as used for our data to detect species-specific DEGs. Up-/down-regulated genes from these data were used for enrichment with our NeuN-/OLIG2- DEGs gene set.
Transcription factors and RNA-binding proteins enrichment.
The transcription factors list was downloaded from http://humantfs.ccbr.utoronto.ca/download.php (87). The RNA-binding proteins list was downloaded from http://rbpdb.ccbr.utoronto.ca/(88).
SZ cell-specific data.
Differential expression analysis of cell-type expression between nuclei obtained from brain tissue derived from SZ cases and controls (CTL) was generated from GSE107638 (32). Briefly, counts were normalized using cpm with the edgeR package in R (62). Genes with no reads in either SZ or CTL samples were further removed. Normalized data were assessed for effects from known biological covariates (diagnosis, age, gender, and hemisphere), technical variables related to sample processing (RIN, Brain Bank, PMI), and technical covariates related to surrogate variation (SV). Age and PMI were converted to categorical variables (named “AgeClass” and “PmiClass”). SVs were calculates using SVA based on “be” method with 100 iterations (63). For the differential expression analysis, we used the lmTest with “robust” parameter and ebayes functions in limma package in R (89) fitting all of the covariates reported. Significant DEGs were categorized with |log2(FC)| > 0.3 and FDR < 0.01 for both NeuN and OLIG2 cell-type SZ vs. CTL analyses (Dataset S5). Detailed information, methods, and analysis are available at https://github.com/konopkalab/Schizophrenia_CellType.
Gene set enrichment.
Gene set enrichment applied for primate DEGs as shown in Fig. 1I, PsychENCODE and SZ cell-type DEGs as shown in Fig. 4 A–D, and transcription factors/RNA-binding proteins as shown in SI Appendix, Fig. S3 was performed using a Fisher’s exact test in R with the following parameters: alternative = “greater,” conf.level = 0.99, simulate.p.value = TRUE, B = 1000. We reported ORs and Benjamini–Hochberg adjusted P values (FDR). Enrichment was further confirmed with a hypergeometric test in R.
Deconvolution.
The human middle temporal gyrus single nuclei RNA-seq data were downloaded from Allen Brain Institute web portal (http://celltypes.brain-map.org/rnaseq/human) (20) and analyzed as we have previously done (32).
Availability of data and material.
The NCBI GEO accession number for the human data reported in this manuscript is GSE107638. Nonhuman primate raw data are deposited with accession number GSE123936.
Code availability.
Codes to support the DGE analysis, WGCNA, and shiny applications for data visualizations are available at https://github.com/konopkalab/Primates_CellType.
Supplementary Material
Acknowledgments
We thank the donors and their families for the tissue samples used in these studies. We thank Angela Mobley of the Flow Cytometry Facility and Vanessa Schmid of the Next Generation Sequencing Core of the University of Texas (UT) Southwestern Medical Center for technical support. G.K. is a Jon Heighten Scholar in Autism Research at UT Southwestern. This work was supported by the Uehara Memorial Foundation to N.U.; the Japan Society for the Promotion of Science (JSPS) Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers (S2603) to N.U., K.T., S.B. and G.K.; the National Chimpanzee Brain Resource, NIH R24NS092988, the NIH National Center for Research Resources P51RR165 (superceded by the Office of Research Infrastructure Programs/OD P51OD11132) to T.M.P.; the National Science Foundation (SBE-131719) to S.V.Y.; the James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition – Scholar Award to G.K.; and the National Institute of Mental Health (MH103517), to T.M.P., G.K., and S.V.Y. Human tissue samples were obtained from the NIH NeuroBioBank (The Harvard Brain Tissue Resource Center, funded through HHSN-271-2013-00030C, the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center; and the University of Miami Brain Endowment Bank) and the UT Neuropsychiatry Research Program (Dallas Brain Collection). Nonhuman primate tissue samples were obtained from Yerkes National Primate Research Center (P51OD11132).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (human data, accession no. GSE107638; nonhuman primate data, accession no. GSE123936).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907982116/-/DCSupplemental.
References
- 1.Gabi M., et al. , No relative expansion of the number of prefrontal neurons in primate and human evolution. Proc. Natl. Acad. Sci. U.S.A. 113, 9617–9622 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preuss T. M., “The human brain: Evolution and distinctive features” in On Human Nature, Tibayrenc M., Ayala F. J., Eds. (Academic Press, San Diego, 2017), chap. 8, pp. 125–149. [Google Scholar]
- 3.Sousa A. M. M., Meyer K. A., Santpere G., Gulden F. O., Sestan N., Evolution of the human nervous system function, structure, and development. Cell 170, 226–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khaitovich P., Enard W., Lachmann M., Pääbo S., Evolution of primate gene expression. Nat. Rev. Genet. 7, 693–702 (2006). [DOI] [PubMed] [Google Scholar]
- 5.King M. C., Wilson A. C., Evolution at two levels in humans and chimpanzees. Science 188, 107–116 (1975). [DOI] [PubMed] [Google Scholar]
- 6.Liu X., et al. , Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Res. 22, 611–622 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sousa A. M. M., et al. , Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konopka G., et al. , Human-specific transcriptional networks in the brain. Neuron 75, 601–617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rilling J. K., van den Heuvel M. P., Comparative primate connectomics. Brain Behav. Evol. 91, 170–179 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Miller D. J., et al. , Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 16480–16485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donahue C. J., Glasser M. F., Preuss T. M., Rilling J. K., Van Essen D. C., Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc. Natl. Acad. Sci. U.S.A. 115, E5183–E5192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mighdoll M. I., Tao R., Kleinman J. E., Hyde T. M., Myelin, myelin-related disorders, and psychosis. Schizophr. Res. 161, 85–93 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Herculano-Houzel S., The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 62, 1377–1391 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Pelvig D. P., Pakkenberg H., Stark A. K., Pakkenberg B., Neocortical glial cell numbers in human brains. Neurobiol. Aging 29, 1754–1762 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Fields R. D., et al. , Glial biology in learning and cognition. Neuroscientist 20, 426–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voineskos A. N., et al. , Oligodendrocyte genes, white matter tract integrity, and cognition in schizophrenia. Cereb. Cortex 23, 2044–2057 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teffer K., Semendeferi K., Human prefrontal cortex: Evolution, development, and pathology. Prog. Brain Res. 195, 191–218 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Fu X., et al. , Rapid metabolic evolution in human prefrontal cortex. Proc. Natl. Acad. Sci. U.S.A. 108, 6181–6186 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Y., Matevossian A., Huang H. S., Straubhaar J., Akbarian S., Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 9, 42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boldog E., et al. , Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci. 21, 1185–1195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berto S., Nowick K., Species-specific changes in a primate transcription factor network provide insights into the molecular evolution of the primate prefrontal cortex. Genome Biol. Evol. 10, 2023–2036 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somel M., et al. , MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 20, 1207–1218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doan R. N., Shin T., Walsh C. A., Evolutionary changes in transcriptional regulation: Insights into human behavior and neurological conditions. Annu. Rev. Neurosci. 41, 185–206 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Hardingham G. E., Pruunsild P., Greenberg M. E., Bading H., Lineage divergence of activity-driven transcription and evolution of cognitive ability. Nat. Rev. Neurosci. 19, 9–15 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Dark C., Homman-Ludiye J., Bryson-Richardson R. J., The role of ADHD associated genes in neurodevelopment. Dev. Biol. 438, 69–83 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Barley K., Dracheva S., Byne W., Subcortical oligodendrocyte- and astrocyte-associated gene expression in subjects with schizophrenia, major depression and bipolar disorder. Schizophr. Res. 112, 54–64 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Haroutunian V., et al. , Myelination, oligodendrocytes, and serious mental illness. Glia 62, 1856–1877 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Srivastava S., Bhatia M. S., Bhargava S. K., Kumari R., Chandra S., A diffusion tensor imaging study using a voxel-based analysis, region-of-interest method to analyze white matter abnormalities in first-episode, treatment-naïve major depressive disorder. J. Neuropsychiatry Clin. Neurosci. 28, 131–137 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Tønnesen S., et al. , White matter aberrations and age-related trajectories in patients with schizophrenia and bipolar disorder revealed by diffusion tensor imaging. Sci. Rep. 8, 14129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandal M. J., et al. ; PsychENCODE Consortium , Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendizabal I., et al. , Cell type-specific epigenetic links to schizophrenia risk in the brain. Genome Biol. 20, 135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glatt S. J., Cohen O. S., Faraone S. V., Tsuang M. T., Dysfunctional gene splicing as a potential contributor to neuropsychiatric disorders. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 156B, 382–392 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quesnel-Vallières M., Weatheritt R. J., Cordes S. P., Blencowe B. J., Autism spectrum disorder: Insights into convergent mechanisms from transcriptomics. Nat. Rev. Genet. 20, 51–63 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Reble E., Dineen A., Barr C. L., The contribution of alternative splicing to genetic risk for psychiatric disorders. Genes Brain Behav. 17, e12430 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Miyata S., Hattori T., Shimizu S., Ito A., Tohyama M., Disturbance of oligodendrocyte function plays a key role in the pathogenesis of schizophrenia and major depressive disorder. BioMed Res. Int. 2015, 492367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis K. L., et al. , White matter changes in schizophrenia: Evidence for myelin-related dysfunction. Arch. Gen. Psychiatry 60, 443–456 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Gibson E. M., et al. , Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science 344, 1252304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y., et al. , Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature 487, 443–448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chamberlain K. A., Sheng Z. H., Mechanisms for the maintenance and regulation of axonal energy supply. J. Neurosci. Res. 97, 897–913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lima Caldeira G., Peça J., Carvalho A. L., New insights on synaptic dysfunction in neuropsychiatric disorders. Curr. Opin. Neurobiol. 57, 62–70 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Mohan H., et al. , Dendritic and axonal architecture of individual pyramidal neurons across layers of adult human neocortex. Cereb. Cortex 25, 4839–4853 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obermayer J., et al. , Lateral inhibition by Martinotti interneurons is facilitated by cholinergic inputs in human and mouse neocortex. Nat. Commun. 9, 4101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hofman M. A., Evolution of the human brain: When bigger is better. Front. Neuroanat. 8, 15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin J. A., et al. , A novel role for oligodendrocyte precursor cells (OPCs) and Sox10 in mediating cellular and behavioral responses to heroin. Neuropsychopharmacology 43, 1385–1394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nave K. A., Ehrenreich H., Myelination and oligodendrocyte functions in psychiatric diseases. JAMA Psychiatry 71, 582–584 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Steele C. J., Zatorre R. J., Practice makes plasticity. Nat. Neurosci. 21, 1645–1646 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Tomassy G. S., Dershowitz L. B., Arlotta P., Diversity matters: A revised guide to myelination. Trends Cell Biol. 26, 135–147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zatorre R. J., Fields R. D., Johansen-Berg H., Plasticity in gray and white: Neuroimaging changes in brain structure during learning. Nat. Neurosci. 15, 528–536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen X., et al. , Novel schizophrenia risk factor pathways regulate FEZ1 to advance oligodendroglia development. Transl. Psychiatry 7, 1293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akizu N., et al. , Biallelic mutations in SNX14 cause a syndromic form of cerebellar atrophy and lysosome-autophagosome dysfunction. Nat. Genet. 47, 528–534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang Y., et al. , LRRK2 and parkin immunoreactivity in multiple system atrophy inclusions. Acta Neuropathol. 116, 639–646 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Hakak Y., et al. , Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 98, 4746–4751 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rash B. G., et al. , Gliogenesis in the outer subventricular zone promotes enlargement and gyrification of the primate cerebrum. Proc. Natl. Acad. Sci. U.S.A. 116, 7089–7094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu Y., et al. , Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi J. S., et al. , ChIP-seq and RNA-seq methods to study circadian control of transcription in mammals. Methods Enzymol. 551, 285–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao H., et al. , CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casper J., et al. , The UCSC genome browser database: 2018 update. Nucleic Acids Res. 46, D762–D769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smedley D., et al. , BioMart–Biological queries made easy. BMC Genomics 10, 22 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D., The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J., Bardes E. E., Aronow B. J., Jegga A. G., ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Falcon S., Gentleman R., Using GOstats to test gene lists for GO term association. Bioinformatics 23, 257–258 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Paradis E., Schliep K., Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Page S. L., Chiu Ch., Goodman M., Molecular phylogeny of Old World monkeys (Cercopithecidae) as inferred from gamma-globin DNA sequences. Mol. Phylogenet. Evol. 13, 348–359 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Page S. L., Goodman M., Catarrhine phylogeny: Noncoding DNA evidence for a diphyletic origin of the mangabeys and for a human-chimpanzee clade. Mol. Phylogenet. Evol. 18, 14–25 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Elango N., Lee J., Peng Z., Loh Y. H., Yi S. V., Evolutionary rate variation in Old World monkeys. Biol. Lett. 5, 405–408 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yi S., Ellsworth D. L., Li W. H., Slow molecular clocks in Old World monkeys, apes, and humans. Mol. Biol. Evol. 19, 2191–2198 (2002). [DOI] [PubMed] [Google Scholar]
- 72.Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium; Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium , Genomic dissection of bipolar disorder and Schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Psychiatric GWAS Consortium Bipolar Disorder Working Group , Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 43, 977–983 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wray N. R., et al. ; eQTLGen; 23andMe; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium , Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Estrada K., et al. , Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 44, 491–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schunkert H., et al. ; Cardiogenics; CARDIoGRAM Consortium , Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morris A. P., et al. ; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium , Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Autism Spectrum Disorders Working Group of The Psychiatric Genomics Consortium , Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savage J. E., et al. , Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davies G., et al. , Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 9, 2098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okbay A., et al. ; LifeLines Cohort Study , Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jansen I. E., et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Grove J., et al. ; Autism Spectrum Disorder Working Group of the Psychiatric Genomics Consortium; BUPGEN; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; 23andMe Research Team , Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Leeuw C. A., Mooij J. M., Heskes T., Posthuma D., MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gautier L., Cope L., Bolstad B. M., Irizarry R. A., affy–Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004). [DOI] [PubMed] [Google Scholar]
- 86.Dannemann M., Lachmann M., Lorenc A., ‘maskBAD’–A package to detect and remove Affymetrix probes with binding affinity differences. BMC Bioinformatics 13, 56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lambert S. A., et al. , The human transcription factors. Cell 175, 598–599 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Cook K. B., Kazan H., Zuberi K., Morris Q., Hughes T. R., RBPDB: A database of RNA-binding specificities. Nucleic Acids Res. 39, D301–D308 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ritchie M. E., et al. , Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.