Significance
The capabilities for invasion and metastasis underlie the mortality and morbidity of most forms of human cancer. Currently, there are no effective therapies specifically targeting these cancer phenotypes, in part due to the paucity of dominant mutations that induce them, and indeed losses of suppressors of invasion and metastasis are increasingly recognized as determinants, posing challenges for drug development. Our results implicate epigenetic gene regulation mediated by elevated expression of distinct microRNAs in orchestrating invasion and metastasis, evidently by abrogating distinctive suppressor mechanisms. Therefore, targeting such microRNAs holds promise as a strategy to combat malignant cancers with epigenetically disrupted tumor suppressor mechanisms.
Keywords: cancer, metastasis, microRNAs, Acvr1c/ALK7, PanNETs
Abstract
MicroRNA-mediated gene regulation has been implicated in various diseases, including cancer. This study examined the role of microRNAs (miRNAs) during tumorigenesis and malignant progression of pancreatic neuroendocrine tumors (PanNETs) in a genetically engineered mouse model. Previously, a set of miRNAs was observed to be specifically up-regulated in a highly invasive and metastatic subtype of mouse and human PanNET. Using functional assays, we now implicate different miRNAs in distinct phenotypes: miR-137 stimulates tumor growth and local invasion, whereas the miR-23b cluster enables metastasis. An algorithm, Bio-miRTa, has been developed to facilitate the identification of biologically relevant miRNA target genes and applied to these miRNAs. We show that a top-ranked miR-137 candidate gene, Sorl1, has a tumor suppressor function in primary PanNETs. Among the top targets for the miR-23b cluster, Acvr1c/ALK7 has recently been described to be a metastasis suppressor, and we establish herein that it is down-regulated by the miR-23b cluster, which is crucial for its prometastatic activity. Two other miR-23b targets, Robo2 and P2ry1, also have demonstrable antimetastatic effects. Finally, we have used the Bio-miRTa algorithm in reverse to identify candidate miRNAs that might regulate activin B, the principal ligand for ALK7, identifying thereby a third family of miRNAs—miRNA-130/301—that is congruently up-regulated concomitant with down-regulation of activin B during tumorigenesis, suggestive of functional involvement in evasion of the proapoptotic barrier. Thus, dynamic up-regulation of miRNAs during multistep tumorigenesis and malignant progression serves to down-regulate distinctive suppressor mechanisms of tumor growth, invasion, and metastasis.
Many cancer patients will succumb to their disease due to metastasis, underscoring the imperative to develop antimetastatic therapies (1). During this complex process, cancer cells invade both into adjacent tissue and into blood vessels, from which they can subsequently extravasate into other organs, and eventually begin proliferating to form macrometastases (2–4). A majority of cancer cells in a tumor typically fail to complete this process, emphasizing the significant barriers encountered and the capabilities necessarily acquired to circumvent them. While various genetic aberrations of oncogenes and tumor suppressor genes have been associated with tumorigenesis, there is currently a dearth of recurrent driver gene mutations specifically implicated in metastasis (5). Notably, recent studies have shown that genetic polymorphisms and epigenetic regulators, including chromatin modifiers and miRNAs, can play important roles during the “metastatic cascade” (6–9).
MicroRNAs are small noncoding RNAs ∼22 nt in length that are able to posttranscriptionally repress the expression of mRNAs, and therefore regulate diverse signaling pathways and biological processes (10, 11). After their biogenesis, the mature strand of a miRNA, along with an Argonaute protein, form the miRNA-induced silencing complex (miRISC), which is responsible for the translational silencing and mRNA decay of its target genes. The pairing between the miRNA seed region (miRNA nucleotides 2 through 7) and a target gene’s complementary sites (miRNA-response elements [MREs], usually located in the 3′ UTR) is mediated by simple Watson–Crick pairing; therefore, the specificity of the miRNA is determined by the length of the seed region and the accessibility of the MRE. As a result, miRNAs are predicted to be able to bind to many genes, which potentially allows them to have pleiotropic effects, consequentially posing challenges to determining functionally relevant gene targets (12–14).
MicroRNAs have been implicated in various aspects of cancer progression, such as proliferation, apoptosis, and the epithelial–mesenchymal transition (EMT), and they can either promote or suppress tumorigenesis and metastasis (15–21). Previously we reported that distinct miRNAs are differentially expressed during the progression of tumorigenesis in the RIP1-Tag2 (RT2) mouse model of pancreatic neuroendocrine tumors (PanNETs) (9). A subset of these PanNETs, designated as “met-like primary” tumors (MLPs), are characterized by miRNAs and mRNA transcriptome signatures in common with liver metastases, posing the intriguing hypothesis that PanNET cancer cells acquire the necessary capabilities for metastasis during primary tumorigenesis (9, 22). Our cross-species analysis revealed that 6 miRNAs—miR-132, miR-137, miR-181, miR-23b, miR-27b, and miR-24-1—were similarly up-regulated in human and mouse metastasis-associated MLP tumors (22). In the current study, we have investigated these and other miRNAs and developed an algorithm to identify and associate candidate target genes with the miRNAs that might regulate them. As a result, we revealed “tumor suppressors” that are down-modulated by distinct miRNAs and collectively implicated in circumventing barriers to invasion and metastasis.
Results
MicroRNA-137 Enhances Primary Tumor Progression.
The MLP-associated and metastasis-specific miRNA signatures contains 33 miRNAs—16 up-regulated and 17 down-regulated—compared to noninvasive insulinoma tumors (ITs) (9), of which a subset is similarly altered both in mouse and human MLP PanNETs (SI Appendix, Fig. S1 A and B) (22). In order to prioritize their investigation, we further examined expression of each in both subtypes of primary tumor—IT and MLP—and in liver metastases in the RT2 PanNET model. The results led us to focus on miR-137 and the miR-23b cluster, due to their distinctive patterns of expression in invasive MLP tumors and liver metastases (SI Appendix, Fig. S1 C and D). Notably, the paralog cluster targeting the same MREs—the miR-23a cluster—is also up-regulated in mouse PanNET metastases (SI Appendix, Fig. S1E).
To probe potential functional effects, miR-137 and the miR-23b cluster were overexpressed in the βTC3 IT cell line, using a piggyBac transposon system enabling doxycycline (DOX)-inducible miRNA expression that includes the fluorescent protein mKate along with luciferase (SI Appendix, Fig. S1 F–H). We first examined the effect of miRNA overexpression in experimental primary tumors arising in an orthotopic transplant model of PanNET, involving the injection of βTC3 cells into the pancreas of immunocompromised mice. Initially, we induced expression of miRNAs in vitro by adding DOX to the cell culture media and then kept administering DOX in vivo via DOX-containing chow. Expression of miR-137, but not the miR-23b cluster, accelerated tumor growth, which led to shorter overall survival (Fig. 1A). To examine whether the enhanced tumor growth was consequent to the initial seeding of the cancer cells or subsequent tumor growth, miRNA expression was induced 1 wk after injection of the cells into the pancreas, by which point solid tumors had become established. Once again, up-regulation of miR-137, but not of the miR-23b cluster, reduced survival, reflecting enhanced tumor growth (Fig. 1B), substantiating its ability to promote primary tumorigenesis.
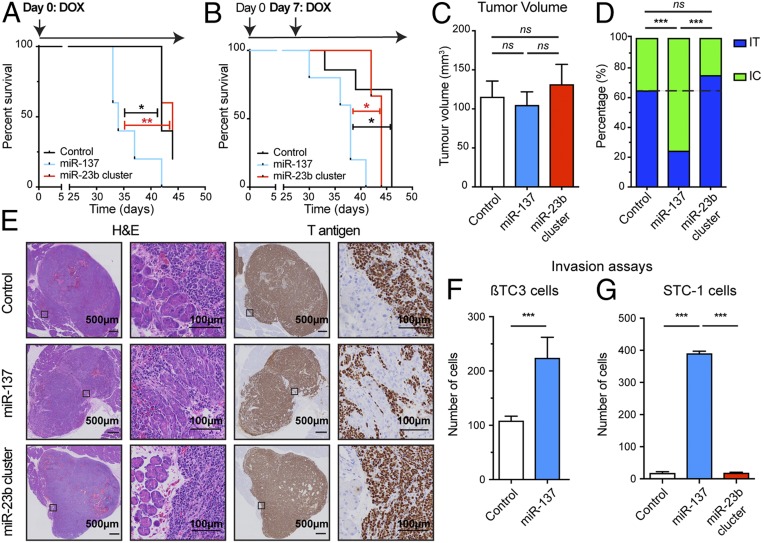
Fig. 1.
MicroRNA-137 reduces survival and increases invasion of PanNET. (A and B) Survival of mice with orthotopic PanNETs elicited by inoculation of control βTC3 cells, or miRNA overexpressing cells. MicroRNA expression was induced either concomitantly (A) or 7 d after injection of the cells into the pancreas parenchyma (B). n = 3 to 7. Log-rank test. *P < 0.05, **P < 0.01. (C) Quantification of tumor volume at the endpoint of the survival studies. (D) Quantification of the distribution between IT and IC tumors. n = 28 to 37. Fisher’s exact test. ns, not significant. ***P < 0.001. (E) H&E staining and immunostaining for the SV40 T antigen (Tag) oncoprotein to reveal cancer cells in tumor tissue sections. (F and G) Effect of miRNA expression on the invasive capabilities of βTC3 (F) and STC-1 (G) in a flow stimulated invasion assay. n = 3 to 6. Student’s t test. ***P < 0.001.
MicroRNA-137 Elicits an Invasive Capability.
We next immunostained a series of tissue sections from similarly sized orthotopic transplant tumors, collected from end-point mice, for the driving SV40 T antigen oncoprotein so as to reveal the cancer cells, and observed that up-regulation of miR-137 increased the proportion of invasive carcinoma (IC; putatively the MLP subtype) versus noninvasive IT tumors, when compared to the control group (Fig. 1 C–E). In contrast, expression of the miR-23b cluster had no impact: the proportions of IT vs. IC/MLP were the same as with the control group (Fig. 1 D and E). A flow-stimulated invasion assay (23) further established the capability of miR-137 to enhance invasiveness of βTC3 cells (Fig. 1F). We also evaluated a second cell line, STC-1, derived from an intestinal carcinoid tumor that developed in the same RT2 mouse model (24), which was similarly engineered to overexpress these miRNAs (SI Appendix, Fig. S1 I and J); miR-137 (but not the miR-23b cluster) again enhanced flow-stimulated invasiveness (Fig. 1G).
These functional analyses collectively reveal that miR-137 is able to enhance progression to invasive carcinomas, in particular by stimulating invasiveness, whereas the miR-23b cluster did not have a discernible effect, consistent with its modest up-regulation in MLP tumors compared to miR-137.
MicroRNA-23b Cluster Enhances Liver Metastasis.
The intriguing observation that the miR-23b cluster was markedly up-regulated in metastasis compared to MLP primary tumors, and its lack of effect on the latter, led us to assess its effects on metastasis. Since analysis of metastasis in mice bearing βTC3 cell-derived orthotopic tumors is not feasible due to severe hypoglycemia consequent to excessive insulin secretion by the cancer cells, we used experimental metastasis models.
Up-regulation of the miR-23b cluster proved to selectively enhance liver metastasis following i.v. inoculation of cancer cells, whereas overexpression of miR-137 had a very modest effect, as scored by the increased number of metastatic foci (Fig. 2A), as well as by higher levels of luciferase (Fig. 2B). Ex vivo fluorescent imaging of the livers immediately after dissection established that the metastatic foci were positive for the mKate reporter expressed along with luciferase in the transposon vector (Fig. 2C), further confirming their identity as transduced cancer cells, as did immunostaining for the SV40 T antigen oncoprotein (Fig. 2D).
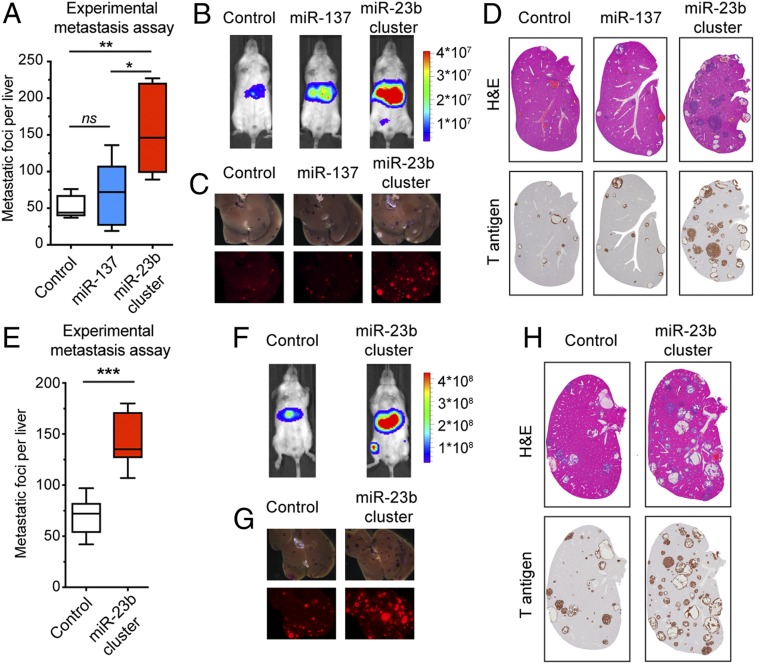
Fig. 2.
Expression of the miR-23b cluster enhances liver metastasis. (A–G) PanNET experimental liver metastasis assays via tail vein injections using control βTC3 cells or miRNA overexpressing cells. (E–H) Intestinal carcinoid liver metastasis assays via tail vein injections using control STC-1 cells, or miRNA overexpressing cells. (A and E) Quantification of metastatic foci in the liver derived from PanNETs (A; n = 5) or intestinal carcinoids (E; n = 7 to 11). (A) One-way ANOVA followed by Holm–Sidak’s multiple comparisons test. ns, not significant. *P < 0.05, **P < 0.01. (E) Student’s t test. ***P < 0.001. (B and F) Bioluminescence imaging before killing (radiance; p/sec/cm2/sr). (C and G) mKate fluorescence imaging of excised livers. (D and H) H&E staining and Tag oncoprotein immunostaining of liver tissue sections.
To further evaluate the prometastatic effect of the miR-23b cluster, we similarly evaluated miRNA-transduced versions of the aforementioned STC-1 cell line. Up-regulation of the miR-23b cluster significantly enhanced the metastatic ability of the STC-1 cells (Fig. 2 E–H). In addition, we assessed the endogenous levels of miR-23b cluster expression in a series of cancer cell lines derived from PanNET tumors arising in the RT2 model that displayed a spectrum of metastatic potential (25) and found that miR-23b and miR-27b were expressed at higher levels in the more highly metastatic cell lines (SI Appendix, Fig. S1 K and L). Notably, both the miR-23b cluster and the miR-23a cluster are similarly up-regulated in high-grade human PanNET tumors (“NF-MLP,” i.e., “nonfunctional,” namely with low-insulin expression) (SI Appendix, Fig. S1 M and N).
Collectively, the results reveal that up-regulation of the miR-23b cluster selectively enhances the metastatic capability of PanNET and intestinal carcinoids without significantly affecting tumor growth and invasion.
MicroRNA-23b Cluster Has a Prosurvival Effect during the Initial Stages of Metastasis.
To further investigate the prometastatic effect of the miR-23b cluster, we varied the timing of DOX-mediated induction of miR-23b cluster expression in βTC3 cells (Fig. 3A). In the first 2 conditions, DOX was added to the media during cell culture before tail vein injection, and we either continued using DOX-containing chow following inoculation into mice (ON/ON), or not, instead using regular chow (ON/OFF). In the remaining 2 conditions, DOX was not added to the media, and it was either supplied immediately after tail vein injection (OFF/ON) or not supplied at all (OFF/OFF). Whereas the ON/OFF condition was prometastatic, phenocopying the ON/ON condition, the OFF/ON was not (Fig. 3 B and C). Quantification of bioluminescence in the liver over time further established that the miR-23b cluster did not affect the expansion phase of metastasis (Fig. 3D and SI Appendix, Fig. S2 A and B), signifying that the miR-23b cluster was important during the initial stages of colonization.
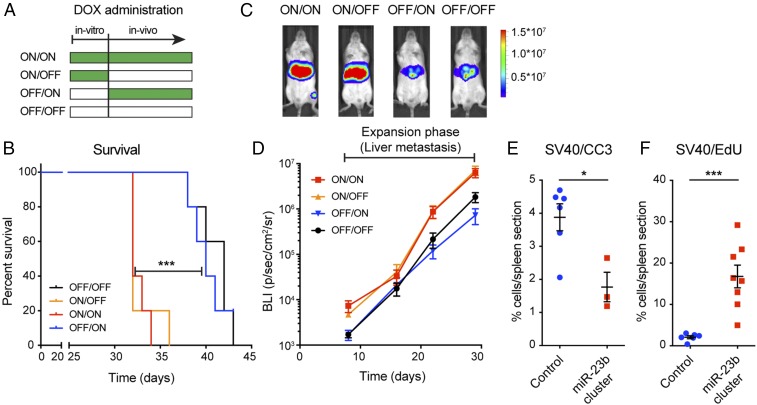
Fig. 3.
Characterization of the prometastatic effects of the miR-23b cluster. (A–D) PanNET experimental liver metastasis assays via tail vein injections using miR-23b cluster overexpressing βTC3 cells with distinctive regimens of DOX-induced expression. (A) DOX administration schedules. (B) Survival of mice variably induced to express the miR-23b cluster. n = 5. Log-rank test. ***P < 0.001. (C) Bioluminescence imaging of representative mice at day 30 for the 4 induction schedules (radiance; p/sec/cm2/sr). (D) Time course of the bioluminescence signal in the liver during the expansion phase. (E and F) PanNET experimental liver metastasis assays via intrahepatic injection using control βTC3 cells or miR-23b cluster overexpressing cells. Quantification of apoptosis (E; Tag+/Cleaved Caspase 3) and proliferation (F; Tag+/EdU) of βTC3 control and miR-23b cluster overexpressing cells, 24 h after intrahepatic injection. n = 3 to 8. Student’s t test. *P < 0.05, ***P < 0.001.
To dissect the prometastatic mechanism, we examined proliferation and apoptosis of control and miR-23b cluster-overexpressing βTC3 cells in the liver 24 h after intrahepatic injection, which efficiently seeds cells into the liver, facilitating characterization of their fate during the initial stages of colonization. We observed that miR-23b cluster overexpression led to decreased apoptosis and increased proliferation compared to control cells (Fig. 3 E and F and SI Appendix, Fig. S2 B and C), whereas there was no difference in the total number of cancer cells lodged in the liver (SI Appendix, Fig. S2C). Thus, we conclude that the miR-23b cluster facilitates cancer cell growth following initial metastatic seeding of the liver.
Development of Bio-miRTa: An Algorithm for Identification of Biologically Relevant miRNA Targets.
MicroRNAs are known to play context-dependent roles, and as such their functionally important gene targets can in principle vary in distinct pathological conditions. Therefore, we developed an algorithm, Bio-miRTa, for the identification of biologically relevant miRNA targets (Fig. 4), which enables the ranking of the miRNA candidate target genes according to the biological process of interest. As outlined below, Bio-miRTa is structured into 2 steps: In the first step, all potential targets of the miRNA(s) of interest are extracted, and in the second, the list is filtered by experimental or descriptive datasets that are related to the biological process of interest, in order to score and rank potential target genes accordingly.
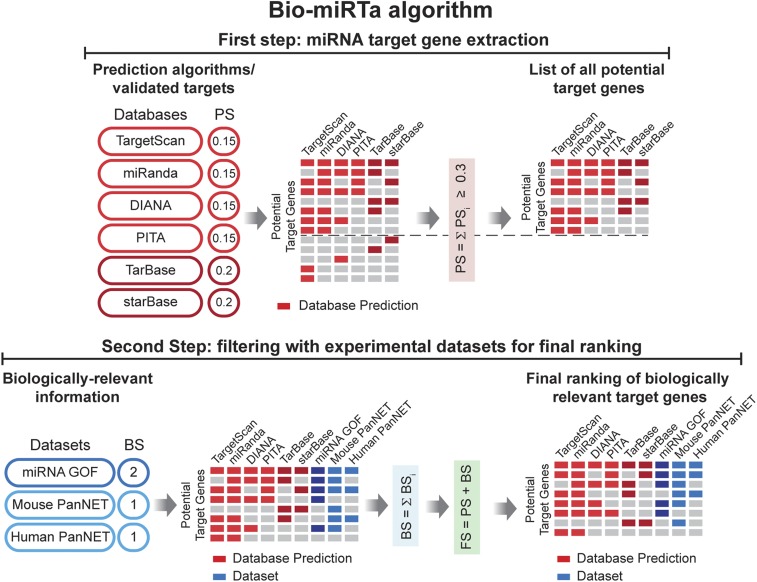
Fig. 4.
Flowchart of the Bio-miRTa algorithm. In the first step the algorithm extracts the miRNA gene targets using information from target prediction algorithms (TargetScan, miRanda, DIANA, and PITA) and databases with experimentally validated targets (TarBase and starBase). A PS is assigned to the potential target genes that appear in the respected database, and those with a cumulative PS < 0.3 are filtered out. In the second step, the algorithm incorporates biologically relevant information provided by the user and assigns a BS accordingly. Finally, it scores and ranks the target genes according to the FS. The details of and rationale for each step are described in the Results and Materials and Methods sections.
In the first step of Bio-miRTa, the algorithm extracts all potential targets of the miRNA(s), regardless of the biological context. During this step, it uses 4 independent target prediction algorithms [TargetScan (26), miRanda (27), DIANA (28), and probability of interaction by target accessibility [PITA] (29)] in which the target genes are primarily predicted based on the presence of an MRE in the target gene, and other algorithm-specific parameters. Since the number of false positive prediction is high in computational algorithms (30, 31), Bio-miRTa also incorporates 2 datasets of experimentally validated miRNA targets [TarBase (32) and starBase (33)] into the prediction pipeline. Each gene predicted as a target of a miRNA by any of the 6 datasets is assigned a prediction score (PS; with values of 0.15 for each prediction algorithm and 0.2 for each experimentally validated dataset). The final PS for a gene is calculated by the sum of PS for each dataset (i.e., PSmax = 1; Fig. 4). Bio-miRTa reports a gene as a potential target of a miRNA if it is presented in at least 2 prediction algorithms, or 1 prediction algorithm and 1 experimentally validated dataset (i.e., PS ≥ 0.3). Notably, the first step of Bio-miRTa is generic to the miRNA, and does not factor in the biological context; therefore, it could be used independently of step 2 to provide a ranked list of all possible targets in principle of miRNA(s) based on their prediction scores (Fig. 4 and Materials and Methods).
In the second step, the algorithm factors in the biological context, by incorporating various datasets provided by the user, which can be grouped into 2 general categories: 1) experimental datasets from functional miRNA perturbations (e.g., miRNA gain of function [GOF] and miRNA loss of function [LOF]), and 2) descriptive datasets comparing relevant states or conditions in experimental mouse models and human transcriptomic data where the candidate miRNA is differentially expressed, and hence its targets could also be differentially expressed. For the assigned biological score (BS) in the aforementioned categories of datasets, we rationalized that the BS of the experimental datasets (i.e., BSED) should be greater than the BS of the descriptive datasets (i.e., BSDD), since in the latter case there could be other complex modulations of target genes, and as such, although relevant, descriptive datasets might provide weaker evidence compared to the functional perturbations of a miRNA(s). Therefore, we assigned BSED = 2 to the first experimental dataset, and BSDD = 1 to the descriptive dataset. In the case of additional datasets, the BS score is increased by 1 for each additional dataset in either category, and then a normalized BS is applied to all datasets in the corresponding category. Finally, Bio-miRTa calculates the final score (FS) by summing the PS from the first step and the BS from the second step, which leads to the final ranking of the candidate target genes (Fig. 4).
To validate the algorithm, we used the transcriptome data from a recent study in which the miR-200 family was inactivated in the RT2 PanNET model (15). Notably, the gene encoding for the transcription factor Zeb1 was predicted by Bio-miRTa as the top target of the miR-200 family (SI Appendix, Fig. S3 and Dataset S1). Zeb1 was shown to be the main gene target in the aforementioned study (15), and moreover, previous studies also implicated the role of the miR-200 family in the regulation of Zeb1 and epithelial-to-mesenchymal transition (34). Other known targets of the miR-200 family, such as Dlc1, Reck, and Ogt, were also identified by the algorithm, as well as a potential gene target, Mbnl1, which plays an important role in pre-mRNA alternative splicing (SI Appendix, Fig. S3).
Using Bio-miRTa to Identify Candidate Target Genes for miR-137 and the miR-23b Cluster.
To assess its utility in identifying relevant target genes, the first step of Bio-miRTa (Fig. 4) was applied to miR-137 and the miR-23b cluster, whose functional importance had been established above (Figs. 1 and 2). In the case of the miR-23b cluster, which is composed of 3 miRNAs, in the first step of Bio-miRTa, the algorithm separately subscored each of the 3 miRNAs that targeted a candidate target gene, and then combined the scores in order to calculate the corresponding PS for the cluster (SI Appendix, SI Materials and Methods and Fig. S4).
For the second step, we applied 1 functional perturbation dataset for each miRNA, and 2 descriptive datasets. The functional miRNA perturbation datasets were composed of significantly down-regulated genes upon miR-137 overexpression (GOF) for the miR-137 inquiry, and significantly down-regulated genes upon miR-23b cluster overexpression (GOF) for the miR-23b cluster inquiry (35). In each analysis the BSED was therefore equal to 2 (Fig. 4). The 2 descriptive datasets contained down-regulated genes in mouse and human MLP tumors and metastatic lesions compared to the lower grade insulinoma/islet tumors (as aforementioned, MLP/metastasis have higher miR-137/miR-23b cluster expression compared to insulinoma/islet tumors). We incorporated these 2 descriptive datasets into both miR-137 and miR-23b cluster target predictions, assigning a BS = 1 for each dataset, resulting in a BSDD = 2, and a maximum combined BS of 4 (Fig. 4).
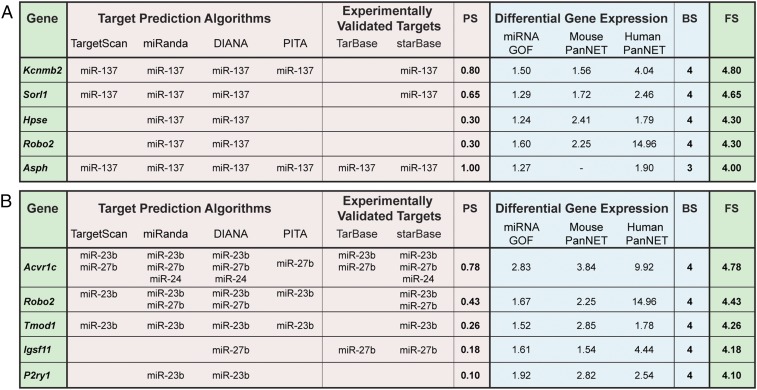
In step 1, we identified 2,340 and 7,563 candidate target genes with PS ≥ 0.3 for miR-137 and the miR-23b cluster, respectively. Notably, this number was substantially reduced by the application of step 2 and the combined scoring of both steps. Particularly, after filtering in only those candidate target genes that appear in the GOF dataset and at least 1 of the 2 descriptive datasets (FS > 3), there were only 20 and 27 candidate target genes for miR-137 and the miR-23b cluster, respectively (Fig. 5 A and B and Datasets S2 and S3).
Fig. 5.
Identification of miRNA gene targets using Bio-miRTa. (A and B) Highly ranked candidate target genes for miR-137 (A) and the miR-23b cluster (B) identified using the Bio-miRTa algorithm. The first column labeled as “gene” shows the candidate target genes sorted according to the FS shown in the last column on the Right. The columns with the headers “target prediction algorithms” and “experimentally validated targets” report the presence of an MRE for the corresponding miRNA in the candidate target gene. The columns with the header “differential gene expression” show the differential gene expression in βTC3 cells after miRNA overexpression in βTC3 cells (miRNA GOF), in mouse IT versus MLP tumors in RT2 mice (mouse PanNETs) and in the human IT versus MLP PanNETs (human PanNETs). The intercalated columns show the PS (based on the presence of MRE according to the first step of the algorithm), BS (based on the differential gene expression), and FS (the sum of the PS and BS scores). The PS scored in the case of the miR-23b cluster is normalized as shown in SI Appendix, Fig. S4. The top 5 genes of each miRNA are shown; Datasets S2 and S3 contain the complete list of ranked genes.
Initially, we focused on candidate target genes of miR-137. Then we turned to investigate the top-ranked miR-23b candidate genes identified by this algorithm, in particular the Tgfβ superfamily receptor Acvr1c (Activin receptor type-1C; commonly known as ALK7), which we have previously reported to be a metastasis suppressor (25).
Using Bio-miRTa in Reverse to Identify Candidate miRNAs Targeting the ALK7 Ligand Activin B.
We previously reported (25) that expression of the principal ALK7 ligand, activin B, encoded by the Inhßb gene, is down-regulated during the early stages of PanNET tumorigenesis; thus we hypothesized that it too might be repressed by miRNAs. Although we lacked a functional perturbation dataset, Bio-miRTa identified a family of 4 similar miRNAs—miR-130a/b and miR-301a/b—predicted to bind a conserved MRE within the 3′ UTR of human and mouse Inhßb; concordantly, these miRNAs proved to be up-regulated during the sequential stages of tumorigenesis in the RT2 PanNET model (SI Appendix, Fig. S5 A–D and Dataset S4). Using luciferase reporter assays, we verified the ability of these miRNAs to bind to the identified MRE within the 3′ UTR of Inhßb mRNA and thereby repress expression of the reporter (SI Appendix, Fig. S5 E and F). Furthermore, the biochemical interaction of miR-301a with Inhβb mRNA has been previously established using the HITS-CLIP assay (36). As such, we infer that activin B expression may also be regulated by dynamic expression of miRNAs during multistep PanNET tumorigenesis, motivating future functional studies.
Notably, this analysis demonstrates that the algorithm can be informative in the absence of a functional (GOF/LOF) dataset involving perturbations in miRNA expression, and that Bio-miRTa can be reversed, so as to identify candidate miRNAs via the MREs present in a particular gene of interest.
MicroRNA-137 Candidate Target Genes Are Associated with Poor Prognosis in Human Cancer Types.
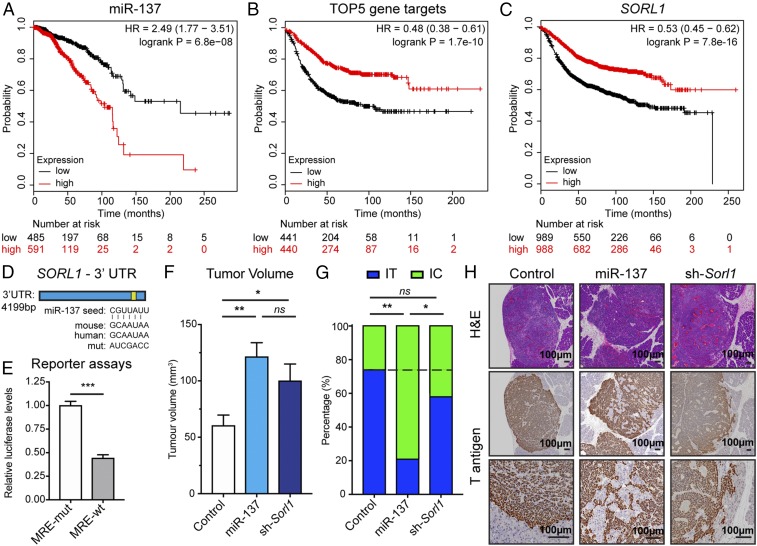
To begin assessing the association of miRNA-137 and its target genes with human cancers, we first audited available public transcriptomic datasets of human breast cancer in The Cancer Genome Atlas (TCGA) and found a clear association of high miR-137 expression with comparatively worse overall survival (OS) in breast cancer (Fig. 6A). Given this association, we further queried available transcriptomic databases for a possible association of putative miR-137 target genes with breast cancer prognosis. The analysis revealed that comparatively low expression of a miR-137 gene signature, composed of the 5 top-ranked miR-137 candidate genes (Fig. 5A), was strongly associated with poorer relapse-free survival (RFS) of breast cancer patients (Fig. 6B), as was the individual expression of each gene (Fig. 6C and SI Appendix, Fig. S6 A–D). Thus, in agreement with a previous report (37), it is plausible that miR-137 enhances invasive tumor growth in human breast cancer, and thereby contributes to progression of the disease. In addition to breast cancer, we assessed the association of miRNA-137 with 20 additional cancer types using TCGA data and found that its expression was associated with poorer OS of patients with head and neck, ovarian, and uterine corpus endometrial cancer, while it was associated with better OS of patients with kidney renal cell and liver hepatocellular carcinoma (SI Appendix, Fig. S7). Future studies will be required to delineate the evidently context-dependent roles of miRNA-137 in these different cancer types.
Fig. 6.
Association of miR-137 and its top-ranked candidate target genes with breast cancer prognosis, and characterization of the Sorl1 target gene in primary PanNET tumors. (A) Association of miR-137 expression with overall survival in a cohort of breast cancer patients including all subtypes. (B and C) Association of an miR-137 gene signature (comprising the top 5 predicted target genes) and 1 of its components, SORL1, with RFS in human breast cancer. (D) The MRE for miR-137 in the 3′ UTR of SORL1. The miRNA seed, the MREs for human and mouse SORL1, as well as the mutated MRE used for the reporter assays, are shown. (E) Luciferase reporter assays. n = 6. Student’s t test. ***P < 0.001. (F–H) Orthotopic PanNET tumor assays using control, miR-137 overexpressing, and Sorl1 knockdown βTC3 cells. The miR-137 overexpression and Sorl1 knockdown were induced by DOX food concomitant with injection of the engineered cells into the pancreas parenchyma, and the mice were killed 30 d after injection. (F) Tumor volumes at the 30-d defined endpoint. n = 6 to 7. One-way ANOVA followed by Holm–Sidak’s multiple comparisons test. *P < 0.05, **P < 0.01. (G) The distribution between IT and IC tumors in the 3 cohorts. n = 14 to 21. Fisher’s exact test. ns, not significant. *P < 0.05, **P < 0.01. (H) H&E staining and immunostaining for the SV40 Tag oncoprotein to reveal cancer cells in tumor tissue sections.
Among the candidate miR-137 target genes (Fig. 5A), we began by assessing Sorl1 (also known as SORLA and LR11), since we found that its expression was comparatively lower in the aggressive triple-negative breast cancer (TNBC; basal) subtype, and that it was associated with shorter time to the first progression of patients with non-small cell lung and gastric cancers (SI Appendix, Fig. S6 E–G). Sorl1 is a member of the “vacuolar protein sorting 10 protein (VPS10P) domain” receptor family responsible for the trafficking and sorting of proteins between the Golgi apparatus, intracellular endosomes, and the cell surface. The mouse and human 3′ UTR of Sorl1 contain 1 conserved MRE for miR-137 (Fig. 6D). To assess the ability of miR-137 to bind to this MRE, we synthesized a fragment of ∼1 kb surrounding the wild-type MRE, as well as a fragment with a mutated MRE (Fig. 6D). We then performed luciferase reporter assays, which revealed that the wild-type MRE decreased luciferase levels by 2-fold compared to the mutated one (Fig. 6E), indicating that miR-137 can bind to the Sorl1 3′ UTR MRE and mediate miRNA-mediated mRNA degradation. The regulation of Sorl1 by miR-137 is also supported by published immunoprecipitation-based biochemical assays (38).
To assess the possible role of Sorl1 in tumor phenotypes, a DOX-inducible piggyBac transposon system was used to knock down Sorl1 in the βTC3 cells (SI Appendix, Fig. S6 H and I). The engineered cells were inoculated (orthotopically) into the pancreas of immunocompromised mice, and the mice were killed 30 d later. We observed that the Sorl1 knockdown increased the tumor burden by ∼2-fold, which is in accordance with the observed tumor-promoting function of miR-137 (Fig. 6F). Tumor sections were immunostained for the driving SV40 T antigen oncoprotein and assessed for the proportion of invasive (IC) versus noninvasive insulinoma-like (IT) tumors. Interestingly, the Sorl1 knockdown had only a minor effect on the invasive phenotype (Fig. 6 G and H), suggesting either that another miR-137 target gene(s) is (are) primarily responsible for its invasion-inducing capability, or that Sorl1 down-regulation acts in a synergistic/additive manner with another target gene.
This pilot study encourages future investigation of the mechanisms and more precise effects of Sorl1 in PanNET and other tumor types, in particular breast cancer, using genetically engineered mouse models. Similar lines of investigation can be envisaged to validate and characterize other target genes for miR-137 as a suppressor(s) of invasive growth; among the candidates are the other top-ranked target genes, Kcnmb2, Hpse, Asph, and Robo2, the latter notably also implicated as a miR-23b target (Fig. 5B).
ALK7 Expression Is Down-Regulated by the miR-23 Cluster.
Three genes stood out as candidate targets of the miR-23b cluster: ALK7, P2ry1, and Robo2 (Fig. 5B). We have recently reported that suppression of ALK7 expression and signaling functionally enhances tumorigenesis and metastasis in PanNETs and breast cancer (25). Using a transposon gene knockdown system (SI Appendix, Fig. S6H), we have now shown that knock down of either P2ry1 or Robo2 can also enhance liver metastasis (SI Appendix, Fig. S8 A–D).
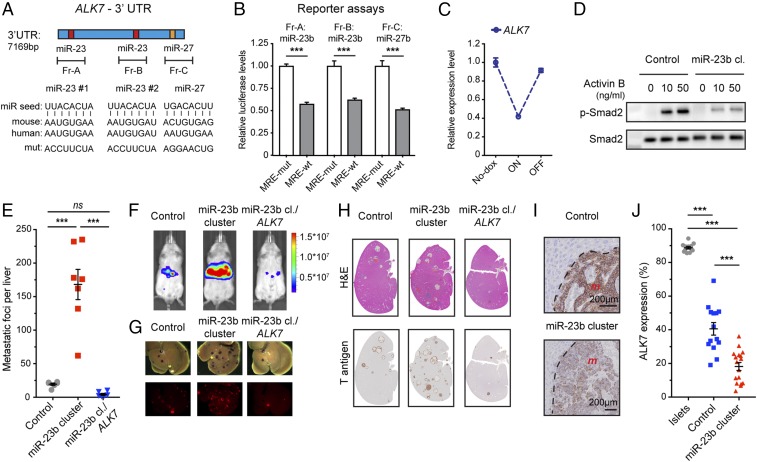
Given the extensive functional validation of ALK7 as a metastasis suppressor (25), we chose to focus on characterizing the potential regulation of ALK7 by the miR-23b cluster. ALK7 is a type I receptor, which upon binding to a ligand, most notably activin B, forms a heterotetramer with a type II receptor (Acvr2a or Acvr2b), resulting in intracellular signaling via phosphorylation of Smad2/3 (SI Appendix, Fig. S9A) (39). Both the mouse and the human 3′ UTRs of ALK7 contain 2 MREs for miR-23b and 1 MRE for miR-27b (Fig. 7A), while for miR-24, there is 1 MRE only in the mouse gene. To assess the ability of miR-23b and miR-27b to bind to their respective MREs in ALK7, we synthesized fragments of ∼1 kb surrounding each MRE and mutationally inactivated the MRE of interest in each fragment (Fig. 7A). Luciferase reporter assays revealed that the presence of a wild-type MRE decreased the luciferase levels around 2-fold in each case, compared to constructs with the mutationally disrupted MRE (Fig. 7B), indicating that miR-23b and miR-27b are able to bind to their respective MREs and mediate miRNA-mediated mRNA degradation. The ability of miR-23 and miR-27 to regulate the expression of ALK7 is further supported by published datasets (40–42) cataloging immunoprecipitation-based biochemical assays of miR-to-MRE interactions, which described the binding of both miRNAs to MREs in the 3′ UTR of ALK7 (SI Appendix, Fig. S9B).
Fig. 7.
ALK7 expression is regulated by miR-23/7b miRNAs during PanNET tumorigenesis. (A) MREs for miR-23b and miR-27b in the 3′ UTR of ALK7. The miRNA seed, the MREs for human and mouse ALK7, as well as the mutated MREs used for the reporter assays, are shown. (B) Luciferase reporter assays for a 3′ UTR reporter gene. n = 6. Student’s t test. ***P < 0.001. (C) Expression of ALK7 in cultured βTC3 cells carrying a tet-regulatable miR-23b cluster transgene following DOX administration to induce miRNA expression and then its withdrawal. (D) Western blotting for phosphorylated Smad2 and total Smad2 in miR-23b cluster-expressing βTC3 cells and control cells, treated as indicated with activin B for 8 h. (E–H) Experimental metastasis assay using βTC3 cells expressing the miR-23b cluster with and without coexpression of the miR23b-insensitive ALK7 ORF, relative to control cells. (E) Quantification of metastatic foci. n = 3 to 8. One-way ANOVA followed by Holm–Sidak’s multiple comparisons test. ns, not significant. ***P < 0.001. (F) Bioluminescence imaging of mice of luciferase-expressing cancer cells before killing (radiance; p/sec/cm2/sr). (G) mKate fluorescence imaging of cancer cells in excised livers. (H) H&E staining and Tag oncoprotein immunostaining of liver sections. (I and J) Representative images and scoring of ALK7 expression (%; total tumor area containing ALK7-positive cells) in liver metastases of control and miR-23b cluster overexpressing βTC3 cells. m, metastasis. n = 12 to 17. One-way ANOVA followed by Holm–Sidak’s multiple comparisons test. ***P < 0.001. Normal pancreatic islets, which express much higher levels (25), are shown for comparison.
As noted above, the “IT-like” βTC3 cell line expresses very low levels of the miR-23b cluster, whereas the “MLP-like” cell line AJ-5257-1 expresses higher levels (SI Appendix, Fig. S1 K and L), similarly to cancer cells in MLP tumors and metastases. Transient transfection of AJ-5257-1 cells with antagomirs to miR-23/27 increased ALK7 expression (SI Appendix, Fig. S9C). Conversely, DOX-induced up-regulation of the miR-23b cluster in βTC3 cells led to decreased ALK7 expression (Fig. 7C and SI Appendix, Fig. S9D). Furthermore, up-regulation of miR-23b suppressed phosphorylation of the ALK7 downstream target Smad2 upon treatment of βTC3 cells with activin B, which is the principal high-affinity ALK7 ligand (Fig. 7D). Finally, to further corroborate the ability of the miR-23b cluster to regulate ALK7, based on our previous study (25), we developed 2 transcriptomic ALK7 signaling pathway signatures: the first includes all genes altered by ALK7 activation in βTC3 cells, whereas the second selectively filters only those genes known to be associated with apoptosis (Dataset S5). Using these mRNA signatures, we observed that forced expression of the miR-23b cluster in βTC3 cells repressed activation of the ALK7 signaling pathway compared to control βTC3 cells when both were treated with activin B so as to induce ALK7 signaling, as indicated by the lower scores for both gene sets (SI Appendix, Fig. S9 E and F). Notably, this repressive effect was comparable to that produced by expression of a dominant-negative form of ALK7 (ALK7-DN; containing a K222R “kinase-dead” mutation) or by ALK7 knockdown in βTC3 cells also treated with activin B (SI Appendix, Fig. S9 E and F); these engineered constructs have been previously described (25).
We next used functional assays involving βTC3 cells engineered with a transposon vector conditionally expressing the miR-23b cluster, with or without the ALK7 open reading frame (ORF) that lacks the 3′ UTR (and hence the miR-23/27 MREs) and is therefore insensitive to the miR-23b cluster (SI Appendix, Fig. S9G). Notably, the capability of the miR-23b cluster to enhance liver metastasis, documented above (Fig. 2), was diminished by coexpression of the miR-23b cluster-insensitive ALK7 ORF (Fig. 7 E–H). Using immunostaining, we observed that ALK7 expression was lower in the metastases overexpressing the miR-23b cluster (Fig. 7 I and J), further establishing its ability to down-regulate the expression of ALK7. Together, these results establish that the members of the miR-23 cluster—miR-23a/b and miR-27a/b—collectively suppress the expression of ALK7. Given the ability of miRNAs to modulate expression of multiple genes, we further queried Bio-miRTa for known ALK7 effector genes containing MREs for the miR-23b cluster, and identified the BH3-only proapoptotic protein Btg2, which we had previously implicated as one of the proteins involved in the ALK7-mediated induction of apoptosis (25). Btg2 has been reported to be a target of miR-27b (43), and indeed we found that Btg2 was down-regulated upon miR-23b cluster expression in βTC3 cells (SI Appendix, Fig. S9H).
Future studies are warranted to assess the potentially distinctive functional effects and potential collaborative activity of ALK7 in combination with the 2 other miR-23b cluster targets, P2ry1 and Robo2, implicated above, as well as others in the signature (SI Appendix, Fig. S1).
Discussion
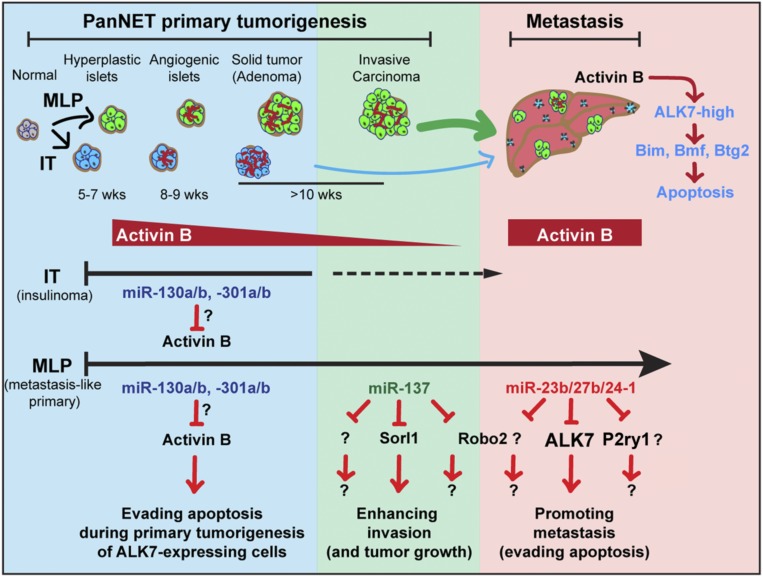
Herein, we investigated miRNAs dynamically regulated during malignant progression in pancreatic neuroendocrine tumors, and thereby have implicated specific miRNAs as modulators of distinctive neoplastic phenotypes: miR-137 enhanced primary tumor growth and invasion, whereas the miR-23b cluster contributed to the successful seeding and colonization of metastatic cancer cells in the liver (Fig. 8). MicroRNAs regulate the expression of multiple genes, which makes it challenging to dissect and understand their roles in complex diseases such as cancer (13, 14, 26). While various computational algorithms have been developed to predict the targets of miRNAs based on the sequence of the miRNA seed and other parameters (26, 28, 29, 44), it has proved challenging to identify and prioritize gene targets of miRNAs that might be important for a particular disease state or biological condition. In this study, we leveraged considerable knowledge and insights acquired over the years studying the RT2 mouse model of PanNETs (9, 22, 45), in concert with the newly developed computational algorithm, Bio-miRTa, to rank candidate targets for particular miRNAs. This strategy led us to candidate genes that are down-regulated by miR-137 or by the miR-23b cluster in the invasive and highly metastatic mouse and human MLP PanNETs, a tumor subtype in which these miRNAs are up-regulated. Importantly, by applying Bio-miRTa we were able to refine the long list of MRE-containing genes more than 100-fold, producing a shortlist of candidate genes that could in principle be screened using unbiased medium throughput approaches, such as CRISPR/Cas9. Notably, the algorithm, Bio-miRTa, can easily be customized for the identification and prioritization of candidate gene targets for miRNAs involved in the progression of other cancer types as well as other diseases, developmental stages, and physiological states, from which short-listed top-ranked candidates could then be further evaluated using functional assays.
Fig. 8.
Schematic diagram illustrating the proposed roles of the implicated miRNAs and their respective gene targets during PanNET tumorigenesis and metastasis.
In previous studies, miR-137 has been reported to have either tumor-promoting or tumor-suppressor capabilities (46). In agreement with our data that implicate miR-137 in promoting invasive growth of PanNETs and associate its elevated expression with worse survival of breast cancer patients, previous studies have shown that miR-137 is up-regulated and promotes invasion in breast, bladder, and non-small cell lung cancer (37, 47, 48). Chang et al. (48) have shown that the promoter of miR-137 contains 4 E-boxes and that its expression in non-small cell lung cancer is regulated by the EMT transcription factor Slug. Congruently, miR-137 is up-regulated in the highly invasive MLP PanNET subtype, which is characterized by an EMT-like transcriptomic phenotype (9, 22). Future studies will be required to identify the mechanisms responsible for the up-regulation of miR-137 in PanNET MLP tumors.
Using publicly available transcriptomic datasets, we show that comparatively low expression of the top 5 candidate miR-137 target genes is associated with poor prognosis of breast cancer patients. The similar association of elevated miR-137 with human breast cancer and mouse PanNET encourages their candidacy as target genes. As a first step, in this study we explored the effect of Sorl1 down-regulation in PanNETs; while known for its role in trafficking of the amyloid precursor protein and its association with Alzheimer’s disease (49), a potential function in cancer progression has not been established. Our data show that down-regulation of Sorl1 enhances tumor growth in a transplant tumor model, and concordantly, comparatively low expression is associated with poorer prognosis of human breast, lung, and gastric cancer. The precise role of Sorl1 and the pathways/functions it might regulate remain to be delineated. Future studies are also warranted to identify and functionally interrogate other miR-137 target genes, in particular the other top 4 candidates Kcnmb2, Hpse, Asph, and Robo2. Notably, the Sorl1 knockdown alone did not phenocopy the effect of miR-137 on invasiveness, implicating other targets as components of its phenotypic effects.
Our results functionally implicating the miR-23b cluster (and potentially its paralog miR-23a cluster) in metastatic colonization is both consistent with and extends upon previous publications. For example, the miR-23b cluster is up-regulated in matched human breast cancer metastasis samples compared to primary tumors, and it promotes experimental lung metastasis (50, 51). Both miR-23a and miR-23b were found to be up-regulated in colorectal liver metastasis compared to primary tumors (52), while miR-23a and miR-27a were shown to be associated with more aggressive gastric tumors and with their metastases (53, 54). Finally, the miR-23a/b cluster was found to be up-regulated in premalignant pancreatic intraepithelial neoplasia (PanIN) lesions as well as in tumor stages of human pancreatic ductal adenocarcinoma (55), was further increased in metastatic lesions (56) and associated with poorer disease-free and overall survival (57).
Herein, and in our recent study (25), we functionally characterized 3 of the identified miR-23b cluster target genes, ALK7, Robo2, and P2ry1, and showed that their knockdowns individually enhanced liver metastasis. Our focus was on ALK7, given its potent and singular effects as a metastasis suppressor (25). That said, the other candidate target genes may be modulating other aspects of the malignant phenotype. Robo2 belongs to the Slit/Robo family of axon guidance molecules, and while our understanding for this family’s role during cancer progression is still imprecise, it is thought that the molecules convey tumor suppressing signals, and accordingly, their expression is down-regulated in many cancers (58, 59). Notably, Robo2 was also 1 of the top 5 miR-137 candidate target genes, and it might also be involved in the invasive phenotype. P2ry1 is a G protein coupled receptor of extracellular ADP whose possible association with cancer is largely unknown (60); our data provide a clue that it too may serve as a barrier to tumor progression. The roles of the Slit/Robo axon guidance molecules as well as of P2ry1 during cancer progression and metastasis warrant future investigation.
While Robo2 and P2ry1 are intriguing, our functional studies highlight the singular importance of suppressing ALK7 signaling during primary tumorigenesis and metastasis in mouse models of PanNET and breast cancer (25). We showed that inactivation of ALK7 promotes metastasis to the lungs, brain, and liver, by enabling cancer cells to escape apoptosis, which is otherwise triggered by activin B secreted by various cells in these tissue sites, including endothelial cells. In the present study, we have demonstrated that the miR-23b cluster is a key repressor of ALK7 expression. Functional perturbation analysis reveals that ALK7 is a key miR-23b target gene, since rescue experiments completely abolished the prometastatic effect of the up-regulated miR-23b cluster.
In the aforementioned report (25) we also implicated activin B/ALK7 signaling as a barrier during the early stages of PanNET tumorigenesis as well as in metastasis, wherein the cell of origin, the islet beta cell, expresses activin B as well as ALK7. Activin B was observed to be down-regulated concomitant with neoplastic progression to proliferative and angiogenic precursor lesions and in turn nascent solid tumors, after which ALK7 was down-regulated during malignant progression (25). Using our newly developed algorithm for associating miRNAs with candidate genes in reverse, we found that a family of miRNAs—miR-130a/b and miR-301a/b—is potentially responsible for down-regulation of activin B during the early stages of tumorigenesis. These miRNAs are up-regulated progressively, beginning at the hyperplastic/dysplastic islet progenitor stage, and their expression continues to increase in subsequent stepwise transitions to more aggressive lesions, congruent with down-regulation of activin B. Future functional studies analogous to those presented herein for miR-137 and the miR-23b cluster are warranted to test the hypothesis that the observed up-regulation of these miRNAs during multistep tumorigenesis serves to repress activin B expression and thereby foster neoplastic progression. Interestingly, this family of miRNAs has been previously implicated in regulation of the canonical Tgfß signaling pathway (61, 62).
Collectively, the results presented herein implicate a set of distinctive miRNAs that are up-regulated during tumorigenesis and progression to invasive and metastatic phenotypes (Fig. 8). A series of biologically relevant gene targets of these miRNAs have been identified and initially characterized. While mutations and other genetic aberrations have proven to be drivers of the initial stages of cell transformation to a cancerous state, less is known about the subsequent events that mediate invasion and metastasis. Our results align with other recent studies implicating epigenetic mechanisms, in particular miRNA-mediated gene regulation, in the specification of malignant phenotypes (6). Elucidation of the upstream regulatory mechanisms governing their expression along with further mechanistic validation of the target genes they suppress may enable the design of new therapeutic strategies.
Materials and Methods
Cell Lines, Culture Conditions, and Cell Culture Assays.
The βTC3, STC-1, and 293-T cell lines were cultured in DMEM media containing 10% (vol/vol) FBS. The AJ-5257-1 was cultured in DMEM-F12 media containing 10% (vol/vol) FBS, 1% (vol/vol) insulin/transferrin/selenium, 4 µg/mL hydrocortisone, and 5 µg/mL mouse EGF. Cell lines were maintained in a 5% CO2 incubator at 37 °C. Cell lines were tested to exclude Mycoplasma contamination. The generation of stable cell lines, gene knockdown, treatment with cytokines, reporter assays, as well as plasmid generation, are described in detail in SI Appendix, SI Materials and Methods.
Western Blotting and Immunostaining.
Western blots and immunostaining were performed as has been previously described (25). Details are outlined in SI Appendix, SI Materials and Methods.
Mouse mRNA Sequencing and Differential Expression Analysis.
RNA sequencing was done as has been previously described (63, 64); details are outlined in SI Appendix, SI Materials and Methods.
Animal Studies.
Animal studies were performed as we have previously described (25), according to protocols approved by the veterinary authorities of the Canton Vaud according to Swiss law on animal protection with the authorization VD3214 (to D.H.). Details are outlined in SI Appendix, SI Materials and Methods.
The Bio-miRTa Algorithm.
Bio-miRTa has been designed to identify biologically relevant miRNA targets. The algorithm consists of 2 main steps: 1) extracting all potential targets of the miRNA(s), and 2) incorporating experimental data provided by the user to score and rank the target genes. The output of the pipeline is a list of genes scored and ranked based on the relevance to the unique biological system that is examined. A scheme of the pipeline is shown in Fig. 4 and SI Appendix, Fig. S4. More details about the algorithm are described in SI Appendix, SI Materials and Methods.
The Bio-miRTa package is available on the Hanahan laboratory website: https://www.epfl.ch/labs/hanahan-lab/data-and-tools/.
Survival Analysis.
Kaplan–Meier (KM) survival analysis was used to assess the relationship of mIR-137 and its candidate target genes expression level with survival using KM-plotter (65). For miR-137, the sample cohort was stratified according to the best cutoff using false discovery rate (FDR) < 5% (66). In each analysis, the sample cohort was stratified into 4 quartiles based on expression. The patients in the first and fourth quartiles were categorized as the “high” and “low” expression, respectively.
Quantification and Statistical Analysis.
Statistical analysis was performed and graphics were produced using R version 3.0.1 or Prism version 7.0c.
Data Availability.
RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131887).
Biological Materials Availability.
All plasmids and cell lines codes are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
We thank M. De Palma (Swiss Institute for Experimental Cancer Research–Swiss Federal Institute of Technology Lausanne [EPFL]) and M. Delorenzi (Swiss Institute of Bioinformatics [SIB]) for insightful comments on the manuscript; M. Mina (SIB) for reviewing the code for Bio-miRTa; S. Lamy and P. Magliano for technical support; J. Dessimoz (histology core facility–EPFL) for assistance with immunostaining; P. L. Sylvain and B. Mangeat (gene expression core facility–EPFL) for assistance with RNA-Seq; N. Zangger (SIB) for assistance with GEO submission, and members of the Hanahan laboratory for fruitful discussions. This work was supported by research grants from the Swiss National Science Foundation to D.H. and by Canadian Institutes of Health Research and Human Frontier Science Program Organization fellowships to I.P.M.
Footnotes
The authors declare no competing interest.
Data deposition: The RNA sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131887). The Bio-miRTa R package source code is deposited at Hanahan Lab website and accessible via https://www.epfl.ch/labs/hanahan-lab/data-and-tools/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913307116/-/DCSupplemental.
References
- 1.Steeg P. S., Targeting metastasis. Nat. Rev. Cancer 16, 201–218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert A. W., Pattabiraman D. R., Weinberg R. A., Emerging biological principles of metastasis. Cell 168, 670–691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massagué J., Obenauf A. C., Metastatic colonization by circulating tumour cells. Nature 529, 298–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaffer C. L., Weinberg R. A., A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Makohon-Moore A. P., et al. , Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat. Genet. 49, 358–366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel S. A., Vanharanta S., Epigenetic determinants of metastasis. Mol. Oncol. 11, 79–96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun M. G. H., Mao J.-H., Chiu C. W., Balmain A., Hanahan D., Polymorphic genetic control of tumor invasion in a mouse model of pancreatic neuroendocrine carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 17268–17273 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDonald O. G., et al. , Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 49, 367–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson P., et al. , MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 23, 2152–2165 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim V. N., MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6, 376–385 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Bartel D. P., Metazoan MicroRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidigal J. A., Ventura A., The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 25, 137–147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krützfeldt J., Poy M. N., Stoffel M., Strategies to determine the biological function of microRNAs. Nat. Genet. 38 (suppl.), S14–S19 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Thomas M., Lieberman J., Lal A., Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 17, 1169–1174 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Title A. C., et al. , Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 9, 4671 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ventura A., Jacks T., MicroRNAs and cancer: Short RNAs go a long way. Cell 136, 586–591 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicoloso M. S., Spizzo R., Shimizu M., Rossi S., Calin G. A., MicroRNAs–The micro steering wheel of tumour metastases. Nat. Rev. Cancer 9, 293–302 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Baranwal S., Alahari S. K., miRNA control of tumor cell invasion and metastasis. Int. J. Cancer 126, 1283–1290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pencheva N., Tavazoie S. F., Control of metastatic progression by microRNA regulatory networks. Nat. Cell Biol. 15, 546–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng Y., Croce C. M., The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 1, 15004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lujambio A., et al. , A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. U.S.A. 105, 13556–13561 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadanandam A., et al. , A cross-species analysis in pancreatic neuroendocrine tumors reveals molecular subtypes with distinctive clinical, metastatic, developmental, and metabolic characteristics. Cancer Discov. 5, 1296–1313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Hanahan D., Hijacking the neuronal NMDAR signaling circuit to promote tumor growth and invasion. Cell 153, 86–100 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Rindi G., et al. , Development of neuroendocrine tumors in the gastrointestinal tract of transgenic mice. Heterogeneity of hormone expression. Am. J. Pathol. 136, 1349–1363 (1990). [PMC free article] [PubMed] [Google Scholar]
- 25.Michael I. P., et al. , ALK7 signaling manifests a homeostatic tissue barrier that is abrogated during tumorigenesis and metastasis. Dev. Cell 49, 409–424.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Agarwal V., Bell G. W., Nam J.-W., Bartel D. P., Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, 101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John B., et al. , Human MicroRNA targets. PLoS Biol. 2, e363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paraskevopoulou M. D., et al. , DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 41, W169–W173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E., The role of site accessibility in microRNA target recognition. Nat. Genet. 39, 1278–1284 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Pinzón N., et al. , microRNA target prediction programs predict many false positives. Genome Res. 27, 234–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fridrich A., Hazan Y., Moran Y., Too many false targets for MicroRNAs: Challenges and pitfalls in prediction of miRNA targets and their gene ontology in model and non-model organisms. Bioessays 41, e1800169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karagkouni D., et al. , DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 46, D239–D245 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J.-H., Liu S., Zhou H., Qu L.-H., Yang J.-H., starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42, D92–D97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korpal M., Kang Y., The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 5, 115–119 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michael I. P., Zangger N., Hanahan D., Analysis of gene expression from βTC3 cells of Rip-Tag2 PanNET mouse model upon miR-137 and miR-23b cluster doxycycline (DOX) induction. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131887. Deposited 13 September 2017.
- 36.Pillai M. M., et al. , HITS-CLIP reveals key regulators of nuclear receptor signaling in breast cancer. Breast Cancer Res. Treat. 146, 85–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying X., Sun Y., He P., MicroRNA-137 inhibits BMP7 to enhance the epithelial-mesenchymal transition of breast cancer cells. Oncotarget 8, 18348–18358 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xue Y., et al. , Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massagué J., How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1, 169–178 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Kishore S., et al. , A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods 8, 559–564 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Gottwein E., et al. , Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10, 515–526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boudreau R. L., et al. , Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron 81, 294–305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L., et al. , MiR-27a-3p functions as an oncogene in gastric cancer by targeting BTG2. Oncotarget 7, 51943–51954 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Betel D., Koppal A., Agius P., Sander C., Leslie C., Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanahan D., Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature 315, 115–122 (1985). [DOI] [PubMed] [Google Scholar]
- 46.Mahmoudi E., Cairns M. J., MiR-137: An important player in neural development and neoplastic transformation. Mol. Psychiatry 22, 44–55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiu Y., et al. , MicroRNA-137 upregulation increases bladder cancer cell proliferation and invasion by targeting PAQR3. PLoS One 9, e109734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Chang T.-H., et al. , Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett. 402, 190–202 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Rogaeva E., et al. , The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 39, 168–177 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin L., et al. , Prooncogenic factors miR-23b and miR-27b are regulated by Her2/Neu, EGF, and TNF-α in breast cancer. Cancer Res. 73, 2884–2896 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ell B., et al. , The microRNA-23b/27b/24 cluster promotes breast cancer lung metastasis by targeting metastasis-suppressive gene prosaposin. J. Biol. Chem. 289, 21888–21895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vychytilova-Faltejskova P., et al. , Genome-wide microRNA expression profiling in primary tumors and matched liver metastasis of patients with colorectal cancer. Cancer Genomics Proteomics 13, 311–316 (2016). [PubMed] [Google Scholar]
- 53.An J., et al. , MiR-23a in amplified 19p13.13 loci targets metallothionein 2A and promotes growth in gastric cancer cells. J. Cell. Biochem. 114, 2160–2169 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Zhang Z., Liu S., Shi R., Zhao G., miR-27 promotes human gastric cancer cell metastasis by inducing epithelial-to-mesenchymal transition. Cancer Genet. 204, 486–491 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Humeau M., et al. , Salivary MicroRNA in pancreatic cancer patients. PLoS One 10, e0130996 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu G., et al. , MicroRNA-23a promotes pancreatic cancer metastasis by targeting epithelial splicing regulator protein 1. Oncotarget 8, 82854–82871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frampton A. E., et al. , MicroRNAs cooperatively inhibit a network of tumor suppressor genes to promote pancreatic tumor growth and progression. Gastroenterology 146, 268–77.e18 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Gara R. K., et al. , Slit/Robo pathway: A promising therapeutic target for cancer. Drug Discov. Today 20, 156–164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu F., Ma Y., Zhang J., Qin F., Fu L., Function of Slit/Robo signaling in breast cancer. Front. Med. 9, 431–436 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Barańska J., Czajkowski R., Pomorski P., P2Y1Receptors–Properties and functional activities. Adv. Exp. Med. Biol. 1051, 71–89 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Jacobsen A., et al. , Analysis of microRNA-target interactions across diverse cancer types. Nat. Struct. Mol. Biol. 20, 1325–1332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu L., et al. , The oncogenic role of microRNA-130a/301a/454 in human colorectal cancer via targeting Smad4 expression. PLoS One 8, e55532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendrikx S., et al. , Endothelial calcineurin signaling restrains metastatic outgrowth by regulating Bmp2. Cell Rep. 26, 1227–1241.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Micheletti R., et al. , The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 9, eaai9118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Györffy B., et al. , An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Lánczky A., et al. , miRpower: A web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 160, 439–446 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131887).