Significance
While the possibilities of living systems are theoretically near-infinite, the reality of biology at any instant is bounded. Here, we demonstrate how the discovery of functional information encoded in natural DNA sequences can be formally completed. Using bacteriophage øX174 as a model system, we identify and disrupt all conserved but unknown open reading frames, demonstrating by experiment that no cryptic functions remain to be discovered. Our proof by synthesis definitively affirms that the øX174 protein-coding annotations are complete and showcases how genomics and synthetic biology can be combined to operationally complete the genetics of natural living systems.
Keywords: synthetic biology, synthetic genomics, reverse genomics, gene discovery, cleanomics
Abstract
We develop a method for completing the genetics of natural living systems by which the absence of expected future discoveries can be established. We demonstrate the method using bacteriophage øX174, the first DNA genome to be sequenced. Like many well-studied natural organisms, closely related genome sequences are available—23 Bullavirinae genomes related to øX174. Using bioinformatic tools, we first identified 315 potential open reading frames (ORFs) within the genome, including the 11 established essential genes and 82 highly conserved ORFs that have no known gene products or assigned functions. Using genome-scale design and synthesis, we made a mutant genome in which all 11 essential genes are simultaneously disrupted, leaving intact only the 82 conserved but cryptic ORFs. The resulting genome is not viable. Cell-free gene expression followed by mass spectrometry revealed only a single peptide expressed from both the cryptic ORF and wild-type genomes, suggesting a potential new gene. A second synthetic genome in which 71 conserved cryptic ORFs were simultaneously disrupted is viable but with ∼50% reduced fitness relative to the wild type. However, rather than finding any new genes, repeated evolutionary adaptation revealed a single point mutation that modulates expression of gene H, a known essential gene, and fully suppresses the fitness defect. Taken together, we conclude that the annotation of currently functional ORFs for the øX174 genome is formally complete. More broadly, we show that sequencing and bioinformatics followed by synthesis-enabled reverse genomics, proteomics, and evolutionary adaptation can definitely establish the sufficiency and completeness of natural genome annotations.
Ongoing improvements in DNA sequencing tools have allowed researchers to begin to catalog the full diversity of genetic information in nature (1, 2). However, assigning biological functions to the so-revealed sequence data has progressed less quickly and surely. For example, genomes are automatically annotated using computational methods that leverage what is known of conserved molecular mechanisms and incorporate training data from an increasing number of annotated genomes. Nevertheless, accurate prediction of protein-coding open reading frames (ORFs) remains challenging (3). One particular challenge is that, typically, only a small subset of apparent ORFs actually encode proteins. Moreover, the quantitative scoring of detailed sequence characteristics for ORFs appears as a continuum, from ORFs that do not encode proteins to full-fledged protein-coding ORFs (4).
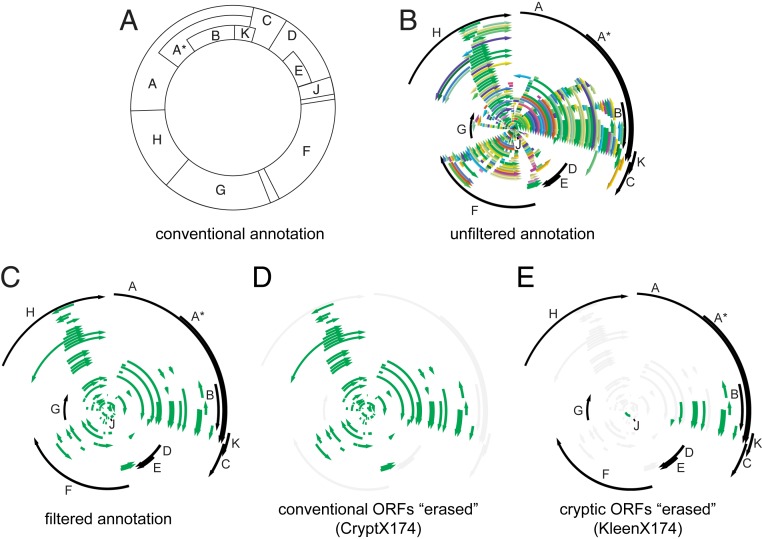
Bacteriophage øX174 has been useful as a model system for genetics research, from before the sequencing era through the present. The number of identified protein-coding ORFs in øX174 increased as methods were developed and applied. For example, successive forward saturating genetic screens enabled identification of 10 protein-coding ORFs in the absence of genome sequence information (5–10). Sequencing of the 5,386 nucleotide øX174 genome (11) and later comparison to the related G4 phage genome enabled identification of one additional protein-coding ORF (12). Today, the øX174 genome is still recognized as encoding only these 11 proteins (Fig. 1A) (13). In contrast, an unfiltered view of the øX174 genome reveals up to 315 ORFs. While most of these ORFs are presumably not protein-coding (Fig. 1B), it is not absolutely certain that every protein-coding ORF in the øX174 genome has been discovered.
Fig. 1.
Designing genomes encoding only essential or only cryptic ORFs. (A) Contemporary genetic map of øX174. Lettered boxes represent the 11 established protein-coding ORFs. (B) The øX174 genome with 315 ORFs, each 60 bp or longer and starting with an ATG, GTG, or TTG codon (various colors), plus the 11 established protein-coding ORFs (black). (C) Filtered annotation of the øX174 genome with ORFs; 11 previously identified protein-coding ORFs (black) and 82 cryptic ORFs of unknown protein-coding status (green). (D) The cryptX174 genome design with the 11 established ORFs disrupted (gray), and 82 cryptic ORFs of unknown protein-coding status (green). (E) The kleenX174 genome design showing disruption of 71 cryptic ORFs (gray), 11 previously identified protein-coding ORFs (black), and remaining 11 cryptic ORFs of unknown protein-coding status (green).
De novo synthesis of the øX174 and other genomes is becoming routine (14, 15) and has already helped show when genes are essential (16), when gene overlaps are nonessential (17), and, in general, that the “genomes encoding natural biological systems can be systematically redesigned and built anew in service of scientific understanding or human intention” (18). Practical applications of genome-scale reverse genetics approaches are also being pioneered, most notably in vaccine development (19, 20). Synthesis has also been used to reveal and better understand more-subtle puzzles ranging from retrotransposon activity (21) to whether the amount of conserved information encoded in a natural DNA sequence exceeds the known functions assigned to the sequence (22).
Thus, and taken together, we envisioned a more formal, systematic, and broadly generalizable approach for determining whether all ORFs in a given genome have been discovered. Rather than searching for individual phenotypes associated with each remaining potential cryptic ORF, we start by accepting a presumption that all cryptic ORFs have, individually and collectively, no functions whatsoever. Then, by using reverse genomics to systematically generate a derivative genome designed to test the starting presumption, we purposefully and efficiently identify whether any further functions remain to be discovered or, if none are found, definitively declare that no such functions remain unknown. While the starting presumption may be widely accepted—otherwise, any still-hidden genes would have been found already by established methods—the presumption itself seems never to have been tested by direct experiment. More importantly, if we can formally demonstrate that no further functional information is encoded in a natural genome, albeit with the simplest model genome as a first example and under limited experimental conditions, then we can demonstrate a method by which genetics, as a discovery science, can approach “completeness” in both an empirical and formal sense (23).
Results
Identifying Conserved ORFs in øX174.
To directly test the completeness of the annotation of the bacteriophage øX174 genome, we compared 23 Bullavirinae genomes to identify whether any highly conserved protein-coding ORFs could be detected. We focused on protein-coding ORFs, since studies of øX174 mRNA have not revealed any noncoding RNA genes (24). We further focused on evolutionarily conserved ORFs, since they are more likely to encode functions compared to nonconserved ORFs (4).
We first filtered the set of all possible ORFs to include only those beginning with one of the 3 most common start codons in the Escherichia coli (E. coli) host (ATG, GTG, and TTG) and preceded by an identifiable Shine−Dalgarno motif (25, 26). Our analysis identified a set of 83 conserved ORFs shared between the øX174 genome and at least one other Bullavirinae genome. Eleven of these identified ORFs were the known protein-coding ORFs of the Bullavirinae subfamily (A, A*, B, C, D, E, F, G, H, J, and K). We designated the remaining set of 72 ORFs as the cryptic ORFs of øX174, because they lack prior evidence of protein expression and function.
We ranked each of the 83 identified ORFs (11 known plus 72 cryptics) by the number of times each was detected by computational analysis of the 23 genomes studied, finding a range of conservation among the set (SI Appendix, Figs. S1 and S2). Eight of the 11 known protein-coding ORFs clustered as the highest scoring, due to their detection by all computational tools in a majority of the 23 searched genomes (SI Appendix, Fig. S3). Known protein-coding ORFs A*, E, and K had much lower scores similar to those of 8 cryptic ORFs (2, 13, 15, 29, 31, 46, 46, and 57), due to their detection by only some of the analysis methods (SI Appendix, Fig. S3). We manually added 10 more cryptic ORFs that are known to be conserved across the PhiX174microvirus genus (SI Appendix, Fig. S2) (27), leading to 82 candidate cryptic ORFs total (Fig. 1C) (GenBank accession no. MN385565 (28)).
Design and Characterization of a Genome Encoding only Cryptic ORFs.
We designed a variant øX174 genome in which the start codons initiating translation of the 11 known essential protein-coding ORFs are simultaneously disrupted. We called the resulting genome “cryptX174,” as it should only express proteins from cryptic ORFs (Fig. 1D). Our goals in making cryptX174 were to test the validity of the established start codons and to construct a template that contains all ORFs except for the 11 known protein-coding ORFs; we wanted a genome whose design better supports detection of peptides that may be expressed at very low levels. Practically, the cryptX174 design changes the ATG start codons of protein-coding ORFs A, A*, B, C, D, E, F, G, H, J, and K to either ATA or ACG (SI Appendix, Table S1 and GenBank accession no. MF426915 (29)); specific alternate codons were selected to have the least impact on the amino acids coded in the other 5 reading frames.
We tested the viability of the cryptX174 design by in vitro construction followed by transfection into host E. coli C cells. We found that transfection of the cryptX174 genome did not produce observable plaques. The cryptX174 nonviability suggests that a disruption in at least one of the essential protein-coding ORFs cannot be compensated for by an upstream or downstream in-frame start codon.
We next sought to determine whether any cryptic ORFs are expressed. Given that the cryptX174 genome did not encode viable phage, we used cell-free transcription and translation to measure protein production directly. We generated linear genomes from both the wild-type and cryptX174 templates, adding a terminal T7 promoter and terminator to each. We used mass spectrometry to measure cell-free protein production from both the wild-type and cryptX174 templates. We identified peptides from 8 of the 11 known protein-coding ORFs, as well as the small cryptic ORF 75, which is only 23 amino acids in length (MSIITPKRKVLRMSVQDCWRPPL). Peptides from cryptic ORF 75 were produced from both wild-type øX174 and cryptX174 templates, and the N-terminal ORF 75 peptide (fMSIITPK) was observed unprocessed with an N-formylmethionine modification still intact (Table 1). N-formylmethionine is a marker of translation initiation in E. coli that is usually cleaved off quickly in vivo (30); the presence of a N-formylmethionine modification on the ORF 75 N-terminal methionine gave us confidence that this peptide arose from ORF 75-specific translation initiation and was not an internal peptide encoded by another ORF. N-formylmethionine peptides produced from genes A, D, and H were also observed, supporting the established start sites for these genes (Table 1).
Table 1.
Peptides observed by mass spectrometry following cell-free expression from wild-type and synthetic genome templates
| Genome templates | ||||||
| Protein | Peptide sequence† | No. of spectra | Notes | WT øX174 | cryptX174 | kleenX174 |
| 75 | fMSIITPK | 22 | N-terminal formylmethionine | + | + | — |
| A | fMQGVEFDNGDMYVDGHK | 3 | N-terminal formylmethionine | — | — | + |
| A | WFLENEK | 7 | + | — | — | |
| A or A* | VTVADVLAAQPVTTNLLK | 4 | + | — | + | |
| B | RDEIEAGK | 5 | — | — | + | |
| C | None detected | — | — | — | ||
| D | fMSQVTEQSVRFQTALASIK | 33 | N-terminal formylmethionine | + | — | + |
| E | None detected | — | — | — | ||
| F | EIQYLNAK | 3 | + | — | + | |
| G | LVLTSVTPASSAPVLQTPK | 7 | + | + | — | |
| G | EIICLQPLK | 8 | — | — | + | |
| H | FGAIAGGIASALAGGAMSK | 9 | + | + | — | |
| H | fMFGAIAGGIASALAGGAMSK | 11 | N-terminal formylmethionine | + | + | — |
| H | LLDLVGLGGK | 11 | + | — | — | |
| H | MQLDNQK | 2 | + | — | — | |
| H | SAIQGSNVPNPDEAAPSFVSGAMAK | 5 | + | + | — | |
| J | GARLWYVGGQQF | 31 | + | — | — | |
| K | LLLCDLSPSTNDSVKN | 5 | + | — | + | |
øX174-specific peptides observed from shotgun mass spectrometry measurement of proteins produced in a Protein synthesis Using Recombinant Elements (PURE) expression reaction. Only peptides with P value ≤ 0.05 and with ≥2 observed spectra are reported. The protein search database contained both E. coli and øX174 protein sequences corresponding to the appropriate phage genome (WT øX174, cryptX174, or kleenX174).
Design and Characterization of a Genome “Cleaned” of Cryptic ORFs.
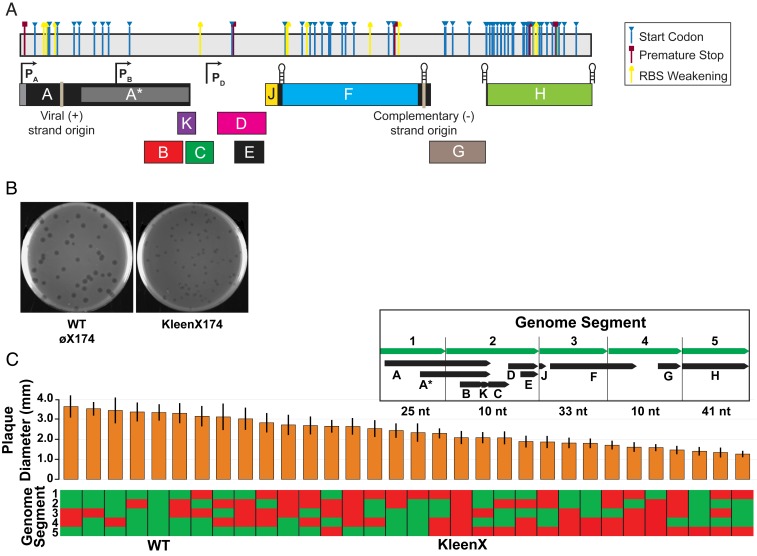
We next designed a variant øX174 genome in which as many cryptic ORFs are simultaneously disrupted as possible. To do so, we introduced silent mutations that either 1) changed the start codon to any codon but ATG, GTG, or TTG, 2) weakened the putative ribosome binding site (RBS) by reducing the frequency of A and G nucleotides, or 3) introduced a premature stop codon near the 5′-end of the ORF. In all cases, we did not alter the protein coding sequence for any of the 11 known øX174 genes. By this approach, we were able to design disruptions for 71 of the 82 cryptic ORFs, including ORF 75 (Dataset S1). We named the resulting genome “kleenX174” (Fig. 1E); the kleenX174 genome differs in only 120 positions from the wild type and maintains the same GC content (Fig. 2A and GenBank accession no. MF426914 (31)).
Fig. 2.
A “clean” øX174 genome in which 71 cryptic ORFs are simultaneously disrupted is viable but has reduced fitness. (A) Linear depiction of kleenX174 genome showing the locations and modes of cryptic ORF disruption; see Dataset S1 for detailed information. (B) Plaques of wild-type and kleenX174 phage. 85 mm diameter plates. (C) Plaque diameter of wild-type/kleenX174 chimeras, arranged from largest average plaque size to smallest. Vertical bars represent 1 SD from n = 50 plaque measurements. Each chimeric phage consists of 5 genome segments chosen from a mixture of wild-type genome segment (green) and kleenX174 modified genome segments (red). (Inset) The boundaries of the 5 genome segments, protein-coding ORFs found in each segment, and total number of nucleotide differences between wild-type øX174 and kleenX174 genome sequences in each segment.
We constructed the kleenX174 genome by in vitro construction followed by transfection into host E. coli C cells, which, in this case, resulted in plaques (Fig. 2B). We found that kleenX174 plaques had a smaller diameter (2.1 ± 0.3 mm) compared to wild-type plaques (3.3 ± 0.4 mm). Smaller plaque sizes generally imply slower growth rates and lower fitness (32).
kleenX174 Chimera Design and Phenotypic Measurements.
To map the kleenX174 growth defect, we made chimeric genomes by sectioning the øX174 genome into 5 segments and assembling 32 chimeras containing either the wild-type or kleenX174 sequence for each segment. We used plaque sizes for each chimeric phage to identify which of the 120 synonymous mutations within the kleenX174 genome design might contribute to a reduced plaque size phenotype. Measured diameters of the resulting plaques from each chimeric genome showed that segment 5, encompassing all of gene H, was implicated as the loci within kleenX174 most responsible for its small-plaque phenotype (Fig. 2C).
Identification of Proteins Produced from kleenX174.
We next wanted to identify the proteins expressed from the kleenX174 genome. We used cell-free expression to produce proteins from linearized kleenX174 template and measured proteins via mass spectrometry. We identified peptides from 5 of the 11 known øX174 genes (Table 1). We did not detect ORF 75 peptides; the start codon of ORF 75 is disrupted in kleenX174 (ATG > ACG). We observed N-formylmethionine peptides produced from genes A and D (Table 1), further supporting the annotated start sites for these genes. We did not detect H protein peptides from kleenX174 template but did from both cryptX174 and wild-type templates. We hypothesized that the lack of detectable H protein peptides from kleenX174 template along with the low fitness of chimeric phage containing kleenX174 segment 5 might both be due to decreased protein H production (Fig. 2C).
Evolutionary Adaptation of kleenX174 Reveals a Functionally Important Codon.
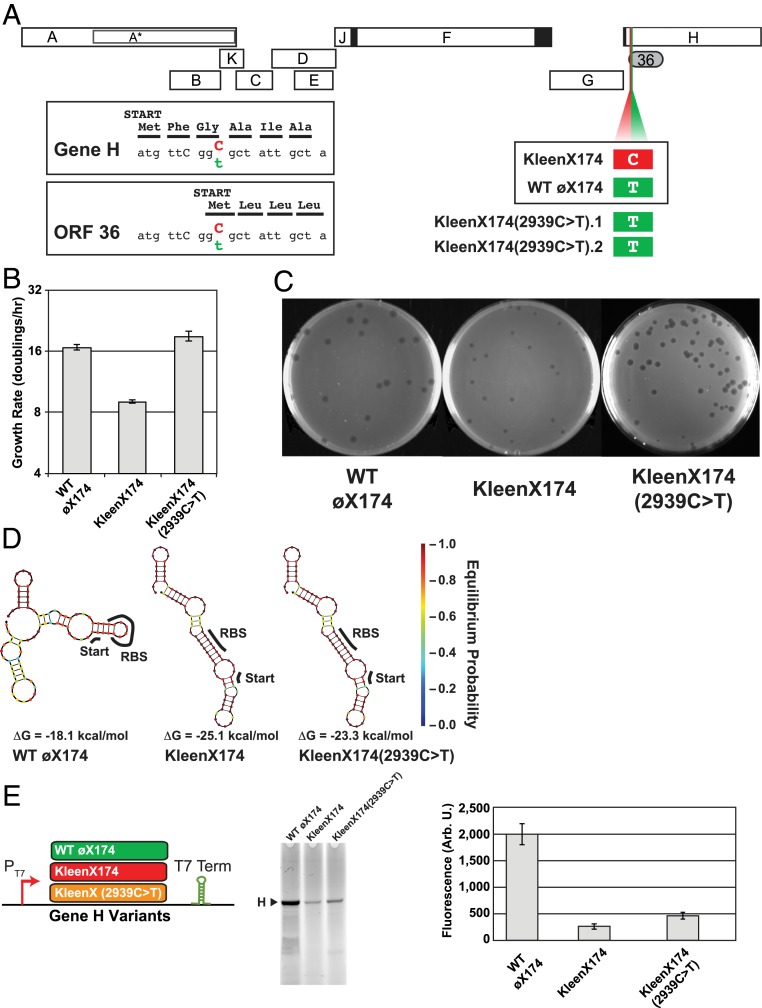
Refactored genomes can serve as a starting point for experimental adaptions that reveal design problems or novel functional genetic elements (33). Thus, to better understand the kleenX174 growth defect, we passaged the phage for 48 generations, selecting for faster-growing mutants. Following this process, sequencing indicated that one change, kleenX174(2939C > T), was most abundant in both bulk culture and individually sequenced plaques (Fig. 3A). Moreover, we observed the 2939C > T mutation in repeated serial passage experiments. We isolated pure kleenX174(2939C > T) phage stock and measured the effect of the 2939C > T mutation on growth rates. We found that the growth rate and plaque size of the kleenX174(2939C > T) mutant was restored to that of wild type (Fig. 3 B and C).
Fig. 3.
Evolutionary adaptation of kleenX174 to high-growth rate results in a reversion (2939C > T) and increased gene H expression. (A) The mutation 2939C > T observed in 2 independent adaptation experiments both restores the putative start codon of cryptic ORF 36 (GCG > GTG) and silently changes the third codon of gene H (GGC > GGT, Gly > Gly). (B) Evolved kleenX174(2939C > T) grows faster than ancestral kleenX174 in liquid culture (population doublings per hour). (C) The kleenX174(2939C > T) mutation also recovers plaque size. (D) RNA structure predictions of gene H sequence variants from wild-type øX174, kleenX174, and mutant kleenX174(2939C > T). NUPACK lowest-energy RNA structures generated from an 83-nt window surrounding the gene H start codon. (E) Protein H production from kleenX174(2939C > T) is increased compared to ancestral kleenX174 genome. Protein H produced from synthetic dsDNA templates (Datasets S2–S4) containing either wild-type øX174, kleenX174, or kleenX174(2939C > T) gene H sequence plus 20-bp upstream sequence identical to genome background, flanked by T7 promoter (PT7) and terminator sequences (T7 Term). PURExpress reactions run with 0.8 nM template were separated on SDS/PAGE followed by fluorescence detection of BODIPY-FL tagged lysine incorporated into proteins produced during the transcription/translation reaction. Error bars represent 1 SD (n = 3).
The 2939C > T mutation is located within segment 5 from the chimera experiment and appears to both revert the start codon of cryptic ORF 36 (from GCG to GTG) and, in a different reading frame, cause a synonymous glycine mutation (from GGC to GGT) in the third codon of gene H (Fig. 3A). GGC is used more frequently than GGT in E. coli (0.35 versus 0.27), suggesting this substitution might modulate gene H translation rate.
To explore the effect of the 2939C > T mutation on gene H mRNA structure, we performed RNA folding simulations of the 5′-ends of gene H sequence variants. We found that the kleenX174 gene H structure is predicted to be −7 kcal/mol (28%) more stable than the wild type and has a different predicted base-pairing pattern (Fig. 3 D, Left and Middle), while the kleenX174(2939C > T) mutant retains the altered RNA base-pairing structure of kleenX174 but should be less stable (Fig. 3 D, Right). We also found that the predicted gene H mRNA structural stability in the kleenX174 design is outside the range predicted using the gene H sequences from all 23 Bullavirinae genomes (SI Appendix, Fig. S7). Higher mRNA stability around the start codon has been implicated in decreased protein expression (34).
To test the hypothesis that the small plaque phenotype of kleenX174 is due to decreased gene H protein production, we created 3 synthetic versions of gene H matching those of wild-type øX174, kleenX174, and kleenX174(2939C > T). We measured H protein production from each template in a cell-free expression system (Fig. 3E). We found that H protein production from a kleenX174 template was only a fraction of wild-type levels (13%) and that the 2939C > T mutation almost doubles the amount of H protein produced (23%) (Fig. 3E).
Discussion
We definitively determined, by perturbation design, direct measurements, and evolutionary adaptation, that the conventional annotation of functional genes in the øX174 genome is complete. Specifically, we found that 71 of 82 highly conserved cryptic ORFs within the øX174 genome are simultaneously dispensable for growth at wild-type rates under the conditions tested and do not encode detectable amounts of protein. Even the 5 cryptic ORFs (2, 29, 31, 46, and 57) that have conservation scores greater than known protein-coding ORFs can be disrupted without apparent phenotypic effect (SI Appendix, Fig. S1). We acknowledge that 11 of 82 conserved cryptic ORFs cannot be disrupted without also altering one or more known essential genes and were therefore not tested by ORF disruption directly. However, for each of these cryptic ORFs, no peptides specific to their expression were detected by mass spectrometry. Disruption of one cryptic ORF (75) for which expressed peptides were detected by mass spectrometry had no observed impact on phenotype or fitness under the conditions tested.
Evolutionary adaptation for faster-growing mutants derived from a synthetic genome devoid of most conserved cryptic ORFs revealed a consistent reversion that partially restored wild-type translation rates for a known essential gene (H) and fully restored phage fitness under laboratory conditions. Gene H is a DNA pilot protein that is present at 12 copies per capsid (13) and needed for genome injection into host cells (35). The rarer glycine codon in the N termini of wild-type gene H could be important for slowing translation if needed to ensure correct folding, timing of synthesis during infection (36), or level matching between B and H proteins during capsid assembly (37). A similar effect was reported in yeast containing a recoded PRE4, producing a growth defect from reduced Pre4 protein levels resulting from a more stable structure at the 3′ end of the recorded ORF (38).
We note that the so-selected reversion also restores a potential GTG start codon for cryptic ORF 36 (Fig. 3). The ORF 36 start codon and sequence is well conserved across 19 of the 23 genomes analyzed (SI Appendix, Figs. S4 and S5). However, despite the presence of an upstream RBS, strong start codon, and strong family conservation, we did not observe any peptides from ORF 36 in our experiments and conclude that ORF 36 does not produce protein.
From a methods perspective, many of the proteins produced via the cell-free expression system had intact N-terminal formylmethionine residues (Table 1). The Protein synthesis Using Recombinant Elements (PURE) transcription and translation system used in this work appears to lack the peptide deformylase activity necessary to remove N-terminal formyl groups (39). Such residues serve as hallmarks of translation initiation in bacteria but are typically quickly removed in vivo, thereby masking true translation initiation sites (40). Established methods such as ribosome footprinting (41) identify translation initiation sites by isolating ribosomes in the vicinity of mRNA start codons. However, detection of formylmethionine on peptide N termini provides direct biochemical evidence of translation initiation. The PURE system therefore shows significant promise as a tool for systematically mapping translation initiation, especially compared to methods that require peptide enrichment (42) or the use of knockout strains and translation inhibitors (43).
Here, we evaluated the information encoded within the øX174 genome as it existed when isolated from nature and since propagated and characterized under typical laboratory conditions. Any latent information that might contribute to future functions or to fitness in response to environmental changes or random mutations is not accounted for. For example, work in yeast has shown that, over evolutionary timescales, nongenic ORFs can be stably maintained and promoted to protein-expressing ORFs (4). Nevertheless, for a genome as it exists at a point in a lineage and within a specific environmental context, we have exhaustively characterized the ORF encoding capacity.
Going forward, it is easy to suggest that Mesoplasma florum, E. coli, Saccharomyces cerevisiae or other well-studied or model minimal organisms may serve as useful next genomes to consider formally completing (15, 44–46). However, extending the approaches developed here to such organisms will be challenging. Improved tools and methods will be essential. For example, we envision that improved cell-free expression systems combined with high-throughput generation and testing of “simplons” (resynthesized linear fragments of DNA designed to only encode a simplified set of annotated functions) may serve a key role. A suite of extracts representing a range of environmental conditions, as well as extracts whose composition are designed to recapitulate key branch points in the phylogenetic history of an organism, could serve to evaluate which known or cryptic ORFs are expressed under what conditions. Such data could then guide which cryptic ORFs, if any, warrant focused study in vivo.
So-informed, future work should also anticipate that completing more-complex genomes will require management of large numbers of quasi-essential genes (16, 47). Conditional knockdown screens using CRISPR interference (48, 49) might identify synthetic lethal combinations to be avoided. Genome sections would be replaced section by section with disrupted cryptic ORFs using homologous recombination assisted by CRISPR-Cas or integrase systems (50–53). Practitioners would follow progress via phenotypic assays and established frameworks (38), eventually leading to complete annotations of cleaned genomes (i.e., “cleanomes”).
In conclusion, we note again that the genomes encoding natural living systems are finite in length; the rate at which novel functional genetic information is “written” into genomes is limited by how quickly mutations can be selected for and fixed within competing populations; and the pace at which encoded functional genetic information is “lost” from genomes is greater than zero, as determined by spontaneous mutation rates and functional selection frequencies. Taken together, these 3 facts—1) an information storage system of finite capacity, 2) a finite rate of information encoding, and 3) a nonzero rate of information loss—would seem to practically bound the amount of functional information that can be encoded in any natural genome at any point in time. Thus, the science of discovering and understanding functional genetic information encoded in natural living systems (i.e., genetics) should itself be formally finite and bounded. Herein, we have explored such a finite framing by starting from the presumption that the existing discovery science for one natural genome had been completed. Although the understanding for almost all other natural living systems is now very far from complete, we submit for consideration that the underlying framing should nevertheless hold, and that the science of genetics may progress to completion more quickly by more systematically complementing classical forward discovery-based approaches with reverse approaches that definitively demonstrate what does not matter. For engineers or others interested in refining and repurposing natural genetic sequences, we note that the generalizable methods applied here can collectively serve to “validate” DNA by demonstrating definitively what molecules are encoded in any given DNA sequence.
Methods
Bacterial Strains, Growth Conditions, and Plaque Assays.
The bacterial strain E. coli C (ATCC 13706) grown at 37 °C in phage lysogeny broth (LB) was used throughout (54). Transformed E. coli C cells were regrown for 1.5 h in phage LB followed by mixing 10 µL of transformed cell serial dilutions with 200 µL of untransformed E. coli C grown overnight to saturation. The mixed cells were plated using the double-layer agar plate method (15-mL bottom-layer 1.2% agar, 5-mL top-layer 0.7% agar) with either phage LB or Tryptone/KCl (TK) agar plates (17, 55). Plates were incubated at 37 °C for 16 to 18 h before visualization. Plaque sizes were measured using Adobe Photoshop CS6 Analysis->Measurement command from images of plates containing between 40 and 100 plaques.
ORF Prediction.
Four established gene prediction tools were used in combination to find ORFs in the øX174 genome: Gene Locator and Interpolated Markov ModelER (GLIMMER) (56), GeneMark (57) using the GeneMark.hmm PROKARYOTIC (version 2.10b) algorithm, EasyGene (58), and Prodigal (59). We analyzed genomes from 23 different Bullavirinae virus subfamily phages from 3 different genera: the PhiX174microvirus phages: WA11 (DQ079895.1), WA10 (DQ079894.1), WA4 (DQ079893.1), S13 (M14428.1), øX174 (NC_001422.1), NC56 (DQ079892.1), NC51 (DQ079891.1), NC41 (DQ079890.1), NC37 (DQ079889.1), NC16 (DQ079888.1), NC11 (DQ079887.1), NC7 (DQ079886.1), NC5 (DQ079885.1), NC1 (DQ079884.1), ID45 (DQ079883.1), ID22 (DQ079881.1), and ID1 (DQ079880.1); the G4microviruses: ID18 (NC_007856.1), G4 (NC_001420.1), and ID2 (NC_007817.1); and the alpha3microviruses: alpha3 (NC_001330.1), St-1 (NC_012868.1), and WA13 (NC_007821.1). These genome sequences were analyzed for ORFs initiating from the strongest 3 start codons in E. coli: ATG, GTG, and TTG. In addition to computationally identified ORFs, we also identified 10 ORFs by reference to past literature that manually predicted potential protein-producing ORFs in the øX174 genome based on RBS location upstream of ATG start codons (27).
Phage Genome Design and Construction.
All known commercial øX174 preparations have sequences that differ from the canonical Sanger øX174 sequence used as the basis for our kleenX174 design (14). Thus, we built from scratch the wild-type øX174 genome corresponding to the original Sanger 1977 sequence, as defined in GenBank (11). Specifically, we downloaded the NC_001422.1 sequence, split it into 5 approximately equal-sized pieces with 60-base pair (bp) overhangs, synthesized, assembled using in vitro homologous recombination (60), and transformed directly into the E. coli C host strain (61, 62) to recover viable phage.
To design the kleenX174 genome (GenBank accession no. MF426914.1), the wild-type øX174 sequence was modified so that 71 of 82 predicted cryptic ORFs would be disrupted (Dataset S1). We constrained genome modifications to only incorporate silent changes, whereby the amino acid sequence of any overlapping known øX174 protein-coding ORF remained the same. We first attempted to modify the start codon of each cryptic ORF, as we reasoned doing so would give the highest probability of successfully disrupting ORFs while minimizing the number of nucleotides altered. However, if a start codon could not be so altered, we instead changed the RBS of the gene to reduce A and G base frequency, or modified in-frame bases to create a premature stop codon as close to the start codon as possible. In 11 cases, no silent changes could be made without also disrupting a known ORF, and so these cryptic ORFs were left unchanged; such cases were concentrated in the area of the genome containing the overlapping essential genes A, A*, B, and K.
The kleenX174 genome was synthesized in 5 parts with 60-bp overhangs, assembled using in vitro homologous recombination (60), and transformed directly into the E. coli C host strain (61, 62).
To design and build the cryptX174 genome (GenBank accession no. MF426915.1), we analyzed the start codons of all known øX174 protein-coding ORFs to determine nucleotide changes that could be made to mutate each to another codon that was not ATG, while minimizing the impact on all overlapping upstream and downstream codons in the 5 remaining reading frames. The so-specified cryptX174 genome was synthesized in 3 parts with 60-bp overhangs, assembled using in vitro homologous recombination followed by transformation directly into E. coli C cells.
Chimeric Genome Design and Construction.
The Sanger 1977 wild-type øX174 and kleenX174 genomes were computationally split into 5 approximately equal-sized segments with joints between segments placed in locations that did not have any modifications from the wild-type sequence. The areas around the synthetic DNA joints had this requirement so that they could be swapped seamlessly with the corresponding area of the wild-type genome during construction of chimeric genomes. Assembly of the 32 chimeric genomes was planned using the Teselagen web application (63). Five wild-type and 5 kleenX174 parts were amplified using primers specified by Teselagen (SI Appendix, Table S2) and assembled from 30 fmol of each of the 5 pieces using in vitro homologous recombination. Assembled genomes were transformed directly into chemically competent E. coli C.
Creating Linear Genome Templates for Cell-Free Expression and Mass Spectrometry.
We linearized the genomes just following the stop codon of gene H and added 88 bp of the H/A-intergenic region and start of gene A to the 3′-end of each template. These modifications ensured that each linear genome contained uninterrupted sequences for all predicted ORFs (SI Appendix, Fig. S2). We added 8.6 nM of each genome and 1 U/µL Murine RNase inhibitor to the PURExpress expression system (New England Biolabs). Each genome was run in a separate reaction using either 12C/14N-, 13C/14N-, or 13C/15N-lysine (Thermo Scientific Pierce) at 25 °C for 16 h. All 3 reactions were pooled and digested with Lys-C, cleaned up, and run on nanoLC-2D and LTQ-Orbitrap Velos (ThermoFisher) followed by analysis with Byonic v2.0-25 software. The peptide search database contained all E. coli protein sequences pooled with the 11 known øX174 proteins and the 82 cryptic ORFs identified in this work.
Evolutionary Selection of kleenX174 Phage.
To subject the kleenX174 phage to evolutionary selection for increased growth rate, we followed an established adaptation protocol (54). Briefly, E. coli C was grown overnight at 37 °C in phage LB then split 1/100 into 25 mL of phage LB and regrown to A600 = 0.5, followed by the addition of 104 kleenX174 phage (multiplicity of infection (MOI) = 2e−5). The infected culture was shaken for 40 min (∼3 generations), and then chloroform was added to a final concentration of 4% (vol/vol). The culture was then centrifuged at 15,000 × g for 15 min, and the supernatant was removed to another tube, titered to determine phage concentration, and used to infect a fresh batch of E. coli cells at A600 = 0.5. After 16 passages (∼48 generations), the lysate was used as template in rolling circle amplification (RCA) reactions and Sanger-sequenced using a set of primers to cover the entire øX174 genome (SI Appendix, Table S2). The lysate was also plated, and isolated plaques were extracted into water and used as template for RCA, followed by Sanger sequencing.
Protein H Production from Synthetic DNA Templates.
Double-stranded synthetic DNA templates corresponding to the wild-type øX174, kleenX174, and kleenX174(2939C > T) sequences were designed and synthesized (IDT) with added 5′ T7 promoter and 3′ T7 terminators, as recommended by NEB in the PURExpress kit. Cell-free in vitro expression reactions were run with 0.8 nM template at 37 °C for 2 h under standard conditions with 1 U/µL Murine RNase inhibitor (NEB) and FluoroTect BODIPY-FL labeled Lysine (Promega). Cell-free reactions were processed with RNase A (0.1 mg/mL) to remove unreacted transfer RNA, then run on 12% Bis-Tris SDS/PAGE using 2-(N-Morpholino)ethanesulfonic acid (MES) buffer (Life Technologies). Fluorescence was visualized using a Typhoon 9410 (GE) scanner with laser settings of Blue2 488-nm BP 520 normal sensitivity. Band volumes were calculated using ImageJ v1.49 (64).
mRNA Structure Predictions.
RNA folding simulations were performed on an 83-nt window around the start codon of each analyzed ORF using the Nucleic Acid Package (NUPACK) web server with default settings (65). Lowest-energy structures were reported. MUltiple Sequence Comparison by Log-Expectation (MUSCLE) (66) multiple sequence alignments were also performed using the European Bioinformatics Institute (EMBL-EBI) web servers (https://www.ebi.ac.uk/Tools/msa/muscle/).
Data Availability.
All data generated or analyzed during this study are available from the corresponding authors on reasonable request. Sequences of wild-type øX174 annotated with the cryptic ORFs (accession no. MN385565), kleenX174 (accession no. MF426914), and cryptX174 (accession no. MF426915) are available via National Center for Biotechnology Information’s GenBank.
Supplementary Material
Acknowledgments
We acknowledge Jerome Bonnet, Pakpoom Subsoontorn, Monica Ortiz, Anwar Sunna, Ariel Hecht, Tom Williams, Heinrich Kroukamp, Jeff Glasgow, Marc Salit, Arend Sidow, and Joe Jacobson for helpful discussions and feedback. We acknowledge Ryan Leib and Chris Adams at the Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry (https://mass-spec.stanford.edu/) for assistance in protein analysis. This work was supported by students enrolled in the Stanford University Bioengineering Department Research Experience for Undergraduates program and the San Mateo High School Biotechnology Career Pathway Internship Program. P.R.J. was supported by a Canadian Natural Sciences and Engineering Research Council Postdoctoral Fellowship (PDF-388725-2010) and Macquarie University’s Molecular Sciences Department, Faculty of Science, and Deputy Vice Chancellor (Research). Additional support was provided by the Stanford/National Institute of Standards and Technology Joint Initiative for Metrology in Biology (https://jimb.stanford.edu/) and a gift from Agilent Technologies, Inc.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Nucleotide Database at https://www.ncbi.nlm.nih.gov/nuccore via accession nos. MN385565, MF426914, and MF426915.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905990116/-/DCSupplemental.
References
- 1.Carlson R., The pace and proliferation of biological technologies. Biosecur. Bioterror. 1, 203–214 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Landenmark H. K. E., Forgan D. H., Cockell C. S., An estimate of the total DNA in the biosphere. PLoS Biol. 13, e1002168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatusova T., et al. , “Prokaryotic genome annotation pipeline” in The NCBI Handbook (National Center for Biotechnology Information, Bethesda, MD, 2013), pp. 175–188. [Google Scholar]
- 4.Carvunis A. R., et al. , Proto-genes and de novo gene birth. Nature 487, 370–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchison C. A. I., Bacteriophage PhiX174: Viral Genes and Functions, PhD thesis, California Institute of Technology, Pasadena, CA (2003). (1969).
- 6.Burgess A. B., Denhardt D. T., Studies on phiX174 proteins. I. Phage-specific proteins synthesized after infection of Escherichia coli. J. Mol. Biol. 44, 377–386 (1969). [DOI] [PubMed] [Google Scholar]
- 7.Jeng Y., Gelfand D., Hayashi M., Shleser R., Tessman E. S., The eight genes of bacteriophages phi X174 and S13 and comparison of the phage-specified proteins. J. Mol. Biol. 49, 521–526 (1970). [DOI] [PubMed] [Google Scholar]
- 8.Mayol R. F., Sinsheimer R. L., Process of infection with bacteriophage phiX174. XXXVI. Measurement of virus-specific proteins during a normal cycle of infection. J. Virol. 6, 310–319 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benbow R. M., Hutchison C. A., Fabricant J. D., Sinsheimer R. L., Genetic map of bacteriophage phiX174. J. Virol. 7, 549–558 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linney E. A., Hayashi M. N., Hayashi M., Gene A of X174. 1. Isolation and identification of its products. Virology 50, 381–387 (1972). [DOI] [PubMed] [Google Scholar]
- 11.Sanger F., et al. , Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265, 687–695 (1977). [DOI] [PubMed] [Google Scholar]
- 12.Tessman E. S., Tessman I., Pollock T. J., Gene K of bacteriophage phi X 174 codes for a nonessential protein. J. Virol. 33, 557–560 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fane B. A., et al. , “PhiX174, the Microviridae” in The Bacteriophages, Abedon S. T., Calendar R. L., Eds. (Oxford University Press, 2005), chap. 11, pp. 129–145. [Google Scholar]
- 14.Smith H. O., Hutchison C. A. 3rd, Pfannkoch C., Venter J. C., Generating a synthetic genome by whole genome assembly: ϕX174 bacteriophage from synthetic oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 100, 15440–15445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson S. M., et al. , Design of a synthetic yeast genome. Science 355, 1040–1044 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Hutchison C. A., 3rd, et al. , Design and synthesis of a minimal bacterial genome. Science 351, aad6253 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Jaschke P. R., Lieberman E. K., Rodriguez J., Sierra A., Endy D., A fully decompressed synthetic bacteriophage øX174 genome assembled and archived in yeast. Virology 434, 278–284 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Chan L. Y., Kosuri S., Endy D., Refactoring bacteriophage T7. Mol. Syst. Biol. 1, 2005.0018 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dormitzer P. R., et al. , Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci. Transl. Med. 5, 185ra68 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Desselberger U., Reverse genetics of rotavirus. Proc. Natl. Acad. Sci. U.S.A. 114, 2106–2108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han J. S., Boeke J. D., A highly active synthetic mammalian retrotransposon. Nature 429, 314–318 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Schneider T. D., Stormo G. D., Excess information at bacteriophage T7 genomic promoters detected by a random cloning technique. Nucleic Acids Res. 17, 659–674 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stent G. S., The Coming of the Golden Age: A View of the End of Progress (Natural History Press, Garden City, NY, 1969). [Google Scholar]
- 24.Hayashi M., Aoyama A., Richardson D., Hayashi M., “Biology of the bacteriophage øX174” in The Bacteriophages, Calendar R., Ed. (Plenum Press, New York, NY, 1988), vol. 2, pp. 1−55. [Google Scholar]
- 25.Hecht A., et al. , Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Res. 45, 3615–3626 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firnberg E., Labonte J. W., Gray J. J., Ostermeier M., A comprehensive, high-resolution map of a gene’s fitness landscape. Mol. Biol. Evol. 31, 1581–1592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godson G. N., Fiddes J. C., Barrell B. G., Sanger F., “Comparative DNA sequence analysis of the G4 and phiX174 genomes” in The Single-Stranded DNA Phages (Cold Spring Harbor Press, Plainview, NY, 1978), vol. 8, pp. 51–86. [Google Scholar]
- 28.Jaschke P. R., Dotson G. A., Hung K., Liu D., Endy D.. Sequence data for Escherichia virus phiX174, complete genome. NCBI Nucleotide Database. https://www.ncbi.nlm.nih.gov/nuccore/MN385565. Deposited 18 September 2019. [Google Scholar]
- 29.Jaschke P. R., Dotson G. A., Hung K., Liu D., Endy D.. Sequence data for Synthetic Enterobacteria phage CryptX174, complete genome. NCBI Nucleotide Database. https://www.ncbi.nlm.nih.gov/nuccore/MF426915. Deposited 06 September 2019. [Google Scholar]
- 30.Meinnel T., Mechulam Y., Blanquet S., Methionine as translation start signal: A review of the enzymes of the pathway in Escherichia coli. Biochimie 75, 1061–1075 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Jaschke P. R., Dotson G. A., Hung K., Liu D., Endy D.. Sequence data for Synthetic Enterobacteria phage KleenX174, complete genome. NCBI Nucleotide Database. https://www.ncbi.nlm.nih.gov/nuccore/MF426914. Deposited 1 October 2019. [Google Scholar]
- 32.Abedon S. T., Yin J., Bacteriophage plaques: Theory and analysis. Methods Mol. Biol. 501, 161–174 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Springman R., Molineux I. J., Duong C., Bull R. J., Bull J. J., Evolutionary stability of a refactored phage genome. ACS Synth. Biol. 1, 425–430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodman D. B., Church G. M., Kosuri S., Causes and effects of N-terminal codon bias in bacterial genes. Science 342, 475–479 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Sun L., et al. , Icosahedral bacteriophage ΦX174 forms a tail for DNA transport during infection. Nature 505, 432–435 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Plotkin J. B., Kudla G., Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 12, 32–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherwa J. E. Jr, Young L. N., Fane B. A., Uncoupling the functions of a multifunctional protein: The isolation of a DNA pilot protein mutant that affects particle morphogenesis. Virology 411, 9–14 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Mitchell L. A., et al. , Synthesis, debugging, and effects of synthetic chromosome consolidation: synVI and beyond. Science 355, eaaf4831 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Shimizu Y., et al. , Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Laursen B. S., Sørensen H. P., Mortensen K. K., Sperling-Petersen H. U., Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingolia N. T., et al. , Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8, 1365–1379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald L., Beynon R. J., Positional proteomics: Preparation of amino-terminal peptides as a strategy for proteome simplification and characterization. Nat. Protoc. 1, 1790–1798 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Spector S., Flynn J. M., Tidor B., Baker T. A., Sauer R. T., Expression of N-formylated proteins in Escherichia coli. Protein Expr. Purif. 32, 317–322 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Fredens J., et al. , Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolisnychenko V., et al. , Engineering a reduced Escherichia coli genome. Genome Res. 12, 640–647 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baby V., et al. , Inferring the minimal genome of Mesoplasma florum by comparative genomics and transposon mutagenesis. mSystems 3, e00198–e00117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuzmin E., et al. , Systematic analysis of complex genetic interactions. Science 360, eaao1729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rousset F., et al. , Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genet. 14, e1007749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang T., et al. , Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat. Commun. 9, 2475 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang K., et al. , Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ostrov N., et al. , Design, synthesis, and testing toward a 57-codon genome. Science 353, 819–822 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Si T., et al. , Automated multiplex genome-scale engineering in yeast. Nat. Commun. 8, 15187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campa C. C., Weisbach N. R., Santinha A. J., Incarnato D., Platt R. J., Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods 16, 887–893 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Rokyta D. R., Abdo Z., Wichman H. A., The genetics of adaptation for eight microvirid bacteriophages. J. Mol. Evol. 69, 229–239 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fane B. A., Hayashi M., Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage phi X174. Genetics 128, 663–671 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L., Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Besemer J., Borodovsky M., GeneMark: Web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33, W451–W454 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsen T. S., Krogh A., EasyGene–A prokaryotic gene finder that ranks ORFs by statistical significance. BMC Bioinformatics 4, 21 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Warren D. J., Preparation of highly efficient electrocompetent Escherichia coli using glycerol/mannitol density step centrifugation. Anal. Biochem. 413, 206–207 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Chung C. T., Niemela S. L., Miller R. H., One-step preparation of competent Escherichia coli: Transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 86, 2172–2175 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hillson N. J., Rosengarten R. D., Keasling J. D., j5 DNA assembly design automation software. ACS Synth. Biol. 1, 14–21 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zadeh J. N., et al. , NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are available from the corresponding authors on reasonable request. Sequences of wild-type øX174 annotated with the cryptic ORFs (accession no. MN385565), kleenX174 (accession no. MF426914), and cryptX174 (accession no. MF426915) are available via National Center for Biotechnology Information’s GenBank.