Significance
Despite studies have shown that mutant p53 acquires its GOF by suppressing p63 and p73, the hypothesis has not been tested in vivo and the underlying mechanism is not fully understood. Here, by using mutant p53-R270H knockin and TAp63/p73-deficient mouse models, we showed that KI mutant p53-R270H antagonizes p63/p73 to shorten mouse life span and to promote T lymphoblastic lymphomagenesis. Mechanistically, we showed that mutant p53 collaborates with the Notch1 intracellular domain (NICD) to abrogate p63/p73-mediated repression of HES1 and ECM1. As a result, HES1 and ECM1 are overexpressed, leading to lymphomagenesis. Thus, we identify a context where mutant p53 contributes to pathogenesis of T lymphoblastic lymphoma by collaborating with the Notch1 pathway to hijack the p63/p73-regulated transcriptional program.
Keywords: mutant p53, GOF, p63/p73, Notch1, T-ALL
Abstract
p53 is the most frequently mutated gene in human cancers and mutant p53 has a gain of function (GOF) that promotes tumor progression and therapeutic resistance. One of the major GOF activities of mutant p53 is to suppress 2 other p53 family proteins, p63 and p73. However, the molecular basis is not fully understood. Here, we examined whether mutant p53 antagonizes p63/p73-mediated tumor suppression in vivo by using mutant p53-R270H knockin and TAp63/p73-deficient mouse models. We found that knockin mutant p53-R270H shortened the life span of p73+/− mice and subjected TAp63+/− or p73+/− mice to T lymphoblastic lymphomas (TLBLs). To unravel the underlying mechanism, we showed that mutant p53 formed a complex with Notch1 intracellular domain (NICD) and antagonized p63/p73-mediated repression of HES1 and ECM1. As a result, HES1 and ECM1 were overexpressed in TAp63+/−;p53R270H/− and p73+/−;p53R270H/− TLBLs, suggesting that normal function of HES1 and ECM1 in T cell activation is hyperactivated, leading to lymphomagenesis. Together, our data reveal a previously unappreciated mechanism by which GOF mutant p53 hijacks the p63/p73-regulated transcriptional program via the Notch1 pathway.
The tumor suppressor p53 plays a central role in maintaining genome integrity and is often referred to as the “guardian of the genome” (1, 2). The most prominent role of p53 is to function as a transcription factor to regulate a series of target genes involved in cell cycle arrest, DNA repair, and apoptosis (3). The importance of p53 in tumor suppression is underscored by the fact that more than half of human cancers contain a mutation or deletion of the TP53 gene.
As the most frequently mutated gene in human cancers, most of p53 mutations are missense mutations and clustered within the central DNA binding domain, such as hotspot mutations R175H, R248W, and R273H. There are mainly 2 types of p53 mutants: conformational mutants, such as R175H, that alter the structure of DNA binding domain and contact mutants, such as R248W or R273H, that attenuate the ability of mutant p53 to bind to DNA (4). As a result, these p53 mutants are deficient in DNA binding and therefore lose their tumor suppressor functions (5). Interestingly, some p53 mutants also acquire new and distinct oncogenic properties, generally referred to as “gain of function” (GOF), such as the ability to promote tumor progression and metastasis (6, 7). For example, cell-based assays have demonstrated various oncogenic properties of mutant p53, including increased survival, DNA synthesis, chemoresistance, angiogenesis, as well as invasion and metastasis (8). Moreover, mutant p53 knockin (KI) mice exhibit significantly different tumor spectra and high incidence of tumor metastasis when compared with p53-null mice (9, 10). Furthermore, multiple clinical studies have shown that high levels of mutant p53 are correlated with more aggressive tumors, poorer outcomes, and enhanced resistance to chemotherapeutic drugs (11, 12). Thus, understanding mutant p53 GOF may lead to the discovery of drugs with broad anticancer effects.
Several mechanisms have been proposed for mutant p53 GOF, including the ability of p53 mutants to bind and inactivate 2 other p53 family members, p63 and p73 (13–15). Indeed, inactivation of p63/p73 has been linked to the ability of mutant p53 to promote chemoresistance, invasion, and metastasis (8) and thus, is suggested to be one of the key mechanisms of mutant p53 GOF. However, it is not certain whether mutant p53 antagonizes p63/p73 in vivo since most studies are cell-based studies. Moreover, the underlying mechanism by which mutant p53 antagonizes p63/p73 is still not fully understood. Early studies showed that the core domain of mutant p53 is sufficient to interact with p63 or p73 in coimmunoprecipitation assays (15). However, it was later found that the interaction between mutant p53 and p63/p73 depends on the nature of the p53 mutation. For example, p53R175H, a conformational mutant that results in misfolded p53 protein, binds much stronger to both TA/ΔN p63α and TA/ΔN p73α than p53R273H, a contact mutant that only slightly perturbs the wild-type (WT) conformation of the protein (13). These data suggest that in addition to direct interaction, mutant p53 inactivates p63/p73 through another mechanism(s).
The Notch1 receptor plays a pivotal role in development and tissue homeostasis (16). Notch1 contains a modular, single-pass transmembrane domain that transduces signals from neighboring cells to nuclear (17). The canonical Notch signaling starts with the binding of the ligand to the Notch1 receptor, followed by proteolytic cleavages to release the intracellular part of the Notch receptor, also called the Notch intracellular domain (NICD), from the inner membrane. The NICD is then translocated into the nucleus and forms a complex with CSL (CBF1, Suppressor of Hairless, Lag-1), also known as RBPJ, to transactivate a set of targets. Interestingly, the role of Notch1 in cancer is context dependent (18). Notch1 acts as a protooncogene in T cell acute lymphoblastic leukemia (T-ALL) but functions as a tumor suppressor in hepatocellular carcinoma (18). These opposing functions of Notch1 in cancer are likely due to its activation of various targets across different contexts. Nevertheless, very little is known whether Notch1 plays a role in mutant p53 GOF.
In the current study, we set out to examine whether mutant p53 antagonizes p63/p73-mediated tumor suppression by using genetically modified mouse models and if so, the underlying mechanism. Indeed, we found that knockin mutant p53-R270H decreases the median survival of p73+/− mice and promotes T lymphoblastic lymphomagenesis in both p63+/− and p73+/− mice. Mechanistically, we found that mutant p53 cooperates with Notch1 to hijack p63/p73-mediated repression of HES1 and ECM1, critical regulators of T cell activation.
Results
Mutant p53 Antagonizes p63/p73-Mediated Tumor Suppression In Vivo.
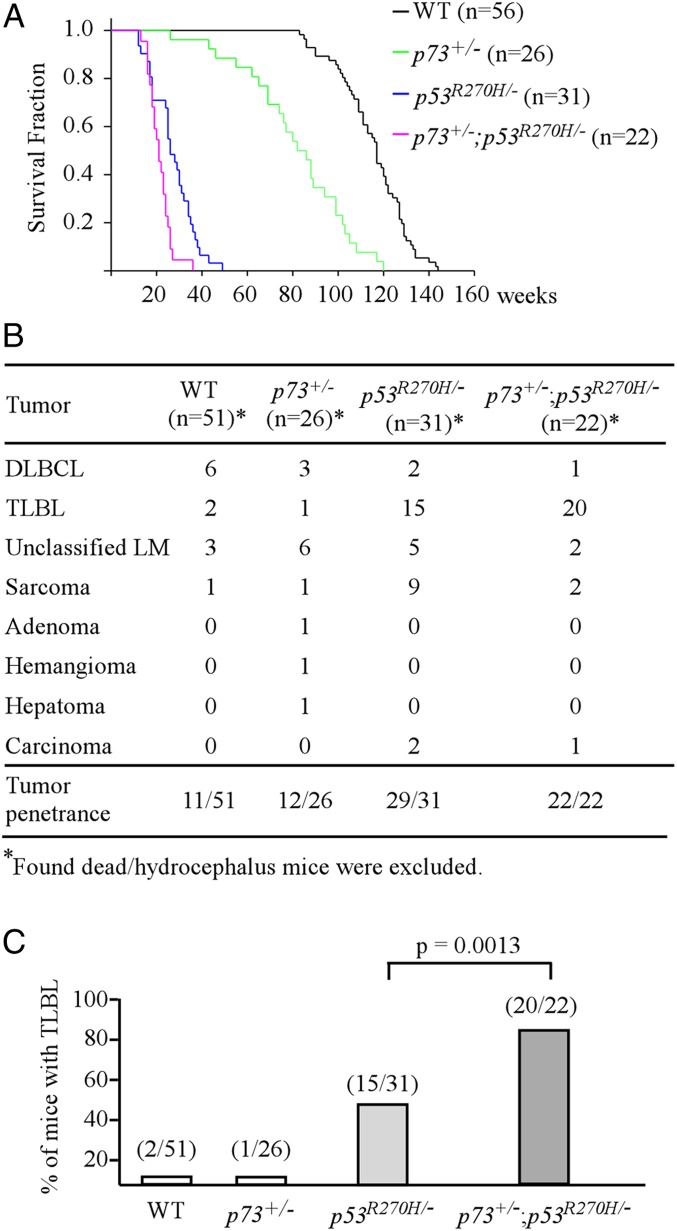
To determine whether mutant p53 antagonizes p73-mediated tumor suppression in vivo, we chose the mutant p53R270H-KI mouse model because: 1) p53R270H/− mice are prone to T lymphoblastic lymphomas (TLBLs) (19), which share similar cytologic and molecular features of human T-ALL; 2) p53-R273H in human (equivalent to R270H in mouse) is a hotspot mutant in hematologic malignancies according to the COSMIC database (https://cancer.sanger.ac.uk/cosmic). Specifically, a cohort of WT, p73+/−, p53R270H/−, and p53R270H/−;p73+/− mice were generated and monitored throughout their life span. To minimize the number of animals used, wild-type mice (n = 56) were adapted from 2 previous studies (19, 20), whereas p53R270H/− mice were adapted from 1 previous study (19). We would like to mention that all of the mice were derived from the same C57BL6 background and maintained at the same facility in the last 5 y. We found that the median life span was 117 wk for wild-type mice, 82 wk for p73+/− mice, 26 wk for p53R270H/− mice, and 21 wk for p53R270H/−;p73+/− mice, respectively (Fig. 1A and SI Appendix, Tables S1, S2, S4, and S5). Statistical analyses indicated that the median life span was significantly different among all 4 groups (Fig. 1A), including p53R270H/− vs. p53R270H/−;p73+/− mice (P = 0.0016 by log-rank test). Next, histopathological analysis was performed and showed that all 4 groups of mice developed spontaneous tumors (Fig. 1B). Tumor penetrance was 21.6% for wild-type mice, 46.2% for p73+/− mice, 93.5% for p53R270H/− mice, and 100% for p73+/−;p53R270H/− mice (Fig. 1B). Statistical analyses indicated that tumor penetrance was significantly higher in p73+/−, p53R270H/−, and p73+/−;p53R270H/− mice as compared to wild-type mice (Fig. 1B). Tumor penetrance in p53R270H/− and p73+/−;p53R270H/− mice was significantly higher than that in p73+/− mice (Fig. 1B). Additionally, knockin mutant p53R270H significantly increased the incidence of TLBLs in p53R270H/− (48.3%) and p73+/−;p53R270H/− (82.6%) mice as compared to wild-type (3.9%) and p73+/− (3.8%) mice (Fig. 1C). Remarkably, p73+/−;p53R270H/− mice were much prone to TLBLs than p53R270H/− mice (P = 0.0013 by Fisher’s exact test) (Fig. 1C).
Fig. 1.
(A) Kaplan–Meier survival curve for wild-type (n = 56), p73+/− (n = 26), p53R270H/− (n = 31), and p73+/−;p53R270H/− (n = 22) mice. The median life span was 117 wk for wild-type mice, 82 wk for p73+/− mice, 26 wk for p53R270H/− mice, and 21 wk for p73+/−;p53R270H/− mice. (B) The tumor spectra of wild-type (n = 51), p73+/− (n = 26), p53R270H/- (n = 31), and p73+/−;p53R270H/− (n = 22) mice. (C) The numbers and percentages of wild-type, p73+/−, p53R270H/−, and p73+/−;p53R270H/− mice with TLBL.
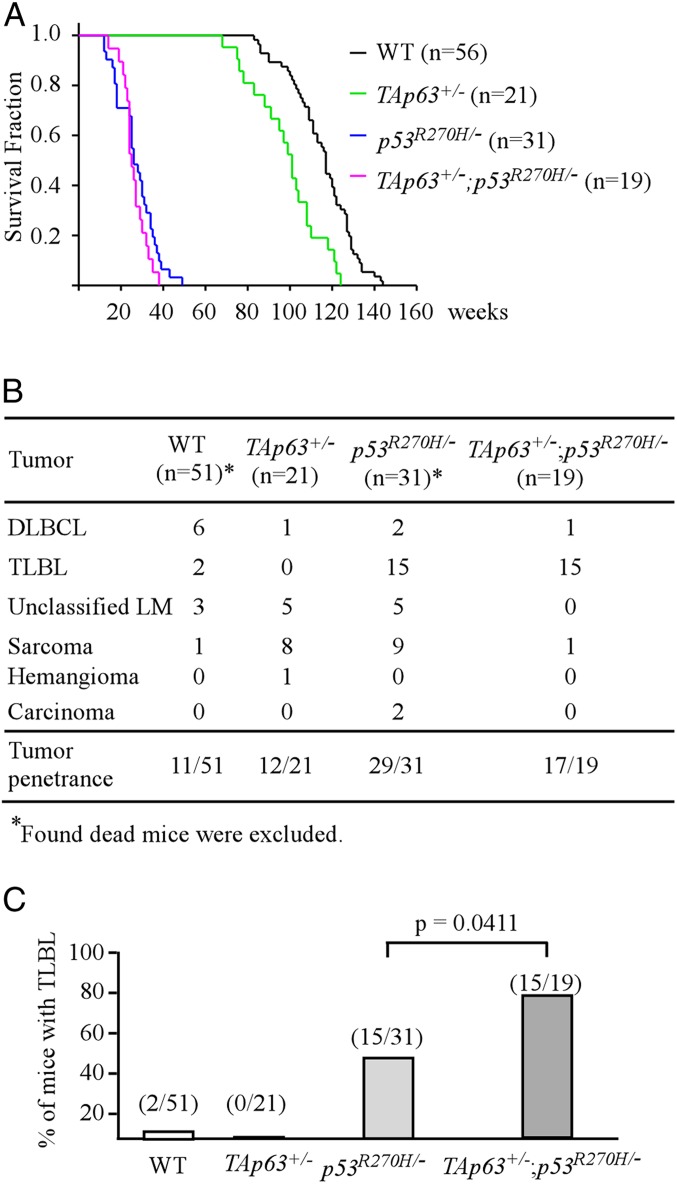
To determine whether mutant p53 can antagonize TAp63-mediated tumor suppression in vivo, a similar approach was taken by monitoring a cohort of WT, TAp63+/−, p53R270H/−, and p53R270H/−;TAp63+/− mice throughout their life span. We would like to mention that the p53R270H/−;TAp63+/− mice were generated for the current study, whereas the TAp63+/− mice were adapted from a previous publication (21). The median life span was 101 wk for TAp63+/− mice, which was significantly shorter than wild-type mice (117 wk) (Fig. 2A). However, there was no significant difference in the survival time between p53R270H/− (26 wk) and p53R270H/−;TAp63+/− (25 wk) mice (P = 0.231 by log-rank test) (Fig. 2A). Moreover, most of the mice developed spontaneous tumors with similar tumor spectra (Fig. 2B). Nevertheless, we found that the incidence of TLBL was markedly increased in p53R270H/−;TAp63+/− mice as compared to that in p53R270H/− mice (78.9% vs. 48.4%; P = 0.0411 by Fisher’s exact test) (Fig. 2C).
Fig. 2.
(A) Kaplan–Meier survival curve for wild-type (n = 56), TAp63+/− (n = 21), p53R270H/− (n = 31), and TAp63+/−;p53R270H/− (n = 19) mice. The median life span was 117 wk for wild-type mice, 101 wk for TAp63+/− mice, 26 wk for p53R270H/− mice, and 25 wk for TAp63+/−;p53R270H/− mice. (B) The tumor spectra of wild-type (n = 51), TAp63+/− (n = 21), p53R270H/− (n = 31), and TAp63+/−;p53R270H− (n = 19) mice. (C) The numbers and percentages of wild-type, p73+/−, p53R270H/−, and p73+/−;p53R270H/− mice with TLBL.
Knockin Mutant p53R270H and TAp63/p73 Deficiency Acts Additively in Enhancing HES1 and ECM1 Expression.
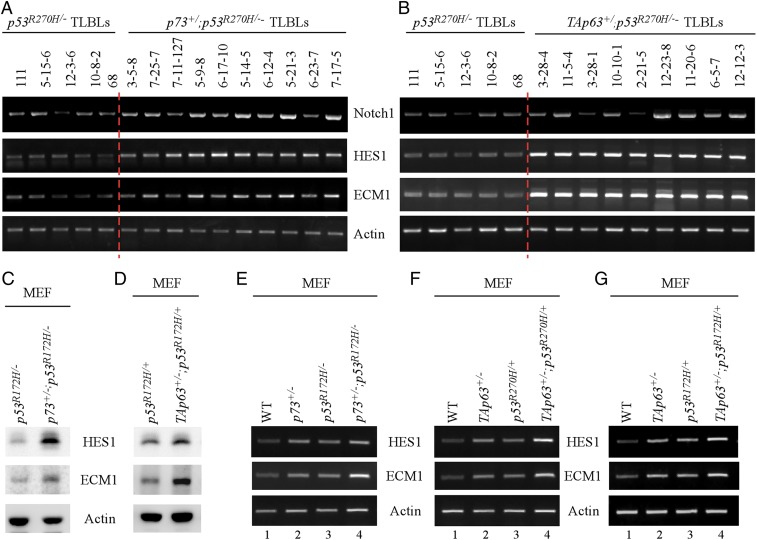
As shown above, the incidence of TLBL was highly increased in both p53R270H/−;p73+/− and p53R270H/−;TAp63+/− mice as compared to that in p53R270H/− mice (Figs. 1C and 2C), suggesting that mutant p53 antagonizes p63/p73 to promote the development of TLBLs. To explore the underlying mechanism, we examined several pathways that can contribute to TLBL, including the Notch1 pathway. We found that compared to p53R270H/− TLBLs, the level of Notch1 transcripts was elevated in most of the p53R270H/−;p73+/− and p53R270H/−;TAp63+/− TLBLs (Fig. 3 A and B, Notch1 panels). These data suggested that activation of the Notch1 pathway contributes to the development of TLBLs in compound mutant mice. Thus, we examined whether p63 or p73 regulates Notch1 expression in human cancer cells. We found that loss of p63 only mildly increased Notch1 transcripts, whereas loss of p73 had almost no effect on Notch1 expression (SI Appendix, Fig. S1 A and B). These data let us speculate that another regulator(s) may contribute to the development of TLBLs in compound mutant mice. In this regard, we searched our RNA-seq data for potential targets of p63/p73 that are regulated by the Notch1 pathway or known to be involved in T cell development. Two potential targets, HES1 and ECM1 (Extracellular matrix protein 1), were chosen for further analysis. HES1, a classic target of Notch1, is essential for T cell development as HES1-knockout (KO) mice show a rudimentary or complete absence of thymus (22). ECM1, a multifunctional protein, was found to play a role in T cell migration and promote angiogenesis in breast cancer (23–25), but has not been identified as a target of Notch1. Indeed, we found that the levels of HES1 and ECM1 transcripts were markedly increased by TAp63/p73 deficiency in p53R270H/− TLBLs (Fig. 3 A and B, HES1 and ECM1 panels). To determine whether p63/p73 represses expression of HES1 and ECM1 in mouse embryonic fibroblasts (MEFs) carrying a mutant p53-R172H, various compound MEFs were generated and showed that the levels of HES1 and ECM1 proteins were much higher in p53R172H/−;p73+/− and p53R172H/+;TAp63+/− MEFs than that in p53R172H/− and p53R172H/+ MEFs, respectively (Fig. 3 C and D). Murine mutant p53-R172H is conserved as R175H in human.
Fig. 3.
(A) The levels of Notch1, HES1, and ECM1 transcripts were examined in TLBLs from p53R270H/− and p73+/−;p53R270H/− mice. The IDs (111, 3-15-6, 12-3-6, 10-8-2, and 68) indicate TLBLs from p53R270H/− mice. The IDs (3-5-8, 7-25-7, 7-11-127, 5-9-8, 6-17-10, 5-14-5, 6-12-4, 5-21-3, 6-23-7, and 7-17-5) indicate TLBLs for p73+/−;p53R270H/− mice. (B) The levels of Notch1, HES1, and ECM1 transcripts were examined in TLBLs from p53R270H/− and TAp63+/−;p53R270H− mice. The IDs (111, 5-15-6, 12-3-6, 10-8-2, and 68) indicate TLBLs from p53R270H− mice. The IDs (3-28-4, 11-5-4, 3-28-1, 10-10-1, 2-21-5, 12-23-8, 11-20-6, 6-5-7, and 12-12-3) indicate the TLBLs from TAp63+/−;p53R270H/− mice. (C) The levels of HES1 and ECM1 proteins were examined in p53R172H/− and p73+/−;p53R172H/− MEFs. (D) The levels of HES1, ECM1, and actin proteins were examined in p53R172H/+ and TAp63+/−;p53R172H/+ MEFs. (E) The levels of HES1, ECM1, and actin transcripts were measured in WT, p73+/−, p53R172H/−, and p73+/−;p53R172H/− MEFs. (F) The levels of HES1, ECM1, and actin transcripts were measured in WT, TAp63+/−, p53R270H/+, and TAp63+/−;p53R270H/+ MEFs. (G) The levels of HES1, ECM1, and actin transcripts were measured in WT, TAp63+/−, p53R172H/+, and TAp63+/−;p53R172H/+ MEFs.
Next, to determine whether mutant p53 antagonizes p63/p73-mediated repression of HES1 or ECM1, several sets of MEFs were generated. We found that the levels of HES1 and ECM1 transcripts were increased by reduced expression of p73 or TAp63 (Fig. 3 E–G, compare lane 1 with 2) or by knockin mutant p53R270H or p53R172H (Fig. 3 F and G, compare lane 1 with 3; SI Appendix, Fig. S1C). Most importantly, the levels of HES1 and ECM1 transcripts were further increased in compound mutant MEFs (p53R172H/−;p73+/−, p53R270H/+;TAp63+/−, p53R172H/+;TAp63+/−) when compared to MEFs with only TAp63/p73 deficiency or mutant p53-KI (Fig. 3 E–G, compare lane 4 with lanes 2 and 3, respectively). Together, these data suggest that mutant p53 antagonizes p63/p73 to enhance HES1 and ECM1 expression.
Mutant p53 Cooperates with Notch1 to Enhance HES1 and ECM1 Expression.
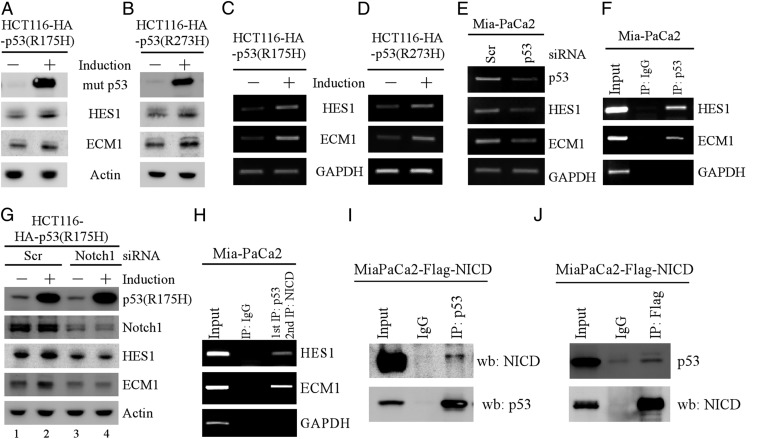
As shown above, knockin mutant p53 enhances HES1 and ECM1 expression in MEFs (Fig. 3 E–G). We thus asked whether HES1 and ECM1 can be regulated by mutant p53 in human cancer cells. To test this, human colorectal HCT116 cells that can inducibly express mutant p53(R175H) or p53(R273H) were used. We showed that the levels of HES1 and ECM1 proteins were increased by both mutant p53(R175H) and p53(R273H) (Fig. 4 A and B). Similarly, ectopic expression of mutant p53(R175H) was capable of increasing expression of HES1 and ECM1 in p53-KO HCT116 cells (SI Appendix, Fig. S2A). By contrast, ectopic expression of wild-type p53 in p53-null HCT116 cells inhibited, whereas knockout of p53 in HCT116 cells increased both HES1 and ECM1 expression (SI Appendix, Fig. S2 B and C). Moreover, we showed that the levels of HES1 and ECM1 transcripts were increased by mutant p53(R175H) or p53(R273H) in HCT116 cells (Fig. 4 C and D), but decreased by knockdown of mutant p53 in Mia-PaCa2 cells that contain mutant p53(R248W) (Fig. 4E). In line with this, luciferase reporter assay showed that both HES1 and ECM1 promoters were activated by mutant p53 (SI Appendix, Fig. S2D). Furthermore, we performed chromatin immunoprecipitation (ChIP) assay and showed that both HES1 and ECM1 promoters were recognized by mutant p53 (Fig. 4F).
Fig. 4.
(A and B) HCT116 cells were uninduced or induced to express HA-tagged mutant p53(R175H) (A) or mutant p53(R273H) (B) for 24 h, followed by Western blot to examine the level of mutant p53, HES1, ECM1, and actin. (C and D) The levels of HES1 and ECM1 transcripts were examined by RT-PCR analysis using cells uninduced or induced to express p53(R175H) (C) or mutant p53(R273H) (D). (E) The levels of HES1 and ECM1 transcripts were examined by RT-PCR analysis in Mia-PaCa2 cells transfected with a scrambled or p53 siRNA for 3 d. (F) ChIP analysis was performed to analyze the binding of mutant p53 to the HES1 and ECM1 promoters. (G) HCT116 cells were transfected with a scrambled or Notch1 siRNA for 3 d, followed with or without induction of mutant p53(R175H) for 24 h. The levels of mutant p53(R175H), Notch1, HES1, ECM1, and actin were analyzed by Western blot analysis. (H) ChIP and reChIP assays were performed with Mia-PaCa2 cells for the co-occupancy of mutant p53 and NICD to the HES1 and ECM1 promoters. DNA–protein complexes were first immunoprecipitated with anti-p53 and then subjected to reChIP analysis using control IgG or anti-NICD antibody. (I and J) Mia-PaCa2 cells were transiently transfected with Flag-tagged NICD and cell lysates were immunoprecipitated with anti-p53 (I) or anti-Flag (J), followed by Western blot analysis with anti-p53 or anti-flag.
Since mutant p53 is deficient in DNA binding, mutant p53 often exerts its GOF through interacting with other proteins (26), which then regulate prosurvival genes. Thus, we sought to identify a mediator of mutant p53 that regulates both HES1 and ECM1 expression. We found a canonical CSL binding site in the ECM1 and HES1 promoters. Previous studies have shown that NICD can form a complex with CSL to transactivate a set of target genes, including HES1 (27). Thus, we examined whether ECM1 can be regulated by Notch1 along with HES1 as a positive control. We showed that the levels of HES1 and ECM1 proteins were decreased by Notch1 knockdown in HCT116, HaCaT, and Mia-PaCa2 cells (SI Appendix, Fig. S3 A–C). Moreover, luciferase assay showed that both HES1 and ECM1 promoters were activated by ectopic NICD (SI Appendix, Fig. S3D). Furthermore, ChIP assay showed that NICD directly bound to the HES1 and ECM1 promoters (SI Appendix, Fig. S3E). These data suggested that ECM1, like HES1, is a target of Notch1. Next, we sought to determine whether Notch1 is involved in mutant p53-dependent induction of HES1 and ECM1. Indeed, we showed that knockdown of Notch1 nearly abrogated increased expression of HES1 and ECM1 by mutant p53 (Fig. 4G, compare lanes 1 and 3 with 2 and 4, respectively). Since NICD and mutant p53 were able to bind to both HES1 and ECM1 promoters (SI Appendix, Fig. S3E and Fig. 4F), we examined whether Notch1 and mutant p53 co-occupy the HES1 and ECM1 promoters by performing ChIP and reChIP assays. Briefly, protein–DNA complexes were first immunoprecipitated with anti-p53, followed by a second immunoprecipitation with anti-NICD antibody. Indeed, we found that both HES1 and ECM1 promoters were detected from NICD–DNA complexes in Mia-PaCa2 cells (Fig. 4H) and in Romas cells (SI Appendix, Fig. S3E), suggesting that NICD and mutant p53 interact on the promoters of HES1 and ECM1. Furthermore, reciprocal IP-Western blot analysis indicated that mutant p53 associated with NICD in Mia-PaCa2 (Fig. 4 I and J) and in Romas cells (SI Appendix, Fig. S3F).
HES1 and ECM1 Are Transcriptionally Repressed by p63 and p73.
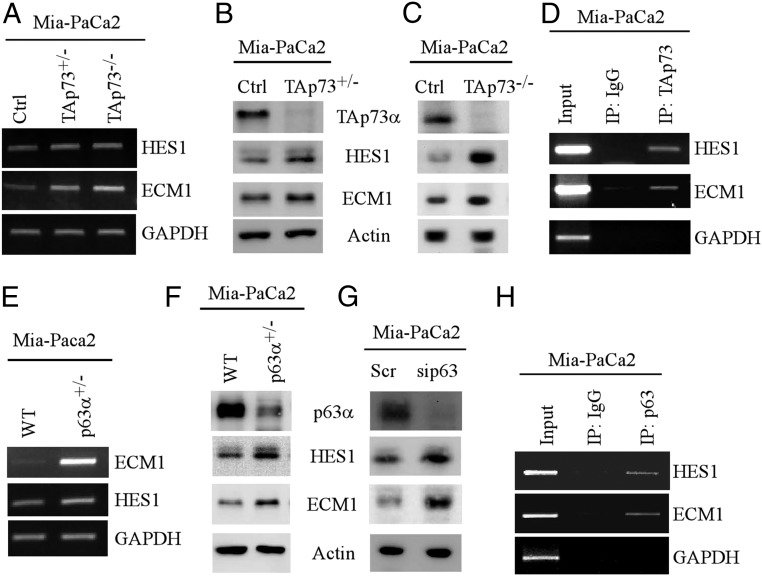
To determine whether p73 transcriptionally represses HES1 and ECM1, H1299 cells that can inducibly express HA-tagged TAp73α were used. We found that the levels of HES1 and ECM1 transcripts and proteins were decreased upon TAp73α induction (SI Appendix, Fig. S4 A and B). To verify that endogenous TAp73 suppresses HES1 and ECM1, CRISPR-Cas9 technology was used to generate stable Mia-PaCa2 cell lines in that one or both TAp73 alleles were deleted. We found that the levels of HES1 and ECM1 transcripts and proteins were much higher in TAp73+/− or TAp73−/− Mia-PaCa2 cells than that in isogenic control Mia-PaCa2 cells (Fig. 5 A–C). Consistently, luciferase reporter assays showed that TAp73α was able to repress both HES1 and ECM1 promoters (SI Appendix, Fig. S4C). Furthermore, ChIP assay showed that endogenous TAp73 directly bound to both HES1 and ECM1 promoters (Fig. 5D). These data suggest that TAp73 acts as a repressor for HES1 and ECM1.
Fig. 5.
(A) The levels of HES1, ECM1, and actin transcripts were measured in isogenic control, TAp73+/−, and TAp73−/− Mia-PaCa2 cells. (B and C) The levels of HES1, ECM1, and actin proteins were measured in isogenic control, TAp73+/− (B), and TAp73−/− (C) Mia-PaCa2 cells. (D) ChIP assays were performed to measure the binding of TAp73 to the HES1 and ECM1 promoters. (E and F) The levels of HES1, ECM1, and actin transcripts (E) and proteins (F) were measured in isogenic control and p63+/− Mia-PaCa2 cells. (G) The levels of HES1, ECM1, and actin proteins were examined in Mia-PaCa2 cells without or without p63 knockdown. (H) ChIP assays were performed to measure the binding of p63 to the HES1 and ECM1 promoters.
To examine whether p63 can repress HES1 and ECM1 expression, MCF7 cells that can inducibly express myc-tagged TAp63α were used. We found that upon induction of TAp63α, the levels of HES1 and ECM1 transcripts and proteins were decreased (SI Appendix, Fig. S5 A and B). By contrast, the levels of HES1 and ECM1 transcripts and proteins were elevated in TAp63+/− Mia-PaCa2 cells as compared to isogenic control Mia-PaCa2 cells (Fig. 5 E and F). Likewise, knockdown of p63 resulted in increased expression of HES1 and ECM1 in Mia-PaCa2 cells (Fig. 5G). Moreover, we showed that TAp63 directly bound to the HES1 and ECM1 promoters in Mia-PaCa2 cells (Fig. 5H) and in MCF7 cells (SI Appendix, Fig. S5C). In line with this, luciferase reporter assays showed that TAp63α was able to repress both HES1 and ECM1 promoters (SI Appendix, Fig. S5D).
The Binding of Mutant p53/Notch1 Complex to the HES1 and ECM1 Promoters Is Enhanced by TAp63/p73 Deficiency.
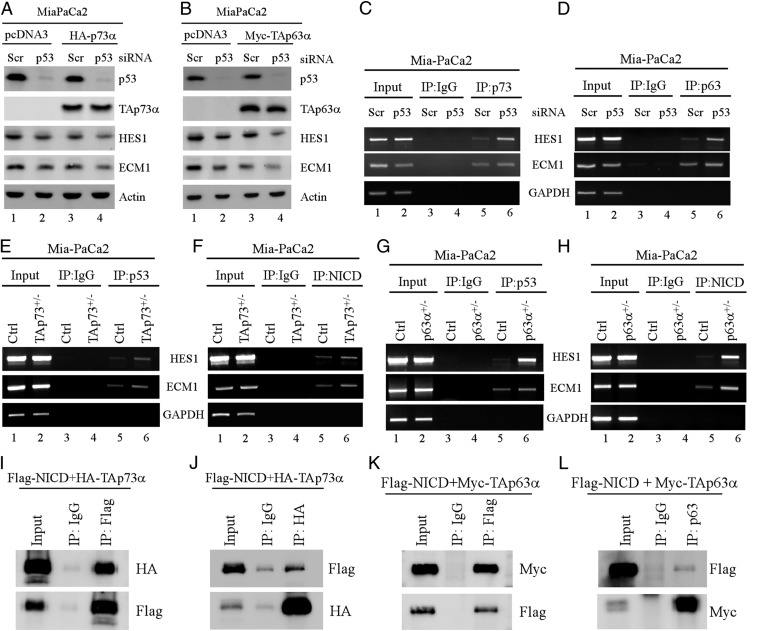
As shown above, our data suggest that mutant p53 and Notch1 increase, whereas p63/p73 repress HES1 and ECM1 expression (Figs. 4 and 5). Interestingly, we also showed that mutant p53 further enhances HES1 and ECM1 expression in p63- and p73-deficient TLBLs and MEFs (Fig. 3), suggesting that mutant p53 antagonizes p63/p73 to enhance HES1 and ECM1 expression. To verify this, Mia-PaCa2 cells were used to knock down endogenous mutant p53 together with or without ectopic expression of p63 or p73. As expected, the levels of HES1 and ECM1 were decreased by knockdown of mutant p53 or by ectopic expression of p63/p73 (Fig. 6 A and B, compare lane 1 with 2 or 3). Importantly, HES1 and ECM1 expression was found to be further decreased upon knockdown of mutant p53 and simultaneous overexpression of p63 or p73 (Fig. 6 A and B, compare lanes 2 and 3 with 4). Consistently, we also showed that the binding of p63 or p73 to the HES1 and ECM1 promoter was enhanced by knockdown of mutant p53 (Fig. 6 C and D, compare lane 5 with 6). These data suggest that mutant p53 can antagonize p63/p73 to facilitate HES1 and ECM1 expression. Next, to further explore the mechanism, we determined whether mutant p53 and NICD compete with p63/p73 to bind to the HES1 and ECM1 promoters. To address this, ChIP assays were performed with isogenic control and p73+/− Mia-PaCa2 cells. We found that the binding of mutant p53 and NICD to both HES1 and ECM1 promoters was increased in p73+/− Mia-PaCa2 cells as compared to isogenic control Mia-PaCa2 cells (Fig. 6 E and F, compare lane 5 with 6). Similarly, we found that the binding of mutant p53 and NICD to both HES1 and ECM1 promoters was increased in p63+/− Mia-PaCa2 cells as compared to isogenic control cells (Fig. 6 G and H, compare lane 5 with 6). Thus, we speculated whether NICD, like mutant p53, interacts with p63/p73 and subsequently prevents the bindings of p63 and p73 to the HES1 and ECM1 promoters. To test this, reciprocal IP analyses were performed to examine whether p63/p73 associated with NICD. Indeed, we found that TAp73α interacted with NICD in both p53-KO HCT116 cells (Fig. 6 I and J) as well as in H1299 cells (SI Appendix, Fig. S6A). Similarly, TAp63α also associated with NICD in p53-KO HCT116 cells (Fig. 6 K and L) as well as in Romas cells (SI Appendix, Fig. S6B). Together, these data suggested that mutant p53 antagonizes p63/p73 to enhance HES1 and ECM1 expression by forming complex with Notch1, which then prevent p63/p73 from binding to the HES1 and ECM1 promoters.
Fig. 6.
(A and B) Mia-PaCa2 cells were transiently transfected with a scrambled or p53 siRNA for 48 h, followed by transient transfection of a control pcDNA3 vector or a vector expressing HA-tagged TAp73α (A) or Myc-tagged TAp63α for 18 h. Cell lysates were collected and subjected to Western blot analysis to detect the levels of p53, TAp73α (A), TAp63α (B), HES1, ECM1, and actin. (C and D) Mia-PaCa2 cells were transiently transfected with a scrambled or p53 siRNA for 3 d, followed by ChIP assays to examine the binding of endogenous p73 (C) or p63 (D) to the HES1 and ECM1 promoters. (E–H) ChIP assays were performed with isogenic control, TAp73+/− (E and F), p63+/− (G and H) Mia-PaCa2 cells to examine the binding of mutant p53 (E and G) or NICD (F and H) to the HES1 and ECM1 promoters. (I and J) p53-KO HCT116 cells were cotransfected with plasmids expressing Flag-tagged NICD or HA-tagged TAp73a, followed by immunoprecipitation with anti-Flag (I) and anti-HA (J). The immunocomplexes were examined by Western blot with anti-Flag or anti-HA. (K and L) MCF7 cells expressing Myc-tagged TAp63α were transiently transfected with Flag-tagged NICD, followed by immunoprecipitation with anti-Flag (K) and anti-p63 (L). The immunocomplexes were examined by Western blot with anti-Flag or anti-Myc.
Discussion
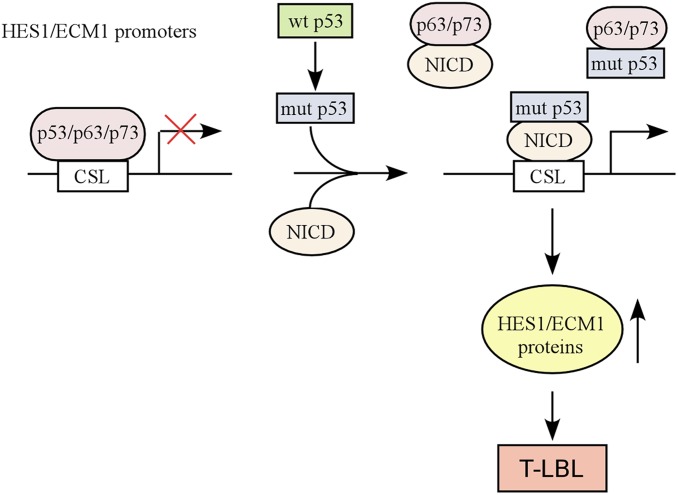
Numerous studies have confirmed that mutant p53 acquires GOF, but the basis for GOF is not fully understood. As mutant p53 is generally considered to be deficient in DNA binding, it is suggested that mutant p53 exerts GOF activities through interactions with other proteins, including 2 other p53 family proteins, p63 and p73 (13, 15). In the current study, we set out to examine whether mutant p53 antagonizes p63/p73-mediated tumor suppression in vivo. By generating various compound mutant mice, we showed that p53R270H/−;p73+/− mice have a reduced life span as compared to p53R270H/− mice (Fig. 1A). Importantly, we found that KI mutant p53R270H markedly enhanced the incidence of TLBL in both TAp63- and p73-deficient mice (Figs. 1C and 2C). Mechanistically, we showed that mutant p53 forms a complex with NICD and subsequently may prevent p63 and p73 from binding to the promoters of HES1 and ECM1, both of which are critical regulators of T cell development. Thus, we have identified a context where mutant p53 contributes to the pathogenesis of T-ALL by collaborating with the Notch1 pathway to hijack the p63/p73-regulated transcriptional program. A model was proposed in Fig. 7 to elucidate the interplay among mutant p53, NICD, p63, and p73.
Fig. 7.
A model to elucidate the interplay among mutant p53, NICD, p63, and p73.
Since p53R270H/−;p73+/− and p53R270H/−;TAp63+/− mice are prone to TLBLs (Figs. 1C and 2C), we were unable to observe other types of tumors. As mutant p53 exerts its GOF in a context-dependent manner, it will be interesting to use a conditional mutant p53 knockin mouse model or a targeted somatic mutant p53 knockin mouse model to further examine how GOF mutant p53 antagonizes p63/p73-mediated tumor suppression. We would also like to mention that p53+/−;p63+/− and p53+/−;p73+/− mice were found to have a reduced survival and increased metastatic rate as compared to p53+/−mice (28). These data suggest that loss of p63/p73 cooperates with loss of wild-type p53 to promote tumorigenesis. As wild-type p53 and p63/p73 represses Notch1 signaling (SI Appendix, Fig. S2 B and C and Fig. 5), it is possible that elevated Notch1 signaling contributes to progression of tumors with loss of p53/p63/p73, which warrants further investigation.
Several mutant p53 binding partners have been identified (26). In this study, we identified Notch1 as a binding partner of mutant p53 that contributes to mutant p53 GOF (Fig. 4). Our data indicate that mutant p53 collaborates with NICD to enhance HES1 and ECM1 expression and subsequently contributes to the pathogenesis of TLBLs or T-ALLs. These results suggest that GOF mutant p53 has a cross-talk with Notch1 signaling and regulates a series of oncogenic events for cancer progression, metastasis, and angiogenesis. Indeed, aberrant Notch1 signaling and p53 mutation were found to be associated with poor prognosis in chronic lymphocytic leukemia (CLL) (29) and breast cancer (30). However, a recent study showed that in acute myeloid leukemia (AML), mutant p53 does not lead to neomorphic GOF activities but instead drives clonal selection through a dominant-negative (DN) effect (31). Since Notch1 mutations are relatively rare in AML (32), these data are not necessarily contradictory to our observations. Thus, further studies are warranted to examine whether the mutant p53-Notch1 cross-talk contributes to progression of these cancers. Moreover, considering that Notch1 inhibitors are currently being tested in clinical trials for patients with advanced or metastatic cancer (33), it is worthwhile to determine whether the mutant p53-Notch1 cross-talk is a major target of Notch1 inhibitors. Notably, we found that unlike mutant p53, wild-type p53 represses HES1 and ECM expression (SI Appendix, Fig. S2 B and C), suggesting that wild-type p53 antagonizes Notch1 signaling. Consistent with this, p53 inhibits Notch1 expression and activation in several thymoma lines (34). However, wild-type p53 was also found to activate Notch1 expression in Ewing’s sarcoma (35) as well as in primary human keratinocytes (36, 37), suggesting that wild-type p53 regulates Notch1 in a context-dependent manner. These data suggest that a complex network exists among Notch1, wild-type, and mutant p53. Thus, further dissecting this network would help us better understand the role of the p53 and Notch pathways in cancer development.
Similar to p53, multiple levels of cross-regulations have been found between p63 and Notch1. Due to the usage of different promoters, TP63 can be expressed as TAp63 isoforms that contain an N-terminal transcription-activating (TA) domain and as ΔNp63 isoforms that lack TA domain but contain a unique ΔN activation domain (38). ΔNp63α is the major isoforms expressed in keratinocytes (39). Interestingly, a negative cross-talk has been found between ΔNp63α and Notch1 in stratified epithelia, wherein ΔNp63α and Notch1 suppress each other’s expression (40, 41). In this study, we found that TAp63 strongly represses HES1 and ECM1 expression in normal MEFs and various human cancer cells (Figs. 3 and 5), suggesting that this repression could be a common event. On the other hand, the Notch1 ligands, JAG1 and JAG2, were found to be induced by TAp63 (42). These data indicate that TAp63 can both activate and repress the Notch1 pathway in a context-dependent manner.
In this study, we showed that mutant p53 forms a complex with NICD and abrogates p73-mediated repression of HES1 and ECM1 (Figs. 4–6), suggesting that the p73-Notch1 pathway plays a role in the pathogenesis of human T-ALL. Indeed, activating Notch1 mutations are detected in more than half of human T-ALL (43). Additionally, p73 is found to be inactivated in human T-ALL due to promoter hypermethylation (44, 45). Thus, it would be interesting to test whether activation of p73 together with Notch1 inhibitors can be exploited as a strategy for the treatment of T-ALL and possibly, other hematological malignancies.
In sum, we provide evidence that mutant p53 acquires GOF by antagonizing p63/p73-mediated tumor suppression via Notch1 signaling. We also provide an insight into a promising strategy to treat tumors harboring mutant p53 by disrupting the cross-talk among mutant p53, Notch1, and p63/p73.
Materials and Methods
Mice.
p73+/− mice were generated by the Mouse Biology Program, University of California at Davis. p53+/−, p53R172H/+, and p53R270H/+ mice were obtained from The Jackson Laboratory as described previously (46). TAp63+/− mice were generated by E. R. Flores’ laboratory as previously described (47). All animals and use protocols were approved by the University of California at Davis Institutional Animal Care and Use Committee.
MEF Isolation.
MEFs are isolated from 12.5 to 13.5 postcoitum (p.c.) mouse embryos as described previously (48). To generate WT, p73+/−, p53R172H/−, and p73+/−; p53R172H/− MEFs, p73+/−;p53+/− mice were bred with p53R172H/+. To generate WT, TAp63+/−, p53R172H/+, p53R270H/+, TAp63+/−; p53R172H/+, and TAp63+/−;p53R270H/+ MEFs, TAp63+/− mice were mated with p53R172H/+ or p53R172H/+ mice. MEFs were cultured in DMEM supplemented with 10% FBS (HyClone), 55 μM β-mercaptoethanol, and 1× nonessential amino acids (NEAA) solution (Cellgro).
Cell Culture and Cell Line Generation.
HCT116, p53−/−HCT116, H1299, MCF7, and Mia-PaCa2 cells and their derivatives were cultured in DMEM (Invitrogen) supplemented with 10% FBS (HyClone). HCT116 cells that can inducibly express HA-tagged p53(R175H) or p53(R273H) under the control of the tetracycline-regulated promoter were generated as previously described (49). To induce p53(R175H) or p53(R273H) expression, tetracycline (1 μg/mL) was added to the medium. H1299 cells that can inducibly express HA-tagged TAp73α and MCF7 cells that can inducibly express Myc-tagged TAp63α were generated as previous described (50). To induce TAp73α or TAp63α expression, tetracycline were removed from the medium. To generate TAp73+/− and TAp73−/− Mia-PaCa2 cell lines by CRISPR-Cas9, pSpCas9(BB)-2A-Puro vector expressing TAp73 sgRNA#1 (5′-CTT CCC CAC GCC GGC CTC CG -3′) and sgRNA#2 (5′-TCA AAC GTG GTG CCC CCA TC-3′) were transfected into Mia-PaCa2 cells. To generate p63a+/− Mia-PaCa2 cell lines, pSpCas9(BB)-2A-Puro vector expressing p63α sgRNA (5′-TGG ACT ATT TCA CGA CCC AG-3′) was transfected into Mia-PaCa2 cells. The cells were selected with puromycin and each individual clone was confirmed by sequencing and Western blot analysis.
Immunoprecipitation and Western Blot Analysis.
These assays were performed as previously described (51). For immunoprecipitation, cell lysates were incubated with 1 μg of antibody or control IgG overnight. The immunocomplexes were brought down by protein A/G beads and subjected to Western blot analysis. For Western blot analysis, cell lysates, prepared using 2× SDS sample buffer, were separated in 8 to 12% SDS/PAGE gel, transferred to a nitrocellulose membrane, and probed with indicated antibodies. The antibodies used in this study were as follows: anti-actin (Sigma), anti-HA (Covance), anti-ECM1 (Santa Cruz Biotechnology), anti-NICD (Cell Signaling), anti-Notch1 (Santa Cruz Biotechnology), anti-HES1 (Cell Signaling), anti-Flag (Sigma), anti-TAp73 (Bethyl), and anti-p63 (customized).
Plasmids.
NICD expression vectors were obtained from Addgene. pcDNA3 vectors expressing myc-tagged TAp63α and or HA-tagged TAp73α were generated as previously described (52).
RNA Interference.
Scrambled siRNA (GGC CGA UUG UCA AAU AAU U) and siRNAs against p53 (5′-CAC CUU GAU CCA GCG GAC UUA-3′) or p63 (5′-CGA CAG UCU UGU ACA AUU U-3′) were purchased from Dharmacon (Chicago, IL). For siRNA transfection, RNAiMax (Life Technology) was used according to the user’s manual.
RNA Isolation and RT-PCR.
Total RNA was isolated with TRIzol reagent as described (51). cDNA was synthesized with reverse transcriptase (Promega) and used for RT-PCR. The PCR program used for amplification was 1) 94 °C for 5 min, 2) 94 °C for 45 s, 3) 60 °C for 45 s, 4) 72 °C for 30 s and 5) 72 °C for 10 min. From steps 2 through 4, the cycle was repeated from 22 to 30 times depending on the targets amplified. The sequences of primers are listed in SI Appendix, Table S7.
ChIP Assay and Sequential ChIP-reChIP Assay.
ChIP assay was performed as previously described (53). To test mutant p53 and Notch1 association on the promoters, a ChIP-reChIP assay was performed as described (53). Briefly, chromatin extracts were incubated overnight at 4 °C with control IgG or antibody against protein of interest. In the Chip-reChip assay, the first Chip was performed with anti-p53, which was then “reChipped” with anti-NICD and control IgG. The primers used for the Chip assays are listed in SI Appendix, Table S7.
Histological Analysis.
Mouse tissues were fixed in 10% (wt/vol) neutral buffered formalin, routinely processed, and embedded in paraffin blocks. Tissue sections (8 μm) were sectioned and stained with H&E.
Data Availability.
All data are included in the article and in SI Appendix. Materials such as reagents and protocols are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We are grateful for the TAp63-KO mouse model provided by Dr. Elsa R. Flores at Moffitt Cancer Center. This work is supported in part by National Institutes of Health R01 grants (CA081237 and CA195828).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913919116/-/DCSupplemental.
References
- 1.Vogelstein B., Lane D., Levine A. J., Surfing the p53 network. Nature 408, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Vousden K. H., Prives C., Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Harms K., Nozell S., Chen X., The common and distinct target genes of the p53 family transcription factors. Cell. Mol. Life Sci. 61, 822–842 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosh R., Rotter V., When mutants gain new powers: News from the mutant p53 field. Nat. Rev. Cancer 9, 701–713 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Ko L. J., Prives C., p53: Puzzle and paradigm. Genes Dev. 10, 1054–1072 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Oren M., Rotter V., Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect. Biol. 2, a001107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller P. A., Vousden K. H., Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 25, 304–317 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freed-Pastor W. A., Prives C., Mutant p53: One name, many proteins. Genes Dev. 26, 1268–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olive K. P., et al. , Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Lang G. A., et al. , Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872 (2004). [DOI] [PubMed] [Google Scholar]
- 11.He C., Li L., Guan X., Xiong L., Miao X., Mutant p53 gain of function and chemoresistance: The role of mutant p53 in response to clinical chemotherapy. Chemotherapy 62, 43–53 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Brachova P., Thiel K. W., Leslie K. K., The consequence of oncomorphic TP53 mutations in ovarian cancer. Int. J. Mol. Sci. 14, 19257–19275 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stindt M. H., et al. , Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene 34, 4300–4310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Como C. J., Gaiddon C., Prives C., p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol. Cell. Biol. 19, 1438–1449 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaiddon C., Lokshin M., Ahn J., Zhang T., Prives C., A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol. Cell. Biol. 21, 1874–1887 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson E. R., Sandberg R., Lendahl U., Notch signaling: Simplicity in design, versatility in function. Development 138, 3593–3612 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Aster J. C., Pear W. S., Blacklow S. C., The varied roles of notch in cancer. Annu. Rev. Pathol. 12, 245–275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lobry C., Oh P., Aifantis I., Oncogenic and tumor suppressor functions of notch in cancer: It’s NOTCH what you think. J. Exp. Med. 208, 1931–1935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H. J., et al. , Ninjurin 1 has two opposing functions in tumorigenesis in a p53-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 114, 11500–11505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., et al. , Ferredoxin reductase is critical for p53-dependent tumor suppression via iron regulatory protein 2. Genes Dev. 31, 1243–1256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y., et al. , The Rbm38-p63 feedback loop is critical for tumor suppression and longevity. Oncogene 37, 2863–2872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita K., et al. , The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 13, 1203–1210 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z., et al. , ECM1 controls T(H)2 cell egress from lymph nodes through re-expression of S1P(1). Nat. Immunol. 12, 178–185 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Lee K. M., et al. , ECM1 regulates tumor metastasis and CSC-like property through stabilization of β-catenin. Oncogene 34, 6055–6065 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Han Z., et al. , Extracellular matrix protein 1 (ECM1) has angiogenic properties and is expressed by breast tumor cells. FASEB J. 15, 988–994 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Kim M. P., Lozano G., Mutant p53 partners in crime. Cell Death Differ. 25, 161–168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarriault S., et al. , Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18, 7423–7431 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flores E. R., et al. , Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7, 363–373 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Zenz T., et al. , TP53 mutation and survival in chronic lymphocytic leukemia. J. Clin. Oncol. 28, 4473–4479 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Reedijk M., et al. , High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 65, 8530–8537 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Boettcher S., et al. , A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 365, 599–604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tohda S., NOTCH signaling roles in acute myeloid leukemia cell growth and interaction with other stemness-related signals. Anticancer Res. 34, 6259–6264 (2014). [PubMed] [Google Scholar]
- 33.Massard C., et al. , First-in-human study of LY3039478, an oral Notch signaling inhibitor in advanced or metastatic cancer. Ann. Oncol. 29, 1911–1917 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Laws A. M., Osborne B. A., p53 regulates thymic Notch1 activation. Eur. J. Immunol. 34, 726–734 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Ban J., et al. , EWS-FLI1 suppresses NOTCH-activated p53 in Ewing’s sarcoma. Cancer Res. 68, 7100–7109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefort K., et al. , Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 21, 562–577 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yugawa T., et al. , Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 27, 3732–3742 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helton E. S., Zhu J., Chen X., The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J. Biol. Chem. 281, 2533–2542 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Dotto G. P., Crosstalk of Notch with p53 and p63 in cancer growth control. Nat. Rev. Cancer 9, 587–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen B. C., et al. , Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 20, 1028–1042 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yugawa T., et al. , DeltaNp63alpha repression of the Notch1 gene supports the proliferative capacity of normal human keratinocytes and cervical cancer cells. Cancer Res. 70, 4034–4044 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Sasaki Y., et al. , The p53 family member genes are involved in the Notch signal pathway. J. Biol. Chem. 277, 719–724 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Ferrando A. A., The role of NOTCH1 signaling in T-ALL. Hematology (Am. Soc. Hematol. Educ. Program) 353–361 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawano S., et al. , Loss of p73 gene expression in leukemias/lymphomas due to hypermethylation. Blood 94, 1113–1120 (1999). [PubMed] [Google Scholar]
- 45.Alexandrova E. M., Moll U. M., Role of p53 family members p73 and p63 in human hematological malignancies. Leuk. Lymphoma 53, 2116–2129 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., et al. , Genetic ablation of Rbm38 promotes lymphomagenesis in the context of mutant p53 by downregulating PTEN. Cancer Res. 78, 1511–1521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su X., et al. , TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 5, 64–75 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., et al. , Translational repression of p53 by RNPC1, a p53 target overexpressed in lymphomas. Genes Dev. 25, 1528–1543 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J., Xu E., Chen X., TAp73 protein stability is controlled by histone deacetylase 1 via regulation of Hsp90 chaperone function. J. Biol. Chem. 288, 7727–7737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu G., Chen X., The C-terminal sterile alpha motif and the extreme C terminus regulate the transcriptional activity of the alpha isoform of p73. J. Biol. Chem. 280, 20111–20119 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Dohn M., Zhang S., Chen X., p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20, 3193–3205 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Shu L., Yan W., Chen X., RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 20, 2961–2972 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qian Y., Jung Y. S., Chen X., Differentiated embryo-chondrocyte expressed gene 1 regulates p53-dependent cell survival versus cell death through macrophage inhibitory cytokine-1. Proc. Natl. Acad. Sci. U.S.A. 109, 11300–11305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article and in SI Appendix. Materials such as reagents and protocols are available from the corresponding author upon reasonable request.