Significance
The evolution of new metabolic pathways has been a driver of diversification from the last universal common ancestor 3.8 billion y ago to the present. Bioinformatic evidence suggests that many pathways were assembled by recruiting promiscuous enzymes to serve new functions. However, the processes by which new pathways have emerged are lost in time. We have little information about the environmental conditions that fostered emergence of new pathways, the genome context in which new pathways emerged, and the types of mutations that elevated flux through inefficient new pathways. Experimental laboratory evolution has allowed us to evolve a new pathway and identify mechanisms by which mutations increase fitness when an inefficient new pathway becomes important for survival.
Keywords: experimental evolution, serendipitous pathway, promiscuity, pyridoxal 5′-phosphate, metabolism
Abstract
PdxB (erythronate 4-phosphate dehydrogenase) is expected to be required for synthesis of the essential cofactor pyridoxal 5′-phosphate (PLP) in Escherichia coli. Surprisingly, incubation of the ∆pdxB strain in medium containing glucose as a sole carbon source for 10 d resulted in visible turbidity, suggesting that PLP is being produced by some alternative pathway. Continued evolution of parallel lineages for 110 to 150 generations produced several strains that grow robustly in glucose. We identified a 4-step bypass pathway patched together from promiscuous enzymes that restores PLP synthesis in strain JK1. None of the mutations in JK1 occurs in a gene encoding an enzyme in the new pathway. Two mutations indirectly enhance the ability of SerA (3-phosphoglycerate dehydrogenase) to perform a new function in the bypass pathway. Another disrupts a gene encoding a PLP phosphatase, thus preserving PLP levels. These results demonstrate that a functional pathway can be patched together from promiscuous enzymes in the proteome, even without mutations in the genes encoding those enzymes.
Evolution of novel enzymes and metabolic pathways has been a primary driver of organismal diversity on Earth. Reconstructions of the genome of the last universal common ancestor suggest that core metabolic pathways, particularly biosynthetic pathways (1–3), had evolved by 3.8 billion y ago. As ecosystems became more complex and new carbon sources became available, myriad degradative pathways capable of supporting heterotrophic metabolisms evolved. Selective pressures to manipulate the environment and compete for resources fostered emergence of pathways for synthesizing complex natural products. Evolution of novel pathways continues in the present due to the introduction of anthropogenic chemicals, including pesticides, pharmaceuticals, solvents, textile dyes, and uncounted numbers of chemicals used in industry and household products.
Bioinformatic evidence suggests that metabolic pathways evolve by recruitment of enzymes that have a promiscuous ability to catalyze a newly important reaction (4–6). “Promiscuous” activities—which we define as adventitious secondary activities that are physiologically irrelevant (7)—are extremely common. A 2,3-dihydroxybenzoate decarboxylase from Fusarium oxysporum acts on 15 other substrates (8). The bacterial RebH tryptophan halogenase acts on 44 out of 93 tested aromatic compounds (9). Most members of the HAD superfamily of phosphatases hydrolyze multiple substrates; some hydrolyze >140 (10). Even inefficient promiscuous activities can accelerate reactions by orders of magnitude (11). Consequently, if the environment changes and a promiscuous activity becomes important for fitness, it can serve as a starting point for evolution of a new enzyme. The potential for assembly of novel pathways using the resources in a given proteome is unknown, but likely considerable. For example, Escherichia coli has ∼1,600 enzymes (12). If we make a modest guess that enzymes have on average 10 promiscuous activities, there could be 16,000 potential reactions whose circuitry could be rewired to generate novel pathways.
We term pathways constructed from promiscuous activities “serendipitous pathways” (SPs) to denote their fortuitous nature. SPs differ from “underground metabolism,” which refers to leaks in the metabolic network due to promiscuous enzyme reactions (13). SPs, in our view, represent previously underground pathways for which flux has been raised to physiologically significant levels by mutations. Thus, the enzymatic reactions in an SP have become physiologically relevant and are no longer promiscuous.
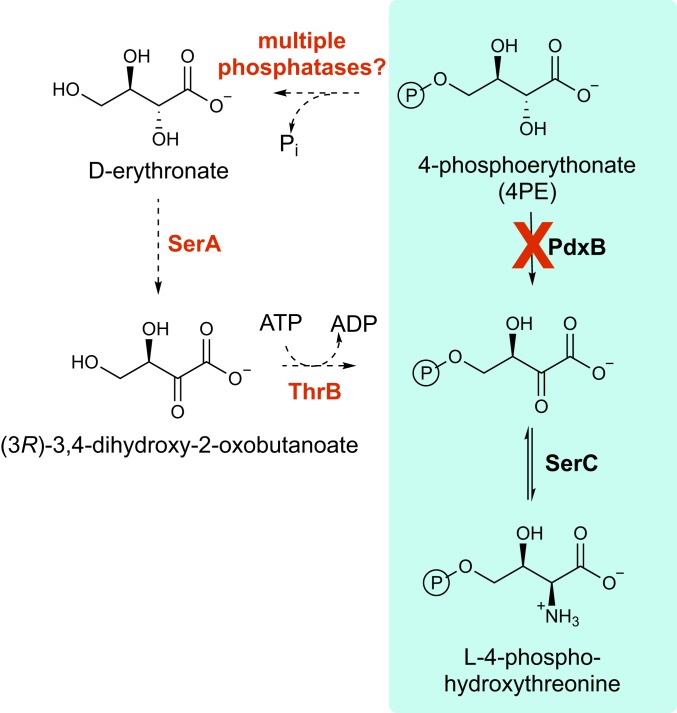
Here we report elucidation of a 4-step SP that restores synthesis of pyridoxal 5′-phosphate (PLP) in a strain of E. coli that lacks pdxB. PLP is an essential cofactor; it is required for the transaminases that convert α-keto acids to α-amino acids, as well as a number of epimerases, decarboxylases, racemases, aldolases, deaminases, lyases, and aminomutases (14). Due to the critical roles of many of these enzymes, a strain of E. coli that lacks PdxB (erythronate 4-phosphate dehydrogenase; Fig. 1) would not be expected to grow on minimal media.
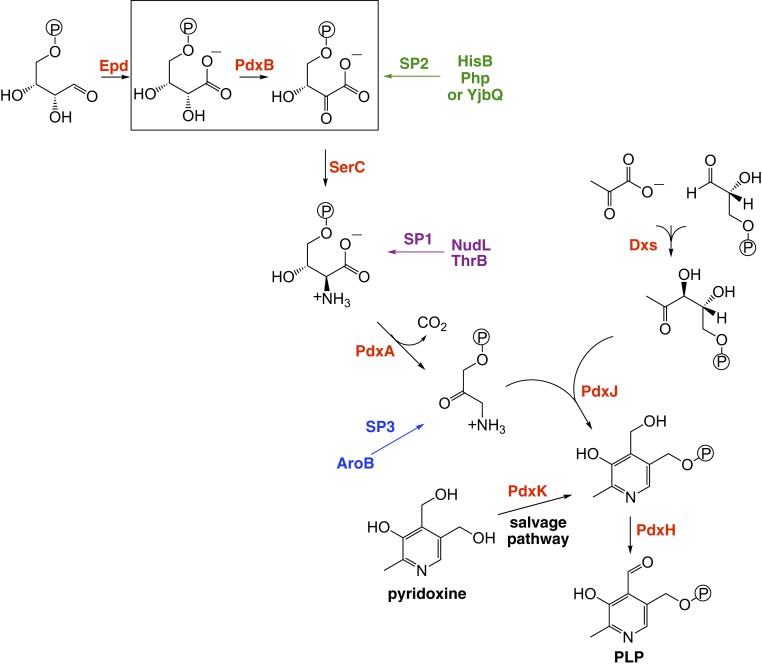
Fig. 1.
The pathway for synthesis of PLP in E. coli. PLP can also be synthesized from pyridoxine by a salvage pathway. Enzymes involved in the canonical synthesis pathway and the salvage pathway are highlighted in red. The reaction catalyzed by PdxB is boxed. The points at which 3 SPs facilitated by overexpression of individual enzymes feed into the pathway are indicated by colored arrows. The sequence of reactions in SP1 is shown in Fig. 2. The sequences of reactions in SP2 and SP3 are not known. Overexpression of PdxA may facilitate an SP or just pull material through SP1 and/or SP2.
We previously showed that overexpression of any one of 7 different genes (yeaB [recently renamed nudL], thrB, hisB, php, yjbQ, aroB, or pdxA) restores growth of a strain lacking pdxB (15). The proteins encoded by these genes do not catalyze the missing enzymatic reaction. Rather, overexpression enables synthesis of PLP by SPs that divert a metabolite from elsewhere in the metabolic network and convert it to a metabolite downstream of the step catalyzed by PdxB.
In the first SP we characterized, SP1 (Fig. 2), NudL, a putative CoA pyrophosphohydrolase (16), hydrolyzes 3-phosphohydroxypyruvate to 3-hydroxypyruvate (3HP). The 3HP undergoes nonenzymatic decarboxylation to produce glycolaldehyde. LtaE, a low-specificity threonine aldolase whose physiological function is unknown, catalyzes the condensation of glycolaldehyde and glycine to a mixture of 4-hydroxythreonine and 4-hydroxy-allo-threonine. Finally, a promiscuous activity of homoserine kinase (ThrB) phosphorylates 4HT, generating 4-phosphohydroxythreonine, an intermediate in the normal PLP synthesis pathway. Overexpression of nudL increases flux into the pathway, whereas overexpression of thrB pulls material through the pathway by coupling the thermodynamically unfavorable aldol condensation of glycine and glycolaldehyde to an exergonic phosphorylation reaction. Genetic studies suggest that at least 2 other SPs can be patched together using the resources in the E. coli proteome; these pathways patch into the normal PLP synthesis pathway at different points (Fig. 1) (15).
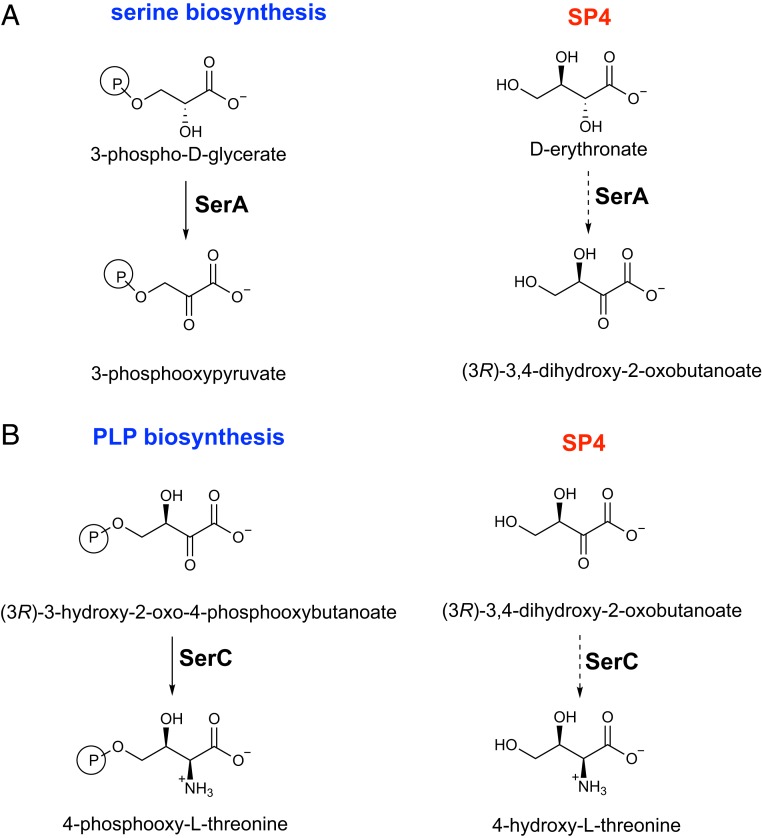
Fig. 2.
Two SPs that restore PLP synthesis in the absence of PdxB. The previously described SP1 (15) is facilitated by overexpression of either nudL or thrB. SP4, which was elucidated in this work, operates in an evolved ∆pdxB strain (JK1). Cofactors are shown only for reactions in SPs. Red denotes promiscuous enzymes that have been recruited to catalyze reactions in SP1 and/or SP4.
The existence of SPs for PLP synthesis was revealed by overexpression of individual genes to levels that may not be achievable by physiological processes (15). In this work, we have asked whether more physiologically relevant processes can restore growth of the ΔpdxB strain. We were able to evolve strains that grow robustly on glucose after only 150 generations and accumulation of 3 to 6 mutations. We identified a new SP (SP4) in strain JK1 that converts the substrate for PdxB to an intermediate downstream of the blocked step in the pathway via 4 reactions catalyzed by promiscuous enzymes (Fig. 2). We have identified the mechanisms by which mutations in 4 genes observed in the adapted strains improve fitness. Deletions or frameshifts eliminate the activity of YbhA, a PLP phosphatase, thus preventing destruction of PLP. Mutations in 3 other genes (gapA, pgl, and serA) enhance the newly needed activity of SerA in SP4, but by 4 different mechanisms.
Results and Discussion
Evolution of the ∆pdxB Strain in M9/Glucose.
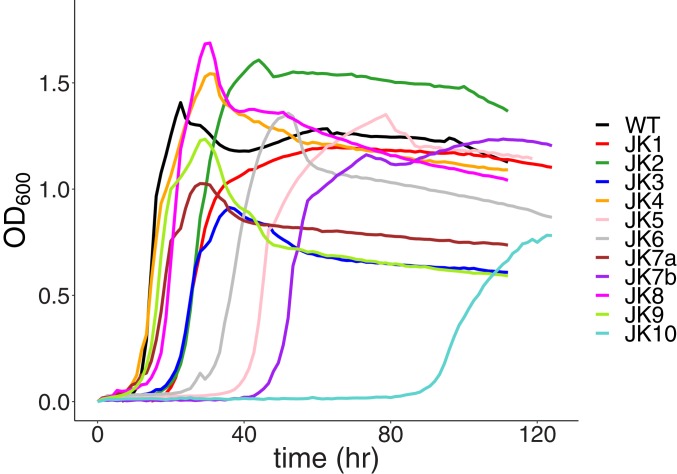
A single colony of a ΔpdxB::kan strain of E. coli BW25113 was grown to an optical density at 600 nm (OD600) of 1.0 in Luria broth (LB) containing kanamycin. The cells were washed 5 times with phosphate-buffered saline (PBS) to remove residual nutrients and inoculated into 6 flasks containing 25 mL M9/glucose (0.4% wt/vol) to an initial OD600 of 0.001. The flasks were incubated at 37 °C with shaking. Slight turbidity was visible after 8 to 10 d (OD600 0.02 to 0.04 for 3 flasks and <0.02 for the other 3), likely due to very low flux through one or more SPs for PLP synthesis. At that point, each culture was split into 2 separate flasks. Serial dilutions (1:100) were carried out when the cultures reached midlog phase (OD600 = ∼0.1) for up to 50 d (SI Appendix, Fig. S1). Two lineages died out early in the experiment. Fig. 3 shows the growth of evolved strains isolated at the end of the experiment. All of the strains grew quite well after a variable lag phase; growth rates ranged between 0.17 and 0.43 h−1. For comparison, the growth rate of wild-type E. coli was 0.58 h−1 (SI Appendix, Fig. S2 and Table S1).
Fig. 3.
Growth of wild-type and adapted clones of ∆pdxB E. coli on M9/glucose (0.4%) at 37 °C. JK7a and JK7b refer to 2 colonies from the final JK7 population. Growth curves for 2 to 4 replicates of each strain were consistent, but only one is shown for each strain for clarity.
Parallelism in Mutational Targets Reveals Adaptive Mutations.
At the conclusion of the experimental evolution, cultures were spread on M9/glucose plates and 11 colonies (2 from lineage 7 and 1 from each of the other lineages) were selected for whole-genome sequencing. Table 1 summarizes the genes in which mutations were found; Dataset S1 provides details about each mutation. Notably, most of the mutations were deletions or insertions that would have caused loss of function. Point mutations observed in gapA and serA also caused loss of function (discussed further below).
Table 1.
Genes in which mutations were found in adapted clones
| Strain | ybhA | pgl | gapA | serA | ettA | purF | ilvH | rpoS | rpe | nadR | pykF | rng | gltB | sdhA | livH | pyrE | rho | lon | rpoC | rpmG | ypjA |
| JK1 | D | D | N | F | N | ||||||||||||||||
| JK2 | D | D | N | I | N | D | F | ||||||||||||||
| JK3 | D | D | N | N | N | ||||||||||||||||
| JK4 | F | I | I | N | X | ||||||||||||||||
| JK5 | D | D | N | D | N | N | D | ||||||||||||||
| JK6 | N | I | N | ||||||||||||||||||
| JK7a | D | D | N | N | N | F | |||||||||||||||
| JK7b | D | D | N | N | F | X | |||||||||||||||
| JK8 | N | N | N | F | D | N | |||||||||||||||
| D | |||||||||||||||||||||
| JK9 | D | D | N | N | I | ||||||||||||||||
| JK10 | N | N | N | F | I |
N, nonsynonymous; F, frameshift; D, deletion; I, insertion; X, intergenic.
Several genes were affected by mutations in multiple lineages. Mutational parallelism—the observation that mutations occurred in the same gene after evolution of replicate populations—is a strong indication that these mutations are adaptive (17–20). The beneficial effects of these mutations could stem from a variety of mechanisms, including prevention of PLP hydrolysis, compensation for low levels of PLP, improvements in PLP synthesis, or simply improvements in growth on glucose as a sole carbon source. Potential mechanisms for these effects will be discussed below.
Identification of a Novel SP for PLP Synthesis.
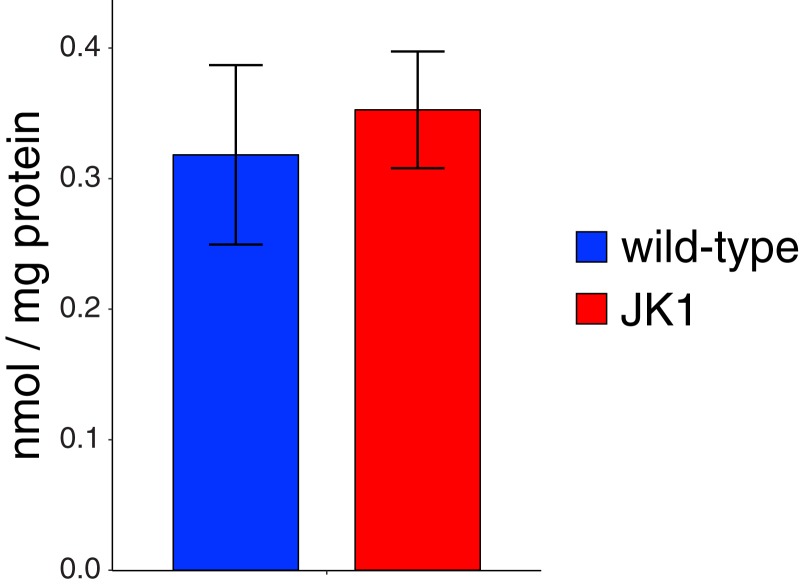
The restoration of growth in the evolved strains begs the question of how PLP synthesis has been restored. We have identified a novel SP that reconstitutes PLP synthesis in strain JK1. Notably, this pathway produces wild-type levels of PLP (Fig. 4).
Fig. 4.
Concentrations of PLP in extracts of JK1 and wild-type E. coli cells grown in M9/glucose (0.4%).
To determine whether JK1 uses the previously discovered SP1 (Fig. 2) to reconstitute PLP synthesis, we deleted ltaE and thrB. Deletion of ltaE does not impair growth on M9/glucose (0.4%) (SI Appendix, Fig. S3A). Deletion of thrB abolishes growth, even when threonine is added to compensate for ThrB’s normal function in threonine synthesis (Fig. 5A). However, addition of both threonine and pyridoxine (which can be converted to PLP via a salvage pathway) allows ∆thrB JK1 to grow, indicating that ThrB is required for PLP synthesis. These data suggest that PLP synthesis in JK1 results from production of 4-hydroxythreonine by an unknown SP, followed by phosphorylation of 4-hydroxythreonine, which we previously showed to be a robust promiscuous activity of ThrB ([kcat/Km] = 4.8 × 103 M−1⋅s−1) (15). Consistent with this hypothesis, deletion of pdxA, which catalyzes the conversion of 4-phosphohydroxythreonine to the next intermediate in the normal PLP synthesis pathway, prevents growth of JK1 on glucose unless pyridoxine is added (Fig. 5B).
Fig. 5.
Effect of deleting (A) thrB, (B) pdxA, (C) serA, and (D) serC on growth of JK1 on M9/glucose (0.4%). The shaded regions surrounding each growth curve represent 1 SD for 2 to 4 replicates. thr, l-threonine; ser, l-serine; gly, glycine; pyr, pyridoxine; arom, l-phenylalanine, l-tyrosine, l-tryptophan, p-aminobenzoate, p-hydroxybenzoate, and 2,3-dihydroxybenzoate. Growth curves for JK1 were not consistent from day to day for undetermined reasons. All growth curves in each panel were taken from the same plate.
A multitude of potential SPs might produce 4HT in JK1. After consideration of many possibilities, we chose to investigate the pathway shown as SP4 in Fig. 2. This SP was appealing because it requires only 3 steps, each of which is a common enzymatic transformation, and the proposed intermediates resemble normal metabolites and thus might be substrates for promiscuous enzymes. Further, as described in the following paragraphs, we could identify good candidates for catalysis of each of the steps.
Two phosphatases that hydrolyze 4PE—heptose 1,7-bisphosphate phosphatase (GmhB) and phosphoglycolate phosphatase (Gph)—were previously identified in a high-throughput substrate profiling screen (10). Deletion of both gmhB and gph impaired growth of JK1, but growth was not improved by addition of pyridoxine, indicating that the growth impairment was due to loss of the native function of one or both proteins and not to a role in PLP synthesis (SI Appendix, Fig. S2B). E. coli has more than 40 other phosphatases; 1 or more of these may catalyze sufficient hydrolysis of 4PE to enable PLP synthesis via SP4. Although we were not able to identify the phosphatase(s) responsible for hydrolysis of 4PE, we detected erythronate in both wild-type E. coli and JK1 (Dataset S2), demonstrating that this reaction takes place in vivo.
The second step in SP4 is oxidation of erythronate to (3R)-3,4-dihydroxy-2-oxobutanoate. Three lines of evidence suggested that 3-phosphoglycerate dehydrogenase (SerA) might catalyze this reaction. First, SerA normally catalyzes oxidation of an alcohol alpha to a carboxylate in 3-phosphoglycerate and thus might have promiscuous activity with the structurally similar erythronate (Fig. 6A). Second, mutations in serA were detected in several adapted clones (Table 1). Third, growth of JK1 is inhibited by serine (SI Appendix, Fig. S4). SerA, the first enzyme in the serine biosynthesis pathway, is subject to feedback inhibition by serine. If SerA is required for PLP synthesis, the presence of serine in the medium would be expected to interfere with SP4.
Fig. 6.
Native (Left) and promiscuous (Right) reactions catalyzed by (A) SerA and (B) SerC. (The stereochemistry of the native and promiscuous reactions of SerC is assumed to be the same.)
SerA in fact has an inefficient ability to oxidize erythronate (Table 2). Further, deletion of serA prevents growth of JK1 on M9/glucose, even when serine is added to compensate for loss of SerA’s essential function in serine synthesis (Fig. 5C). However, growth is restored when both serine and pyridoxine are added to the medium. Together, the in vitro and in vivo evidence indicate that SerA has been recruited to serve as an erythronate dehydrogenase in JK1.
Table 2.
Kinetic parameters for wild-type and mutant versions of SerA
| Strain (mutation) | kcat,3PG, s−1 | Km,3PG, mM | kcat/Km,3PG, M−1⋅s−1 | kcat/Km,erythronate*, M−1⋅s−1 | Specificity† | Ki,serine, µM |
| Wild-type | 7.7 ± 1.1 | 4.7 ± 1.1 | 1.6 ± 0.3 × 103 | 0.07 ± 0.01 | 2.3 × 104 | 7.0 ± 1.9 |
| JK6 (insertion of MV after I381) | 7.7 ± 0.2 | 1.7 ± 0.1 | 4.5 ± 0.2 × 103 | 0.34 ± 0.02 | 1.3 × 104 | >2,000 |
| JK7b (I304L) | 1.5 ± 0.2 | 3.2 ± 0.9 | 4.7 ± 0.9 × 102 | 0.12 ± 0.01 | 3.9 × 103 | 14.7 ± 3.5 |
| JK8 (Q371P ∆T372) | 10.5 ± 0.3 | 0.2 ± 0.0 | 4.3 ± 0.2 × 104 | 6.64 ± 0.34 | 6.5 × 103 | >2,000 |
| JK9 (G377C) | 16.1 ± 1.0 | 1.1 ± 0.2 | 1.4 ± 0.1 × 104 | 0.37 ± 0.02 | 3.8 × 104 | >2,000 |
Errors are SDs.
It was not possible to saturate SerA with up to 6 mM erythronate for most of the enzymes, so only values for kcat/Km are given. For the I304L variant found in JK8, kcat = 0.014 ± 0.001 s−1 and Km = 2.0 ± 0.3 mM.
Specificity is defined as (kcat/Km,3PG)/(kcat/Km,erythronate).
The third step in SP4 is transamination of (3R)-3,4-dihydroxy-2-oxobutanoate to 4-hydroxythreonine. SerC is an attractive candidate for catalysis of this reaction because 1 of its 2 physiological substrates, (3R)-3-hydroxy-2-oxo-4-phosphooxybutanoate, differs from (3R)-3,4-dihydroxy-2-oxobutanoate only by the presence of a phosphoryl group on carbon 4 (Fig. 6B). We assayed the ability of SerC to convert 4-hydroxythreonine to (3R)-3,4-dihydroxy-2-oxobutanoate (the opposite direction of the reaction in SP4) because (3R)-3,4-dihydroxy-2-oxobutanoate is not commercially available. SerC does indeed have weak activity with 4-hydroxythreonine; kcat is 2.0 ± 0.1 × 10−5 s−1, Km is 2.6 ± 0.7 mM, and kcat/Km is 7.4 ± 1.0 × 10−3 M−1⋅s−1. For comparison, kcat/Km for the reaction with the native substrate 4-phosphohydroxythreonine is 727 M−1⋅s−1 (21).
Testing the putative role of SerC in SP4 is complicated because adding serine to the medium to compensate for loss of SerC’s function in serine synthesis would inhibit SerA, which is required for SP4. Consequently, we introduced a serine-insensitive allele of serA (serA*, which encodes N364A SerC) (22) into JK1 prior to deleting serC. This strategy was only partially successful, as preliminary experiments showed that the ∆serC serA* JK1 strain grew poorly in M9/glucose even when both serine and pyridoxine were added to the medium. High concentrations (100 mM) of serine inhibit growth of wild-type E. coli (23, 24) due to inhibition of homoserine dehydrogenase and aspartokinase (25, 26) and impaired synthesis of aromatic amino acids (23). Although we added only 10 mM serine to the medium, the ∆serC serA* JK1 strain may be more susceptible to serine toxicity than wild-type E. coli. We solved the problem by supplementing the medium with 2 mM glycine, which can be converted to serine but is less toxic. We also added 6 aromatic compounds (l-phenylalanine, l-tyrosine, l-tryptophan, p-hydroxybenzoate, p-aminobenzoate, and 2,3-dihydroxybenzoate) because serC is upstream of aroA in an operon, and serC deletion may perturb expression of aroA, which is required for synthesis of aromatic compounds. Fig. 5D shows that ∆serC serA* JK1 is unable to grow on the supplemented M9/glucose unless pyridoxine is added, confirming that SerC is required for PLP synthesis.
Elimination of an Alternative SP.
We also considered an alternative SP (Fig. 7) that, like SP4, requires SerA, ThrB, and SerC, but in this case ThrB phosphorylates (3R)-3,4-dihydroxy-2-oxobutanoate, and SerC serves its canonical function in PLP synthesis. Distinguishing between these 2 SPs requires determining whether ThrB phosphorylates (3R)-3,4-dihydroxy-2-oxobutanoate or 4-hydroxythreonine. We assessed ThrB activity by measuring conversion of adenosine 5′-triphosphate (ATP) to adenosine 5′-diphosphate (ADP) by 31P-NMR in reaction mixtures containing ThrB, ATP, and erythronate and NAD+. In this reaction mixture, 9% of the ATP was converted to ADP and inorganic phosphate after 24 h at 37 °C (SI Appendix, Fig. S5). This formation of ADP is due to hydrolysis of ATP in the active site of ThrB in the absence of a second substrate. A comparable amount was converted to ADP when SerA was included to convert erythronate to (3R)-3,4-dihydroxy-2-oxobutanoate, indicating that (3R)-3,4-dihydroxy-2-oxobutanoate is not a substrate for ThrB. However, when SerC was included to convert (3R)-3,4-dihydroxy-2-oxobutanoate to 4-hydroxythreonine, 14% of the ATP was turned over to ADP, suggesting that 5% of the ATP had been used by ThrB to phosphorylate 4-hydroxythreonine, as proposed for SP4. These data rule out the pathway shown in Fig. 7.
Fig. 7.
An alternative SP that involves SerA, ThrB, and SerC in a different order than in SP4.
Some Adaptive Mutations Likely Improve Growth on Glucose as a Sole Carbon Source.
Mutations are clearly responsible for elevating flux through SP4 and restoring PLP synthesis to wild-type levels. However, some of the adaptive mutations may simply improve growth on glucose as a sole carbon source. Mutations that likely fall into this category will be discussed before we address the more interesting mutations that impact PLP levels.
Previous studies have reported a number of mutations that occur frequently during evolution of E. coli in minimal media. Mutations in pykF and nadR, which occurred in 2 lineages, occurred during long-term adaption of E. coli in glucose (27). Two strains acquired mutations in rpoS, which encodes the alternative sigma factor σS, the master regulator of the general stress response. One causes an early frameshift; the other changes Leu177 to Arg. Null mutations in rpoS are common during experimental evolution (28, 29). JK1 acquired a mutation in rpoC, which encodes the β′ subunit of RNA polymerase. Mutations in rpoC are frequently observed during experimental evolution (30–36) and have pleiotropic effects that improve growth rate on minimal media (32).
Two strains acquired mutations in rng, which encodes a ribonuclease involved in 16S ribosomal RNA maturation and messenger RNA (mRNA) turnover. These nonsynonymous mutations most likely cause loss of function. Deletion of rng stabilizes a number of mRNAs, including several encoding glycolytic enzymes (glucose 6-phosphate isomerase, triose phosphate isomerase, and enolase) (37). It is plausible that increased production of these enzymes might enhance growth on glucose.
Some Adaptive Mutations Likely Enhance Fitness by Compensating for Low Levels of PLP Early in Adaptation.
Nine of 11 strains acquired mutations affecting ybhA; most were large deletions or frameshift mutations (Table 1 and SI Appendix, Fig. S5). YbhA, a member of the haloacid dehalogenase-like superfamily of phosphatases, hydrolyzes both PLP and fructose 1,6-bisphosphate (38). If PLP synthesis in the adapted strains is inefficient early in adaptation, loss of a PLP phosphatase would certainly be advantageous.
Three strains acquired nonsynonymous mutations in IlvH, the regulatory subunit of acetohydroxyacid synthase (AHAS) III. AHAS III is 1 of 2 isozymes required for early steps in branched-chain amino acid synthesis in E. coli K12 strains. IlvH is necessary for full activity of AHAS III and also confers sensitivity to feedback inhibition by branched-chain amino acids. The residues affected by mutations (Ala36, Ser13, and Ile30) are located at the dimer interface in or near the predicted effector binding site. The Ala36Val change we observed in JK5 as well as mutations affecting Gly14 (immediately adjacent to Ser13) are known to abolish feedback inhibition by valine (39). Since later steps in branched-chain amino acid synthesis require PLP-dependent enzymes (IlvA and either IlvE or TyrB), enhancing the upstream AHAS III activity may help push material through branched-chain amino acid synthesis pathways during the early stages of evolution when levels of active IlvA, IlvE, and TyrB may be suboptimal due to limited availability of PLP.
How Do Mutations Improve PLP Synthesis?
The most interesting mutations are those that improve flux through SP4. Such mutations might increase the level of an enzyme in SP4 or of its novel substrate. Alternatively, they might decrease the level of the native substrate for an enzyme in SP4, thereby diminishing its ability to competitively inhibit the newly needed SP4 reaction.
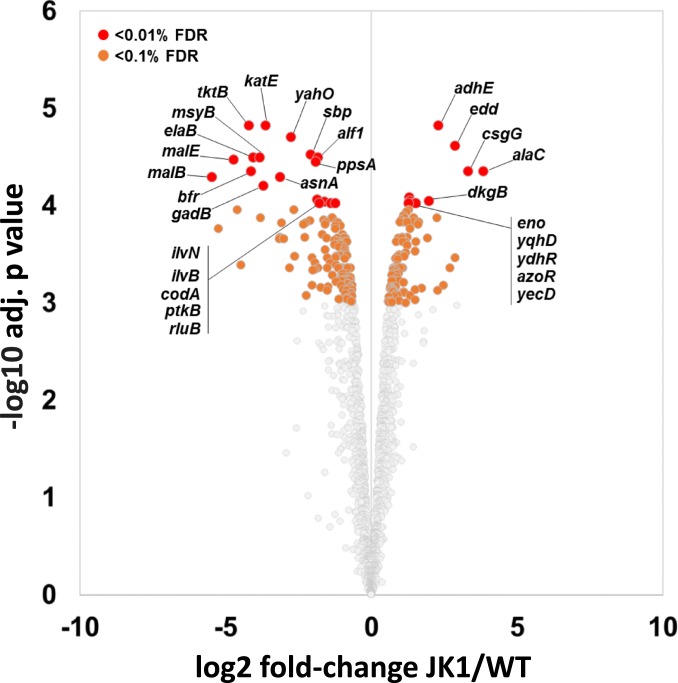
We analyzed the proteomes of JK1 and wild-type E. coli to determine whether SP4 enzymes are more abundant in JK1. The proteome of JK1 is significantly different from that of wild-type E. coli; 271 proteins are more abundant in JK1, and 303 are less abundant (Fig. 8 and Dataset S3). These global changes may be due to the mutations in rpoC and rpoS. The effects of rpoS deletion on the proteome (40) and of rpoC mutations on the transcriptome (32) have previously been investigated. However, the presence of mutations in both rpoS and rpoC in JK1 makes it difficult to attribute specific changes to one or the other of these mutations.
Fig. 8.
Volcano plot comparing adjusted P value and log2 fold change of 1,790 proteins in JK1 compared to wild-type E. coli grown in M9/glucose (0.4%). FDR, false discovery rate.
Levels of SerC and ThrB, which catalyze novel reactions in SP4, are increased by 64% and 40%, respectively, in JK1. The level of SerA is unchanged. The modest changes in SerC and ThrB seem unlikely to be solely responsible for the success of SP4.
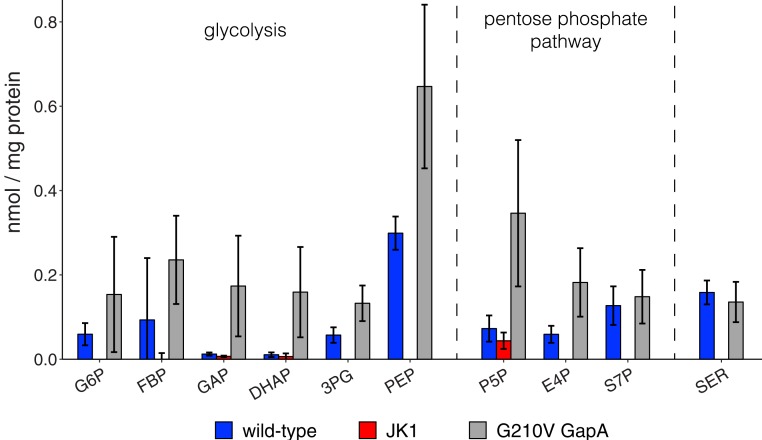
We measured the concentrations of 23 metabolites in cell lysates using targeted metabolomics. Levels of 4PE and erythronate are similar in wild-type and JK1 cells (Dataset S2); thus, material is not pushed through SP4 by an increase in the level of the 4PE precursor of SP4 or erythronate. However, the data reveal 2 striking changes that should increase the rate of erythronate oxidation by SerA. First, 3-phosphoglycerate (3PG), the native substrate for SerA, is undetectable in JK1 (Fig. 9). The decreased 3PG concentration should decrease inhibition of erythronate oxidation by SerA in SP4. Second, serine is also undetectable in JK1. The low level of serine in JK1 should decrease feedback inhibition of SerA and ensure that it is available for PLP synthesis.
Fig. 9.
Concentrations of selected metabolites in extracts of JK1, wild-type, and G210V GapA E. coli cells grown in M9/glucose (0.4%). G6P, glucose 6-phosphate; FBP, fructose 1,6-bisphosphate; GAP, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; 3PG, 3-phosphoglycerate; PEP, phosphoenolpyruvate; P5P, pentose 5-phosphates; E4P, erythrose 4-phosphate; S7P, sedulose 7-phosphate; SER, serine. Error bars represent SD.
We initially hypothesized that the dramatic decrease in 3PG concentration in JK1 was due to a mutation in gapA, which encodes glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Fig. 10). JK1 has a gapA mutation that changes Gly210 to Val; 9 other adapted strains have a mutation that changes Leu158 to Gln. Gly210 is located near the active site; Leu158 is buried in the interior of the protein at a distance from the active site (SI Appendix, Fig. S7). Changing Gly210 to Val decreases kcat/Km for glyceraldehyde 3-phosphate by 8-fold (Table 3). In contrast, changing Leu158 to Gln does not affect catalytic activity but decreases melting temperature by 6.1 °C (from 72.6 ± 0.2 to 66.5 ± 0.4 °C). GAPDH activity in cell lysates is decreased by 80% in JK1 and by 60% in JK2, which carries the L158Q gapA (SI Appendix, Fig. S8). Similar decreases are observed when these mutations are introduced into wild-type E. coli.
Fig. 10.
Proteins and metabolites in JK1 that change by >2-fold relative to wild-type E. coli. Transketolase and transaldolase reactions in the pentose phosphate pathway are indicated by purple and blue arrows, respectively. Particularly important changes discussed in the text are highlighted in red. TktB (the minor transketolase) and TalA levels are decreased by 19- and 4-fold, respectively, in JK1, but levels of TktA, the major transketolase, and TalB are unchanged. Thus, adequate transaldolase and transketolase activities should be available to support the pentose phosphate pathway reactions.
Table 3.
Values for kcat/Km,GAP for wild-type and mutant versions of GapA
| Enzyme | kcat/Km,GAP*, M−1⋅s−1 |
| Wild-type | 6.2 ± 0.4 × 104 |
| G201V | 7.6 ± 0.8 × 103 |
| L158Q | 7.2 ± 0.5 × 104 |
We were not able to saturate the enzymes at up to 2 mM GAP, so only kcat/Km,GAP is reported.
We hypothesized that decreased GAPDH activity might enhance PLP synthesis by decreasing the level of the downstream metabolite 3PG. To test this hypothesis, we introduced the G210V gapA mutation into wild-type E. coli. However, the metabolome of JK1 is considerably different from that of the G210V gapA mutant strain (Fig. 9). Thus, the low level of 3PG in JK1 cannot be attributed solely to the G210V gapA mutation.
An important clue to the low 3PG concentration in JK1 is the loss of 6-phosphogluconolactonase (Pgl), the second enzyme in the oxidative pentose phosphate pathway (Fig. 10) due to a 3,812-bp deletion that removes ybhA (discussed above) as well as 140 bp at the 5′ end of pgl (SI Appendix, Fig. S6). Similar deletions were found in 6 other adapted strains. Loss of Pgl should substantially decrease flux through the oxidative pentose phosphate pathway (although may not abolish it entirely since 6-phosphogluconolactone may hydrolyze nonenzymatically). Consistent with this expectation, levels of erythrose 4-phosphate and sedulose 7-phosphate are greatly diminished in JK1. In the absence of Pgl, ribose for nucleotide synthesis, ribulose for flavin synthesis, and erythrose 4-phosphate for PLP and aromatic amino acid synthesis must be produced from glycolytic intermediates by the transketolase and transaldolase reactions highlighted in blue and purple in Fig. 10. Notably, all of these reactions require GAP. We attribute the low level of 3PG in JK1 to the combination of diversion of GAP into the pentose phosphate pathway necessitated by loss of Pgl and the decreased activity of GAPDH, which will diminish flux into the lower part of glycolysis. An additional contributing factor may be the 4-fold decrease in the level of PpsA (phosphoenolpyruvate synthetase) (Dataset S3), which should decrease flux from pyruvate back through reactions in the lower glycolytic pathway. The altered balance between the opposing activities of PpsA and the pyruvate kinases PykF and PykA would be expected to deplete phosphoenolpyruvate (PEP), as well as the upstream intermediates, 2-phosphoglycerate (2PG) and 3PG. Consistent with this hypothesis, PEP is undetectable in JK1 cells (Fig. 9). Further, deletion of ppsA in wild-type E. coli also decreases the level of PEP (41).
The ability of SerA to oxidize erythronate in SP4 can also by enhanced by mutations in serA itself. Mutations in serA occurred in 4 strains, although not in JK1. The locations of the amino acids affected by these changes are shown in SI Appendix, Fig. S9. Three (an insertion of MV after Ile381, a change of Gln370 to Pro with concomitant deletion of Thr371, and a change from Gly377 to Cys) are near the serine binding site in the regulatory subunit. Each of these mutations abolishes feedback inhibition of the enzyme by serine (Table 2), which will maintain active SerA for oxidation of erythronate in SP4 even if serine levels are adequate. The fourth mutation changes Ile304 to Leu in the hinge between the regulatory and catalytic domains. This mutation diminishes the inhibition constant, Ki, for serine inhibition by 2-fold, and the kcat/Km for 3PG by 3.5-fold. However, kcat/Km for erythronate is increased by 1.7-fold. Both the decreased feedback inhibition by serine and the altered substrate specificity should improve SerA’s ability to oxidize erythronate in SP4.
Conclusions
Although bioinformatic evidence supports the notion that novel pathways have evolved by patchwork recruitment of promiscuous enzymes to serve new functions, we lack empirical data about the process by which new pathways emerge. We have little or no knowledge of the resources available in the proteome when a new pathway arose, how the environment shaped the evolutionary possibilities, and how mutations enabled an underground pathway to rise to a physiologically significant level of function. Laboratory evolution allows us to address these important questions.
We were able to evolve a novel 4-step SP in E. coli in only 150 generations. The ease with which SP4 was assembled can be attributed to 3 factors. First, the intermediates in SP4 resemble the native substrates for SerA, SerC, and ThrB. Second, growth on glucose as a sole carbon source requires serine and threonine synthesis, so these enzymes will be active. If the medium contained these amino acids, SerA would be inactive due to feedback inhibition by serine, and ThrB expression would be low due to attenuation of the threonine operon. Finally, a single deletion encompassing some or all of ybhA and pgl appears to confer 2 beneficial effects, diminishing the concentration of 3PG and thus inhibition of erythronate oxidation in SP4 and preserving PLP by destroying an enzyme that hydrolyzes PLP.
Enhancing oxidation of erythronate by SerA appears to be a critical factor for elevating flux through SP4. In theory, this could be accomplished by 6 different mechanisms: 1) increasing the concentration of SerA; 2) increasing the concentration of erythronate; 3) changing the specificity of SerA to improve its ability to oxidize erythronate; 4) decreasing the concentration of 3PG, the native substrate for SerA, to decrease inhibition of the newly needed reaction in SP4; 5) decreasing the sensitivity of SerA to feedback inhibition by serine; and 6) decreasing the concentration of serine. We observed 4 of these mechanisms in our analyses of the adapted strains. In our most intensively studied strain, JK1, enhanced conversion of erythronate to (3R)-3,4-dihydroxy-2-oxobutanoate in SP4 can be attributed to dramatic decreases in the concentrations of 3PG and serine.
The emergence of an SP in response to an environmental challenge is contingent upon the availability of suitable promiscuous enzymes. An initial SP may not be a particularly elegant solution to the problem. SP4, for example, involves hydrolysis of a phosphate ester at the beginning of the pathway, and then rephosphorylation at the end of the pathway, costing one more ATP than the standard PLP synthesis pathway. Further, SP4 requires a PLP-dependent enzyme (SerC) to make PLP. Interestingly, many extant pathways have suboptimal features. For example, in E. coli, PLP synthesis normally requires a PLP-dependent enzyme (SerC), thiamin diphosphate synthesis requires a thiamin diphosphate-dependent enzyme (Dxs), and adenine nucleotide synthesis requires ATP. Pathways in which the product is required for a step in the pathway itself are problematic; if the product is depleted, no more can be synthesized. Other metabolic pathways produce unstable intermediates that decompose readily or cause toxicity. The first step in pentachlorophenol degradation in Sphingobium chlorophenolicum generates tetrachlorobenzoquinone (42), a potent alkylating agent that must be sequestered by a protein–protein interaction to prevent depletion of glutathione and damage to macromolecules (43). The pathway for 1,2-propanediol degradation in Salmonella generates toxic propionaldehyde, which is prevented from harming cellular macromolecules by sequestration of the pathway enzymes in a protein microcompartment (44).
Even a poor SP may allow a microbe to survive and reproduce until a better solution is found. New SPs may be refined by mutations, particularly if recruitment of a promiscuous enzyme to serve a new function prevents optimal tuning of fluxes toward 2 different products. For example, feedback inhibition of SerA when serine levels are adequate will impair PLP synthesis in JK1. This adaptive conflict could be solved by gene duplication and divergence to generate a new allele encoding a dedicated erythronate dehydrogenase that is no longer subject to feedback inhibition by serine. Similarly, duplication and divergence of thrB could produce an allele dedicated to SP4 that is no longer subject to transcriptional attenuation in the presence of threonine.
Remarkably, mutations in genes encoding SP4 enzymes were uncommon in the adapted strains. Mutations occurred only in serA, and in only 4 of 11 adapted strains. JK1, which we studied in detail, has no mutations in genes encoding enzymes in SP4. Further, the emergence of SP4 in JK1 was enabled entirely by loss-of-function mutations, which are more readily achieved than gain-of-function mutations. These findings demonstrate that emergence of a SP does not necessarily require mutations in genes encoding enzymes in the SP but can result from mutations that alter levels of proteins and metabolites elsewhere in the metabolic network in ways that indirectly improve flux through the novel SP.
Materials and Methods
Strains.
Strains used for this work are listed in SI Appendix, Table S1. E. coli K12 BW25113 (designated the wild-type herein) and a strain in which pdxB is replaced with a kanamycin resistance gene (designated the ΔpdxB strain) were obtained from the Keio collection (45). Cultures were routinely grown in LB medium (46) at 37 °C with kanamycin, chloramphenicol, ampicillin, or anhydrotetracycline as required. Cultures for adaptation and analysis of growth rate were grown in M9 minimal medium (46) containing 0.4% (wt/vol) glucose.
Evolution of the ∆pdxB Strain.
The ∆pdxB strain was grown in LB to an OD600 of 0.5. The cells were harvested by centrifugation at 13,000 × g at room temperature. The cell pellet was washed 5 times with ice-cold PBS. The washed cells were inoculated into 6 parallel cultures (25 mL) of M9 medium (46) containing 0.4% (wt/vol) glucose at an initial OD600 of 0.001. The cultures were grown at 37 °C with shaking. When visible turbidity developed, aliquots of each culture were inoculated into 2 separate flaks. Serial transfers were carried out at mid- to late-log phase for 1 mo. Daily measurements of log2(A/A0) were used to estimate the number of generations per day. Whole-genome sequencing of multiple clones at the end of the experiment confirmed the presence of kan in place of pdxB, suggesting that none of the cultures had been taken over by contaminating bacteria.
Growth Tests.
Freezer stocks of cell cultures were spread onto LB plates and incubated overnight at 37 °C. A single colony was inoculated into 3 mL of LB medium and the culture was incubated overnight at 37 °C. A 2-µL aliquot of the overnight culture was used to inoculate 3 mL of M9/glucose (0.4%) containing supplements as needed. (For the ∆serA strain, 10 mM l-serine was added. For the ∆serC strain, 2 mM glycine, 500 µM l-phenylalanine, 250 µM l-tyrosine, 200 µM l-tryptophan, 6 µM p-aminobenzoate, 6 µM p-hydroxybenzoate, and 50 µM 2,3-dihydroxybenzoate were added. For the ∆thrB strain, 1 mM threonine was added.) After overnight incubation at 37 °C, a 1-mL aliquot was subjected to centrifugation at 16,000 × g for 1 min. The pellet was washed 5 times with 0.5 mL of ice-cold PBS. The final pellet was suspended in 1 mL of M9/glucose (0.4%) and diluted to give an OD600 of 0.01. Ten microliters of this suspension were inoculated into 90 µL of M9/glucose with supplements as indicated in the text in individual wells of a 96-well plate. The plates were incubated in a Varioskan (Thermo) plate reader at 37 °C with 1 min of shaking every 5 min. The absorbance at 600 nm was measured every 20 min. The data were smoothed using the TRIMMEAN function of Excel by discarding the 2 lowest and 2 highest values from a moving window of 9 data points and then taking the arithmetic mean of the remaining values.
Enzyme Expression and Purification.
Construction of expression plasmids and methods for expressing and purifying SerA, SerC, ThrB, d-2-hydroxyglutarate dehydrogenase, and electron transfer flavoprotein AB are described in SI Appendix.
Enzyme Assays.
Details of enzyme assays are provided in SI Appendix.
Measurement of Melting Temperatures for Wild-Type and Variant Versions of GAPDH.
Melting temperatures were determined using a Protein Thermal Shift Dye Kit (Applied Biosystems) in a Step One Real Time PCR Systems instrument (Applied Biosystems).
Metabolomics and Proteomics.
Procedures for sample preparation and metabolomics and proteomics analyses are described in SI Appendix.
Data Availability.
All data are available within the paper and supplementary material.
Supplementary Material
Acknowledgments
This work was supported by grant R01GM083285 from the National Institutes of Health to S.D.C., NNA15BB04A from the National Aeronautics and Space Administration Astrobiology Institute to S.D.C. and V.S.C., and Defense Advanced Research Projects Agency cooperative agreement 13-34-RTA-FP-007 to W.M.O.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915569116/-/DCSupplemental.
References
- 1.Weiss M. C., Preiner M., Xavier J. C., Zimorski V., Martin W. F., The last universal common ancestor between ancient Earth chemistry and the onset of genetics. PLoS Genet. 14, e1007518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ouzounis C. A., Kunin V., Darzentas N., Goldovsky L., A minimal estimate for the gene content of the last universal common ancestor–Exobiology from a terrestrial perspective. Res. Microbiol. 157, 57–68 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Koonin E. V., Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1, 127–136 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Noda-Garcia L., Liebermeister W., Tawfik D. S., Metabolite-enzyme coevolution: From single enzymes to metabolic pathways and networks. Annu. Rev. Biochem. 87, 187–216 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Teichmann S. A., et al. , The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. J. Mol. Biol. 311, 693–708 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Chothia C., Gough J., Vogel C., Teichmann S. A., Evolution of the protein repertoire. Science 300, 1701–1703 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Copley S. D., An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 40, 72–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X., et al. , Biochemical characterization and substrate profiling of a reversible 2,3-dihydroxybenzoic acid decarboxylase for biocatalytic Kolbe-Schmitt reaction. Enzyme Microb. Technol. 113, 37–43 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Andorfer M. C., et al. , Understanding flavin-dependent halogenase reactivity via substrate activity profiling. ACS Catal. 7, 1897–1904 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang H., et al. , Panoramic view of a superfamily of phosphatases through substrate profiling. Proc. Natl. Acad. Sci. U.S.A. 112, E1974–E1983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien P. J., Herschlag D., Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6, R91–R105 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Freilich S., et al. , The complement of enzymatic sets in different species. J. Mol. Biol. 349, 745–763 (2005). [DOI] [PubMed] [Google Scholar]
- 13.D’Ari R., Casadesús J., Underground metabolism. BioEssays 20, 181–186 (1998). [DOI] [PubMed] [Google Scholar]
- 14.John R. A., Pyridoxal phosphate-dependent enzymes. Biochim. Biophys. Acta 1248, 81–96 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Kim J., Kershner J. P., Novikov Y., Shoemaker R. K., Copley S. D., Three serendipitous pathways in E. coli can bypass a block in pyridoxal-5′-phosphate synthesis. Mol. Syst. Biol. 6, 436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLennan A. G., The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieberman T. D., et al. , Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat. Genet. 43, 1275–1280 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenaillon O., et al. , The molecular diversity of adaptive convergence. Science 335, 457–461 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Kvitek D. J., Sherlock G., Whole genome, whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLoS Genet. 9, e1003972 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenaillon O., et al. , Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165–170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drewke C., et al. , 4-O-phosphoryl-L-threonine, a substrate of the pdxC(serC) gene product involved in vitamin B6 biosynthesis. FEBS Lett. 390, 179–182 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Al-Rabiee R., Zhang Y., Grant G. A., The mechanism of velocity modulated allosteric regulation in D-3-phosphoglycerate dehydrogenase. Site-directed mutagenesis of effector binding site residues. J. Biol. Chem. 271, 23235–23238 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Tazuya-Murayama K., et al. , Effect of L-serine on the biosynthesis of aromatic amino acids in Escherichia coli. J. Nutr. Sci. Vitaminol. (Tokyo) 52, 256–260 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Neumann S., Vladimirov N., Krembel A. K., Wingreen N. S., Sourjik V., Imprecision of adaptation in Escherichia coli chemotaxis. PLoS One 9, e84904 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costrejean J. M., Truffa-Bachi P., Threonine-sensitive homoserine dehydrogenase and aspartokinase activities of Escherichia coli K12. Kinetic and spectroscopic effects upon binding of serine and threonine. J. Biol. Chem. 252, 5332–5336 (1977). [PubMed] [Google Scholar]
- 26.Hama H., Sumita Y., Kakutani Y., Tsuda M., Tsuchiya T., Target of serine inhibition in Escherichia coli. Biochem. Biophys. Res. Commun. 168, 1211–1216 (1990). [DOI] [PubMed] [Google Scholar]
- 27.Woods R., Schneider D., Winkworth C. L., Riley M. A., Lenski R. E., Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 103, 9107–9112 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Notley-McRobb L., King T., Ferenci T., rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184, 806–811 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferenci T., The spread of a beneficial mutation in experimental bacterial populations: The influence of the environment and genotype on the fixation of rpoS mutations. Heredity 100, 446–452 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Cheng K. K., et al. , Global metabolic network reorganization by adaptive mutations allows fast growth of Escherichia coli on glycerol. Nat. Commun. 5, 3233 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Lee Y. H., Nam K. H., Helmann J. D., A mutation of the RNA polymerase β′ subunit (rpoC) confers cephalosporin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 57, 56–65 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad T. M., et al. , RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. U.S.A. 107, 20500–20505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wytock T. P., et al. , Experimental evolution of diverse Escherichia coli metabolic mutants identifies genetic loci for convergent adaptation of growth rate. PLoS Genet. 14, e1007284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knöppel A., et al. , Genetic adaptation to growth under laboratory conditions in Escherichia coli and Salmonella enterica. Front. Microbiol. 9, 756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conrad T. M., et al. , Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 10, R118 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandberg T. E., et al. , Evolution of Escherichia coli to 42 °C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol. 31, 2647–2662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke J. E., Kime L., Romero A D., McDowall K. J., Direct entry by RNase E is a major pathway for the degradation and processing of RNA in Escherichia coli. Nucleic Acids Res. 42, 11733–11751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuznetsova E., et al. , Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 281, 36149–36161 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Kaplun A., et al. , Structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from Escherichia coli. J. Mol. Biol. 357, 951–963 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Patten C. L., Kirchhof M. G., Schertzberg M. R., Morton R. A., Schellhorn H. E., Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272, 580–591 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Ishii N., et al. , Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science 316, 593–597 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Dai M., Rogers J. B., Warner J. R., Copley S. D., A previously unrecognized step in pentachlorophenol degradation in Sphingobium chlorophenolicum is catalyzed by tetrachlorobenzoquinone reductase (PcpD). J. Bacteriol. 185, 302–310 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yadid I., Rudolph J., Hlouchova K., Copley S. D., Sequestration of a highly reactive intermediate in an evolving pathway for degradation of pentachlorophenol. Proc. Natl. Acad. Sci. U.S.A. 110, E2182–E2190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury C., Sinha S., Chun S., Yeates T. O., Bobik T. A., Diverse bacterial microcompartment organelles. Microbiol. Mol. Biol. Rev. 78, 438–468 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T., et al. , Construction of Escherichia coli K12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J., Russell D. W., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, ed. 3, 2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available within the paper and supplementary material.