Significance
In all eukaryotic cells, protein and lipid cargoes are transported between organelles in membrane-bound vesicles. These vesicles, generated by the assembly of protein coats, are captured at their destination organelle by tethering factors. How coats and tethering factors cooperate to ensure proper cargo delivery is poorly understood. Here, we combine X-ray crystallography with biochemical and functional studies to elucidate how the tethering factor Dsl1 uses a tryptophan-containing motif to bind to the vesicle coat protein α-COP. A second tryptophan-containing motif, present within α-COP itself, is critical for coat assembly. Competition between these 2 motifs suggests that Dsl1 preferentially captures partially coated COPI vesicles.
Keywords: membrane trafficking, COPI, vesicle coat, tethering complex, Dsl1
Abstract
Coat protein I (COPI)-coated vesicles mediate retrograde transport from the Golgi to the endoplasmic reticulum (ER), as well as transport within the Golgi. Major progress has been made in defining the structure of COPI coats, in vitro and in vivo, at resolutions as high as 9 Å. Nevertheless, important questions remain unanswered, including what specific interactions stabilize COPI coats, how COPI vesicles recognize their target membranes, and how coat disassembly is coordinated with vesicle fusion and cargo delivery. Here, we use X-ray crystallography to identify a conserved site on the COPI subunit α-COP that binds to flexible, acidic sequences containing a single tryptophan residue. One such sequence, found within α-COP itself, mediates α-COP homo-oligomerization. Another such sequence is contained within the lasso of the ER-resident Dsl1 complex, where it helps mediate the tethering of Golgi-derived COPI vesicles at the ER membrane. Together, our findings suggest that α-COP homo-oligomerization plays a key role in COPI coat stability, with potential implications for the coordination of vesicle tethering, uncoating, and fusion.
Vesicles mediate cargo transport in eukaryotic cells, providing a critical infrastructure for protein maturation and turnover, organelle homeostasis, and more specialized processes such as neurotransmitter release. Proteinaceous vesicle coats select cargo and reshape the membrane of donor organelles to bud off coated vesicles (1–3). As a vesicle matures, the coat is shed, exposing the underlying membrane. Finally, the vesicle delivers its cargo by fusing its membrane with that of a target organelle (or the plasma membrane), a process driven by the energetically favorable assembly of membrane-bridging SNARE protein complexes (4).
In addition to coat proteins and SNAREs, vesicle traffic also requires organelle-specific tethering factors (5–7). These large protein complexes are thought to mediate the initial, reversible attachment between a vesicle and its target membrane, and are presumed to promote SNARE assembly and membrane fusion indirectly, by increasing vesicle dwell time, or directly, by binding to SNAREs. Early studies suggested that tethering factors capture vesicles by recognizing Rab GTPases on the vesicle membrane (8, 9). Subsequently, 2 tethering factors that mediate retrograde trafficking in the early secretory pathway, the Dsl1 and conserved oligomeric Golgi (COG) complexes, were found to bind to coat protein I (COPI) (10–13). This discovery led to the suggestion that tethering factors may in some cases recognize coated vesicles.
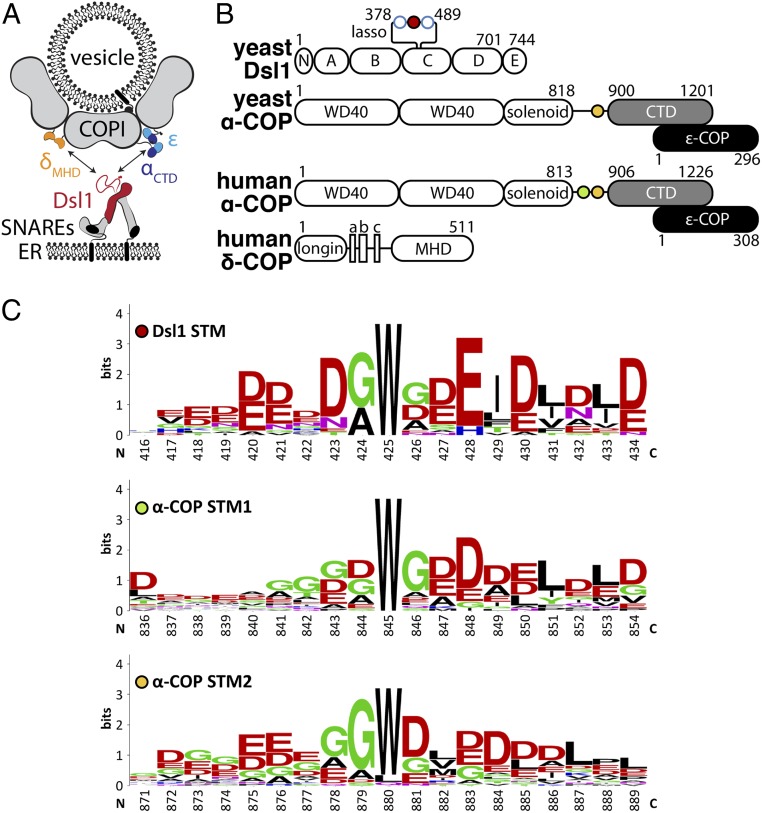
The yeast Dsl1 multisubunit tethering complex has served as a model system for studying tether–coat interactions (10, 14–16). The Dsl1 complex is essential for retrograde Golgi-to-endoplasmic reticulum (ER) traffic mediated by COPI vesicles. X-ray structures and negative-stain electron microscopy revealed that the complex consists of 2 legs connected by a flexible hinge (Fig. 1A). Near the ends of the 2 legs are binding sites for ER-anchored SNARE proteins, presumably accounting for the stable association of the Dsl1 complex with the surface of the ER (17). Near the hinge, the yeast Dsl1 complex contains a low-complexity segment (the so-called “lasso”) with closely spaced binding sites for 2 COPI subunits, α-COP and δ-COP (10, 11) (Fig. 1 A and B). These observations lead to the hypothesis that yeast COPI vesicles, originating at the Golgi, become tethered at the ER through the direct binding of the Dsl1 lasso to the vesicle coat.
Fig. 1.
STM sequences in Dsl1 and α-COP. (A) The Dsl1 tethering factor binds to ER-localized SNAREs (Bottom) and, via its Dsl1 subunit (red), to 2 domains of the COPI vesicle coat, αCTD (blue) and δMHD (orange). (B) The domain structures of the proteins in this study are shown schematically. The C-terminal domain (CTD) of α-COP (gray) forms a stable complex with ε-COP (black). Singleton tryptophan motifs (STMs) are depicted as filled circles, while open blue circles represent di-tryptophan motifs (see SI Appendix, Fig. S1A for details). (C) Sequence logos of STMs found in Dsl1 and α-COP (full aligned sequences are shown in SI Appendix, Fig. S1). Dsl1 numbering is relative to S. cerevisiae, whereas α-COP numbering is relative to Homo sapiens.
Recent cryoelectron tomography (cryo-ET) of COPI-coated liposomes, combined with crystal structures or homology models of the coat subunits, led to a quasiatomic model of the assembled mammalian COPI coat (18–20). The building block of the COPI coat is the triad, itself composed of 3 COPI heteroheptamers. Triads tile the curved membrane surface, connected by 4 different types of intertriad linkages (19). Interestingly, the 4 linkage types each involve either α-COP or δ-COP, the 2 lasso-binding COPI subunits noted above. However, whereas the overall resolution of the COPI coat structure was 9 Å, the resolution of the linkage regions was lower, between 15 and 17 Å (20). At this lower resolution, the detailed nature of the intertriad interactions could not be discerned, nor was it clear whether they are compatible with lasso binding.
Previous work had established the structural basis for the interaction between the Dsl1 lasso and δ-COP (21); we therefore set out to characterize the interaction between the Dsl1 lasso and α-COP. Our biochemical, structural, and in vivo functional experiments reveal the central role of a single tryptophan residue, embedded within the acidic lasso, in binding to the C-terminal domain (CTD) of α-COP. Furthermore, α-COP itself contains a lasso-like sequence (11). We find that this sequence mediates α-COP homo-oligomerization in vitro and appears to stabilize the COPI coat in vivo. Competition between Dsl1•α-COP and α-COP•α-COP interactions may direct the Dsl1 tethering complex toward partially uncoated COPI vesicles.
Results
Crystallization of α-COP Bound to Lasso-Like Sequences.
The Dsl1 tethering complex interacts with COPI using a long, flexible loop within its Dsl1 subunit (10, 11) (Fig. 1 A and B). This loop, known as the lasso, exhibits low sequence complexity, a predominance of acidic residues, and several conserved tryptophan residues (Fig. 1C and SI Appendix, Fig. S1A). The lasso binds directly to both δ-COP and α-COP. The interaction with δ-COP involves di-tryptophan motifs within the lasso (10, 11), which bind with ∼10 μM affinity to the μ-homology domain of δ-COP (δMHD) (21). The interaction with α-COP involves a different lasso region containing a single isolated tryptophan residue (10, 11, 22) (Fig. 1 B and C). This “singleton tryptophan motif” (STM) binds to the CTD of α-COP (αCTD), but neither the structural basis nor the physiological significance of the STM•αCTD interaction has been elucidated.
Structures of Bos taurus and Saccharomyes cerevisiae αCTD, each in a complex with the small ε-COP subunit, were reported previously (22, 23). In an attempt to characterize how αCTD recognizes the Dsl1 lasso, we conducted extensive cocrystallization trials using αCTD•ε from various yeasts together with STM-containing peptides derived from the Dsl1 lasso. Although these efforts yielded a new crystal form of S. cerevisiae αCTD•ε, and thereby a higher resolution structure (SI Appendix, Table S1), the lasso peptides in all cases either prevented crystallization or were excluded from the crystal lattice. An alternative approach was suggested by the observation that α-COP itself contains a lasso-like STM (or, in many species, 2 STMs) immediately N-terminal to the CTD (11) (Fig. 1 B and C and SI Appendix, Fig. S1B). We therefore conducted additional crystallization trials, this time using slightly longer fragments of α-COP that included an STM. We reasoned that this strategy would at a minimum guarantee that an STM was present in any crystals that might form. After screening constructs derived from several species, we were ultimately successful in visualizing the STM•CTD interaction using human α-COP.
Human α-COP contains tandem STMs, STM1 and STM2 (Fig. 1 B and C and SI Appendix, Fig. S1B). We separately crystallized 2 constructs, one containing only STM1 (αSTM1-CTD•ε) and one containing only STM2 (αSTM2-CTD•ε) (Fig. 2A). Both constructs crystallized in the same space group, with 2 α•ε monomers in the asymmetric unit (SI Appendix, Fig. S2A). The corresponding structures were determined by molecular replacement and refined to 3.0- and 3.1-Å isotropic resolution, respectively. In each case, electron density was observed for only one STM per asymmetric unit. After reprocessing the diffraction data using an anisotropic resolution cutoff of 2.7 Å (24), we were able to build 12 and 9 ordered residues in the STM1 and STM2 structures, respectively, with the latter consisting of 2 discontinuous fragments (Fig. 2A and SI Appendix, Fig. S2 C and D). The C-terminal regions of STM1 and STM2 are each sandwiched in a crystal contact (SI Appendix, Fig. S2B) and appear to be partially disordered, whereas the tryptophan and flanking residues display unambiguous electron density and are therefore the focus of our analysis below.
Fig. 2.
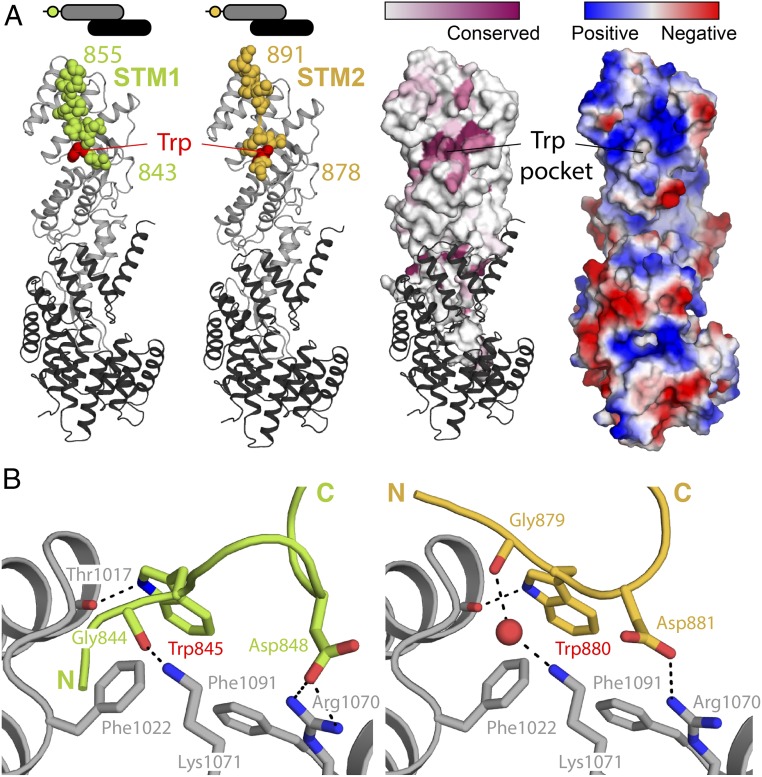
Structural basis of STM1/STM2 recognition by human αCTD•ε. (A) Crystal structures of human αSTM1-CTD•ε (Left) and αSTM2-CTD•ε (Right). The αCTD•ε heterodimer is oriented with αCTD (gray) at the Top and ε-COP (black) at the Bottom. In each case, the STM (spheres) belongs to a symmetry-related molecule (see text and Fig. 3A). On the Right, the surface of αCTD is colored by amino acid conservation or by electrostatic potential, highlighting the conserved, nonpolar cavity that accommodates the singleton tryptophan residue. (B) αCTD binds STM1 and STM2 through a combination of nonpolar interactions, hydrogen bonds, and salt bridges. The side chains of the STM1 and STM2 singleton tryptophan residues are superimposable within experimental error.
Structural Basis for αCTD•STM Binding.
STM1 and STM2 both bind to the same highly conserved site on αCTD (Fig. 2A). As expected based on the acidic nature of the STMs, the binding site displays a strongly positive electrostatic surface potential (Fig. 2A). A hydrophobic pocket accommodates the singleton tryptophan residue, either Trp845 (STM1) or Trp880 (STM2) (Fig. 2B). The main-chain carbonyl of αCTD residue Thr1017 provides a hydrogen-bonding partner for the indole nitrogen of the tryptophan side chain. Adjacent to the tryptophan-binding pocket, 2 conserved basic residues, Arg1070 and Lys1071, appear to play key roles in peptide recognition. Arg1070 forms a salt bridge with the side chain of Asp848 (STM1) or Asp881 (STM2), while Lys1071 forms a charge-stabilized hydrogen bond with the main-chain carbonyl of Gly844 (STM1) or, via a bridging water, Gly879 (STM2).
Based on the specific side-chain interactions common to both structures, we infer that Trp845/880 and Asp848/881 contribute to the binding motif recognized by αCTD. In STM2, Trp880 is adjacent to Asp881, whereas in STM1 Trp845 is separated by 2 intervening residues from Asp848. Consistently, close examination reveals that Dsl1 lasso sequences and α-COP lasso-like sequences contain 1 of the 2 consensus patterns, W[D/E] or Wxx[D/E], with x signifying any residue (SI Appendix, Fig. S1 A and B). Interestingly, variable spacing between key residues—in this case, W and D/E—is also observed for other COPI–ligand interactions, including δ-COP binding to di-tryptophan (Wx1–6W) motifs (21) and α- and β′-COP binding to di-lysine motifs (25). In addition to the conserved W(xx)[D/E] sequence, STMs in Dsl1 and α-COP are highly enriched in acidic residues on either side of the singleton tryptophan (Fig. 1C). Although their positioning relative to the tryptophan is not conserved, these acidic residues likely contribute to the stability of the interaction through electrostatic complementarity with the positive surface surrounding the tryptophan binding pocket (Fig. 2A).
STM-Mediated α-COP Oligomerization In Vitro.
A striking feature of the αSTM2-CTD•ε structure is that α-COP residues 891 and 906—that is, the last ordered residue in STM2 and the first ordered residue in CTD—lie at opposite ends of the molecule, separated by a distance of more than 100 Å. Since it is impossible for the intervening 15 residues to span this distance, the STM that we observe bound to the CTD must be contributed by a symmetry-related molecule—that is, in trans. Indeed, in our crystals, residue 891 is only 21 Å from residue 906 of the nearest symmetry mate (Fig. 3A). In contrast, STM1 is separated from CTD by 51 residues in the native sequence, which would allow it to bind either in cis or trans.
Fig. 3.
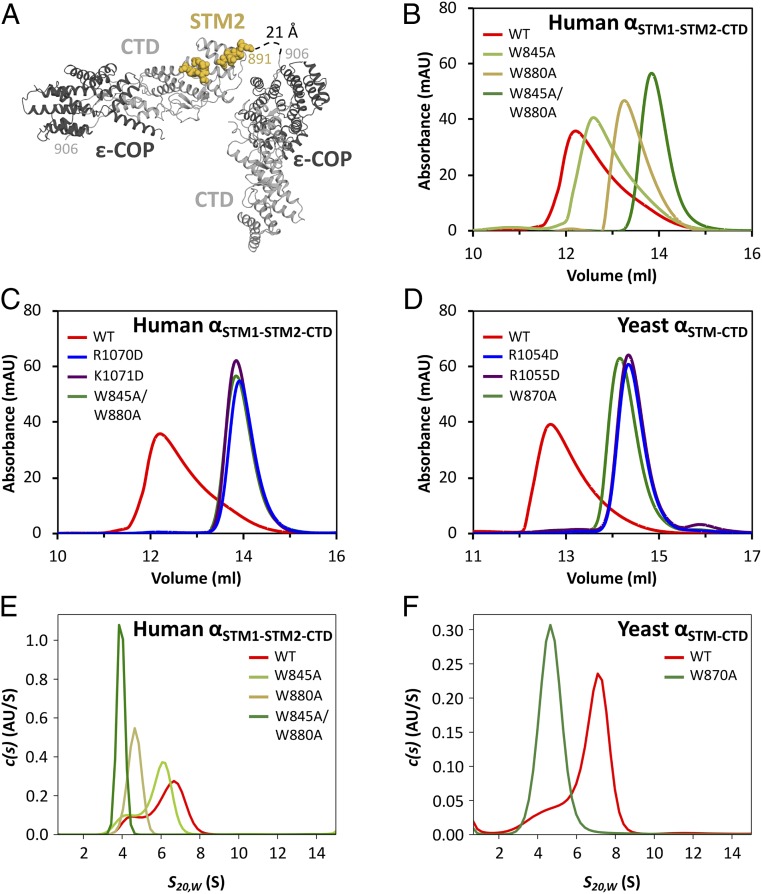
The STM•CTD interaction drives α-COP homo-oligomerization. (A) In the human αSTM2-CTD•ε crystal structure, the bound STM sequence is contributed by a symmetry-related molecule. (B) Size exclusion chromatography was used to analyze wild-type human αSTM1-STM2-CTD•ε, as well as mutants with one or both singleton tryptophans replaced with alanine. (C) Charge-reversal mutations in the STM binding site disrupt homo-oligomerization to the same extent as mutation of both singleton tryptophans. (D) Size exclusion chromatography was used to analyze wild-type yeast αSTM-CTD•ε, as well as the corresponding tryptophan replacement and charge reversal mutants. (E) Sedimentation velocity analytical ultracentrifugation of wild-type and mutant human αSTM1-STM2-CTD•ε confirmed that the STM•CTD interaction is required for homo-oligomerization. (F) Sedimentation velocity analytical ultracentrifugation of wild-type and mutant yeast αSTM-CTD•ε confirmed that the STM•CTD interaction is required for homo-oligomerization.
To test whether STM1 or STM2 mediate trans interactions among α-COP monomers in solution, we began with human α-COP containing both STMs and the CTD, again stabilized by ε-COP. This construct, denoted αSTM1-STM2-CTD•ε, displayed an asymmetric size exclusion chromatography profile consistent with an equilibrium mixture of various homo-oligomers (Fig. 3B). Disrupting the interaction between the STMs and the CTD, either by substituting both Trp845 and Trp880 with alanine (W845A/W880A) or by reversing the charges of CTD residues Arg1070 or Lys1071 (R1070D or K1071D), abolished homo-oligomerization (Fig. 3 B and C). The latter results confirm that the residues on either side of the tryptophan-binding pocket are critical for interacting with STMs. Single W845A and W880A mutants displayed an intermediate degree of oligomerization (Fig. 3B). W880A affected oligomerization more than W845A (Fig. 3B), suggesting that mammalian αCTD has a higher affinity for STM2 than for STM1.
S. cerevisiae α-COP, like many other fungal α-COPs, contains only one STM (Fig. 1B and SI Appendix, Fig. S1B), which resembles human STM2. Nonetheless, a construct containing both the STM and the CTD, yeast αSTM-CTD•ε, displayed robust homo-oligomerization (Fig. 3D). Replacing the tryptophan with alanine (W870A) abolished oligomerization. Oligomerization was also abolished by R1054D or R1055D (equivalent to human R1070D and K1071D). These results demonstrate that yeast α-COP uses the same site on its CTD to recognize the STM, and that the ability of this interaction to mediate α-COP oligomerization is broadly conserved.
We also characterized α-COP homo-oligomerization using sedimentation velocity analytical ultracentrifugation. At low micromolar concentrations—similar to those used for gel filtration—human αSTM1-STM2-CTD•ε sedimented as a broad peak, consistent with a mixture of different size species, and its s value was significantly larger than the 4.2 S expected for a monomer (Fig. 3E and SI Appendix, Fig. S3 A and G). Since the s value increased with protein concentration, we concluded that it did not represent the precise stoichiometry of a single species, but rather the relative abundance of multiple species (SI Appendix, Fig. S3 A–D, and G). Replacing either singleton tryptophan with alanine (W845A or W880A) shifted the size distribution toward monomer, while replacing both tryptophans abolished homo-oligomerization entirely. Analogous behavior was observed for yeast αSTM-CTD•ε, which upon mutation of the singleton tryptophan (W870A) shifted from a mixture of oligomeric species to pure monomer (Fig. 3F). At high concentrations, the apparent size of both the human (SI Appendix, Fig. S3 A–D) and yeast (SI Appendix, Fig. S3 E and F) proteins approached an asymptotic value of 8 S, corresponding approximately to α•ε trimers (SI Appendix, Fig. S3G). Consistent with this interpretation, when we combined mutants to cap assembly at a dimer, the resulting complex sedimented much more slowly than wild-type oligomers (SI Appendix, Fig. S4).
STM-Mediated α-COP Oligomerization In Vivo.
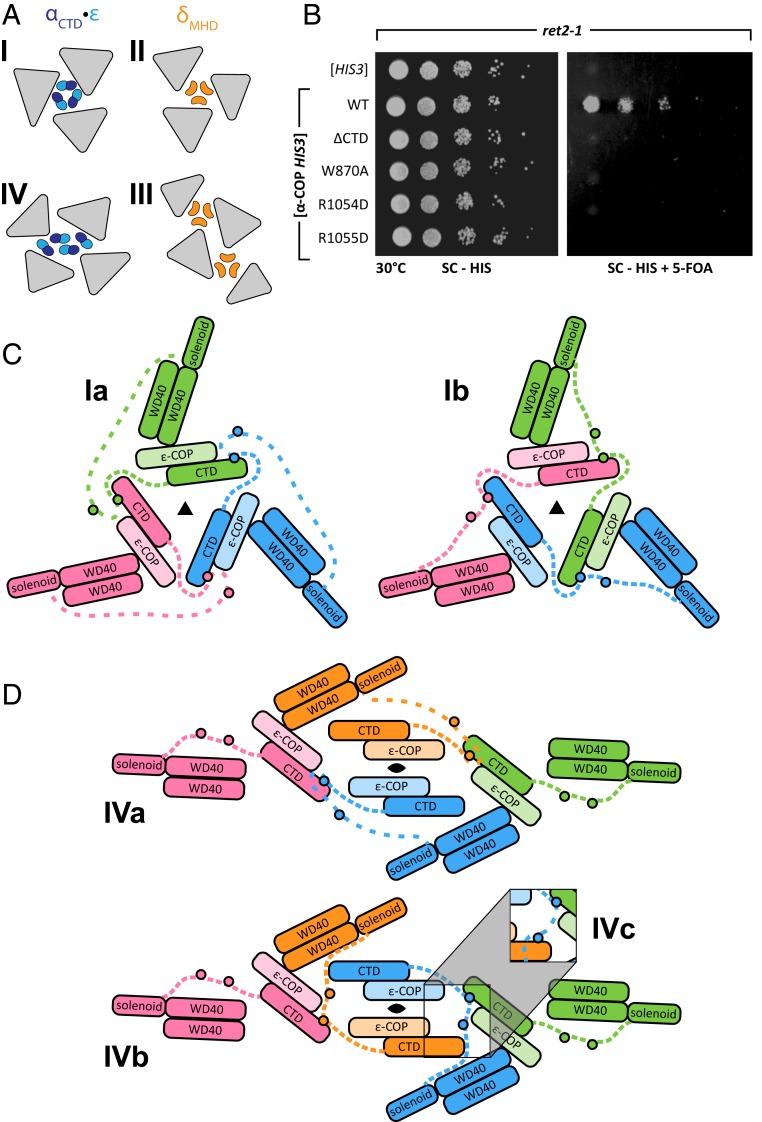
We next asked whether STM•CTD mediated interactions between α-COP monomers might play a role in COPI coat assembly in vivo. As noted above, COPI triads interact with one another via 4 different types of linkage. Two of these (linkages I and IV) involve αCTD•ε, while the other 2 (linkages II and III) involve δMHD (Fig. 4A) (19, 20). Surprisingly, yeast lacking ε-COP, lacking δMHD, or even lacking both are viable (26). This could suggest either that none of the linkages are essential (20) or, alternatively, that linkages I and IV are sufficient for viability and can form in the absence of ε-COP.
Fig. 4.
The role of STM•CTD interactions in COPI linkage formation. (A) Four linkages are observed in the COPI reconstruction (20). Linkages I and II join 3 triads (gray triangles) and linkages III and IV join 4 triads. Trimers of δMHD (orange) are found in linkages II and III, and trimers or tetramers of αCTD (dark blue) and ε (cyan) are found in linkages I and IV, respectively. (B) In ret2-1 yeast, α-COP mutations that disrupt binding between the STM and CTD cause lethality upon 5-FOA counterselection against wild-type α-COP (Right). Maintenance of the α-COP allele plasmid, which confers histidine prototrophy, is confirmed by the plating control (Left). ret2-1 yeast produce a truncated form of δ-COP lacking a major portion of the μ-homology domain and are presumed to be unable to form linkages II and III. (C) Schematic representation of linkage I. Within each α-COP (colored in either red, green, or blue), an intrinsically disordered linker (dashed lines) containing 2 STMs (small circle) connects αCTD to the remainder of the protein. Models Ia and Ib assume that each αCTD binds to an STM2. Model Ib represents the minimal extension of the solenoid–STM2 connector. (D) The 2-fold symmetric linkage IV connects 4 triads. Two types of αCTD•ε are present: peripheral (red and green) and central (orange and blue). Model IVa (Top) assigns the central αCTD•ε units to the closest triad, whereas model IVb (Bottom) swaps central αCTD•ε units to minimize the distance spanned by the solenoid–STM2 connector. In models IVa and IVb, only pairs of triads are connected by STM•CTD binding. Model IVc (Inset), which includes STM1 binding, connects all 4 triads.
To test these ideas, we used the yeast strain ret2-1, which carries a nonsense mutation resulting in a truncated δ-COP μ-homology domain (26, 27) and should therefore be unable to form either type II or type III linkages. A plasmid shuffling strategy enabled us to replace wild-type α-COP with variants that abolished homo-oligomerization in vitro, including the STM mutant W870A and the CTD mutants R1054D and R1055D. While none of these mutations caused a growth defect in otherwise wild-type yeast (SI Appendix, Fig. S5A), all 3 were lethal in the ret2-1 mutant background (Fig. 4B and SI Appendix, Fig. S5B). These results imply that αCTD and δMHD stabilize functionally redundant linkages. Moreover, they strongly suggest that the stability of linkages I and IV depends on the interaction in trans between the STM and CTD of α-COP.
STM-Mediated α-COP Oligomerization and COPI Coat Structure.
Is it plausible, given our current structural understanding of the mammalian COPI coat (19, 20), that linkages I and IV are mediated by STM•CTD binding between α-COP monomers? The key to answering this question is to consider the path taken by the α-COP segment connecting the α-solenoid and the CTD (Fig. 1B). This linker, which contains the STM(s), anchors αCTD•ε to the body of the triad. Because of its presumed flexibility, the linker is missing from the quasiatomic models of linkages I and IV that were proposed by Dodonova et al. (20). Furthermore, the limited resolution of the cryo-ET density in the linkage regions (∼15 Å), while sufficient for positioning the αCTD•ε units, cannot reveal whether they are bound to an STM. Nonetheless, as explained below, the added constraint that STM•CTD interactions are critical for coat stability affords insights into the linker paths and thereby the architectures of linkages I and IV.
Linkage I connects 3 triads and displays approximately 3-fold symmetry (20). The core of linkage I is formed by 3 αCTD•ε units arranged head-to-tail with their STM binding sites facing outward. To model the paths of the linkers, we began by assuming that each αCTD•ε binds to an STM2; STM2 was chosen because of its universal conservation and greater role in homo-oligomerization. Without altering the quasiatomic model of Dodonova et al. (20), it was possible to route each linker such that STM2 bound to the αCTD•ε unit of the counterclockwise-related triad (Fig. 4C, model Ia). An alternative model (model Ib) was even better, in that the distance spanned by the solenoid–STM2 connector was reduced from 1.9 to 1.0 Å per residue, bringing it in line with other flexible linkers within the COPI coat (SI Appendix, Table S2). Model Ib is related to model Ia by a cyclic exchange of αCTD•ε units in a clockwise direction around the approximate 3-fold axis (Fig. 4C). A third model (not illustrated in Fig. 4C), involving a counterclockwise exchange of αCTD•ε units, stretches the connectors even further than model Ia. Overall, although unable to rule out the alternatives entirely, we strongly favor model Ib.
Linkage IV is more complicated, connecting 4 triads arranged with 2-fold symmetry. This gives rise to nonequivalent pairs of central and peripheral triads, linkers, and αCTD•ε units (Fig. 4 A and D). The 4 αCTD•ε units are arranged with the STM binding sites facing inward. The peripheral STMs (red and green in Fig. 4D) are too far away to reach any of the 4 αCTD•ε units. Focusing therefore on the 2 central linkers (blue and orange in Fig. 4D), 2 possible paths can be considered (models IVa and IVb). In both models, the central STM2s bind to a peripheral αCTD•ε unit. Model IVb, which features an exchange of central αCTD•ε units, minimizes the distance traversed by the solenoid–STM2 connector (SI Appendix, Table S2). Model IVb is also attractive because, in organisms containing 2 STMs, linkage IV may be further stabilized by STM1•αCTD•ε binding (Fig. 4D, model IVc; SI Appendix, Table S2). Taken together, our analyses suggest that STM•CTD interactions, likely accompanied by the exchange of αCTD•ε units among triads (models Ib and IVb/c), are responsible for stabilizing linkages I and IV and, thereby, the COPI coat.
Vesicle Tethering via the Dsl1 Lasso STM.
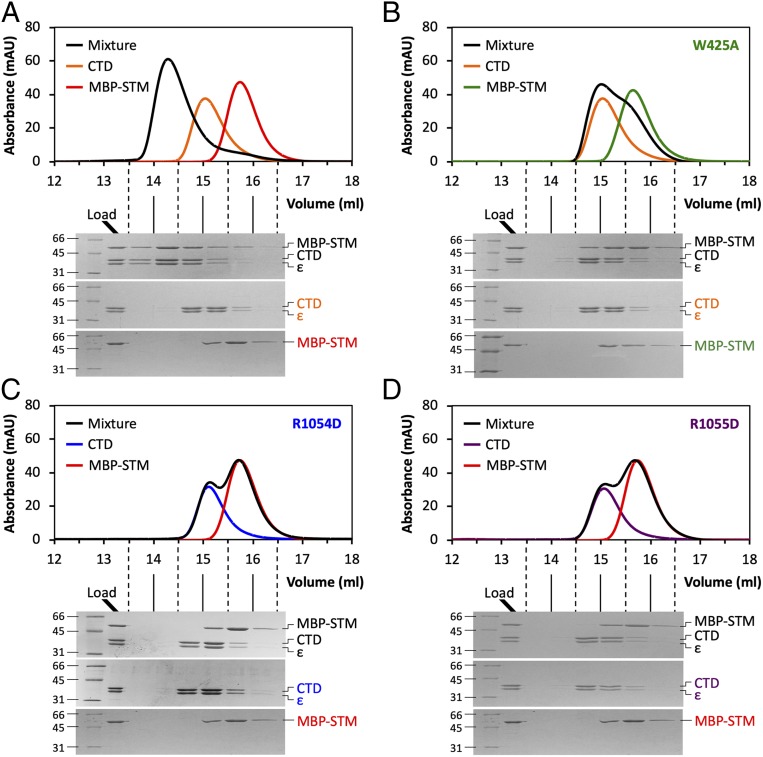
The results presented so far establish that STM sequences within the α-COP linker drive oligomerization by binding in trans to a conserved site on αCTD. We next sought to confirm that the Dsl1 lasso, via its STM, binds to this same site. MBP- and GST-Dsl1STM, fusion proteins containing a 31-residue segment of the S. cerevisiae lasso centered on the singleton tryptophan (Trp425), both formed complexes with S. cerevisiae αCTD•ε that were stable during size exclusion chromatography (Fig. 5A and ref. 22). The equilibrium dissociation constant for MBP-Dsl1STM:αCTD•ε binding, as judged by isothermal titration calorimetry, was ∼2 μM (SI Appendix, Fig. S6A). The analogous yeast MBP-αSTM fusion protein bound to αCTD•ε with comparable affinity (∼4 μM; SI Appendix, Fig. S6B). The interaction between MBP-Dsl1STM and αCTD•ε was abolished by replacing the singleton tryptophan with alanine (W425A; Fig. 5B) and by the CTD charge reversal mutations R1054D and R1055D (Fig. 5 C and D). Thus, the same conserved site on αCTD binds with similar affinity to STMs present in the Dsl1 lasso and in α-COP itself. Although we were unsuccessful in obtaining a crystal structure of the Dsl1 lasso bound to αCTD•ε, we note that like human STM1 it contains the motif Wxx[D/E] (Fig. 1C). We speculate, therefore, that the bound conformation of the lasso may resemble that of STM1.
Fig. 5.
The Dsl1 lasso binds to the same conserved site on αCTD. (A) Size exclusion chromatography and SDS/PAGE demonstrate that yeast αCTD•ε forms a stable 1:1 complex with MBP-Dsl1410–440. In separate experiments, αCTD•ε, MBP-Dsl1410–440, or an equimolar mixture of the 2 (preincubated 1 h at 4 °C) was loaded onto the column. (B) Complex formation between yeast αCTD•ε and MBP-Dsl1410–440W425A is greatly diminished, as the elution profile of the mixture resembles the sum of the individual profiles. The data presented for αCTD•ε alone are identical to those presented in panel (A). (C) Complex formation between αCTDR1054D•ε and MBP-Dsl1410–440 is greatly diminished, as the elution profile of the mixture is indistinguishable from the sum of the individual profiles. The data presented for MBP-Dsl1410–440 alone are identical to those presented in panel (A). (D) As in C, but for αCTDR1055D•ε.
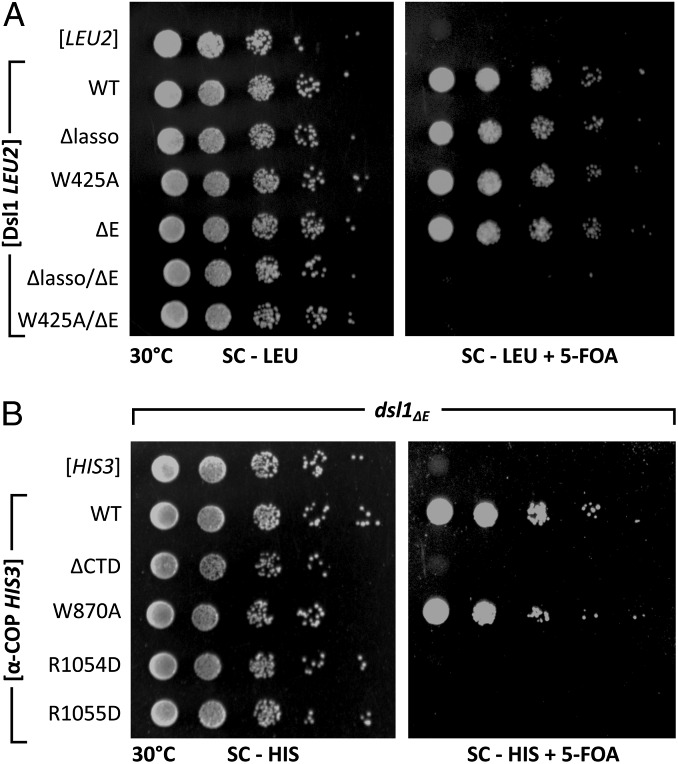
To test the physiological importance of the lasso•αCTD interaction, we again turned to plasmid shuffling experiments in yeast. To render the lasso essential, we disabled a genetically redundant mechanism by deleting the C-terminal E domain of Dsl1 (21, 28) (Fig. 1B). The precise role of the E domain is not known, although recent findings suggest that it might involve Rab binding (29). As reported previously, replacing wild-type Dsl1 with Dsl1Δlasso (an internal deletion of the entire lasso) or Dsl1ΔE (a truncation removing the entire E domain) did not cause discernable growth defects, but replacing wild-type Dsl1 with Dsl1Δlasso/ΔE was lethal (21, 28) (Fig. 6A). Strikingly, in the Dsl1ΔE sensitized background, the single tryptophan substitution W425A was also lethal (Fig. 6A and SI Appendix, Fig. S7A). Likewise, α-COP mutations that disrupted lasso binding in vitro, ranging from αCTD deletion to single residue substitutions (R1054D or R1055D), were lethal in the Dsl1ΔE background (Fig. 6B and SI Appendix, Fig. S7B). As expected for mutations abolishing a binding site for both vesicle tethering and for coat stability, α-COP R1054D and R1055D mutations were synthetic lethal with both Dsl1ΔE and ret2-1. The separability of these 2 functions, however, is demonstrated by the fact that the α-COP W870A mutation shows no synthetic defect in combination with Dsl1ΔE (Fig. 6B and SI Appendix, Fig. S7B). Thus, in a suitably sensitized background, the lasso plays an essential role requiring binding between the Dsl1 STM and αCTD. Taken together, these findings suggest that the ability of Dsl1ΔE to bind to αCTD is required for COPI-dependent transport to the ER.
Fig. 6.
Binding of the Dsl1 lasso to αCTD is essential in sensitized yeast. (A) Alleles of Dsl1, provided on a leucine prototrophy plasmid, were assayed for viability upon loss of a wild-type Dsl1 plasmid (Right). A plating control was included to quantify leucine plasmid maintenance (Left). (B) Alleles of α-COP, provided on a histidine prototrophy plasmid, were assayed for viability in combination with the dsl1ΔE allele (Right). A plating control was included to quantify histidine plasmid maintenance (Left).
Discussion
Unlike COPII and clathrin-coated vesicles, which feature inner adaptor and outer cage layers, COPI vesicles have a single-layered coat created by the en bloc recruitment of COPI heteroheptamers onto Golgi membranes (2, 3). Three heteroheptamers form a triad, the roughly triangular assembly that tiles the vesicle surface. Triads interact with one another, in sets of 3 or 4, via linkages involving either αCTD•ε (linkages I and IV) or δMHD (linkages II and III) (Fig. 4A). Here, we provide evidence that linkages I and IV are mediated by intermolecular interactions between the linker region and the CTD of α-COP. These α-COP•α-COP interactions are driven by the ability of αCTD to bind flexible, acidic segments containing singleton tryptophan residues.
Our crystal structures revealed that STMs in the α-COP linker bind to the most highly conserved region on the surface of αCTD. The STM-binding site features a complementary pocket to accommodate the singleton tryptophan, surrounded by a large number of basic residues, 2 of which make conserved interactions with residues flanking the tryptophan. Binding could be eliminated by changing the singleton tryptophan to alanine or by making charge-reversal mutations in the key basic residues. Either strategy was effective in abolishing α-COP oligomerization in vitro. The charge-reversal mutations, however, also abolished the ability of αCTD to bind the Dsl1 lasso STM (see below). Thus, tryptophan-to-alanine mutants afford the most selective means of eliminating a given STM interaction in vivo. Strikingly, single tryptophan-to-alanine mutations in the α-COP linker or the Dsl1 lasso displayed the same synthetic lethalities as deleting the entire αCTD or lasso, respectively.
Although cryo-ET places αCTD•ε at the center of linkages I and IV, ε-COP is not an essential gene in yeast (30). Similarly, δMHD is located at the center of linkages II and III but can be deleted without apparent consequence (26). Most surprisingly of all, yeast were viable in the absence of both ε-COP and δMHD (26), a finding that cast the functional significance of the 4 COPI linkages into doubt. Here, we show that replacing the singleton tryptophan of α-COP with alanine is lethal in yeast that lack a functional δMHD. We conclude that linkages I and IV, while they do not require ε-COP, do require STM•CTD interactions between α-COP monomers. Moreover, despite some functional redundancy, the COPI linkages play an essential role in yeast, presumably by stabilizing the COPI coat.
Our results argue that COPI vesicles, like COPII and clathrin-coated vesicles, are stabilized by lateral interactions among the elements of the coat. Nonetheless, these lateral interactions are of strikingly different natures. Clathrin cages are stabilized by the intertwining of leg segments (31), whereas COPII cages are stabilized by the interaction of rod-like Sec13•31 heterotetramers at 4-way vertices (32). COPI coats, by contrast, display pleomorphic morphologies in vitro and in vivo, with lateral interactions of 4 different types (Fig. 4A) (18–20, 33). Linkages II and III appear to be stabilized by the formation of, respectively, 1 or 2 homotrimers of δMHD units (Fig. 4A), although the detailed structural basis for formation of these homotrimers has not yet been elucidated. Linkages I and IV, by contrast, display 2 fundamentally different geometries, each involving αCTD•ε. The αCTD•ε units used in the formation of linkages I and IV are connected to the body of the COPI heteroheptamer by long, intrinsically disordered linkers. These linkers, by binding adjacent αCTD•ε units, play a critical and to our knowledge unprecedented role in stabilizing linkages I and IV. The short length of the STM2–CTD connector (SI Appendix, Table S2) serves to hold the αCTD•ε units in close proximity. This construction strategy allows for some flexibility in the linkages, which could be especially important for vesicles tiled by invariant triads. Moreover, it allows the formation of 2 different linkage geometries from a single type of intermolecular interaction (Fig. 4).
The COPI coat, while it helps drive vesicle budding and serves as a target for Dsl1-mediated tethering, represents a barrier to SNARE assembly and membrane fusion. Intriguingly, the Dsl1 lasso recognizes precisely those COPI domains—namely, δMHD and αCTD—that are most centrally involved in stabilizing higher-order COPI structure. To better understand this apparent connection between coat assembly and vesicle tethering, we focused on the second of these interactions. In fact, the lasso•αCTD interaction appears to be the more important of the 2 since, unlike the lasso•δMHD interaction (21), its elimination in sensitized (dsl1ΔE) yeast is lethal. As we have shown above, the lasso STM and the α-COP linker STM compete for the same site on αCTD. We therefore propose that tethering is directed toward “disengaged” αCTD•ε units; that is, those that are not mediating linkages I/IV. A disengaged αCTD•ε unit is of course free to bind to the lasso because its STM-binding site is unoccupied. A disengaged αCTD•ε unit is also likely to be flexibly connected to the remainder of the coat—by means of the intrinsically disordered linker—instead of being confined to the center of a linkage, enabling it to embark on excursions away from the vesicle membrane and explore a larger volume of the cytosol. A fraction of the αCTD•ε units will be disengaged in a fully assembled coat containing a mixture of linkages I–IV. Proportionally more will be disengaged in a partially disassembled coat. In particular, uncoated regions(s) will be ringed by disengaged αCTD•ε units. Tethering partially coated vesicles by their coat edges would seem to be advantageous in promoting SNARE assembly and fusion, and we propose that it is the main mode of Dsl1 action.
Relatively little is known about COPI vesicle uncoating in vivo, or about its temporal and mechanistic relationship to vesicle tethering. Our work suggests that partially coated COPI vesicles, which appear to be common in vivo (33), are the preferred targets of Dsl1 lasso binding. Nevertheless, a great many questions remain to be answered. Does tethering affect uncoating, or do tethers simply monitor the uncoating status of the vesicle? Both COPI and the Dsl1 lasso possess multiple binding sites for the other—how does this multivalency modulate vesicle tethering? Does Dsl1 also capture soluble COPI, and if so, how does this presumably nonproductive interaction compete with recognition of vesicles? What is the fate of a fully uncoated COPI vesicle? These and other questions await future investigation.
Materials and Methods
Protein Expression and Purification.
A DNA fragment corresponding to the yeast α-COP STM and CTD (residues 860–1201) was amplified from S. cerevisiae S288C genomic DNA by PCR and cloned directionally into pETDuet-1 coexpression vector (Novagen) using the NdeI/XhoI restriction sites. The corresponding full-length yeast ε-COP was then amplified and inserted using the BamHI/SacI restriction sites in-frame with the N-terminal His6 tag. The human heterodimeric complex was cloned using a similar strategy, amplifying from T47D cDNA and inserting a DNA fragment corresponding to the α-COP CTD plus STM1 and STM2 (residues 835–1226) into the NdeI/XhoI restriction sites and full-length ε-COP into the BamHI/NotI restriction sites. Dsl1 and α-COP peptides were cloned using the BamHI/NotI restriction sites into a modified pQLink vector backbone containing an N-terminal His7-MBP tag (34). Point mutations and deletions were generated using a modified QuikChange protocol (35).
Expression plasmids were transformed into chemically competent BL21-Codon Plus (DE3)-RIL cells (Agilent) for all yeast constructs and, due to toxicity, C43 (DE3) (Lucigen) cells for human constructs. Protein expression was carried out in high-salt LB media (Sigma), inducing at OD600 ∼ 0.6 with 1 mM isopropyl β-D-1-thiogalactopyranoside, and 18 h of continued shaking at 15 °C. Cells were pelleted and resuspended in lysis buffer containing HBS (20 mM Hepes, pH 7.5, 150 mM NaCl) supplemented with 5 mM β-mercaptoethanol, 1 mM phenymethylsulfonyl fluoride, and 10 μg/mL DNase I. The cell suspension was lysed using a cell disrupter (Avestin) and clarified by centrifugation for 30 min at 37,000 × g. The lysate was then bound to His60 SuperFlow Ni-NTA resin (Clontech) and eluted using an imidazole step gradient. The eluate was applied to an HR MonoQ 10/100 column (Pharmacia) equilibrated with a buffer containing HBS supplemented with 1 mM reduced DTT and eluted with a linear salt gradient (150–500 mM NaCl). Peak fractions were concentrated and loaded onto a Superdex 200 16/600 column (GE Healthcare) equilibrated with HBS supplemented with 1 mM DTT. Fractions of sufficient purity were pooled, concentrated to between 10 and 40 mg/mL, and flash-cooled for storage at −80 °C.
Crystallization and Structure Determination.
For crystallization of human αSTM2-CTD•ε, 1 μL of α-COP870–1226•His6-ε-COP at 8 mg/mL formulated in a buffer containing HBS supplemented with 1 mM DTT was mixed with 1 μL of well solution containing 100 mM sodium citrate pH 6.0, 1% (vol/vol) Tacsimate, pH 5.0 (Hampton), 9% (wt/vol) polyethylene glycol 3350, and 5 mM DTT in hanging-drop format. Crystals appeared after 1 d as cubes 0.1 mm in size. Crystals were cryoprotected by transferring to a solution containing 10 mM Hepes, pH 7.5; 50 mM sodium citrate, pH 5.75; 75 mM NaCl; 0.5% (vol/vol) Tacsimate, pH 5.0; 12% (wt/vol) polyethylene glycol 3350; and 30% (vol/vol) ethylene glycol; and flash-cooled in liquid nitrogen. Data were collected at the NSLS-II AMX beamline.
Crystallization of human αSTM1-CTD•ε was similar, with the following minor changes. Human α-COP835–1226,∆854–887•His6-ε-COP was formulated at 6 mg/mL and crystallized in well solution containing 100 mM sodium citrate, pH 6.0; 0.5% (vol/vol) Tacsimate, pH 5.0; 11% (wt/vol) polyethylene glycol 3350; and 5 mM DTT. Crystals were cryoprotected in a solution containing 10 mM Hepes, pH 7.5; 50 mM sodium citrate, pH 6.0; 75 mM NaCl; 0.25% (vol/vol) Tacsimate, pH 5.0; 13% (wt/vol) polyethylene glycol 3350; and 30% (vol/vol) ethylene glycol. Data were collected at the NSLS-II FMX beamline.
Data were indexed, integrated, and scaled using XDS (36). Initial phases were determined by molecular replacement from the bovine heterodimer (23) (PDB ID code 3MKR) using Phaser (37). Structural refinement was then performed through iterative cycles of de novo model building in COOT (38) and refinement in Phenix.refine (39). To improve interpretability of the maps during model building, particularly in the peptide binding site, an anisotropic cutoff was applied to the data using the StarANISO server (24). Both isotropic and anisotropic datasets were refined.
In the process of screening for cocrystallization conditions for yeast αCTD•ε and Dsl1 peptides, a previously unreported, higher resolution yeast αCTD•ε crystal form was identified. One microliter of yeast α-COP900–1201•His6-ε-COP at 8 mg/mL formulated in a buffer containing HBS supplemented with 1 mM DTT was mixed with 1 μL of well solution containing 100 mM Tris, pH 8.5; 200 mM Li2SO4; 15% (wt/vol) polyethylene glycol 3350; and 25% (vol/vol) glycerol in hanging-drop format. Crystals appeared after 1 mo as long rod clusters. Single rods were dissociated from the cluster and cryoprotected in a solution containing 100 mM Tris, pH 8.5; 200 mM Li2SO4; 23% (wt/vol) polyethylene glycol 3350; and 25% (vol/vol) glycerol; and flash-frozen in liquid nitrogen. Data were collected at the Cornell High Energy Synchrotron Source (CHESS) F1 beamline.
Data were processed by the HKL suite (40) and phased using the previously published yeast heterodimer structure (22) (PDB ID code 3MV2) as a search model for molecular replacement. Refinement was performed as above.
Gel Filtration Assays.
Proteins were combined in a total volume of 200 μL and final concentrations of 10 μM (for binding assays) or 20 μM (for homo-oligomerization assays). Mixtures were incubated for 1 h at 4 °C, then loaded onto a Superdex 200 Increase 10/300 column (GE Healthcare) preequilibrated with HBS supplemented with 1 mM DTT. Complex formation was confirmed by analysis of eluted proteins using SDS/PAGE followed by Coomassie Blue staining.
Analytical Ultracentrifugation.
Proteins were dialyzed against 4 L of buffer containing HBS supplemented with 1 mM DTT using 10-kDa molecular weight cutoff dialysis cassettes (Pierce). Except where indicated, proteins were assayed between 6 and 9 μM. Sedimentation velocity experiments were performed using a Beckman model Optima XL-A instrument equipped with a 4-place An-60 Ti rotor. SV-AUC runs used 2-channel Epon, charcoal-filled centerpieces containing 435-µL samples and 450-µL buffer references. Sedimentation velocity boundary movement was measured at a speed of 42,000 rpm at 20 °C using a step size of 0.003 cm, a delay time of 0 s, and a total of 70 scans. Samples were monitored at 280 nm with a requirement for the starting absorbance to be between 0.3 and 1.0. Temperature-corrected partial specific volumes, densities, and viscosities were calculated using Sednterp (version 1.08) (41). Model-independent analysis for determining heterogeneity of samples was carried out using Sedfit (version 14.4) (42) using continuous sedimentation distribution, c(s), with maximum entropy regularization. The s and D parameters required to solve the Lamm equation were calculated with the approximation that all species in solution for a given sample have similar signal-average frictional coefficients, f/f0. The distributions were plotted using the Gussi interface (version 1.0.3) implemented in Sedfit (version 14.4) (42, 43). Theoretical s values were calculated according to ref. 44.
Isothermal Titration Calorimetry.
Yeast proteins were exchanged into HBS supplemented with 1 mM DTT using a Bio-Spin 6 desalting column (Bio-Rad), and then back-diluted to the appropriate concentration. MPB-Dsl1410–440 or MBP-α-COP855–885 was loaded into the titration syringe, while α-COP900–1201•His6-ε-COP heterodimer was loaded into the sample cell. Experiments were performed using a MicroCal PEAQ-ITC (Malvern) at 25 °C. Data were analyzed using the MicroCal PEAQ-ITC Analysis Software package (Malvern).
Yeast Methods.
A diploid strain heterozygous for the deletion of α-COP, marked with the KanMX cassette, was purchased from the essential gene knockout collection (Dharmacon). Full-length α-COP, flanked by 500 bases upstream of the coding sequence, was cloned from S288C gDNA and inserted into the pRS416 yeast shuttle vector (45) using the BamHI/NotI restriction sites. Diploids were transformed with the URA3 α-COP covering vector (46), and then sporulated in sporulation media containing 0.3% (wt/vol) potassium acetate and 0.02% (wt/vol) raffinose at 23 °C for 7 d. After dissection of tetrads, haploids containing a chromosomal deletion for α-COP covered by the URA3 vector were screened using genetic markers. Haploids were mated to a strain bearing the δ-COP W385* mutation (also known as ret2-1) or Dsl1 H702* mutation (also known as Dsl1∆E) marked with NatMX (28) and sporulated as above. Double-mutant progeny were identified after dissection using markers for antibiotic resistance and uracil prototrophy. The Dsl1 deletion strain with covering plasmid has been previously described (15).
For viability experiments, strains were transformed with pRS413 plasmids containing the appropriate alleles of α-COP or pRS415 plasmids containing the appropriate alleles of Dsl1. Transformants were then grown in synthetic complete leucine or histidine drop-out liquid culture overnight, back-diluted to an OD of 0.2, serially diluted in 10-fold increments, then spotted onto histidine or leucine drop-out plates respectively, supplemented with 0.1% (wt/vol) 5-fluoroorotic acid (GoldBio), where indicated.
Modeling of COPI Linkage Structures.
Human αCTD•ε•STM2 was fit using least-squares regression to αCTD of linkage I (ID code 5NZT, chain A) and both unique αCTDs of linkage IV (ID code 5NZV, chains B and H) (20) using Pymol, and through-space distances were measured between sequences as listed (SI Appendix, Table S2). Alternative positions for the α-COP peripheral solenoid in linkage IV were determined by fitting the COPI triad structure (ID code 5NZR) to chain H.
Figures.
Sequence alignments were performed using ClustalW (47) and rendered using JalView (48). Sequence logos were created using WebLogo (49). Structures were rendered in PyMol (Schrodinger), with imported conservation scores from ConSurf (50), where applicable. Cartoons and figure assemblies were created in Illustrator (Adobe).
Data Availability.
Atomic coordinates and structure factors can be accessed using PDB ID codes 6TZT, 6U3V, and 6U3W (51–53).
Supplementary Material
Acknowledgments
We thank Gena Whitney, Christopher Teng, and Ajibike Lapite for the kind gifts of cDNA and plasmids; and Philip Jeffrey, Angela Chan, Liz Gavis, Alexei Korennykh, Sabine Petry, Mark Rose, Venu Vandavasi, Ginger Zakian, and members of the F.M.H. laboratory for helpful advice and discussion. The Princeton Biophysics and Macromolecular Crystallography core facilities provided essential assistance with isothermal titration calorimetry and X-ray crystallography, respectively. This work was supported by National Science Foundation (NSF) Grant MCB-1243656 (R.F.) and by National Institutes of Health (NIH) Grants T32GM007388 (S.M.T.), F31GM12676 (S.M.T.), and R01GM071574 (F.M.H.). This research used the AMX and FMX beamlines of the National Synchrotron Light Source II, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract DE-SC0012704. The Life Science Biomedical Technology Research Resource, which supports AMX and FMX, is primarily supported by the NIH, National Institute of General Medical Sciences (NIGMS), through a Biomedical Technology Research Resource P41 grant (P41GM111244), and by the DOE Office of Biological and Environmental Research (KP1605010). This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the NSF and the NIH/NIGMS under NSF Award DMR-1332208, using the Macromolecular Diffraction at the CHESS (MacCHESS) facility, which is supported by Award GM-103485 from the NIH, through its NIGMS.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6TZT, 6U3V, and 6U3W).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909697116/-/DCSupplemental.
References
- 1.Jackson L. P., Kümmel D., Reinisch K. M., Owen D. J., Structures and mechanisms of vesicle coat components and multisubunit tethering complexes. Curr. Opin. Cell Biol. 24, 475–483 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Béthune J., Wieland F. T., Assembly of COPI and COPII vesicular coat proteins on membranes. Annu. Rev. Biophys. 47, 63–83 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Arakel E. C., Schwappach B., Formation of COPI-coated vesicles at a glance. J. Cell Sci. 131, jcs218347 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Südhof T. C., Rothman J. E., Membrane fusion: Grappling with SNARE and SM proteins. Science 323, 474–477 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu I. M., Hughson F. M., Tethering factors as organizers of intracellular vesicular traffic. Annu. Rev. Cell Dev. Biol. 26, 137–156 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Hong W., Lev S., Tethering the assembly of SNARE complexes. Trends Cell Biol. 24, 35–43 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Ungermann C., Kümmel D., Structure of membrane tethers and their role in fusion. Traffic 20, 479–490 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Cai H., Reinisch K., Ferro-Novick S., Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12, 671–682 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Stenmark H., Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Andag U., Schmitt H. D., Dsl1p, an essential component of the Golgi-endoplasmic reticulum retrieval system in yeast, uses the same sequence motif to interact with different subunits of the COPI vesicle coat. J. Biol. Chem. 278, 51722–51734 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Zink S., Wenzel D., Wurm C. A., Schmitt H. D., A link between ER tethering and COP-I vesicle uncoating. Dev. Cell 17, 403–416 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Suvorova E. S., Duden R., Lupashin V. V., The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157, 631–643 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zolov S. N., Lupashin V. V., Cog3p depletion blocks vesicle-mediated Golgi retrograde trafficking in HeLa cells. J. Cell Biol. 168, 747–759 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi A., Ren Y., Jeffrey P. D., Hughson F. M., Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex. Nat. Struct. Mol. Biol. 16, 114–123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren Y., et al. , A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell 139, 1119–1129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeter S., Beckmann S., Schmitt H. D., Coat/tether interactions-exception or rule? Front. Cell Dev. Biol. 4, 44 (2016). Erratum in: Front. Cell Dev. Biol.4, 90 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraynack B. A., et al. , Dsl1p, Tip20p, and the novel Dsl3(Sec39) protein are required for the stability of the Q/t-SNARE complex at the endoplasmic reticulum in yeast. Mol. Biol. Cell 16, 3963–3977 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faini M., et al. , The structures of COPI-coated vesicles reveal alternate coatomer conformations and interactions. Science 336, 1451–1454 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Dodonova S. O., et al. , VESICULAR TRANSPORT. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349, 195–198 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Dodonova S. O., et al. , 9Å structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments. eLife 6, e26691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suckling R. J., et al. , Structural basis for the binding of tryptophan-based motifs by δ-COP. Proc. Natl. Acad. Sci. U.S.A. 112, 14242–14247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsia K. C., Hoelz A., Crystal structure of alpha-COP in complex with epsilon-COP provides insight into the architecture of the COPI vesicular coat. Proc. Natl. Acad. Sci. U.S.A. 107, 11271–11276 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee C., Goldberg J., Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell 142, 123–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tickle I. J., et al. , STARANISO (Global Phasing Ltd., Cambridge, United Kingdom, 2018). http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi. Accessed 27 July 2019.
- 25.Ma W., Goldberg J., Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J. 32, 926–937 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arakel E. C., Richter K. P., Clancy A., Schwappach B., δ-COP contains a helix C-terminal to its longin domain key to COPI dynamics and function. Proc. Natl. Acad. Sci. U.S.A. 113, 6916–6921 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cosson P., Démollière C., Hennecke S., Duden R., Letourneur F., Delta- and zeta-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 15, 1792–1798 (1996). [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers J. V., McMahon C., Baryshnikova A., Hughson F. M., Rose M. D., ER-associated retrograde SNAREs and the Dsl1 complex mediate an alternative, Sey1p-independent homotypic ER fusion pathway. Mol. Biol. Cell 25, 3401–3412 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu D., et al. , Rab18 promotes lipid droplet (LD) growth by tethering the ER to LDs through SNARE and NRZ interactions. J. Cell Biol. 217, 975–995 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duden R., Kajikawa L., Wuestehube L., Schekman R., epsilon-COP is a structural component of coatomer that functions to stabilize alpha-COP. EMBO J. 17, 985–995 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fotin A., et al. , Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432, 573–579 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Stagg S. M., et al. , Structural basis for cargo regulation of COPII coat assembly. Cell 134, 474–484 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bykov Y. S., et al. , The structure of the COPI coat determined within the cell. eLife 6, e32493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker R. W., et al. , A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science 349, 1111–1114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H., Naismith J. H., An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Schuck P., Zhao H., Editorial for the special issue of methods “modern analytical ultracentrifugation.” Methods 54, 1–3 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Dam J., Velikovsky C. A., Mariuzza R. A., Urbanke C., Schuck P., Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s). Biophys. J. 89, 619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stafford W. F., 3rd, Boundary analysis in sedimentation transport experiments: A procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 203, 295–301 (1992). [DOI] [PubMed] [Google Scholar]
- 44.Schuck P., Zhao H., Sedimentation Velocity Analytical Ultracentrifugation: Interacting Systems (CRC Press, Taylor and Francis Group, Boca Raton, FL, 2018). [Google Scholar]
- 45.Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P., Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Gietz R. D., Schiestl R. H., High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Sievers F., et al. , Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., Barton G. J., Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashkenazy H., et al. , ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Travis S., et al. , Roles of singleton tryptophan motifs in COPI coat stability and vesicle tethering. Protein Data Bank. https://www.rcsb.org/structure/6TZT. Deposited 30 July 2019. [DOI] [PMC free article] [PubMed]
- 52.Travis S., et al. , Roles of singleton tryptophan motifs in COPI coat stability and vesicle tethering. Protein Data Bank. https://www.rcsb.org/structure/6U3V. Deposited 13 August 2019. [DOI] [PMC free article] [PubMed]
- 53.Travis S., et al. , Roles of singleton tryptophan motifs in COPI coat stability and vesicle tethering. Protein Data Bank. https://www.rcsb.org/structure/6U3W. Deposited 13 August 2019. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors can be accessed using PDB ID codes 6TZT, 6U3V, and 6U3W (51–53).