Significance
Identifying sites in the human genome that are extraordinarily sensitive to ultraviolet radiation (UV) would make it easier to measure a person’s past UV exposure and thus predict that person’s future skin cancer risk. This paper reports that human melanocytes, responsible for skin and hair color, contain over 2,000 such genomic sites that are up to 170-fold more sensitive than the average site. These sites occur at specific locations near genes, so may let UV radiation drive direct changes in cell physiology rather than act through rare mutations.
Keywords: UV, cyclobutane pyrimidine dimer, melanoma, ETS, TOP tract
Abstract
If the genome contains outlier sequences extraordinarily sensitive to environmental agents, these would be sentinels for monitoring personal carcinogen exposure and might drive direct changes in cell physiology rather than acting through rare mutations. New methods, adductSeq and freqSeq, provided statistical resolution to quantify rare lesions at single-base resolution across the genome. Primary human melanocytes, but not fibroblasts, carried spontaneous apurinic sites and TG sequence lesions more frequent than ultraviolet (UV)-induced cyclobutane pyrimidine dimers (CPDs). UV exposure revealed hyperhotspots acquiring CPDs up to 170-fold more frequently than the genomic average; these sites were more prevalent in melanocytes. Hyperhotspots were disproportionately located near genes, particularly for RNA-binding proteins, with the most-recurrent hyperhotspots at a fixed position within 2 motifs. One motif occurs at ETS family transcription factor binding sites, known to be UV targets and now shown to be among the most sensitive in the genome, and at sites of mTOR/5′ terminal oligopyrimidine-tract translation regulation. The second occurs at A2–15TTCTY, which developed “dark CPDs” long after UV exposure, repaired CPDs slowly, and had accumulated CPDs prior to the experiment. Motif locations active as hyperhotspots differed between cell types. Melanocyte CPD hyperhotspots aligned precisely with recurrent UV signature mutations in individual gene promoters of melanomas and with known cancer drivers. At sunburn levels of UV exposure, every cell would have a hyperhotspot CPD in each of the ∼20 targeted cell pathways, letting hyperhotspots act as epigenetic marks that create phenome instability; high prevalence favors cooccurring mutations, which would allow tumor evolution to use weak drivers.
DNA photoproduct induction and repair rates are generally similar across the genome, broadly modified by chromatin state (1–6). Yet the biological effects of photoproducts are often localized to specific genes or regulatory regions (7–9). Are oncogenic and other biological events driven only by rare, stochastically located DNA lesions that affect the cell via rare mutations followed by phenotypic selection, or also by outlier genomic sequences hypersensitive to environmental agents and damaged often? Identifying ultraviolet (UV) sensitive sites would also facilitate a practical application: Measurement of personal past UV exposure as a predictor of future skin cancer risk.
The chief UV photoproduct is the cyclobutane pyrimidine dimer (CPD), created in DNA when an UV photon covalently joins 2 adjacent pyrimidines (dipyrimidine or PyPy sites) (10). CPDs cause oncogenic mutations in cells of the skin and later activate signaling pathways that drive single mutant cells to clonally expand (11–17). It is known that large genes are preferentially inactivated by UV (7); genes typically contain CPD hotspots, sustaining these photoproducts at 10-fold higher frequencies than elsewhere in the gene, especially at methylated CG sites (11, 18, 19), and CPD frequency correlates with mutation frequency in Escherichia coli but not in human cells (11, 20). CPDs have been mapped across the genome at a resolution of 200 to 1,000 nt (5, 6). Single-nucleotide measurements averaged over ETS family transcription factor binding motifs in many genes mutated in melanoma reveal that CPDs are elevated at these positions (21–23). Yet the frequency of CPD induction at single-nucleotide resolution across the entire genome remains unknown, as do hotspots and tissue-dependence.
Here we probed the genome of primary human fibroblasts and melanocytes for genome regions that are outliers for UV sensitivity. We first devised a method, adductSeq, for tagging sites of CPDs or other DNA lesions and using high-throughput DNA sequencing to map the tags at single-base resolution across the genome. “Adduct” is used in its freer sense as a covalent change in DNA structure. We then developed a set of statistical methods, freqSeq, that uses the full capabilities of this physical resolution to quantify an individual site’s overrepresentation relative to a genome average, rather than computationally pooling hundreds or thousands of preselected sites (21, 22, 24, 25). FreqSeq also overcomes confounders that include unannotated collapsed repeats, spontaneous DNA lesions, and the fact that a DNA sequencer lane is always filled, either with desired inserts from a small number of cells or, in untreated or repairing cells, with background noise derived from many cells. We find that human cells contain individual pyrimidine dinucleotides hypersensitive to UV; these sites differ between cell types and lie in biologically significant locations, and untreated melanocytes carry extraordinarily high levels of spontaneous DNA lesions including CPDs.
Methods
AdductSeq.
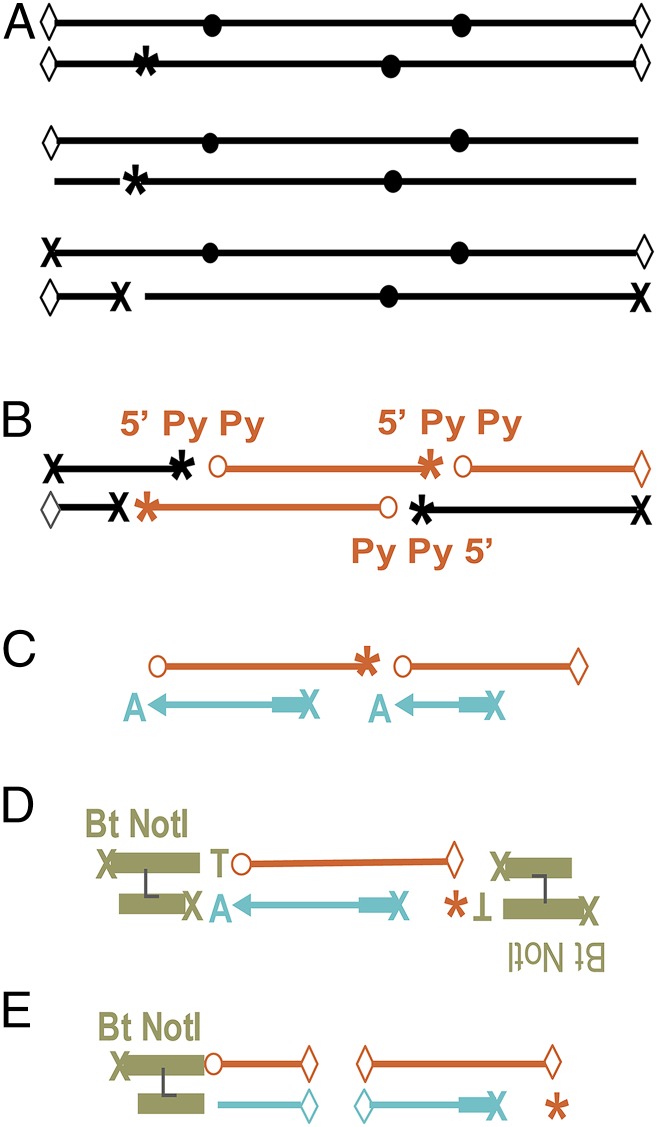
The adductSeq strategy detected DNA adducts at nucleotide resolution across the genome by nicking the DNA at lesion sites with a repair enzyme, tagging the nick with a linker, and mapping the linkers by high-throughput sequencing, in analogy to BLESS used for double-strand breaks (26). The challenge is that, compared to typical DNA sequencing, CPDs are present at <1 per 10 kb after a physiologically relevant UV dose and the particular pyrimidine dinucleotides (PyPy sites) targeted are different in each cell. AdductSeq uses T4 endonuclease V to nick between the 2 PyPy of the CPD, photolyase to remove the CPD, primer extension from a set of semirandom primers to create a double-stranded end at the CPD site for ligation, Covaris shearing to create uniformly sized fragments for library purification, and a biotinylated linker for purification from linkerless fragments (Fig. 1 and SI Appendix, Supplementary Methods). T4 endonuclease V acts by cleaving the glycosidic bond of the 5′ Py of the CPD, creating a baseless site, then nicking 3′ to this abasic site. Preexisting abasic sites are blocked to focus on CPD detection. Primary human fibroblasts and melanocytes were exposed to UVC (254 nm) or narrow-band UVB (nbUVB) (305–315 nm), sometimes allowing time for excision repair, and analyzed for CPDs by adductSeq. Mapped, filtered reads totaled ∼400,000,000 and 700,000,000 for fibroblasts and melanocytes, respectively (SI Appendix, Table S1). These data are publicly available (27).
Fig. 1.
The adductSeq method, illustrated for quantifying CPDs at single-base resolution across the genome. (A) Gently isolated genomic DNA contains shear breaks (◊), abasic sites (*), and, if UV-irradiated, CPDs (●). Treating with USER enzyme nicks at abasic sites and uracils not lying within a CPD. Shear breaks and nicks are 5′ dephosphorylated (X) to suppress detection. (B) Incubation with T4 endonuclease V incises between the 2 PyPy of the CPD, leaving an abasic site at the 5′ Py and a 5′ phosphate before the 3′ Py. Incubation with photolyase reverts the CPD to a normal 3′ Py (o). Sequencing the gold fragments will reveal the CPD locations. (C) Primer extension from a set of semirandom primers creates a double-stranded end at the CPD site, which is given an A extension to facilitate ligation. (D) Ligation to a linker tags the CPD site. (E) Covaris shearing and biotin purification creates uniformly-sized fragments for a high-throughput DNA sequencing library.
FreqSeq.
Whole-genome DNA sequencing is usually not an analytic technique, due to uncharacterized (“collapsed”) repeats, stochastic variations in genome coverage, and differences in DNA loading and sequencing quality between experiments. Because we were searching for rare outliers rather than consensus sequences, it was important to increase statistical resolution to achieve quantification of rare lesions at single-base resolution across the genome. The raw data could be processed in the usual way: Reads were filtered for spurious linker insertions, read quality, and PCR duplicates, and aligned to the hg19 reference human genome using the Burrows–Wheeler alignment algorithm (SI Appendix, Fig. S1). Then, to exclude artifacts as stringently as possible and normalize against collapsed repeats and other spurious signals, we added 2 additional layers of ratio-based analysis that constitute freqSeq (SI Appendix, Supplementary Methods and Fig. S2). Data analysis scripts are publicly available (28).
Results
CPD Mapping Covers the Genome.
Exposing 293T DNA or human fibroblast cells to UVC increased the fraction of reads located at PyPy sites from the expected ∼25% at 0 J/m2 UVC (i.e., residual shear breaks from a large number of cells, distributed equally between PyPy, PuPu, PyPu, and PuPy) to ∼90% at 40 J/m2 (CPDs from a small number of cells, plus shear breaks) (SI Appendix, Table S1). To compare doses, PyPy reads were corrected for the number of initial cells represented on the sequencer lane: PuPu or CA reads were used as a measure of shear breaks independent of preexisting DNA damage, and assumed constant per cell across experiments (SI Appendix, Supplementary Methods). The PyPy/PuPu odds (proportional to CPD density) (SI Appendix, Supplementary Methods) increased monotonically with dose (SI Appendix, Fig. S3A). The pool of sequences in the semirandom primers for second-strand extension accessed the entire mapped human genome, and all regions were ligatable: For each experiment, at each of several window lengths (1 nt to 100 kb), we tallied the number of accepted, mappable reads in that experiment and the number of newly encountered windows containing ≥1 read of any dinucleotide type, where “newly encountered” is a window not seen in any prior experiment. Plotting the cumulative encountered unique windows (as a fraction of the theoretical number in 6,600 Mb of single-strand haploid genome) vs. the cumulative read count yielded a saturation plateau for windows as small as 100 nt (SI Appendix, Fig. S3B). The plateaus occurred at ∼85%, approximately the fraction of the genome that is uniquely mappable. For smaller windows, it is evident that larger experiments would reach a similar plateau. Approximately 90% of mappable 10-nt regions were encountered at least once, so the method is unbiased. Similar plots for individual cell types and dinucleotide types reveal the uniformity of this result as well as the percentage of the genome’s dinucleotide sites that we had probed at this scale of experimentation (SI Appendix, Fig. S3 C and D). In primary melanocytes, the experiments sampled 15% of the mappable PyPy dinucleotides at least once.
Unirradiated Melanocytes Contain Pervasive Apurinic Sites and TG Lesions.
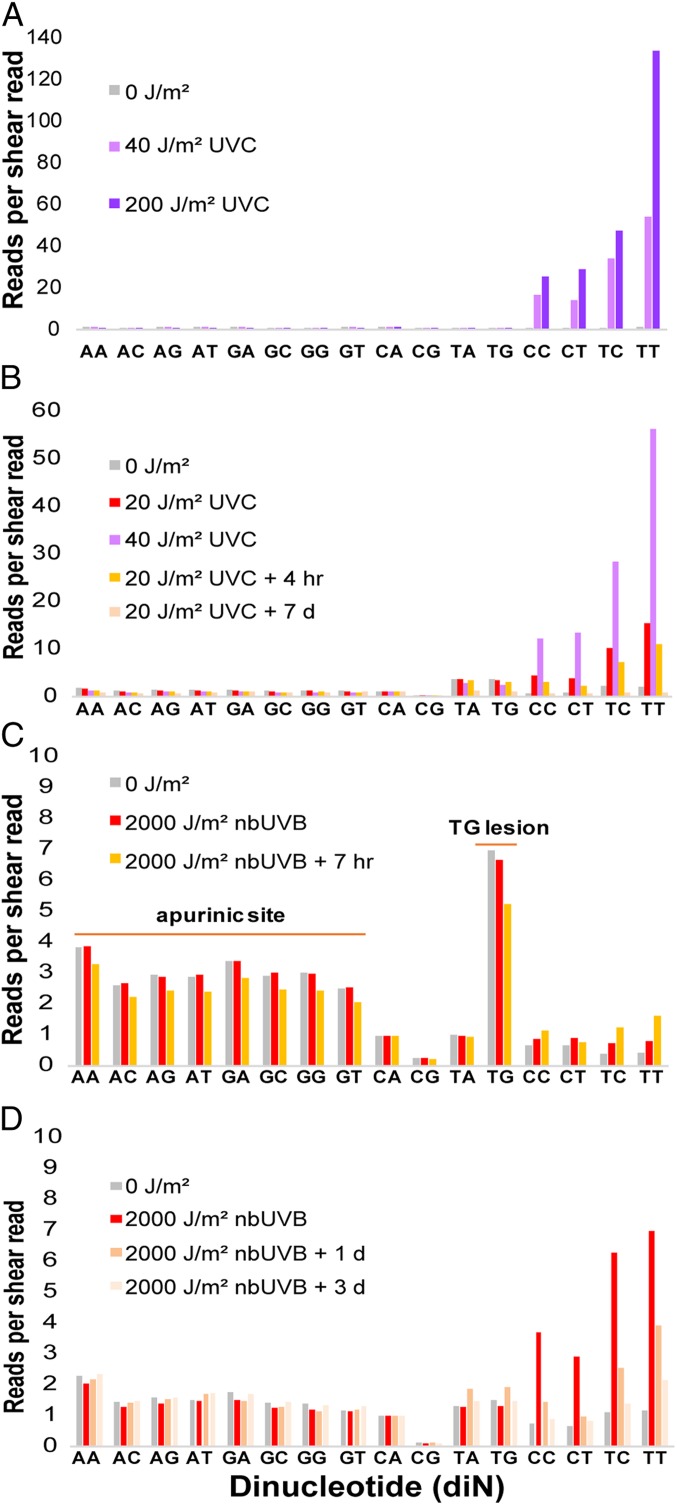
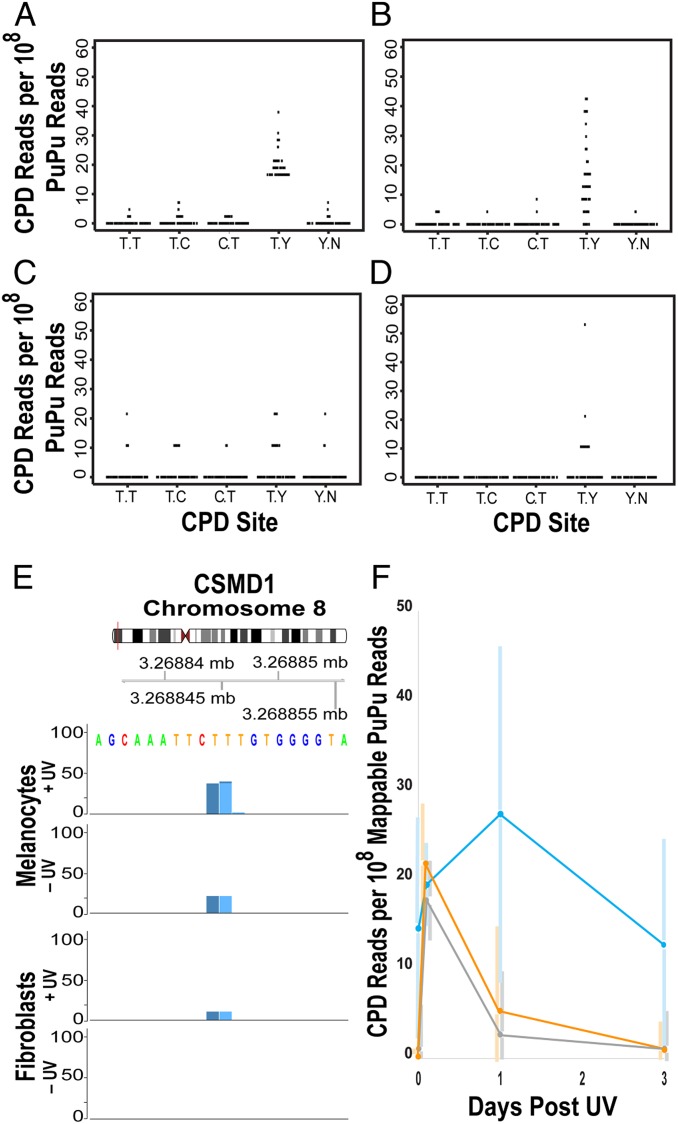
Irradiating naked DNA or human fibroblast cells yielded almost solely CPDs, at the expected relative frequency TT > TC > CT > CC (Fig. 2 A and B). Fibroblasts showed a detectable elevation in lesions at TA and TG sequences without UV; these are not sites of spontaneous purine loss (apurinic sites), which adductSeq would score as a PuN sequence. Melanocytes presented a striking contrast, containing all classes of PuN reads even in unirradiated cells; this level exceeded the number of CPDs created by UV (Fig. 2C). Apurinic sites are plausible because melanin synthesis is a radical-generating process (29) and oxidation of deoxyribose leads to depurination (30). UV-induction of CPDs at PyPy sites was observable above this background. At 7 h after UV exposure, CPDs had continued to increase in melanocytes (Fig. 2C), consistent with the creation of “dark CPDs” by chemiexcitation of melanin (31).
Fig. 2.
High levels of spontaneous apurinic sites and lesions at TG sequences in primary human melanocytes. (A) UVC-irradiation of 293T DNA produces primarily CPDs, which occur at dipyrimidine sites. Reads arising only from shear breaks would give a value of 1.0. (B) Primary human fibroblasts contain some lesions at TA and TG sequences prior to UV exposure. (C) Primary human melanocytes contain high levels of apurinic sites and a lesion at TG sequences, even without irradiation. These exceed the level of CPDs produced by nbUVB irradiation. (D) Preblocking these sites reveals CPD induction by nbUVB and subsequent repair.
The most frequent spontaneous lesion in melanocytes occurred at TG (Fig. 2C). The only known lesion restricted to this tandem DNA sequence is a formamido thymidine, resulting from peroxidation of T followed by migration of the peroxyl radical to G to create the well-known 8-oxo-dG adduct (32, 33); this reaction has been proposed as the true origin of 8-oxo-dG. A formamido pyrimidine can be nicked by some abasic endonucleases, evidently at TG by our T4 endonuclease (34, 35). In test tube experiments, the reaction is even more common at GT sequences, but in adductSeq that lesion would be detected at gTN and so would be aggregated with TT and TC CPDs. Therefore, in subsequent experiments, we prenicked the DNA with the enzyme USER prior to the dephosphorylation step in order to block apurinic sites and TG lesions from being detected by T4 endonuclease (Fig. 2D). We conclude that spontaneous melanin synthesis is capable of damaging and presumably mutating genes leading to melanoma via oxidative DNA damage, which could include the non-UV signature BRAF V600E PuTPu→PuAPu mutation seen in nevi and melanomas. We also conclude that melanocytes are not the ideal cell type to use for genomic dosimeters of a person’s prior UV exposure; keratinocytes may be preferable.
CPD Hyperhotspots.
The size scale of our experiments, ∼1 lane of sequencing and sampling only 15% of PyPy sites once or more, resulted in a genomic average of 0.07 and 0.14 reads per mappable PyPy site in fibroblasts and melanocytes, respectively. Nevertheless, the fibroblast genome contained a remarkable 157 individual PyPy sites having ≥5 recurrent reads; this level of recurrence is 80- to 125-fold greater than expected from the average recurrence rate on a site’s chromosome (Poisson P for recurrence = 10−7 to 10−11) (Dataset S1). Each recurrence’s paired-end sequence differed, ruling out PCR duplicates. Some sites were present thousands of fold more frequently than expected for that CPD recurrence level (SI Appendix, Supplementary Methods). We therefore term these sites “CPD induction hyperhotspots.” Although genes constitute only 27.5% of the human genome, and exons 1.5%, 76% of the hyperhotspots resided in genes or in their 5′UTR/promoter region. Within genes the overrepresentation of hyperhotspots was similar, so evolution has not expunged the most UV-sensitive regions from genes.
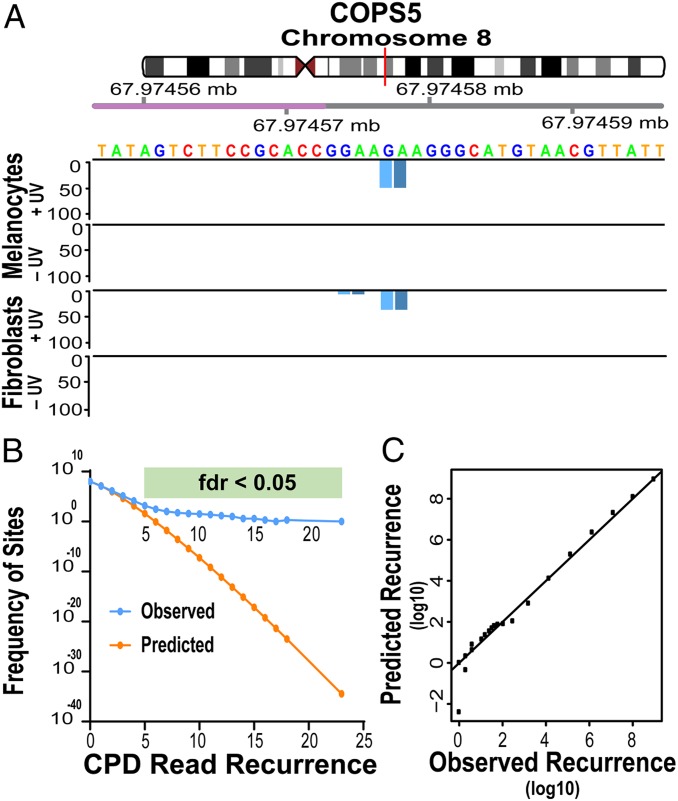
Melanocytes, a 2-fold larger dataset, contained 2,180 PyPy sites having ≥6 reads and 10 sites with over 15 reads (Poisson P = 10−7–10−43); this is 25 to 1036-fold more sites than expected to have that number of recurrent reads (false-discovery rate [FDR] 0.04 to 10−36) (Dataset S2). Jackpot events are ruled out by appearance in multiple experiments (Dataset S3). An example hyperhotspot is shown in Fig. 3A. Of the melanocyte hyperhotspots, 67% resided within genes. A rigorous criterion for discovery of individual hyperhotspots without prior information is FDR ≤ 0.05; there were then 39 in fibroblasts and 2,180 in melanocytes, while only ∼0.1 would have been missed (SI Appendix, Supplementary Methods). Having discovered CPD hyperhotspots on the basis of read recurrence in sequenced DNA regions, it is possible to characterize their CPD occurrence relative to the entire genome: CPDs at the hyperhotspot dinucleotides were generated up to 170-fold more frequently than the genomic average (Datasets S1 and S2 and SI Appendix, Table S3.)
Fig. 3.
CPD hyperhotspots are distinct entities. (A) Genome region containing a hyperhotspot in the promoter of the COPS5 gene. Violet indicates the major transcription start site; tick mark aligns to the base to its right. DNA sequence is for the positive strand, a 5′TC3′ CPD hyperhotspot lies on the negative strand, the 3′ Py shown in light blue is present in the adductSeq read, and the 5′ Py shown in dark blue is deduced from the reference genome. The vertical axis represents a position's normalized PyPy readcount per 108 mappable PuPu reads, the PuPu normalization enabling comparison between UV doses or cell types (see text). Most PyPy sites had no CPDs represented on the sequencer; 2 barely discernable sites correspond to 1 read (normalized to ∼3 for melanocytes, ∼11 for fibroblasts), which could be stochastic. (B) The frequency of sites having a particular CPD (PyPy) read recurrence in melanocytes follows the Poisson distribution appropriate to the chromosomal mean of ∼0.14 reads per site (which will depend on the size of the experiment), up to ∼4 recurrences. (C) The distribution of read recurrence frequencies fits a 3-component Poisson model (see text). Recurrences ≥7 result from the third Poisson distribution.
Hyperhotspots are a distinct entity. The distribution of recurrent PyPy sites in melanocytes fit a 3-component Poisson model (Fig. 3 B and C), in which: 61.0% of the genome’s mappable diPy sites having 0 recurrences were genuine “CPD cold” sites (mean 0); 39.0% were “CPD ordinary” sites with 0 to 6 recurrences (mean 0.286), whose distribution could reflect stochasticity in CPD production rates or stochasticity in representation on the sequencer; and 553 sites with ≥7 recurrences (mean 7.865) (maximum-likelihood Poisson fitted FDR < 0.05). The ≥7 threshold coincided with the transition to distinctive DNA sequence motifs at the hyperhotspot sites (see below). The top 3 hyperhotspots were outliers, even for the 3-component model. The main Poisson distribution of observed sites therefore contains coldspots, average sites, ordinary CPD hotspots on the order of 10× the genomic average, and perhaps hyperhotspots with 6 recurrences in the tail of the distribution due to the mechanisms governing ordinary sites; hyperhotspots with ≥7 recurrences are a novel object not predictable from the photophysics underlying the main Poisson distribution.
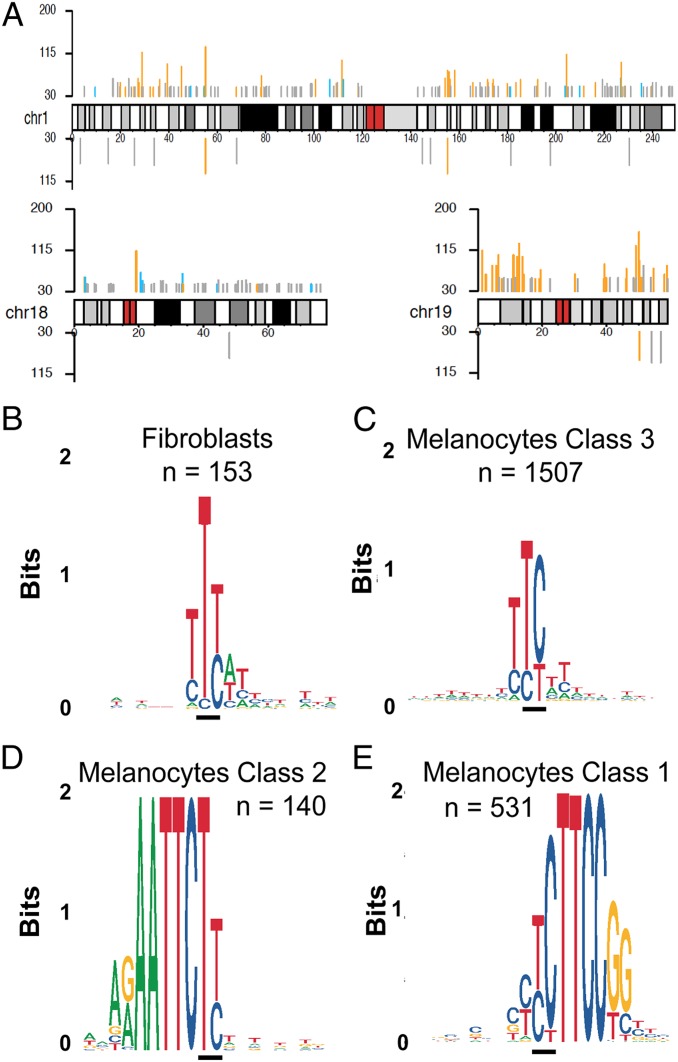
Hyperhotspots were spread across the genome, most frequently near genes and in gene-rich chromosomes (Fig. 4A). The consensus motif for the fibroblast PyPy hyperhotspots was not simply YY, with Y indicating a pyrimidine and underlines indicating the CPD-forming bases, but instead YTY (Fig. 4B). In melanocytes, the majority of hyperhotspots occurred at a DNA sequence motif similar to that in fibroblasts (Fig. 4C); we will refer to the YTY motif as class 3.
Fig. 4.
CPD hyperhotspots occur at DNA sequence motifs in human fibroblasts and melanocytes. (A) Map of CPD hyperhotspots across 3 exemplar chromosomes (chr), including gene-poor chr18 and gene-rich chr19. Melanocyte hyperhotspots are shown above the chromosome map and fibroblast hyperhotspots below. y axis is normalized PyPy readcount per 108 mappable PuPu reads. Hyperhotspots are the most frequently encountered 0.0001% of mappable PyPy sites; the likelihood of a site exhibiting multiple CPDs decreases rapidly according to the Poisson distribution. Gray, blue, and orange bars represent class 3, class 2, and class 1 hyperhotspot classes, respectively. (B) DNA sequence logo for fibroblast hyperhotspots (class 3, 5′YTY3′); black bar indicates the CPD location. (C) Sequence logo for the majority of melanocyte hyperhotspots (class 3, 5′YTY3′). (D) Melanocyte class 2 hyperhotspots (5′(A)2–15TTCTY3′). (E) Melanocyte class 1 hyperhotspots at ETS-like transcription factor binding sites and TOP tracts (5′yYYTTCCg/t3′).
In contrast, most of the top 300 melanocyte hyperhotspots (and 31% of the total [684]) occurred at 1 of 2 precise DNA sequence motifs. The less prevalent motif, termed class 2, was 5′ (A)2–15TTCTY 3′ (Fig. 4D). These sites often featured an additional stretch of A:T base pairs 3′ to the CPD site (Dataset S2). Stacked A bases are known to conduct UV energy down the DNA helix (36), apparently acting as a UV “antenna” for the PyPy site generating the CPD. This motif resembles that seen in prior measurements of CPD hotspots in UV-irradiated DNA and in studies of bacterial and melanoma mutations: A stretch of A:T base pairs lying 5′ to a pyrimidine tract and the CPD typically occurring at the 3′ end of the pyrimidine tract (11, 17, 18, 37). Where the pyrimidine is a C, the UV signature mutation of C→T at a dipyrimidine site can arise (37).
The principal CPD hotspot motif in melanocytes, termed class 1, was 5′ yYYTTCCg/t 3′, with lowercase letters indicating less-stringent parts of the motif (Fig. 4E). A smaller number of CPDs were evident at the TC position. This motif lacks the leading-(A:T) run; the CPDs arose in the 5′ portion of the pyrimidine tract, rather than the typical 3′ end; and the CPD typically contained C despite T being more common in CPDs (38). Fibroblasts only rarely contained either of these motifs (Dataset S1). The Reactome pathway database (https://reactome.org/) was used to identify pathways containing genes having a class 1 hyperhotspot. Of the 18 most-significant pathways (Benjamini–Hochberg adjusted P = 3 × 10−8–2 × 10−6), 14 were composed of proteins involved in ribosomal structure or mRNA translation (Dataset S4), a point revisited below. Proteins in the next most significant group of pathways instead included TP53, RB1, ATR, BRCA1, XPF/ERCC4 and other proteins involved in cell cycle checkpoints, DNA recombination, and DNA repair. These 2 groups are prominent in murine melanomas (39) and overlap via RPS27A, a fusion protein having ribosomal protein S27a at the C terminus and ubiquitin at the N terminus, a common pattern for control by proteasomal degradation.
The hyperhotspot classes present in the 2 cell types were compared more directly by randomly down-sampling reads from the 15,917 nonproblematic sites having ≥5 reads in the larger melanocyte dataset, and comparing the down-sampled melanocyte readcounts to the readcounts at corresponding fibroblast sites. Recurrence was calculated across 40 independent down-samplings and the median used to calculate the CPD ratio. SI Appendix, Fig. S4 shows that melanocytes irradiated with nbUVB contain more CPD hyperhotspots, at higher CPD frequencies, and at a different DNA sequence motif, compared to fibroblasts exposed to an equivalent dose of UVC. Class 1 hyperhotspots dominate only in melanocytes and class 2 hyperhotspots are nearly exclusive to them.
Class 1 CPD Hyperhotspots in 5′UTRs Target Mammalian Target of Rapamycin-Directed Translation.
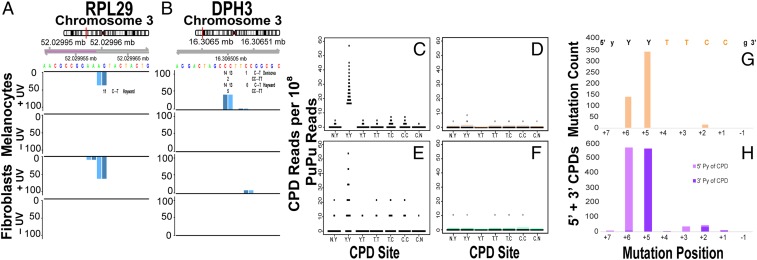
The class 1 hyperhotspot motif appeared in 2 contexts. In 1 context, the C at a yYYTTCCg/t hyperhotspot’s CT or TC aligned at or near a transcriptional start site of a ribosomal protein gene, such as RPL29, RPL34, or RPL37 (40, 41) (Fig. 5A, SI Appendix, Fig. S5A, and Dataset S2). These hyperhotspots reside within “TOP tracts,” 5′ terminal oligopyrimidine tracts that control the mRNA translation rate of growth-related proteins regulated by mammalian target of rapamycin (mTOR), such as those encoding ribosomal proteins and translational elongation factors (42). Ribosomal protein gene TOP tracts contain CTTTCC or CT2–3C1–2 (43). The binding protein is unknown. Most melanomas have activated mTOR pathways (44). The ATR hyperhotspot was also located at a putative TOP tract. In TP53, the hyperhotspot resided at an intronic TOP-like tract for WRAP53 on the other strand, which encodes a functional TP53 antisense transcript and a component of the telomerase holoenzyme (45, 46). The intronic RB1 hyperhotspot was located at a TOP-like tract for the LPAR6 gene on the other strand, coding a G protein-coupled receptor associated with prostate cancer metastasis (47). These hyperhotspots can have biological effects, whatever the binding protein: An eventual rare mutation in DNA can alter the TOP tract’s translational activity (48) and could move the transcription start site; transcriptional mutagenesis [which produces a base change only in the mRNA (49)] would occur sooner and more ubiquitously. The CPD itself can alter translation rates due to CPD-induced alternative splicing (50). The preeminence of ribosome-related genes in the Reactome analysis suggests that the congruence between CPD hyperhotspots and TOP tracts is not fortuitous, and the effect on translation rates deserves detailed investigation. The number of hyperhotspots located at TOP tracts may be larger than evident here because not all transcriptional start sites have been mapped and not all cell types have been examined.
Fig. 5.
UV-sensitive CPD hyperhotspots at YYTTCC are targeted to specific nucleotides and align with recurrent mutations in melanoma. (A) Melanocyte CPD hyperhotspot situated at a TOP tract that regulates the translation rate of RPL29; violet indicates the major transcription start site. The C is the site of 11 UV signature C→T mutations in melanomas and these are recurrent above expectation (41). (B) Melanocyte CPD hyperhotspot at an ETS-like transcription factor binding site in the DPH3 gene promoter, and recurrent mutations in melanomas. This CPD hyperhotspot is present in primary human melanocytes but not in primary human fibroblasts. Black numbers beneath the sequence indicate the location and frequency of C→T mutations observed in 2 collections of melanomas (40, 41). Numbers lying between 2 pyrimidines indicate CC→TT mutations. (C) Melanocyte normalized CPD reads per 108 mappable PuPu reads at and adjacent to the ETS family transcription factor core binding sequence, after UV exposure. Dot indicates a CPD joining the 2 nucleotides; note that each nucleotide appears twice, once as the 5′ Py of a CPD at PyPy and once as the 3′ nucleotide. Means are, left to right, 0.2, 21.8, 0.3, 0.2, 1.1, 0.7, 0.02. (D) Distribution of CPDs across this hyperhotspot motif in the absence of UV exposure. Horizontal bars are 20× mean. (E) Fibroblast CPD recurrence rates at the melanocyte hyperhotspot sites. Means are 0.6, 12.5, 0.4, 0.7, 2.8, 1.6, 0.06. (F) Distribution in fibroblasts in the absence of UV exposure. (G) Distribution of melanoma mutations across the same motif in 52 genes having ≥5 recurrent mutations (all are C→T) (41). The ETS family transcription factor core binding sequence is shown in color; lowercase letters are less stringently required. (H) Distribution of CPD readcounts across this motif in the mutated genes. CPD counts at a mutation position are shown as stacked counts from the CPD in which the mutated position is the 5′Py (light violet) of the CPD and the 3′Py of the CPD (the usual position of a CPD-induced mutation, dark violet).
Class 1 CPD Hyperhotspots in Promoters and 5′UTRs Target ETS-Family Regulated Genes.
In the other context, the YY in yYYTTCCg/t lies outside of, but immediately adjacent to, the TTCC:GGAA core binding motif of the ETS family of transcription factors (24, 40). The 5′ Y does not lie within the ETS family consensus sequence (A/G)(C/T)(T/A)TCCG (51), so these hyperhotspots reflect noncanonical binding sites. The relatively rare YYTTCC variant sequence does bind ETS family members, as shown by University of California, Santa Cruz ENCODE transcription factor binding site tracks. Of the genes that had class 1 CPD hyperhotspots, 94.3% were targets of ETS family transcription factor binding, according to the ENCODE Transcription Factor Targets dataset. ETS family transcription factors are proto-oncogenes, important in the development of neurons and neural crest cells, such as melanocytes. ETS1 is required for melanocyte development; moreover, its inducer, basal fibroblast growth factor (bFGF), is a key component of melanocyte growth medium (52–54). The CPD hyperhotspot in the DPH3 gene promoter is shown in Fig. 5B, which also reveals that this site is not a hyperhotspot in primary human fibroblasts. Additional examples of this motif in individual gene promoters are given in SI Appendix, Fig. S5 B–H. The strong bias for the YY position in these CPD hyperhotspot examples was seen across all instances of the yYYTTCCg/t motif in melanocytes, whereas other dipyrimidines, such as the CC position, were minor targets (Fig. 5 C and D). This CPD positional preference has been noticed before by averaging hundreds of preselected ETS family TTCCG sites, particularly those mutated in melanomas, using TERT-immortalized fibroblasts or melanoma cell lines (21, 22). However, preselection misses hyperhotspots such as at yYYTTCCt (SI Appendix, Fig. S5 I and J). The physical basis at the lesser, TC, CPD site was demonstrated in vitro: The adjacent binding of the ETS1 transcription factor alters the DNA base separation and torsion angle to a conformation that favors CPD formation (21). The present results identify the individual ETS-bound genes that account for this UV sensitivity pattern and show that they underlie the tallest peaks of CPD formation in the normal melanocyte genome.
A prediction of the ETS mechanism is cell-type specificity: Primary fibroblasts should be less enriched for CPD hyperhotspots at this motif. The reasoning is that, although ETS1 mediates extracellular matrix degradation in vivo, primary fibroblasts up-regulate ETS1 only after adding bFGF or upon TERT-immortalization (55–57). In primary fibroblasts, ETS-related CPD hyperhotspots were indeed rare (Fig. 4A and SI Appendix, Fig. S4); only 0.7% of the fibroblast hyperhotspots were located at YYTTCC, and their excess over expected read frequencies was orders-of-magnitude lower than in melanocytes. The cell-type–specific difference in CPD frequencies was largely confined to the YY position in the YYTTCC motif, which was modestly enhanced over neighbors in primary fibroblasts but was a pronounced peak in primary melanocytes (Fig. 5 C and E and SI Appendix, Fig. S6). Only 1% of melanocyte hyperhotspots were also hyperhotspots in fibroblasts. Of the 23 sites that did intersect, 13 occurred at the ETS-like sequence, suggesting that these transcription factors have the same biophysical effects in fibroblasts but are bound to fewer sites.
Mutation-Aligned CPD Hyperhotspots.
In 5′ UTRs, a UV-signature C→T mutation at a TOP tract hyperhotspot would nullify the TOP tract and could decrease or increase the translation rate. Indeed, the CPD hyperhotspot in RPL29 lies at its TOP tract’s critical transcription start site C, located at chr3:52029960; this aligns precisely with recurrent C→T mutations in melanomas that were found to be present more frequently than random expectation (41). No mRNA changes were observed, but none are expected if the mutation acts by altering the translation rate. RPL29 has an important role in tumor angiogenesis (58).
In promoters, Colebatch et al. (24) searched for noncoding tumor mutations that, like TERT mutations, alter transcription factor binding sites. All noncoding mutations present at >10% frequency were located in melanomas, specifically in active promoters within the sequence motif 5′ yYCYTCC 3′. Mutations were C→T, preferentially located at either base of YC, with some mutations at the first C of the CC. This pattern mirrors our CPD distribution in melanocytes and is more focused than the CPD pattern in fibroblasts. Individual genes displaying this 1:1 mapping between CPD hyperhotspots and recurrent mutations are shown in Fig. 5 A and B and SI Appendix, Fig. S5. In analogy to studies of averaged data from just ETS family sites (21, 22), Fig. 5G shows the summed count of yYYTTCCg/t melanoma mutations in a recent compilation (41), for class 1 CPD hyperhotspot sites having an FDR < 1 × 10−10 (readcount >6) and a mutation count >4. The 2 mutation hotspots coincide with the single CPD hyperhotspot in Fig. 5C; the CPDs at position +3 do not lead to mutations because the base is T. To precisely compare mutations to CPD frequencies, note that a mutation in a pyrimidine run could receive contributions (in separate cells) from 2 CPDs, involving the pyrimidine upstream or downstream (Fig. 5H; this happens not to occur in Fig. 5 A and B). The near absence of CPDs at the yY position implies that the YY CPD hyperhotspot supplied the mutations at both 5′ and 3′ pyrimidines. A conclusive demonstration that mutations align with CPDs includes the negative controls (Table 1). While only 0.00095% of yYYTTCCg/t sites having <6 CPD reads showed ≥5 melanoma mutations, 19% of the CPD hyperhotspots did, an enrichment of 20,000-fold (P = 2 × 10−242).
Table 1.
The 20,000-fold enrichment for melanoma promoter mutation sites at class 1 CPD hyperhotspots
| <5 Mutations* | ≥5 Mutations | |
| <6 CPD† | 8,390,959 sites | 80 sites (0.00095%) |
| ≥6 CPD | 281 sites | 64 sites (19%) |
Human melanoma mutation counts at 5′ yYYTTCCGg/t 3′ (41).
Recurrent CPD reads at the YY position of the 5′ yYYTTCCGg/t 3′ motif.
Of 180 mutation clusters found in gene promoters or 5′UTRs of melanomas (24), 22 had >10% mutant fraction in the tumor. Of these, 59% were located at our CPD hyperhotspots (Dataset S2). From another study of significantly recurrent promoter mutations (41), 31% (79 of 251) aligned with yYYTTCCg/t sites that had >2 CPDs in our sampling. A CCTTCCg motif for regulatory region mutations was noted in a third study (25), which in addition showed that chronic UV irradiation of melanoma cells or keratinocytes produced subclonal mutations at the YY positions in DPH3 and RPL13A, both top CPD hyperhotspots. In chronically sun-exposed human skin, the same mutation motif appears in the promoters of these and other mutated genes, such as USMG5/PDCD11 and MRPS31 (figure 1 of ref 25; see also ref. 59). Averaging CPD measurements over preselected ETS family binding sites in fibroblasts had revealed a 16-fold increase in CPDs at the YY position, with a smaller increase at TC (21). FreqSeq in melanocytes let us resolve these motifs at the level of individual genes, revealing that these sites are, in fact, among the most CPD-sensitive base positions in the human genome and acquire CPDs up to 170-fold more frequently than the genomic average.
Phenotypically, ETS binding can either increase or decrease gene expression; for example, it has opposing effects on glycolysis and the citric acid cycle (55). The predominant mutation, C→T at YCTTCC, was not expected to abrogate ETS binding because it does not alter the consensus ETS binding motif (C/T)(A/T)TCCG. Nevertheless, engineering a UV-signature CC→TT mutation into the CCTTCC hyperhotspot of DPH3 led to increased transcription (40). Sequence changes may alter ETS phosphorylation rather than binding (60). The second most-frequent promoter mutation, at YCTTCC, would block ETS binding because the mutated TTTC sequence precludes the core ETS binding motif TTCC. A deeper investigation of the effect of hyperhotspots on gene expression is needed but is nontrivial because differences in specific mRNA isoforms exist between melanocytes and melanomas (61).
CPDs Accumulate at Class 2 Hyperhotspots Prior to UV Exposure.
The existence of hyperhotspots suggests that these sites might be sensitive enough to accumulate CPDs arising in melanocytes without UV, as a consequence of melanogenesis-related redox reactions that chemiexcite melanin and transfer energy to DNA in the dark (31). The situation would be exacerbated at hyperhotspots where nucleotide excision repair was slow. In unirradiated melanocytes, the class 1 ETS-like hyperhotspots did show a UV-like distribution of CPDs across the ETS-like motif, although much weaker than with UV; unirradiated fibroblasts did not (Fig. 5 D and F).
Strikingly, the class 2 hyperhotspots at 5′ (A)2–15TTCTY 3′ showed a pronounced peak at the TY position in unirradiated melanocytes that was nearly as large as after UV exposure (Fig. 6 A and B). This pattern was 5-fold weaker in unirradiated fibroblasts (Fig. 6 C and D) (P = 3.2 × 10−15 by the exact binomial test on PyPy readcounts per mappable PuPu reads). Fig. 6E shows a specific example in the CSMD1 gene, proposed to be a tumor suppressor in melanoma (62). This behavior is consistent with dark CPD formation via excited-state melanin in melanocytes, and also rules out an origin in technical artifacts due to sequence-specific nicking, which would be independent of cell type. These observations in the absence of UV constitute evidence that: 1) Some fibroblast-melanocyte differences reflect cell type rather than UV wavelength, 2) chemiexcitation and dark CPDs occur in unirradiated melanocytes, and 3) CPDs can accumulate in hyperhotspots despite cell proliferation. Correspondingly, the class 2 sites showed an unusual time course after UV exposure (Fig. 6F). Whereas most class 1 and 3 hyperhotspots showed substantial CPD induction by UV and were largely repaired in 24 h, the class 2 sites showed weak immediate CPD induction, continued to incur CPDs after UVB exposure, and showed repair only at 3 d. Melanin fragments are known to intercalate into A:T-rich tracts and block DNA polymerases (63). Intercalation of excited-state melanin fragments would then cause dark CPD formation and the subsequent de-excited fragments could inhibit their removal by excision repair. Sensitivity to dark CPD production and slow repair would direct CPD accumulation to this class of hyperhotspots. The (A)2–15TTCTY hyperhotspot motif does not correspond to a known transcription factor binding site. The TY position is usually TT, so rather than acting by C→T mutation, these motifs are more likely to block DNA replication, alter transcription, or trigger TP53 signaling via CPD-blocked transcription in active genes (14).
Fig. 6.
CPD hyperhotspots at (A)2–15TTCTY accumulate CPDs prior to UV exposure. (A) Melanocyte CPD hyperhotspot recurrence rates per 108 mappable PuPu reads at the class 2 motif, after UV exposure. The mean at the TY location is 25.7. (B) Distribution of CPDs across this hyperhotspot motif in the absence of UV exposure. Mean at TY, 19.2. (C) Fibroblast CPD recurrence rates at the melanocyte hyperhotspot sites. Mean at TY, 3.6. (D) Distribution in fibroblasts in the absence of UV exposure. Mean at TY, 4.2. (E) A class 2 hyperhotspot in the CSMD1 gene. (F) Induction and repair of CPDs in melanocytes at the 3 classes of hyperhotspot motifs. Orange, class 1 yYYTTCCg/t; blue, class 2 (A)2–15TTCTY; and gray, class 3 YTY that are not embedded in 1 of the other 2 motifs. n = 345, 64, and 199, respectively. Data are from sites with readcounts >6 after UV exposure; the similarity of 0 h +UV normalized readcounts between sites is a consequence of the threshold criterion set for hyperhotspots, in comparison to the normalized genome-wide mean of 0.298. Individual sites have different behaviors, especially in class 2; their distribution is indicated by bars spanning 1 SD.
Hyperhotspot-Containing Genes Are Expressed.
The cellular effect of a DNA hyperhotspot requires that the mRNA be expressed, except if it activates a normally unexpressed gene. The role of transcription factors in creating hyperhotspots suggests that the difference in hyperhotspots between fibroblasts and melanocytes will reflect cell-type differences in gene expression. For the majority of the CPD hyperhotspots, expression data in fibroblasts and melanocytes was available (64). The hyperhotspot-containing genes are expressed, with the class 1 hyperhotspots highly expressed (SI Appendix, Fig. S7). That study found hundreds of genes expressed in melanocytes that were below the threshold of 1.0 reads per kilobase per million mapped reads in fibroblasts and keratinocytes. Yet, hyperhotspot-containing genes had similar expression in melanocytes and fibroblasts (albeit biased toward higher expression in melanocytes). Thus, 1) most hyperhotspot-containing genes have a universal, housekeeping-like, expression pattern (as befits their enrichment for genes involved in translation) and 2) mRNA expression level does not explain why hyperhotspots differ between melanocytes and fibroblasts. This equivalence held even at levels below the 1.0 threshold, suggesting that the expression of these genes is tightly regulated. Our gene-set analysis showed the expected highly significant enrichment for H3K27me3 chromatin immunoprecipitation sequencing peaks in the low-expression lower left quadrant of SI Appendix, Fig. S7, and enrichment for H3K27ac (and RNA binding functions, GO:0003723) in the upper right quadrant. Thus, expression per se is primarily governed by histone marks, while cell-type–specific transcription factor binding creates CPD hyperhotspots despite only a modest shift in transcription level.
CPDs in Hyperhotspots Are Frequent Enough to Direct Physiology.
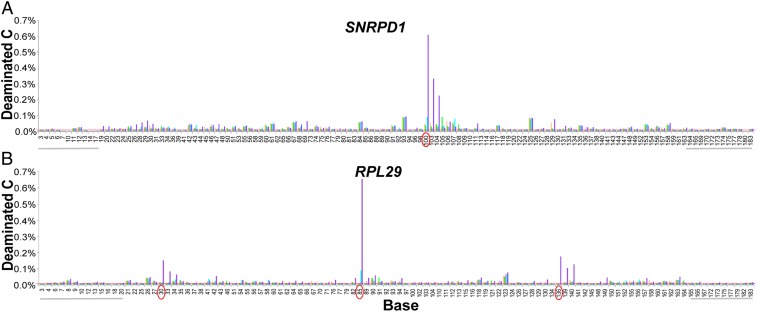
In principle, only DNA fragments bounded by a CPD site are loaded onto the sequencer, so adductSeq does not quantify the absolute CPD frequency. To quantify the absolute, rather than relative, CPD frequencies at hyperhotspots, and to validate their existence using an independent method, we took advantage of the fact that cytosines in CPDs deaminate to uracil 106-fold more rapidly than elsewhere (38, 65). Subsequent photoreactivation of the CPD and PCR amplification with a polymerase that reads U leads to C→T mutations at CPD sites. Next-generation (NextGen) sequencing of amplicons from hyperhotspot regions of RPL29, RPL34, DPH3, COPS5, and SNRPD1 from nbUVB-irradiated primary melanocytes revealed CPD frequencies of 0.25 to 0.7% per genome at individual dipyrimidine sites. CPDs are also frequent in the adjacent positions of the hyperhotspot motif, doubling the figure for the gene carrying a hyperhotspot and thus ranging up to 3% per diploid cell (Fig. 7 and SI Appendix, Fig. S8). These values are, as expected, 50- to 200-fold higher than the genomic average: The genome-wide average induction of C-containing CPDs at these UV doses is 0.01% of C-containing PyPy sites [mass spectrometry (31) for our nbUVB source (11, 38, 66), for UVC]. For the 5 genes tested, the genomic average frequency is below the error rate of NextGen DNA sequencing but it can be estimated by extrapolating the correlation between deamination-based absolute CPD frequency and freqSeq-based relative CPD frequency to x = 1 (SI Appendix, Fig. S8). The average CPD frequency at a C-containing site in these regions was ∼0.003%.
Fig. 7.
At sunburn levels of UVB exposure, each hyperhotspot acquires a CPD in up to 1% of melanocytes. Bar height indicates the percentage of reads having a C→T or G→A substitution. (A) SNRPD1 gene. The hyperhotspot position is circled. (B) RPL29 gene. The 3 hyperhotspot positions are circled. For both genes, CPDs are also apparent in the adjacent positions of the hyperhotspot motif. Color key: tan, no treatment; green, photoreactivation only; pink, heat + photoreactivation; light blue, 2,000 J/m2 nbUVB + photoreactivation; violet, 2,000 J/m2 nbUVB + heat + photoreactivation. Numbering indicates the position of cytosines within the amplicon (on either strand); gray horizontal bars indicate primer locations (and thus DNA sequencer error rate, which accounts for most bars); red horizontal line indicates the average CPD frequency across the 5 regions examined (see text); sites with 5 bars of equal height are presumably due to sequencer errors arising near the end of a 150-nt read.
The genome-wide average CPD frequency is equivalent to 1 CPD per 20,000 nt of single-stranded DNA, with the stochastic locations of these rare CPDs differing in each cell. In contrast, hyperhotspots concentrate CPDs in defined locations within gene promoters and regulatory regions. The impact at the level of metabolic pathways is considered below (Discussion).
Discussion
Hyperhotspots.
The present unsupervised genome-wide measurements reveal the existence of CPD hyperhotspots: At a handful of individual dipyrimidines, CPDs are generated up to 170-fold more frequently than at an average genomic site. One of the 3 motifs determining hyperhotspots is the binding of ETS-like transcription factors, previously shown to be UV-sensitizing (21, 22). Another motif appears related to energy conduction down the π electron stack of A:T bases and intercalation of melanin in these regions. Yet the genome-wide number of class 1 sequence motifs, for example, is over 8 million, much greater than the number satisfying the hyperhotspot FDR threshold we imposed. Additional sequencing would reveal whether CPD creation at these sites is just below the threshold or lacks additional determinants, such as chromatin structure.
ETS1 levels decrease significantly during melanoma progression (60). Correspondingly, our analysis of CPD sequence reads from a recent study of averaged class 1 motif sites in melanoma cell lines (22) showed an absence of the individual hyperhotspots we find in primary melanocytes (SI Appendix, Supplementary Methods, Bioinformatics). This reduction in hyperhotspots resembles the behavior of primary fibroblasts. While the ETS1 decline was not seen in an earlier paper (67), that report used a PCR primer and hybridization probe targeting exon 1 of a rare tissue-specific short isoform that is not expressed in melanocytes. The more common long isoform is expressed in melanocytes but uses an alternative upstream 5′UTR and skips the targeted exon. Thus, Elliott et al. (22) may have found not only an ETS-CPD–mutation relation but also a correlation to ETS physiology.
Fibroblasts were irradiated with UVC and melanocytes with nbUVB to follow prior publications on these cell types, with doses matched on lethality. The differences in CPD hyperhotspot frequency and motif might reflect cell type or wavelength, but the expectation is cell type: 1) Melanocytes but not fibroblasts rely on ETS transcription factors (52–57); 2) at these doses, UVB differs from UVC only in causing a 5-fold increase in the rarer CC CPD and at CG (19, 68, 69), but the hyperhotspots reported here, including yYYTTCCg/t, increased ∼100 fold and do not lie at CG (Fig. 4). An experimental approach to the question is discussed in SI Appendix.
Mutations at Hyperhotspots.
The reason for recurrent mutations in tumors is a topic of intense interest. Conventionally, the question is framed in terms of the effect size of the mutation and the resulting selection for that mutation during tumor evolution. However, those measures of mutation selection are actually measures of mutation recurrence above an expectation level, and thus are blind toward the DNA lesions and polymerase errors that created them. For example, the OncodriveFML algorithm used in Hayward et al. (41) asks whether a mutation disrupts or creates a particular transcription factor binding site in tumors more often than expected from random base substitutions. A hit from this algorithm, therefore, does not reflect the frequency of CPDs that led to the mutation, and is even independent of the CPD’s bias toward C→T base substitutions. The recent proposal (21–23) that recurrent promoter mutations in melanoma are driven primarily by CPD frequency rather than phenotypic selection is strongly supported by our finding that, genome-wide and at multiple individual genes, the promoters and nucleotides recurrently mutated are those with ∼100-fold elevated CPD frequencies at the mutated nucleotide. At these regulatory regions, it is evidently more important to create mutations in many cells than to create a mutation with high effect size in 1 cell. This dichotomy is provocatively similar to the species evolution dichotomy of r- versus K-selection (high offspring production rate versus small numbers of high-survival offspring). High prevalence would increase the probability that 1 of the cells contained a mutation combination conferring a phenotype adapted to the microenvironment; prevalence explores the fitness landscape. The predilection of UV hyperhotspots for growth-related genes and the susceptibility of melanoma cells to loss by immune rejection would seem to favor an r-like high-prevalence/low-survival alternative to “K-selected” strong driver genes that match the fitness landscape unaided.
That said, there is also evidence of mutagenesis-related special properties at these sites. The promoters, 5′ UTRs, and introns involved in hyperhotspots are favorable regions for examining the mutation process: Like tumor-suppressor genes (13), their function can be altered by many base substitutions; consequently, mutations more closely reflect the initial mutagen without distortion by phenotypic selection. In cells exposed to a known UV source, the fraction of classic UV signature mutations, C→T or CC→TT at a dipyrimidine site, is typically 60 to 75%, becoming 90% in repair-defective cells (37). Melanoma mutations in structural genes have this typical level of UV signature mutations (17). Thus, it is striking that 100% of the promoter mutations at the class 1 hyperhotspot motif (22, 24, 25, 40, 41) are UV signature mutations. Therefore, it appears either: 1) The melanoma patient—or melanoma founder cell—had an elevated C→T mutation frequency due to a promoter-specific CPD repair deficit or enhancement of error-prone translesion synthesis at CPDs (70, 71); 2) the hyperhotspot site also favors atypically fast CPD deamination; or 3) these promoter mutations undergo selection on the basis of biophysical interactions between DNA and DNA-binding proteins available with T but not C.
Monitoring Individual Cancer Risk at Hyperhotspots.
The dominant component of a person’s risk of skin cancer is prior UV exposure. If CPDs accumulated in hyperhotspots, these regions could serve as objective sentinels of prior exposure, quantifiable in small skin samples. CPDs are known to persist unrepaired for at least 1 wk in telomeres and heterochromatin of proliferating mammalian cells (8, 72) and for over 1 mo in epidermal stem cells (73). The present study found that, indeed, class 2 hyperhotspot sites had accumulated CPDs in the days or weeks before the experiment began, presumably reflecting internal CPD production by chemiexcitation (31).
To fashion CPD hyperhotspots into genomic dosimeters, it would be important to have sites with a range of sensitivities, so that both low and high UV exposures are quantifiable. This is provided by the 4× range of CPD counts in our hyperhotspot sites, combined with the many sites available. Pragmatically, it is useful that 11 regions contained 2 to 6 hyperhotspots on the same 200-bp amplicon.
Environmental Sensing by Cells at Hyperhotspots.
For a hyperhotspot causing CPDs in 1% of cells at these sunburn-level UVB doses, ∼20 of the ∼2,000 hyperhotspots in melanocytes would have a CPD in any cell. Given that hyperhotspots are concentrated in ∼20 pathways, every pathway will have been hit once. Physiology could therefore be altered in a defined way immediately upon UV exposure, without waiting for rare mutations. For example, UV exposure of the class 1 TOP tract sites would lead to immediate activation or inhibition of mTOR pathways. The fact that most melanomas have activated mTOR pathways (44) suggests that such changes can become permanent. CPDs at hyperhotspots would then be epigenetic marks, as has been proposed for 8-oxo-dG in the redox-sensitive G quadruplexes of gene promoters (74–76). One can speculate that evolution did not eliminate CPD hyperhotspots because they serve a purpose as sensors for environmental exposure.
Supplementary Material
Acknowledgments
We thank Dr. R. Halaban and A. Bacchiocchi, Specimen Resource Core, Yale Specialized Programs of Research Excellence (SPORE) in Skin Cancer, for primary fibroblasts and melanocytes; Dr. A. Narayan (Yale) for experimental advice; Drs. A. Sancar and C. Selby (University of North Carolina) for photolyase; and I. Tikhonova and C. Castaldi (Yale Center for Genome Analysis) for assistance with Illumina library preparation and sequencing. This study was supported by the Yale SPORE in Skin Cancer, National Cancer Institute Grant 2 P50 CA121974 Project 1 (to D.E.B.); and National Institute of Environmental Health Sciences Grant 1 R01 ES030562 (to D.E.B.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE137226). Data analysis scripts are available on GitHub at https://github.com/sameetmehta/freqseq.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907860116/-/DCSupplemental.
References
- 1.Surrallés J., Ramírez M. J., Marcos R., Natarajan A. T., Mullenders L. H., Clusters of transcription-coupled repair in the human genome. Proc. Natl. Acad. Sci. U.S.A. 99, 10571–10574 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanders M. H., Bates S. E., Wilbur B. S., Holmquist G. P., Repair rates of R-band, G-band and C-band DNA in murine and human cultured cells. Cytogenet. Genome Res. 104, 35–45 (2004). [DOI] [PubMed] [Google Scholar]
- 3.Zheng C. L., et al. , Transcription restores DNA repair to heterochromatin, determining regional mutation rates in cancer genomes. Cell Rep. 9, 1228–1234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han C., Srivastava A. K., Cui T., Wang Q. E., Wani A. A., Differential DNA lesion formation and repair in heterochromatin and euchromatin. Carcinogenesis 37, 129–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J., Adebali O., Adar S., Sancar A., Dynamic maps of UV damage formation and repair for the human genome. Proc. Natl. Acad. Sci. U.S.A. 114, 6758–6763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García-Nieto P. E., et al. , Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 36, 2829–2843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKay B. C., et al. , Regulation of ultraviolet light-induced gene expression by gene size. Proc. Natl. Acad. Sci. U.S.A. 101, 6582–6586 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rochette P. J., Brash D. E., Human telomeres are hypersensitive to UV-induced DNA damage and refractory to repair. PLoS Genet. 6, e1000926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg R. A., The Biology of Cancer (Garland Science, New York, ed. 2, 2014), 876 pp. [Google Scholar]
- 10.Friedberg E. C., et al. , DNA Repair and Mutagenesis (ASM Press, Washington, DC, ed. 2, 2005). [Google Scholar]
- 11.Brash D. E., Seetharam S., Kraemer K. H., Seidman M. M., Bredberg A., Photoproduct frequency is not the major determinant of UV base substitution hot spots or cold spots in human cells. Proc. Natl. Acad. Sci. U.S.A. 84, 3782–3786 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brash D. E., et al. , A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. U.S.A. 88, 10124–10128 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegler A., et al. , Mutation hotspots due to sunlight in the p53 gene of nonmelanoma skin cancers. Proc. Natl. Acad. Sci. U.S.A. 90, 4216–4220 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brash D. E., et al. , The DNA damage signal for Mdm2 regulation, Trp53 induction, and sunburn cell formation in vivo originates from actively transcribed genes. J. Invest. Dermatol. 117, 1234–1240 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Zhang W., et al. , UVB-induced apoptosis drives clonal expansion during skin tumor development. Carcinogenesis 26, 249–257 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein A. M., Brash D. E., Jones P. H., Simons B. D., Stochastic fate of p53-mutant epidermal progenitor cells is tilted toward proliferation by UV B during preneoplasia. Proc. Natl. Acad. Sci. U.S.A. 107, 270–275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krauthammer M., et al. , Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 44, 1006–1014 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brash D. E., Haseltine W. A., UV-induced mutation hotspots occur at DNA damage hotspots. Nature 298, 189–192 (1982). [DOI] [PubMed] [Google Scholar]
- 19.Ikehata H., Ono T., Significance of CpG methylation for solar UV-induced mutagenesis and carcinogenesis in skin. Photochem. Photobiol. 83, 196–204 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Kunala S., Brash D. E., Excision repair at individual bases of the Escherichia coli lacI gene: Relation to mutation hot spots and transcription coupling activity. Proc. Natl. Acad. Sci. U.S.A. 89, 11031–11035 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao P., et al. , ETS transcription factors induce a unique UV damage signature that drives recurrent mutagenesis in melanoma. Nat. Commun. 9, 2626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott K., et al. , Elevated pyrimidine dimer formation at distinct genomic bases underlies promoter mutation hotspots in UV-exposed cancers. PLoS Genet. 14, e1007849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts S. A., Brown A. J., Wyrick J. J., Recurrent noncoding mutations in skin cancers: UV damage susceptibility or repair inhibition as primary driver? Bioessays 41, e1800152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colebatch A. J., et al. , Clustered somatic mutations are frequent in transcription factor binding motifs within proximal promoter regions in melanoma and other cutaneous malignancies. Oncotarget 7, 66569–66585 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fredriksson N. J., et al. , Recurrent promoter mutations in melanoma are defined by an extended context-specific mutational signature. PLoS Genet. 13, e1006773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crosetto N., et al. , Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Premi S., et al. , AdductSeq data for “Genomic sites hypersensitive to ultraviolet radiation.” Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE137226. Deposited 10 September 2019. [DOI] [PMC free article] [PubMed]
- 28.Kornacker K., et al. , freqSeq: A statistical method to quantify rare DNA lesions at single-base resolution across the genome. GitHub. https://github.com/sameetmehta/freqseq. Deposited 9 October 2019.
- 29.Smit N. P., et al. , Increased melanogenesis is a risk factor for oxidative DNA damage–Study on cultured melanocytes and atypical nevus cells. Photochem. Photobiol. 84, 550–555 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Povirk L. F., Steighner R. J., Oxidized apurinic/apyrimidinic sites formed in DNA by oxidative mutagens. Mutat. Res. 214, 13–22 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Premi S., et al. , Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science 347, 842–847 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Box H. C., Freund H. G., Budzinski E. E., Wallace J. C., Maccubbin A. E., Free radical-induced double base lesions. Radiat. Res. 141, 91–94 (1995). [PubMed] [Google Scholar]
- 33.San Pedro J. M. N., Greenberg M. M., 5,6-Dihydropyrimidine peroxyl radical reactivity in DNA. J. Am. Chem. Soc. 136, 3928–3936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latham K. A., Lloyd R. S., Delta-elimination by T4 endonuclease V at a thymine dimer site requires a secondary binding event and amino acid Glu-23. Biochemistry 34, 8796–8803 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Bourdat A. G., Gasparutto D., Cadet J., Synthesis and enzymatic processing of oligodeoxynucleotides containing tandem base damage. Nucleic Acids Res. 27, 1015–1024 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nordlund T. M., Sequence, structure and energy transfer in DNA. Photochem. Photobiol. 83, 625–636 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Brash D. E., UV signature mutations. Photochem. Photobiol. 91, 15–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Setlow R. B., Carrier W. L., Pyrimidine dimers in ultraviolet-irradiated DNA’s. J. Mol. Biol. 17, 237–254 (1966). [DOI] [PubMed] [Google Scholar]
- 39.Ferguson B., et al. , Different genetic mechanisms mediate spontaneous versus UVR-induced malignant melanoma. eLife 8, e42424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denisova E., et al. , Frequent DPH3 promoter mutations in skin cancers. Oncotarget 6, 35922–35930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayward N. K., et al. , Whole-genome landscapes of major melanoma subtypes. Nature 545, 175–180 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Thoreen C. C., et al. , A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wool I. G., Chan Y. L., Glück A., Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 73, 933–947 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Karbowniczek M., Spittle C. S., Morrison T., Wu H., Henske E. P., mTOR is activated in the majority of malignant melanomas. J. Invest. Dermatol. 128, 980–987 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Mahmoudi S., et al. , Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell 33, 462–471 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Venteicher A. S., et al. , A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ketscher A., et al. , LSD1 controls metastasis of androgen-independent prostate cancer cells through PXN and LPAR6. Oncogenesis 3, e120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dutton-Regester K., et al. , A highly recurrent RPS27 5'UTR mutation in melanoma. Oncotarget 5, 2912–2917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morreall J. F., Petrova L., Doetsch P. W., Transcriptional mutagenesis and its potential roles in the etiology of cancer and bacterial antibiotic resistance. J. Cell. Physiol. 228, 2257–2261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williamson L., et al. , UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell 168, 843–855.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei G. H., et al. , Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 29, 2147–2160 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torlakovic E. E., Bilalovic N., Nesland J. M., Torlakovic G., Flørenes V. A., Ets-1 transcription factor is widely expressed in benign and malignant melanocytes and its expression has no significant association with prognosis. Mod. Pathol. 17, 1400–1406 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Saldana-Caboverde A., et al. ; NISC Comparative Sequencing Program , The transcription factors Ets1 and Sox10 interact during murine melanocyte development. Dev. Biol. 407, 300–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Halaban R., Ghosh S., Baird A., bFGF is the putative natural growth factor for human melanocytes. In Vitro Cell Dev. Biol. 23, 47–52 (1987). [DOI] [PubMed] [Google Scholar]
- 55.Lindvall C., et al. , Molecular characterization of human telomerase reverse transcriptase-immortalized human fibroblasts by gene expression profiling: Activation of the epiregulin gene. Cancer Res. 63, 1743–1747 (2003). [PubMed] [Google Scholar]
- 56.Cho M. C., et al. , Epiregulin expression by Ets-1 and ERK signaling pathway in Ki-ras-transformed cells. Biochem. Biophys. Res. Commun. 377, 832–837 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Hahne J. C., Okuducu A. F., Fuchs T., Florin A., Wernert N., Identification of ETS-1 target genes in human fibroblasts. Int. J. Oncol. 38, 1645–1652 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Jones D. T., et al. , Endogenous ribosomal protein L29 (RPL29): A newly identified regulator of angiogenesis in mice. Dis. Model. Mech. 6, 115–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martincorena I., et al. , Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mattia G., et al. , Constitutive activation of the ETS-1-miR-222 circuitry in metastatic melanoma. Pigment Cell Melanoma Res. 24, 953–965 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z., Pal S., Bi Y., Tchou J., Davuluri R. V., Isoform level expression profiles provide better cancer signatures than gene level expression profiles. Genome Med. 5, 33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang M. R., Wang Y. X., Guo S., Han S. Y., Wang D., CSMD1 exhibits antitumor activity in A375 melanoma cells through activation of the Smad pathway. Apoptosis 17, 927–937 (2012). [DOI] [PubMed] [Google Scholar]
- 63.Geng J., et al. , Bacterial melanin interacts with double-stranded DNA with high affinity and may inhibit cell metabolism in vivo. Arch. Microbiol. 192, 321–329 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Reemann P., et al. , Melanocytes in the skin—Comparative whole transcriptome analysis of main skin cell types. PLoS One 9, e115717 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng W., Shaw B. R., Accelerated deamination of cytosine residues in UV-induced cyclobutane pyrimidine dimers leads to CC→TT transitions. Biochemistry 35, 10172–10181 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Barak Y., Cohen-Fix O., Livneh Z., Deamination of cytosine-containing pyrimidine photodimers in UV-irradiated DNA. Significance for UV light mutagenesis. J. Biol. Chem. 270, 24174–24179 (1995). [DOI] [PubMed] [Google Scholar]
- 67.Rothhammer T., et al. , The Ets-1 transcription factor is involved in the development and invasion of malignant melanoma. Cell. Mol. Life Sci. 61, 118–128 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Douki T., Cadet J., Individual determination of the yield of the main UV-induced dimeric pyrimidine photoproducts in DNA suggests a high mutagenicity of CC photolesions. Biochemistry 40, 2495–2501 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Rochette P. J., et al. , UVA-induced cyclobutane pyrimidine dimers form predominantly at thymine-thymine dipyrimidines and correlate with the mutation spectrum in rodent cells. Nucleic Acids Res. 31, 2786–2794 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kunala S., Brash D. E., Intragenic domains of strand-specific repair in Escherichia coli. J. Mol. Biol. 246, 264–272 (1995). [DOI] [PubMed] [Google Scholar]
- 71.Wigan M., et al. , A UVR-induced G2-phase checkpoint response to ssDNA gaps produced by replication fork bypass of unrepaired lesions is defective in melanoma. J. Invest. Dermatol. 132, 1681–1688 (2012). [DOI] [PubMed] [Google Scholar]
- 72.Bérubé R., Drigeard Desgarnier M. C., Douki T., Lechasseur A., Rochette P. J., Persistence and tolerance of DNA damage induced by chronic UVB irradiation of the human genome. J. Invest. Dermatol. 138, 405–412 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Mitchell D. L., et al. , Effects of chronic low-dose ultraviolet B radiation on DNA damage and repair in mouse skin. Cancer Res. 59, 2875–2884 (1999). [PubMed] [Google Scholar]
- 74.Fleming A. M., Ding Y., Burrows C. J., Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 114, 2604–2609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seifermann M., Epe B., Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark? Free Radic. Biol. Med. 107, 258–265 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Ba X., Boldogh I., 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol. 14, 669–678 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.