Abstract
In 2006 several yeast-like fungi were isolated from apples that showed a postharvest disorder named “white haze.” These strains were morphologically and molecularly assigned to the genus Tilletiopsis. Following the recent reclassification of yeasts in Ustilaginomycotina and the genus Tilletiopsis in particular, species that caused “white haze” disorder were re-identified based on the phylogenetic analysis of five DNA-loci (ITS, LSU, SSU, RPB2, and TEF1) and analysis of D1/D2 domains of the 26S/28S rRNA (LSU). Six novel species belonging to three orders in the Exobasidiomycetes, namely Entyloma belangeri (holotype: CBS 111600; ex-type: DSM 29114) MB 823155, Entyloma davenportii (holotype: CBS 111604; ex-type: DSM 100135) MB 823154, Entyloma elstari (holotype: CBS 111593; ex-type: DSM 29113) MB 823153, Entyloma randwijkense (holotype: CBS 111606; ex-type: DSM 100136) MB 823156, Jamesdicksonia mali (holotype: CBS 111625; ex-type: DSM 29121) MB 823151 and Golubevia heteromorpha (holotype: CBS 111610; ex-type: DSM 100176) MB 823152 are proposed to accommodate these strains. In addition, sequences representing phylogenetically related but yet undescribed fungi were obtained from GenBank in order to show the diversity of Tilletiopsis-like yeast states in Exobasidiomycetes.
Keywords: six new species, Tilletiopsis, apple, white haze, postharvest disorder
Introduction
Species of the genus Tilletiopsis are saprotrophic yeast-like basidiomycete fungi. The name Tilletiopsis was first introduced by Derx (1930) and was chosen to reflect the morphological resemblance of his isolates to species of the smut fungi Tilletia. Eighteen years later, Derx (1948) described the genus without designating a type species (Derx, 1948). Unfortunately, the strain he was working with got lost so that Nyland (1950) selected a morphologically similar strain to serve as the neotype of the type species of the genus Tilletiopsis, which he named Tilletiopsis washingtonensis. Another species with smaller ballistospores was named Tilletiopsis minor, and both yeasts were isolated near Puyallup (Washington, United States) from living and dead plant material (Nyland, 1950). Two years later Tubaki described two additional species and a variety based on spore size and culture morphology, namely Tilletiopsis cremea, Tilletiopsis lilacina, and T. minor var. flava (Tubaki, 1952). In 1972 Gokhale added Tilletiopsis fulvescens, Tilletiopsis albescens and Tilletiopsis pallescens to the genus (Gokhale, 1972). In 1991 the name Tilletiopsis flava was proposed for T. minor var. flava (Boekhout, 1991). The latest described species were Tilletiopsis derxii, Tilletiopsis oryzicola, and Tilletiopsis penniseti (Takashima and Nakase, 2001). The treatment of T. cremea and T. lilacina as synonyms of T. washingtonensis (Boekhout, 1998) was disproved by molecular studies of Hamamoto et al. (2000).
Traditionally, phenotypic, i.e., morphological and physiological, criteria were used to distinguish yeast species. In culture, yeasts of the genus Tilletiopsis are distinguished by tough to soft colonies that are cream, pinkish-cream, pinkish-yellow, yellow-brown or brown in color, with a reticulate, transversely wrinkled, postulate or velutinous surface (Boekhout, 1998, 2011). The micromorphology of species is rather uniform including narrow, septate, hyaline, monokaryotic regularly branched hyphae with retraction septa, and cytoplasm-containing cells that are usually surrounded by lysed cells (Boekhout, 1998, 2011). Ballistoconidia occur in species of Tilletiopsis (Boekhout, 1998, 2011). With the development of identification methods (reviewed in Barnett, 2004; Kurtzman et al., 2011) biochemical characteristics such as Q-10 as major ubiquinone, and cell walls without xylose, positive Diazonium blue B and urease reactions, and the inability to produce starch-like compounds were found to be characteristic for the genus (Boekhout, 1991, 2011; Boekhout et al., 1992). As has been noted by several authors, the morphology of Tilletiopsis spp. resembles that of some smut fungi, such as Entyloma and Tilletia, suggesting possible anamorph-teleomorph relationships between these yeasts and plant parasites in Exobasidiomycetes (e.g., Boekhout, 1987, 1991). Application of ribosomal gene sequencing confirmed the phylogenetic relationships of the genus Tilletiopsis in Exobasidiomycetes, but also indicated that the genus Tilletiopsis is polyphyletic (Boekhout et al., 1995; Takashima and Nakase, 1996). Further studies confirmed that species of Tilletiopsis are placed in three orders, namely Georgefischerales, Entylomalates, and Doassansiales, and some species represent the anamorphic stage of known teleomorphic species (Begerow et al., 2000; Fell et al., 2000; Bauer et al., 2001, 2006; Boekhout, 2011; Wang et al., 2015).
Recent changes introduced by the International Code of Nomenclature for algae, fungi, and plants (ICN, Melbourne Code, 2012; McNeill et al., 2012) discontinued the use of dual nomenclature for naming fungi with a pleomorphic life-cycle and the so-called “One Fungus = One Name” principle (Hawksworth, 2011; Taylor, 2011) was applied to unify nomenclature of sexual and asexual fungi in the phylum Ustilaginomycotina (Wang et al., 2015). In the revision of the genus Tilletiopsis by Wang et al. (2015) most of the described species were reclassified in either new genera (i.e., Robbauera and Golubevia) or already existing teleomorphic genera (i.e., Phragmotaenium and Gjaerumia). Specifically, T. albescens was recombined as Robbauera albescens; T. pallescens as Golubevia pallescens; T. derxii, T. flava, T. fulvescens, and T. oryzicola as Phragmotaenium derxii, Phragmotaenium flavum, Phragmotaenium fulvescens, and Phragmotaenium oryzicola, respectively; T. penniseti and T. minor as Gjaerumia penneseti and Gjaerumia minor, respectively. Only the type species, T. washingtonensis, and the two related species T. cremea and T. lilacina remained in the genus Tilletiopsis. Despite previously supposed close relationships with the genus Entyloma, none of the described Tilletiopsis species was transferred to this genus.
Species of the genus Tilletiopsis sensu lato are ubiquitous and can be found in various environments, and the most frequently reported habitats of these yeasts include diverse, either dead or living, plant material (Boekhout, 1991). Additionally, the propagules have been detected in the air, from where these yeasts can be transferred to flowers, plant surfaces, soils, sewage and deep-sea sediments, seawater and even sea animals (Gokhale, 1972; Shivas and Brown, 1989; Fairs et al., 2010; Li et al., 2011; Mahé et al., 2014; Yue et al., 2015). T. minor is the only known species of this genus, that was reported from clinical specimens and is a possible causing agent of pneumonia and corneal abscess (Ramani et al., 1997; Kechkekian et al., 2007; AL-Zaydani et al., 2014). Some species may have evolved a mycoparasitic lifestyle as has been suggested for T. pallescens and T. albescens (Boekhout, 2011; Begerow et al., 2014). Interestingly, these Tilletiopsis species show potential as biocontrol agents against powdery mildews, especially against the Cucurbitaceae pathogens of Podosphaera (Last, 1970; Kiss, 2003). The active principle seems to be a fatty acid ester, and similar compounds are already known to be secreted by other members of the Ustilaginomycotina, including the commercially available biocontrol agent Pseudozyma flocculosa (Urquhart and Punja, 2002), which was erroneously reclassified in the genus Anthracocystis due to the errors with the type material of this species (Richard R. Bélanger and T. Boekhout, personal communication).
Together with other conventional epiphytes, Tilletiopsis occurs on the surface of apples where its growth matters as a disruptive element in the commercial production and distribution of the fruit (Boekhout et al., 2006). Different species were isolated from apples in several European countries and were ultimately linked to the postharvest disorder named “white haze,” an intensive fungal growth on the apple fruit surface which results in a compromised quality of the fruits (Boekhout et al., 2006; Baric et al., 2010; Weber and Zabel, 2011; Prencipe et al., 2016). A high relative humidity and lower temperatures were suggested as key factors controlling the disorder that appears after Ultra-Low Oxygen storage (Boekhout et al., 2006).
Several potential new species of the genus Tilletiopsis were discovered from apples showing white haze disorder (Boekhout et al., 2006). The identification based on the rDNA sequencing (D1/D2 domains of the LSU and ITS) together with morphological analyses suggested their close relationships with the genus Entyloma and other genera within the Exobasidiomycetes (Boekhout et al., 2006). Because these cultures were not included in the study by Wang et al. (2015) and because the genus Tilletiopsis is currently restricted to the clade containing its generic type T. washingtonensis, we performed a multi-locus phylogenetic analysis to provide a proper placement of these hitherto undescribed species. In addition to the nucleotide sequences determined by Boekhout et al. (2006), we analyzed partial sequences of the ribosomal small subunit (SSU rDNA), as well as fragments of the genes encoding the second largest subunit of RNA polymerase II (RPB2) and the translation elongation factor 1 alpha (TEF1). Herein we describe six new species and indicate possible novel taxa based on the phylogenetic analysis of LSU rDNA sequences from public repositories.
Materials and Methods
All 28 studied strains were isolated by Boekhout et al. (2006) and preserved in the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands (Table 1). Temperature growth tests were performed on MEA, GPYA, and PDA. Physiological tests were performed in liquid culture at 10°C and examined after 1, 2, 3, and 4 weeks. Nucleotide sequences of ITS and LSU of those strains have been published before by Boekhout et al. (2006). Other LSU sequences obtained from NCBI GenBank1 and analyzed here represent potential novel species that were originally identified as members of the genus Tilletiopsis by the authors (Fungsin, 2003; Jindamorakot, 2006; Takashima et al., 2012). Reference sequences were derived from GenBank and from Wang et al. (2015). All sequence and strain accession numbers for the five-loci analysis are listed in Table 2. Clades and strains of the new species are labeled (A–G) after the classification of Boekhout et al. (2006).
TABLE 1.
Strains and GenBank accession numbers of new species, proposed type strains in bold.
| Strain | Proposed name | Clade | ITS | LSU | SSU | RPB2 | TEF1 |
| CBS 111625 | Jamesdicksonia mali | A | AY879279 | AY879274 | LT615037 | LT614983 | LT615009 |
| CBS 111628 | Jamesdicksonia mali | A | AY879281 | AY272007 | LT615038 | LT614984 | LT615010 |
| CBS 111610 | Golubevia heteromorpha | B | AY259058 | AY272003 | LT615026 | LT614973 | LT614999 |
| CBS 111608 | Golubevia heteromorpha | B | AY259056 | AY272001 | LT615024 | LT614971 | LT614997 |
| CBS 111609 | Golubevia heteromorpha | B | AY259065 | AY272011 | LT615025 | LT614972 | LT614998 |
| CBS 111611 | Golubevia heteromorpha | B | AY259060 | AY272005 | LT615027 | LT614974 | LT615000 |
| CBS 111612 | Golubevia heteromorpha | B | AY879275 | AY879270 | LT615028 | LT614975 | LT615001 |
| CBS 111614 | Golubevia heteromorpha | B | AY259079 | AY272032 | LT615029 | LT614976 | LT615002 |
| CBS 111615 | Golubevia heteromorpha | B | AY259053 | AY272025 | LT615030 | LT614977 | LT615003 |
| CBS 111616 | Golubevia heteromorpha | B | AY259049 | AY272022 | LT615031 | LT614978 | LT615004 |
| CBS 111617 | Golubevia heteromorpha | B | AY259050 | AY272023 | LT615032 | LT614979 | LT615005 |
| CBS 111618 | Golubevia heteromorpha | B | AY259078 | AY272031 | LT615033 | LT614980 | – |
| CBS 111620 | Golubevia heteromorpha | B | AY879276 | AY879273 | LT615034 | LT614981 | LT615006 |
| CBS 111621 | Golubevia heteromorpha | B | AY259082 | AY272035 | LT615035 | LT614982 | LT615007 |
| CBS 111741 | Golubevia heteromorpha | B | AY259061 | AY272006 | LT615036 | – | LT615008 |
| CBS 111604 | Entyloma davenportii | E | AY259064 | AY272010 | LT615013 | LT614961 | LT614987 |
| CBS 111607 | Entyloma davenportii | C | AY259051 | AY272024 | LT615011 | LT614959 | LT614985 |
| CBS 111603 | Entyloma davenportii | E | AY259054 | AY272026 | LT615012 | LT614960 | LT614986 |
| CBS 111593 | Entyloma elstari | D | AY259048 | AY272021 | LT615023 | DQ234552 | DQ028593 |
| CBS 111600 | Entyloma belangeri | F | AY259074 | AY272019 | LT615019 | LT614967 | LT614993 |
| CBS 111596 | Entyloma belangeri | F | AY259055 | AY272027 | LT615015 | LT614963 | LT614989 |
| CBS 111597 | Entyloma belangeri | F | AY259070 | AY272015 | LT615016 | LT614964 | LT614990 |
| CBS 111598 | Entyloma belangeri | F | AY259071 | AY272016 | LT615017 | LT614965 | LT614991 |
| CBS 111599 | Entyloma belangeri | F | AY259075 | AY272020 | LT615018 | LT614966 | LT614992 |
| CBS 111601 | Entyloma belangeri | F | AY259072 | AY272017 | LT615020 | LT614968 | LT614994 |
| CBS 111602 | Entyloma belangeri | F | AY259069 | AY272028 | LT615021 | LT614969 | LT614995 |
| CBS 111605 | Entyloma belangeri∗ | G1 | AY259076 | AY272029 | LT615022 | LT614970 | LT614996 |
| CBS 111606 | Entyloma randwijkense | G2 | AY259080 | AY272033 | LT615014 | LT614962 | LT614988 |
∗Provisionally assigned to Entyloma belangeri species complex.
TABLE 2.
Species, strains, and GenBank accession numbers for the five-loci tree.
| Species | LSU | ITS | SSU | RPB2 | TEF1 |
| Entyloma arnoseridis CBS 203.36∗ | DQ645528 | DQ911609 | DQ645529 | DQ645530 | DQ645531 |
| Entyloma calendulae CBS 513.93 | DQ663687 | DQ663689 | DQ663688 | DQ663690 | DQ663691 |
| Entyloma ficariae CBS 480.91∗ | AJ235295 | – | KP322949 | KP323102 | KP323125 |
| Exobasidium gracile DSM 4460∗ | DQ663699 | DQ663700 | DQ785786 | DQ663701 | DQ663703 |
| Gjaerumia minor CBS 543.50T* | AJ235287 | KP322989 | KP322972 | KP323097 | KP323114 |
| Gjaerumia penniseti CBS 110032T* | AB052825 | – | KP322975 | KP323085 | KP323143 |
| Golubevia pallescens CBS 364.85T* | AJ235292 | – | KP322973 | KP323101 | KP323123 |
| Microstroma juglandis CBS 287.63∗ | AF009867 | DQ789988 | DQ789987 | DQ789989 | DQ789991 |
| Phragmotaenium derxii CBS 110078T* | AB052823 | AB045707 | AB045704 | KP323086 | KP323138 |
| Phragmotaenium flavum CBS 401.84T* | AJ235285 | KP322987 | KP322970 | – | KP323126 |
| Phragmotaenium fulvescens CBS 607.83T* | AJ235282 | KP322988 | KP322971 | KF706530 | KF706483 |
| Rhamphospora nymphaeae CBS 172.38∗ | DQ831032 | DQ831034 | DQ831033 | DQ831035 | DQ831036 |
| Robbauera albescens CBS 608.83T* | AJ235289 | KP322986 | KP322968 | KP323095 | KP323127 |
| Tilletia goloskokovii AFTOL-ID 1713∗ | AY818998 | DQ832248 | DQ832247 | DQ832249 | DQ832251 |
| Tilletiaria anomala CBS 436.72T* | AJ235284 | DQ234558 | AY803752 | AY803750 | DQ835991 |
| Tilletiopsis cremea CBS 605.83T* | AJ235279 | AB025690 | KP322969 | KP323108 | KP323129 |
| Tilletiopsis lilacina CBS 435.92T* | AJ235309 | KP322984 | KP322966 | KP323110 | KP323112 |
| Tilletiopsis washingtonensis CBS 544.50T* | AJ235278 | DQ835994 | KP322976 | DQ835995 | DQ835996 |
| Mycosarcoma maydis CBS 504.76∗ | AF453938 | AY854090 | KP322979 | KP323090 | KP323130 |
TSequences from type material; ∗sequences used by Wang et al. (2015).
Genomic DNA was extracted from cultures grown on potato dextrose agar (PDA, BD Difco), using the EZ-10 Spin Column Genomic DNA kit for plant samples (Bio Basic Canada Inc., Markham, Canada). The extractions were performed using the manufacturer’s protocol. For cell disruption a Precellys 24 homogenizer (Bertin Technologies, France) was used at a speed of 6000 rpm for 2 × 40 s. Four gene regions were amplified, namely a part of the ribosomal small subunit (SSU or 18S rRNA gene), the internal transcribed spacer region (ITS) and fragments of the two protein-coding genes encoding the second largest RNA polymerase II subunit (RPB2) and the translation elongation factor 1 alpha (TEF1). Primers ITS1 and ITS4 (White et al., 1990), NS23UCB and NS24UCB (Gargas and Taylor, 1992) were used to amplify SSU, RPB2-6F and RPB2-7R primers to amplify RPB2 (Liu et al., 1999), and EF1-983F and EF1-2218R to amplify TEF1 (Matheny et al., 2007). All primer sequences are provided at the PriMicro Database Project2 (Supplementary Table S1). PCR products were purified with the EZ-10 Spin Column PCR Purification kit (Bio Basic Canada Inc., Markham, Canada) and the FastGene Gel/PCR Extraction kit (Nippon Genetics Europe GmbH, Düren, Germany) following the manufacture’s protocols. Purified PCR amplification products were sequenced by the department of Genome Analytics at the Helmholtz Centre for Infection Research (Braunschweig, Germany), using the same primers as used for the PCR reaction.
Independent alignments and phylogenetic analyses were performed for each locus. Multiple sequence alignments were performed with the nucleotide sequences using MAFFT 7.017 algorithm (Katoh et al., 2002) with default parameters. The resulting alignments were additionally cured with Gblocks (Castresana, 2000; Talavera and Castresana, 2007) allowing smaller final blocks, gap positions within the final blocks, and less strict flanking positions. The following two datasets were used in the phylogenetic analyses: (i) a five-loci dataset (ITS, LSU, SSU, RPB2, and TEF1) was used to analyse the placement of the novel species within the Exobasidiomycetes and (ii) a LSU dataset was used to analyse nucleotide sequences of potential novel species available in public databases. Single-gene alignments corresponding to five-loci were concatenated and the best nucleotide substitution model was determined with MEGA 7.0.14 (Kumar et al., 2016). Phylogenetic relationships were inferred by the maximum likelihood (ML) method based on the general time reversible (GTR + G + I) model for the five-loci dataset and K80 + G + I for the LSU dataset, respectively. Trees were calculated with RAxML 7.2.8 (Stamatakis, 2006) and the PhyML (Guindon and Gascuel, 2003) plugins implemented in Geneious 7.1.4 (Biomatters Ltd., Auckland, New Zealand), followed by 1000 bootstrap replicates. Mycosarcoma (formerly Ustilago) maydis was selected as an outgroup.
Results
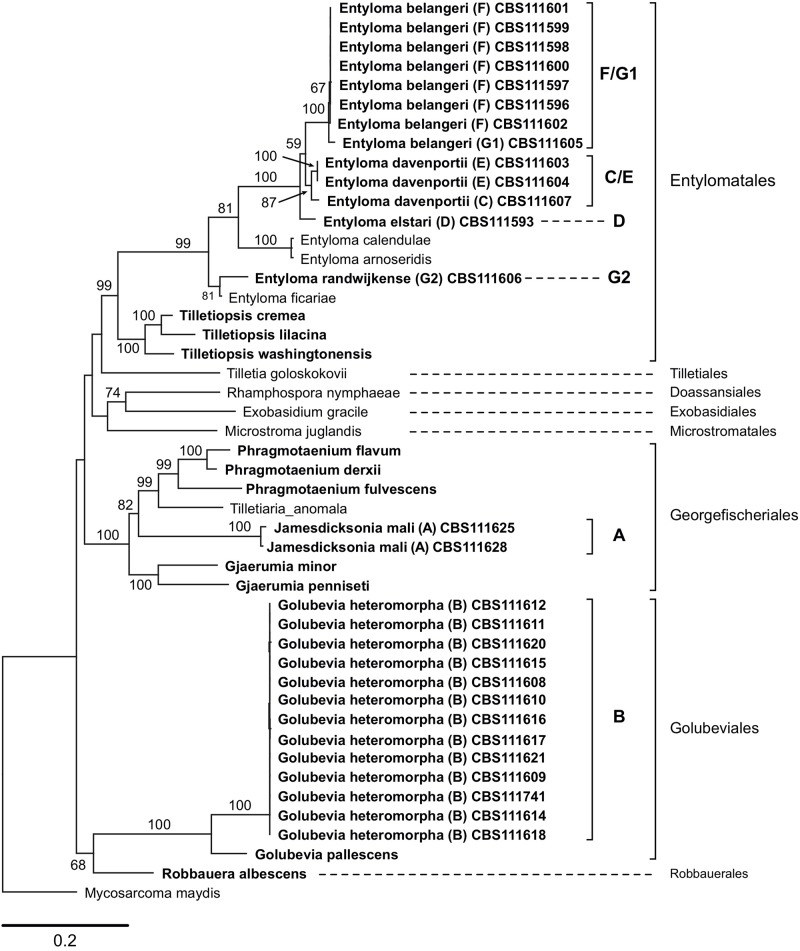
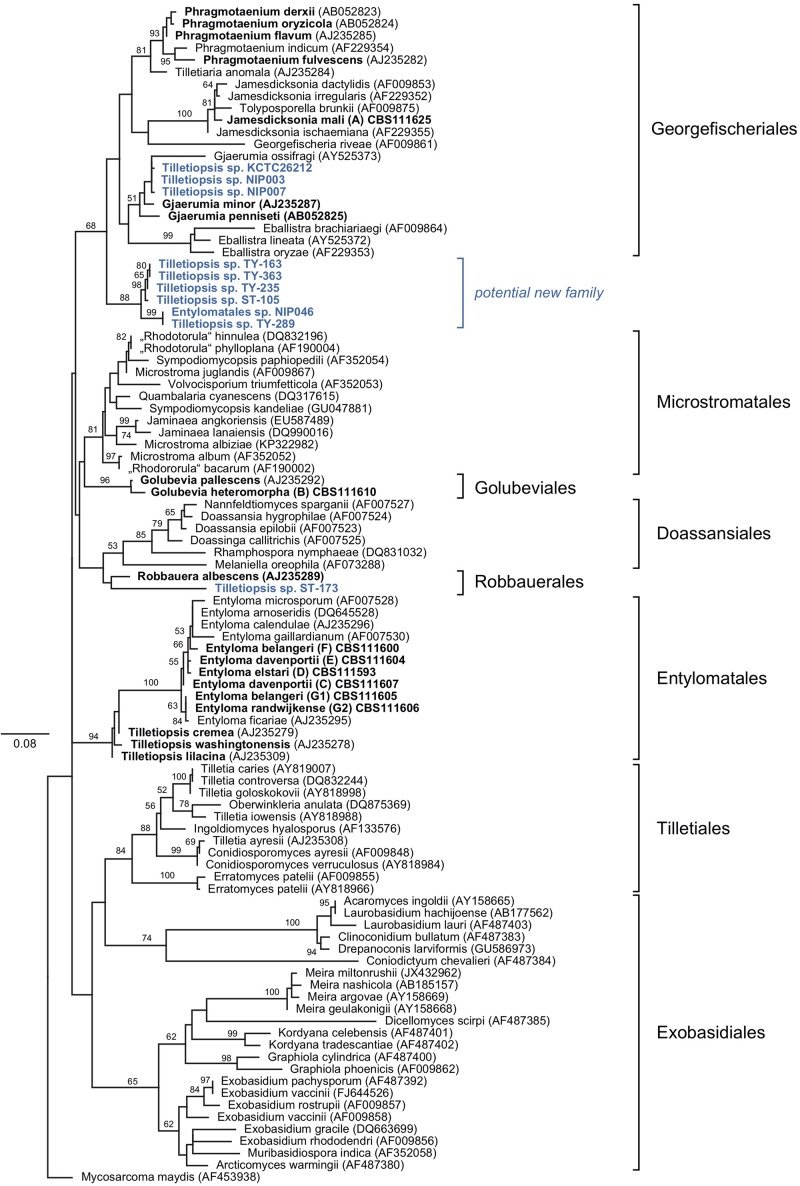
Sequences that were generated in this study, and those obtained from GenBank, reference sequences from Wang et al. (2015) and data published by Boekhout et al. (2006) were used to produce two phylogenetic trees (Figures 1, 2). The five-loci tree provides an overview on the phylogenetic relationships of asexual and sexual taxa in Exobasidiomycetes (Figure 1). The orders Tilletiales, Doassansiales, Microstromatales, and Exobasidiales were represented by a single species to provide a better overview and not to overload the tree. In a taxonomically broad analysis based on LSU rRNA sequences (Figure 2), new species described in this study were represented by a few sequences only. In both analyses, the placement of six new species was resolved. Specifically, they were placed in orders Georgefischeriales, Golubeviales, and Entylomatales. Both trees showed, however, a rather weak support for higher ranks, but the respective families received a good support.
FIGURE 1.
Maximum likelihood analysis of a five-loci alignment combining sequences of the ITS region (including 5.8S rRNA gene), LSU (D1/D2 domains), and SSU rRNA gene, RPB2, and TEF1, showing the relationships of taxa within the Exobasidiomycetes. The numbers on branches are frequencies (>50%) with which a given branch appeared in 1000 bootstrap replications. The scale bars indicate the numbers of expected substitutions accumulated per site. Nucleotide sequences of Tilletiopsis-like fungi are in bold; clade classification (A–G) follows (Boekhout et al., 2006).
FIGURE 2.
Diversity of Tilletiopsis-like fungi determined with Maximum Likelihood analysis of the LSU rRNA gene (D1/D2 domains). The numbers on branches are frequencies (>50%) with which a given branch appeared in 1000 bootstrap replications. The scale bars indicate the numbers of expected substitutions accumulated per site. Nucleotide sequences of Tilletiopsis-like fungi are in bold. Potential new species are given in blue color.
In the genus Entyloma, pairwise sequence comparisons showed 90–98% similarity in the ITS region and 98–100% similarity in LSU. Intraspecific variation of ITS sequences in Entyloma davenportii was 3%. Most strains of Entyloma belangeri showed highly similar ITS (99–100% similarity) and LSU (100% similarity) sequences but nucleotide sequences of strain CBS 111605 showed only 92% similarity in ITS and 98% similarity in LSU. Two strains of Jamesdicksonia mali shared identical LSU sequences, whereas ITS sequences showed 94% similarity. In the genus Tilletiopsis, interspecific pairwise sequence comparisons resulted in 99 and 92–95% similarity values for LSU and ITS sequences, respectively.
In the five-loci phylogenetic analysis, two strains (clade A in Boekhout et al., 2006) were placed in the Georgefischeriales in an intermediate position between the already known Tilletiopsis species, which were recently transferred to the genera Gjaerumia and Phragmotaenium (Wang et al., 2015). This result was also supported by the LSU analysis, where the clade A was placed in the genus Jamesdicksonia with high support (ML: 100%). Clade B represented by 13 strains was placed in the order Golubeviales close to the recently reclassified G. pallescens with high support (ML: 100 and 96% in five-loci and LSU analyses, respectively). The other four new species were placed in the order Entylomatales (clades C/E, F/G1, D, and G2) close to Entyloma ficariae (Figure 1). Despite some observed sequence heterogeneity between strains (9% ITS and 2% LSU), our results suggest that clade F/G1 are likely to represent one new species (ML: 100%). The two other clades C/E and D (one strains) constitute two other novel species (Figure 1). The total intraspecific sequence similarity within clades F/G1 and C/E was 98% (Supplementary Data Sheet S1). The LSU tree supported the placement of these species in Entylomatales, but also showed two conflicts between the trees, namely (i) the placement of strain G1 next to G2 in close relationship to E. ficariae, and (ii) the separation of the clade C/E in the LSU tree (Figure 2). However, the five-loci tree strongly supported the placement of new species within the order Entylomatales, while internal nodes in Entylomatales received no support in the LSU analysis (Figures 1, 2). As several smut and false smut fungi, species of the genus Entyloma are distinguished based on the combination of morphology (e.g., spore measurements), host plant, and genetic information, usually ITS sequences (e.g., Begerow et al., 2002; Boekhout et al., 2006; Lutz and Pia̧tek, 2016). Because plant parasites have been studied by traditional mycologists and phytopathologists, only a few Entyloma species are available in culture and three of them have been used by Wang et al. (2015) in the multi-gene phylogenetic analysis. The lack of cultures and nucleotide sequence data (e.g., sequences of protein-coding genes) from teleomorphic species is hampering a better taxonomic placement in many fungal lineages (Kachalkin et al., 2019), including smuts (e.g., Wang et al., 2015; Begerow et al., 2017). Right now it is impossible to univocally interpret the unexpectedly high variability of ITS sequences among E. belangeri isolates. This result contradicts to some extent current views on species delimitation in the genus Entyloma (e.g., Lutz and Pia̧tek, 2016), but it is important to document that total sequence variability was low (Supplementary Data Sheet S1). In the lack of host data and teleomorph morphology, phylogenetic characters, i.e., results of the multi-gene analysis, are used here to delimit new species. We are in favor of a rather conservative decision to keep strain CBS 111605 provisionally assigned to E. belangeri species complex until more Entyloma specimens and cultures will be sequenced.
In addition to the afore-mentioned new species, sequences obtained from public databases were included in the LSU dataset to provide a broader look on the genetic diversity of fungi morphologically resembling the genus Tilletiopsis. These nucleotide sequences represent potential undescribed species. Our analysis placed a few of these sequences in two orders of the Exobasidiomycetes, namely Georgefischeriales and Robbauerales, and in a new clade that represents a distinct cluster close to the order Georgefischeriales (Figure 2). The sequence of ST-173 (GenBank: DQ404470) was placed close to R. albescens, the only presently known species in the order Robbauerales. Despite low statistical support, this sequence represents most likely the second species in this order. Three sequences retrieved from GenBank (AB726595, AB726598, AF459717) were placed in the Georgefischeriales close to G. penneseti and G. minor, but with little statistical support in the analysis. Sequences of these strains display high pair-wise similarity and likely represent a new species. Six strains formed a clade close to Georgefischeriales (Figure 2). The clade itself received moderate support (ML: 88%), but its relationship with the order Georgefischeriales was not supported (ML: 68%). Our analysis suggests that this new clade likely corresponds to a new family or order. Within the clade sequences NIP046 (AB726628) and TY-289 (AY313022) probably represent one well-supported (ML: 99%) new species, and the sister clade with four more strains (DQ404458, AY313021, AY313023, AY313020) could represent at least one more new species.
From an ecological point of view, it is important to note that the species studied here were involved in a post-harvest disorder on apples, called white haze (Boekhout et al., 2006) that occurred most prominently in the apple harvest season during humid and colder periods and under Low Oxygen storage. As standard growth tests of yeasts are done at 25°C our first attempts were also done at this temperature. However, several of the strains did not grow well or not at all at this temperature. Therefore, we investigated the growth range of the isolates at temperatures ranging from 4 to 30°C on three different media. All isolates tested grew well at 4–15°C, and hence we repeated the growth tests, both carbon and nitrogen compounds, at 10°C. All studied cultures required a longer period of ca. 2 weeks to start growing well, but once growth started quite some visible biomass in the test tubes was accumulated.
Taxonomy
Description of Jamesdicksonia mali Richter, Yurkov, and Boekhout, sp. nov. (MB 823151)
Etymology: The specific epithet mali refers to the plant genus Malus Mill. (Rosaceae), which includes apple trees, the source of isolation.
After 2 weeks at 22°C on PDA, the colonies are tough, wrinkled, dull, covered with slender fascicles, yellow to cream colored, with an entire to slightly ridged margin. Ballistospores are allantoid or cylindrical, 2–3 × 12–19 μm (Figure 3I).
FIGURE 3.
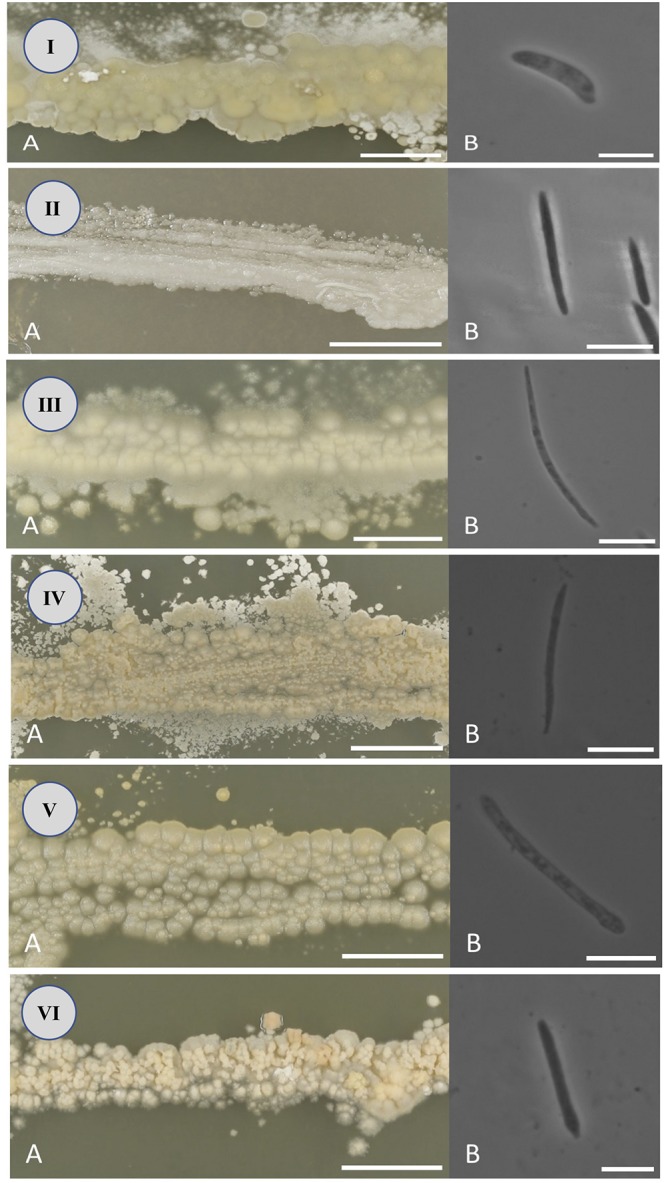
Colony (A) and ballistospore (B) morphology of Jamesdicksonia mali (I), Golubevia heteromorpha (II), Entyloma elstari (III), Entyloma davenportii (IV), Entyloma belangeri (V), and Entyloma randwijkense (VI) on potato dextrose agar (bars: A = 10 mm, B = 10 μm).
Fermentation of glucose absent. Growth on glucose, galactose, D-glucosamine (at 10°C), D-ribose, D-xylose (delayed), L-arabinose, rhamnose (weak or negative), sucrose, maltose, trehalose, cellobiose, arbutin (weak), melibiose, lactose, raffinose, melezitose, inuline, soluble starch, glycerol (weak), erythritol (weak), arabinitol (delayed), glucitol, mannitol, D-gluconate, glucoronate, succinate, and quinic acid. No growth on sorbose, D-arabinose, methyl-alpha-D-glucoside, salicin, ribitol, xylitol, galactitol, inositol, glucono-delta-lactone, 2-keto-gluconate, D-galacturonate, saccharate, lactate, citrate, methanol, ethanol, propane-1,2-diol, butane-2,3-diol, and galactonic acid. Growth on potassium nitrate, sodium nitrite, lysine, and ethylamine (weak or negative). No growth on cadaverine, creatinine, creatinine, D-glucosamine and imidazole. No growth in the presence of 0.01% cycloheximide, in the presence of 10% NaCl and in 50% glucose. Urea hydrolysis and Diazonium Blue B reaction are positive. Optimal growth temperature: 15–22°C. Maximum growth temperature: 25°C; growth at 30°C is weak (on MEA) or negative (on GPYA).
Molecular characteristics (holotype): Nucleotide sequences of ITS, SSU, LSU (D1/D2 domains) rRNA and RPB2 and TEF1 deposited in NCBI/EMBL (GenBank) under the accession numbers AY879279 (ITS), AY879274 (LSU), LT615037 (SSU), LT614983 (RPB2), LT615009 (TEF1).
Deposits: Holotype CBS 111625 isolated from an apple of the cultivar Elstar showing signs of white haze obtained through the Plant Protection Service [Plantenziektenkundige Dienst (PD)] in Wageningen, Netherlands in 1994 (Boekhout et al., 2006), preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Ex-type culture is deposited at the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (DSM 29121).
Strains studied: paratypes CBS 111625 (=DSM 29121), CBS 111628 (=DSM 29122).
Description of Golubevia heteromorpha Boekhout, Richter, and Yurkov, sp. nov. (MB 823152)
Etymology: The specific epithet heteromorpha (Gr. adj. f., hetero – different, morphe shape) refers to a diverse morphological appearance as plate culture on culture medium.
After 2 weeks at 22°C on PDA, the colonies are tough, wrinkled, shiny or dull, covered with slender fascicles, cream colored or whitish, with an entire to slightly ridged margin. Ballistospores are cylindrical, elongate, or lunate, 1–2 × 12–20 μm (Figure 3II).
Fermentation of glucose absent. Growth on glucose, sorbose (sometimes delayed), L-arabinose (sometimes weak), D-arabinose, sucrose, maltose, trehalose, melibiose (delayed), raffinose (delayed), melezitose (delayed), inulin (weak), and soluble starch (delayed). Variable growth on galactose, ribose, xylose, methyl-alpha-D-glucoside, cellobiose, arbutin, lactose, glycerol, erythritol, ribitol, xylitol, arabinitol, glucitol, mannitol, inositol, glucono-delta-lactone, 2-keto-gluconate, succinate, and quinic acid. No growth on D-glucosamine, rhamnose, salicin, galactitol, gluconate, glucoronate, galacturonate, saccharate, lactate, citrate, methanol, ethanol, propane-1,2-diol, butane-2,3-diol, and galactonic acid. Growth on potassium nitrate, sodium nitrite, lysine (variable) and imidazole (variable). No growth on ethylamine, cadaverine, creatine, creatinine, and D-glucosamine. No growth in the presence of 0.01% cycloheximide, in the presence of 10% NaCl and in 50% glucose. Urea hydrolysis and Diazonium Blue B reaction are positive. Optimal growth temperature: 15–22°C. Maximum growth temperature: 20°C; some strains can grow at 25°C on MEA.
Molecular characteristics (holotype): Nucleotide sequences of ITS, SSU, LSU (D1/D2 domains) rRNA and RPB2 and TEF1 deposited in NCBI/EMBL (GenBank) under the accession numbers AY259058 (ITS), AY272003 (LSU), LT615026 (SSU), LT614973 (RPB2), LT614999 (TEF1).
Deposits: Holotype CBS 111610 isolated from an apple of the cultivar Elstar showing signs of white haze obtained through the Plant Protection Service [Plantenziektenkundige Dienst (PD)] in Wageningen, Netherlands in 1994 (Boekhout et al., 2006), preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. Ex-type culture is deposited at the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (DSM 100176).
Strains studied: paratypes CBS 111610 (=DSM 100176), CBS 111608 (=DSM 100175), CBS 111609 (=DSM 29125), CBS 111611 (=DSM 100138), CBS 111612 (=DSM 100177), CBS 111614 (=DSM 100139), CBS 111615 (=DSM 100140), CBS 111616 (=DSM 100141), CBS 111617 (=DSM 100142), CBS 111618 (=DSM 100143), CBS 111620 (=DSM 100144), CBS 111621 (=DSM 100145), CBS 111741 (=DSM 29124).
Description of Entyloma elstari Yurkov, Boekhout, and Richter, sp. nov. (MB 823153)
Etymology: The specific epithet elstari refers to the apple cultivar Elstar, which is the source of isolation.
After 2 weeks at 22°C on PDA, the colonies are tough, slightly wrinkled, dull, pale whitish-tan colored, with a ridged margin. Ballistospores are elongate, falcate or lunate, 0.5–2 × 16–32 μm (Figure 3III).
Fermentation of glucose absent. Growth on glucose, galactose (weak), ribose (delayed), xylose (latent), L-arabinose (delayed), sucrose, maltose, trehalose, methyl-alpha-D-glucoside (latent), cellobiose (delayed), melibiose (latent), lactose (latent), raffinose, melezitose, inulin (weak), soluble starch (weak), glycerol (delayed), erythritol (latent), ribitol (latent), xylitol (latent), arabinitol (latent), glucitol (weak), mannitol (delayed), and inositol (latent). No growth on sorbose, D-glucosamine, D-arabinose, rhamnose, salicin, arbutin, galactitol, 2-keto-gluconate, gluconate, glucoronate, galacturonate, saccharate, lactate, succinate, citrate, methanol, ethanol, propane-1,2-diol, butane-2,3-diol, quinic acid, and galactonic acid. Growth on potassium nitrate and sodium nitrite. No growth on lysine, ethylamine, cadaverine, creatine, creatinine, D-glucosamine, and imidazole. No growth in the presence of 0.01% cycloheximide, in the presence of 10% NaCl and in 50% glucose. Urea hydrolysis and Diazonium Blue B reaction are positive. Optimal growth temperature: 15–22°C. Maximum growth temperature: 25°C; growth at 30°C is weak (on MEA) or negative (on GPYA).
Molecular characteristics (holotype): Nucleotide sequences of ITS, SSU, LSU (D1/D2 domains) rRNA and RPB2 and TEF1 deposited in NCBI/EMBL (GenBank) under the accession numbers AY259048 (ITS), AY272021 (LSU), LT615023 (SSU), DQ234552 (RPB2), DQ028593 (TEF1).
Deposits: Holotype CBS 111593 isolated from an apple of the cultivar “Elstar” showing signs of “white haze” was obtained from an Ultra-Low Oxygen storage room in Netherlands in 1998 (Boekhout et al., 2006), preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. An ex-type culture is deposited at the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (DSM 29113).
Strain studied: CBS 111593 (=DSM 29113).
Description of Entyloma davenportii Yurkov, Richter, and Boekhout, sp. nov. (MB 823154)
Etymology: The specific epithet davenportii is dedicated to R. R. Davenport, in recognition of his contribution to his ecological studies of yeasts on fruits.
After 2 weeks at 22°C on PDA, the colonies are soft, wrinkled or furrowed, dull, whitish-cream colored, with a ridged margin. Ballistospores are elongate, falcate or lunate, 1–2 × 13–24 μm (Figure 3IV).
Fermentation of glucose absent. Growth on glucose, galactose, ribose (sometimes weak), xylose (sometimes weak), L-arabinose, sucrose, trehalose, cellobiose, raffinose, inulin (delayed), melezitose, soluble starch, glycerol (delayed), erythritol, arabinitol (weak), and mannitol (delayed).
No growth on sorbose, D-glucosamine, D-arabinose, rhamnose, maltose, methyl-alpha-D-glucoside, salicin, arbutin, melibiose, lactose, ribitol, xylitol, glucitol, galactitol, inositol, 2-keto-gluconate, gluconate, glucoronate, galacturonate, saccharate, lactate, succinate, citrate, methanol, ethanol, propane-1,2-diol, butane-2,3-diol, quinic acid, and galactonic acid. Growth on potassium nitrate, sodium nitrite, lysine, and creatinine. No growth on ethylamine, cadaverine, creatine, D-glucosamine, and imidazole. No growth in the presence of 0.01% cycloheximide, in the presence of 10% NaCl and in 50% glucose. Urea hydrolysis and Diazonium Blue B reaction are positive. Optimal growth temperature: 15–22°C. Maximum growth temperature: 25°C.
Molecular characteristics (holotype): Nucleotide sequences of ITS, SSU, LSU (D1/D2 domains) rRNA and RPB2 and TEF1 deposited in NCBI/EMBL (GenBank) under the accession numbers AY259064 (ITS), AY272010 (LSU), LT615013 (SSU), LT614961 (RPB2), LT614987 (TEF1).
Deposits: Holotype CBS 111604 isolated from an apple of the cultivar “Elstar” showing signs of “white haze” obtained through the Plant Protection Service [Plantenziektenkundige Dienst (PD)] in Wageningen, Netherlands in 1994 (Boekhout et al., 2006), preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. An ex-type culture is deposited at the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (DSM 100135).
Strains studied: paratypes CBS 111604 (=DSM 100135), CBS 111603 (=DSM 100134), CBS 111607 (=DSM 100137).
Description of Entyloma belangeri Boekhout, Richter, and Yurkov, sp. nov. (MB 823155)
Etymology: The specific epithet belangeri is dedicated to Richard R. Bélanger, who developed Sporodex®, a commercial powdery mildew biocontrol agent, based on P. flocculosa.
After 2 weeks at 22°C on PDA, the colonies are tough, slightly wrinkled, dull, white or cream colored, with a slightly ridged margin. Ballistospores are elongate, cylindrical or allantoid, 1–2 × 18–29 μm (Figure 3V).
Fermentation of glucose absent. Growth on glucose, galactose, ribose (weak), xylose (variable), L-arabinose, sucrose, maltose (variable), trehalose, cellobiose (variable), raffinose, melezitose (variable), inulin (weak, delayed), soluble starch (weak), glycerol (weak), erythritol (weak), ribitol (variable), and mannitol (weak). No growth on sorbose, D-glucosamine, D-arabinose, rhamnose, methyl-alpha-D-glucoside, salicin, arbutin, melibiose, lactose, xylitol, arabinitol, glucitol, galactitol, inositol, glucono-delta-lactone, 2-keto-gluconate, gluconate, glucoronate, galacturonate, saccharate, lactate, succinate, citrate, methanol, ethanol, propane-1,2-diol, butane-2,3-diol, quinic acid, and galactonic acid. Growth on potassium nitrate, sodium nitrite, and imidazole (variable). No growth on lysine, ethylamine, cadaverine, creatine, creatinine, and D-glucosamine. No growth in the presence of 0.01% cycloheximide, in the presence of 10% NaCl and in 50% glucose. Urea hydrolysis and Diazonium Blue B reaction are positive. Optimal growth temperature: 15–22°C. Maximum growth temperature: 25°C growth at 30°C is variable.
Molecular characteristics (holotype): Nucleotide sequences of ITS, SSU, LSU (D1/D2 domains) rRNA and RPB2 and TEF1 deposited in NCBI/EMBL (GenBank) under the accession numbers AY259074 (ITS), AY272019 (LSU), LT615019 (SSU), LT614967 (RPB2), LT614993 (TEF1).
Deposits: Holotype CBS 111600 isolated from an apple of the cultivar “Elstar” showing signs of russetting obtained from the orchard Krabbendijk, Netherlands in 1998 (Boekhout et al., 2006), preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. An ex-type culture is deposited at the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (DSM 29114).
Strains studied: CBS 111600 (=DSM 29114), CBS 111596 (=DSM 100129), CBS 111597 (=DSM 100130), CBS 111598 (=DSM 100131), CBS 111599 (=DSM 100132), CBS 111601 (=DSM 100133), CBS 111602 (=DSM 29115).
Note: strain CBS 111605 (=DSM 29116) has been provisionally assigned to E. belangeri species complex based on results of the multi-gene phylogenetic analysis (Figure 1). Nucleotide sequences of this strain showed 98 and 92% similarity to the type culture in LSU and ITS regions, respectively.
Description of Entyloma randwijkense Richter, Boekhout, and Yurkov, sp. nov. (MB 823156)
Etymology: The specific epithet randwijkense refers to the Dutch village Randwijk, which was the location of the experimental station from which the isolation source (apples) derived.
After 2 weeks at 22°C on PDA, the colonies are tough, flat, furrowed, dull, creamish-tan colored, with an entire margin. Ballistospores are cylindric, allantoid or falcate, 1–2 × 8–22 μm (Figure 3VI).
Fermentation of glucose absent. Growth on glucose, galactose (sometimes weak), L-arabinose (delayed), rhamnose, sucrose, maltose (weak), trehalose, cellobiose (weak), raffinose, melezitose (delayed), inulin (weak), soluble starch (weak), glycerol (latent), erythritol (latent), mannitol (weak), and 2-keto-gluconate (latent).
No growth on sorbose, D-glucosamine, ribose, xylose, D-arabinose, methyl-alpha-D-glucoside, salicin, arbutin, melibiose, lactose, ribitol, xylitol, arabinitol, glucitol, galactitol, inositol, glucono-delta-lactone, gluconate, glucoronate, galacturonate, saccharate, lactate, succinate, citrate, methanol, ethanol propane-1,2-diol, butane-2,3-diol, quinic acid, and galactonic acid. Growth on potassium nitrate (weak), sodium nitrite (weak), lysine (weak), and imidazole. No growth on ethylamine, cadaverine, creatinine, creatinine, and D-glucosamine. No growth in the presence of 0.01% cycloheximide, in the presence of 10% NaCl and in 50% glucose. Urea hydrolysis and Diazonium Blue B reaction are positive. Optimal growth temperature: 15–22°C. Maximum growth temperature: 25°C.
Molecular characteristics (holotype strain): Nucleotide sequences of ITS, SSU, LSU (D1/D2 domains) rRNA and RPB2 and TEF1 deposited in NCBI/EMBL (GenBank) under the accession numbers AY259080 (ITS), AY272033 (LSU), LT615014 (SSU), LT614962 (RPB2), LT614988 (TEF1).
Deposits: Holotype CBS 111606 isolated from an apple obtained from the orchard Wognum II, Netherlands in 1998 (Boekhout et al., 2006), preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. An ex-type culture is deposited at the German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany (DSM 100136).
Strain studied: CBS 111606 (=DSM 100136).
Correction of Tilletiopsis albescens Gokhale, Nova Hedwigia 23: 801 (1972) (MB 324630)
The species T. albescens was described by Gokhale (1972) based on a culture isolated by Cooke from sewage in Libertyville, IL, United States. The type material (culture UBC 926) was clearly designated by the author but the material was deposited not in a single but in two institutions, i.e., in Mycology Herbarium of the University of British Columbia (UBC) and in the American Type Culture Collection (ATCC). However, according to the catalog of the UBC herbarium, the specimen UBC F926 is Lycogala epidendrum, a species of slime mold (accessed on June 6, 2019). The original culture from UBC herbarium was deposited in the CBS culture collection (presently the Westerdijk Fungal Biodiversity Institute) by Bandoni. This culture is permanently preserved in a metabolically inactive state under the number CBS 608.83. In accordance with the Art. 9.2 of the ICN (Shenzhen code, 2017; Turland et al., 2018) we modify the protologue as following:
Deposits: original culture UBC 926 isolated from filamentous organisms from sewage in Libertyville, IL, United States (Gokhale, 1972) was deposited as two isotypes preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute (Utrecht, Netherlands) as CBS 608.83 and in the American Type Culture Collection (Manassas, United States) as ATCC 24343.
Recently, Wang et al. (2015) recombined T. albescens as R. albescens and designated this species as the type species of the genus Robbauera.
Correction of Tilletiopsis pallescens Gokhale (MB 324632) and Validation of Golubevia Q. M. Wang, F. Y. Bai, Begerow, and Boekhout (MB 812694) and Golubevia pallescens (Gokhale) Q. M. Wang, F. Y. Bai, Begerow, and Boekhout (MB 812695)
The species T. pallescens was described by Gokhale (1972) based on a culture isolated from a fruiting body of Sirobasidium sp. collected in Shamoda, Japan. The type material (culture UBC 8007) was clearly designated by the author but the material was deposited not in a single but in two institutions, i.e., in Mycology Herbarium of the University of British Columbia (UBC) and in the American Type Culture Collection (ATCC). However, according to the catalog of the UBC herbarium, the specimen UBC F8007 is Hebeloma glutinosum, a basidiomycete (accessed on June 6, 2019). The original culture from UBC herbarium was deposited in the CBS culture collection (presently the Westerdijk Fungal Biodiversity Institute) by Bandoni. This culture is permanently preserved in a metabolically inactive state under the number CBS 606.83. In accordance with the Art. 9.2 of the ICN (Shenzhen code, 2017; Turland et al., 2018) we modify the protologue as following:
Deposits: original culture UBC 8007 isolated from a fruiting body of Sirobasidium sp. collected in Shamoda, Japan (Gokhale, 1972) was deposited as two isotypes preserved in a metabolically inactive state in the CBS collection of the Westerdijk Fungal Biodiversity Institute (Utrecht, Netherlands) as CBS 608.83 and in the American Type Culture Collection (Manassas, United States) as ATCC 24345.
Recently, Wang et al. (2015) recombined T. pallescens as G. pallescens. However, the basionym T. pallescens was not indicated in the publication. Therefore, this taxonomic combination is invalid according to Art. 41.5 of the ICN (Shenzhen code, 2017; Turland et al., 2018). Here, we provide validation of G. pallescens, the type species of the genus Golubevia.
Golubevia pallescens (Gokhale) Q. M. Wang, F. Y. Bai, Begerow and Boekhout comb. nov. Studies in Mycology 81: 78 (2015) (MB 812695).
Basionym: T. pallescens Gokhale, Nova Hedwigia 23: 805 (1972) (MB 324632).
Golubevia Q. M. Wang, F. Y. Bai, Begerow and Boekhout, Studies in Mycology 81: 78 (2015); MycoBank MB 812694.
Type species: G. pallescens (Gokhale) Q. M. Wang, F. Y. Bai, Begerow and Boekhout, Studies in Mycology 81: 78 (2015); MycoBank MB 812695.
The genus was proposed by Wang et al. (2015) to accommodate a single species T. pallescens. The second species of the genus is described in the present study as G. heteromorpha Boekhout, Richter and Yurkov (MB 823152).
Discussion
In this study, we analyze and formally describe six new species belonging to three orders of the Exobasidiomycetes. These species were resolved in the multigene tree based on five DNA-loci, and in the LSU-based phylogenetic tree. In addition, our analyses included sequences that represent potential new species (Figures 1, 2). Comparing the results from the two analyses, we suggest that it is generally important to enlarge a phylogenetic analysis with additional DNA-loci. As an example, strain G1, which was placed close to G2 in the LSU dataset, clustered with the clade F when four additional loci were included in the analysis. Additional protein-coding gene fragments improved separation of strains and statistical support of the clades, as compared to the LSU-based tree. However, even this five-loci dataset was not sufficient to resolve relationships between the orders in Exobasidiomycetes, as displayed by low bootstrap values.
Asexual yeast and yeast-like states of plant parasites in Ustilaginomycotina are already known for almost 100 years (reviewed in Begerow et al., 2014, 2017). Some of them produce anamorphs in situ (e.g., Ustilago, Entyloma, Anthracoidea, Exobasidium), but the characteristics of these asexual morphs have been neglected from the taxonomy of this group as it was solely based on the characters describing sexual reproduction. Fungi morphologically similar to the anamorphs of Ustilaginomycotina were frequently isolated from sources other than the known host plants, for example soil, air or animal samples (e.g., Boekhout, 2011; Begerow et al., 2014, 2017). Also isolation of such yeasts from healthy and non-host plants were repeatedly reported (Inácio et al., 2008; Begerow et al., 2014, 2017). For a long time these yeasts were described in often large asexual genera like Candida, Rhodotorula, Sporotrichum, and Sterigmatomyces, until a rather stable system of dual naming of sexual and asexual members of Ustilaginomycotina was established (Boekhout, 1998, 2011; Bauer et al., 2001; Begerow et al., 2014, 2017). Most of the yeasts were accommodated in the two genera Pseudozyma (anamorphic Ustilaginaceae) and Tilletiopsis (anamorphic Exobasidiomycetes). Because the nomenclature of plant parasites and yeasts developed independently, big formal genera spanning over taxonomic families and orders were long a workable solution. With the reclassification of teleomorphic species based on cell ultrastructure and DNA-markers (e.g., Bauer et al., 2001), it became obvious that the genera comprising asexual morphs are also polyphyletic (e.g., Fell et al., 2000; Scorzetti et al., 2002) and should be split in the future (e.g., Inácio et al., 2008). After the use of dual nomenclature for the taxonomy of fungi with pleomorphic life cycle was terminated from 2013 (Hawksworth, 2011; Taylor, 2011), it became necessary to come up with a new concept and to revise sexual and asexual taxa together.
The former concept of the genus Tilletiopsis, which was originally based on morphological characteristics, was changing and expanding along with a growing number of species discovered. However, it was still based on the characteristics of the type species (T. washingtonensis), so this genus could not suit a place to accommodate other asexual Exobasidiomycetes, such as Jaminaea (Sipiczki and Kajdacsi, 2009), Sympodiomycopsis (Sugiyama et al., 1991), and Quambalaria (Simpson, 2000). With the introduction of ribosomal DNA sequencing techniques, the genus Tilletiopsis turned out to be polyphyletic, containing species of different orders scattered across the Exobasidiomycetes. This situation was similar to another large anamorphic genus, Pseudozyma, which also served as a “catch-it-all”-genus in the Ustilaginomycetes (Bauer et al., 2001; Boekhout, 1998, 2011; Begerow et al., 2014, 2017). With time, the concept of the genus Tilletiopsis to accommodate all yeast states in Exobasidiomycetes was changed and, consequently, novel genera circumscribed with molecular phylogenetic data were erected, e.g., Jaminaea, Acaromyces (Boekhout, 2003; Sipiczki and Kajdacsi, 2009). These genera may also represent anamorphic stages of plant parasites, as suggested by the recent phylogenetic analyses (Wang et al., 2015), but the lack of nucleotide sequence data from teleomorphic species is hampering a better taxonomic placement. Phylogenetic analyses performed by Wang et al. (2015) and in this study show that the genus Tilletiopsis was polyphyletic. Also, sequences from undescribed species derived from public databases suggest that additional phylogenetic clades comprising Tilletiopsis-like yeasts are to be expected. Some of them might represent a new family or order in the Exobasidiomycetes close to Georgefischeriales. However, phylogenetic placement of these species would require a solid multi-gene analysis in the future.
Asexual yeast states in Exobasidiomycetes are known to produce compounds which are active against other organisms (reviewed in Begerow et al., 2014). Antagonistic reactions toward other fungi were reported for members of genera Acaromyces, Golubevia, Meira, Moesziomyces, Tilletiopsis, and Sympodiomycopsis (reviewed in Begerow et al., 2014). Similarly to a few formerly classified in the genus Tilletiopsis species (e.g., Urquhart and Punja, 2002; Kiss, 2003; Begerow et al., 2014), antagonistic interactions of Golubevia heteromorpha (cited as T. pallescens) against powdery mildew Blumeria graminis f. sp. tritici has been recently reported (Köhl et al., 2019). This yeast proved to be a promising biocontrol agent which significantly reduced the number of powdery mildew pustules (Köhl et al., 2019).
Data Availability Statement
The datasets generated for this study can be accessed from GenBank, provided in Tables 1, 2.
Author Contributions
CR, TB, and AY performed the experiments. TB, MS, and AY designed the experiments. CR, TB, AY, and MS wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Patrick Bork is acknowledged for putting much effort into the project. Evelyne Brambilla and Gabrielle Gresenz (Leibniz Institute DSMZ) are acknowledged for their assistance in the lab. The authors thank Merle Langevoord and Armanda den Exter (Westerdijk Fungal Biodiversity Institute) for helping with the physiology data.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02544/full#supplementary-material
References
- AL-Zaydani I. A., Joseph M. R. P., Suheel A. M., Al-Hakami A. M., Hamid M. E. (2014). Severe pneumonia with a massive pleural effusion in a child caused by Tilletiopsis minor: the first case from Saudi Arabia. Ann. Saudi Med. 35 475–478. 10.5144/0256-4947.2015.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric S., Lindner L., Marschall K., Dalla Via J. (2010). Haplotype diversity of Tilletiopsis spp. causing white haze in apple orchards in Northern Italy. Plant Pathol. 59 535–541. 10.1111/j.1365-3059.2009.02217.x [DOI] [Google Scholar]
- Barnett J. A. (2004). A history of research on yeasts 8: taxonomy. Yeast 21 1141–1193. 10.1002/yea.1154 [DOI] [PubMed] [Google Scholar]
- Bauer R., Begerow D., Oberwinkler F., Piepenbring M., Berbee M. L. (2001). “Ustilaginomycetes,” in The Mycota VII Part B Systematics and Evolution, eds McLaughlin D. J., McLaughlin E. G., Lemke P. A. (Berlin: Springer; ), 7–83. [Google Scholar]
- Bauer R., Begerow D., Sampaio J. P., Weiβ M., Oberwinkler F. (2006). The simple-septate basidiomycetes: a synopsis. Mycol. Prog. 5 41–66. 10.1007/s11557-006-0502-0 [DOI] [Google Scholar]
- Begerow D., Bauer R., Boekhout T. (2000). Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycol. Res. 104 53–60. 10.1017/s0953756299001161 [DOI] [Google Scholar]
- Begerow D., Kemler M., Feige A., Yurkov A. (2017). “Parasitism in yeasts,” in Yeasts in Natural Ecosystems: Ecology, eds Buzzini P., Lachance M. A., Yurkov A. (Cham: Springer; ), 179–210. 10.1007/978-3-319-61575-2_7 [DOI] [Google Scholar]
- Begerow D., Lutz M., Oberwinkler F. (2002). Implications of molecular characters for the phylogeny of the genus Entyloma. Mycol. Res. 106 1392–1399. 10.1017/s0953756202006962 16638136 [DOI] [Google Scholar]
- Begerow D., Schäfer A. M., Kellner R., Yurkov A., Kemler M., Oberwinkler F., et al. (2014). “Ustilaginomycotina,” in The Mycota VII Part A - Systematics and Evolution, 2nd Edn, eds McLaughlin D., Spatafora J. (Berlin: Springer-Verlag; ), 295–329. 10.1007/978-3-642-55318-9_11 [DOI] [Google Scholar]
- Boekhout T. (1987). Systematics of anamorphs of Ustilaginales (smut fungi) - A preliminary survey. Stud. Mycol. 30 137–149. [Google Scholar]
- Boekhout T. (1991). A revision of ballistoconidia-forming yeasts and fungi. Stud. Mycol. 33 1–192. [Google Scholar]
- Boekhout T. (1998). “Tilletiopsis Derx ex Derx,” in The Yeasts, a Taxonomic Study, 4th Edn, eds Kurtzman C. P., Fell J. W. (Amsterdam: Elsevier; ), 848–853. 10.1016/b978-044481312-1/50114-3 [DOI] [Google Scholar]
- Boekhout T. (2003). Novel anamorphic mite-associated fungi belonging to the Ustilaginomycetes: Meira geulakonigii gen. nov., sp. nov., Meira argovae sp. nov. and Acaromyces ingoldii gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 53 1655–1664. 10.1099/ijs.0.02434-0 [DOI] [PubMed] [Google Scholar]
- Boekhout T. (2011). “Tilletiopsis Derx ex Derx (1930),” in The Yeasts: a Taxonomic Study, 5th Edn, eds Kurtzman C. P., Fell J. W., Boekhout T. (Amsterdam: Elsevier Science; ), 2003–2014. 10.1016/b978-0-444-52149-1.00160-9 [DOI] [Google Scholar]
- Boekhout T., Fell J. W., O’Donnell K. (1995). Molecular systematics of some yeast-like anamorphs belonging to the Ustilaginales and Tilletiales. Stud. Mycol. 38 175–183. [Google Scholar]
- Boekhout T., Gildemacher P., Theelen B., Müller W. H., Heijne B., Lutz M. (2006). Extensive colonization of apples by smut anamorphs causes a new postharvest disorder. FEMS Yeast Res. 6 63–76. 10.1111/j.1567-1364.2005.00002.x [DOI] [PubMed] [Google Scholar]
- Boekhout T., Yamada Y., Weijman A. C. M., Roeymans H. J., Batenburg-Van der Vegte W. H. (1992). The significance of coenzyme Q, carbohydrate composition and septal ultrastructure for the taxonomy of ballistoconidia-forming yeasts and fungi. Syst. Appl. Microbiol. 15 1–10. 10.1016/s0723-2020(11)80129-3 [DOI] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Derx H. G. (1930). Étude sur les Sporobolomycètes. Ann. Mycol. 28 1–23. [Google Scholar]
- Derx H. G. (1948). Itersonilia nouveau genre de Sporobolomycètes à mycélium bouclé. Bull Jard Bot Buitenzorg 17 465–472. [Google Scholar]
- Fairs A., Wardlaw A. J., Thompson J. R., Pashley C. H. (2010). Guidelines on ambient intramural airborne fungal spores. J. Investig. Allergol. Clin. Immunol. 20 490–498. [PubMed] [Google Scholar]
- Fell J. W., Boekhout T., Fonseca A., Scorzetti G., Statzell-Tallman A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50 1351–1371. 10.1099/00207713-50-3-1351 [DOI] [PubMed] [Google Scholar]
- Fungsin B. (2003). Taxonomic Studies of Ballistoconidium-Forming Yeasts Found in the Phyllosphere of Thailand. Setagaya: Tokyo University Of Agriculture. [Google Scholar]
- Gargas A., Taylor J. W. (1992). Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18s rDNA from lichenized fungi. Mycologia 84 589–592. 10.1080/00275514.1992.12026182 [DOI] [Google Scholar]
- Gokhale A. A. (1972). Studies on the genus Tilletiopsis. Nov Hedwigia 23 795–809. [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hamamoto M., Tamura M., Nakase T. (2000). Emended descriptions of Tilletiopsis washingtonensis, Tilletiopsis cremea and Tilletiopsis lilacina. Int. J. Syst. Evol. Microbiol. 50 925–930. 10.1099/00207713-50-2-925 [DOI] [PubMed] [Google Scholar]
- Hawksworth D. L. (2011). A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys 1 7–20. 10.5598/imafungus.2011.02.02.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio J., Landell M. F., Valente P., Wang P. H., Wang Y. T., Yang S. H., et al. (2008). Farysizyma gen. nov., an anamorphic genus in the Ustilaginales to accommodate three novel epiphytic basidiomycetous yeast species from America, Europe and Asia. FEMS Yeast Res. 8 499–508. 10.1111/j.1567-1364.2008.00377.x [DOI] [PubMed] [Google Scholar]
- Jindamorakot S. (2006). The Species Diversity of Yeasts in Some Natural Habitats of Thailand. Bangkok: Kasetsart University. [Google Scholar]
- Kachalkin A. V., Turchetti B., Inácio J., Carvalho C., Mašínová T., Pontes A., et al. (2019). Rare and undersampled dimorphic basidiomycetes. Mycol. Prog. 18 945–971. 10.1007/s11557-019-01491-5 [DOI] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechkekian A., Gari-Toussaint M., Gastaud P., Freton A., Gantier J. C. (2007). Keratites due to Tilletiopsis minor post LASIK. J. Mycol. Med. 17 119–121. 10.1016/j.mycmed.2007.03.001 [DOI] [Google Scholar]
- Kiss L. (2003). A review of fungal antagonists of powdery mildews and their potential as biocontrol agents. Pest. Manag. Sci. 59 475–483. 10.1002/ps.689 [DOI] [PubMed] [Google Scholar]
- Köhl J., Goossen-van de Geijn H., Groenenboom-de Haas L., Henken B., Hauschild R., Hilscher U., et al. (2019). Stepwise screening of candidate antagonists for biological control of Blumeria graminis f. sp. tritici. Biol. Control 136:104008 10.1016/j.biocontrol.2019.104008 [DOI] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. P., Fell J. W., Boekhout T., Robert V. (2011). “Methods for isolation, phenotypic characterization and maintenance of yeasts,” in The Yeasts: a Taxonomic Study, 5th Edn, eds Kurtzman C. P., Fell J. W., Boekhout T. (Amsterdam: Elsevier Science; ), 87–110. 10.1016/b978-0-444-52149-1.00007-0 [DOI] [Google Scholar]
- Last F. T. (1970). Factors associated with the distribution of some phylloplane microbes. Netherlands J. Plant Pathol. 76 140–143. 10.1007/bf01974320 [DOI] [Google Scholar]
- Li S. L., Feng S. L., Li Z. T., Xu H., Yu Y. P., Qiao D. R., et al. (2011). Isolation, identification and characterization of oleaginous fungi from the soil of Qinghai Plateau that utilize D-xylose. Afr. J. Microbiol. Res. 5 2075–2081. [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Lutz M., Pia̧tek M. (2016). Phylogenetic placement, DNA barcoding, morphology and evidence for the spreading of Entyloma cosmi, a species attacking Cosmos bipinnatus in temperate climate gardens. Eur. J. Plant Pathol. 145 857–869. 10.1007/s10658-016-0874-1 [DOI] [Google Scholar]
- Mahé S., Rédou V., Le Calvez T., Vandenkoornhuyse P., Burgaud G. (2014). “Fungi in deep-sea environments and metagenomics,” in The Ecological Genomics of Fungi, 1st Edn, ed. Martin F. (Hoboken, NJ: John Wiley & Sons, Inc; ), 325–354. 10.1002/9781118735893.ch15 [DOI] [Google Scholar]
- Matheny P. B., Wang Z., Binder M., Curtis J. M., Lim Y. W., Nilsson R. H., et al. (2007). Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 43 430–451. 10.1016/j.ympev.2006.08.024 [DOI] [PubMed] [Google Scholar]
- McNeill J., Barrie F. R., Buck W. R., Demoulin V., Greuter W., Hawksworth D. L., et al. (2012). International Code of Nomenclature for Algae, Fungi, and Plants (Melbourne Code) Adopted by the Adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. Regnum Vegetabile, 154. Königstein: Koeltz Scientific Books [Google Scholar]
- Nyland G. (1950). The genus Tilletiopsis. Mycologia 42 487–496. 10.1080/00275514.1950.12017853 [DOI] [Google Scholar]
- Prencipe S., Spadaro D., Fruk G., Jemric T. (2016). First report of Tilletiopsis pallescens causing white haze on apple in Croatia. Plant Dis 100 225–225. 10.1094/pdis-05-15-0505-pdn [DOI] [Google Scholar]
- Ramani R., Kahn B. T., Chaturvedi V. (1997). Tilletiopsis minor: a new etiologic agent of human subcutaneous mycosis in an immunocompromised host. J. Clin. Microbiol. 35 2992–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzetti G., Fell J. W., Fonseca A., Statzell-Tallman A. (2002). Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2 495–517. 10.1016/s1567-1356(02)00128-9 [DOI] [PubMed] [Google Scholar]
- Shivas R. G., Brown J. F. (1989). Yeasts associated with fluid in pitchers of Nepenthes. Mycol. Res. 93 96–100. 10.1016/s0953-7562(89)80143-1 [DOI] [Google Scholar]
- Simpson J. A. (2000). Quambalaria, a new genus of eucalypt pathogens. Australas. Mycol 19 57–62. [Google Scholar]
- Sipiczki M., Kajdacsi E. (2009). Jaminaea angkorensis gen. nov., sp. nov., a novel anamorphic fungus containing an S943 nuclear small-subunit rRNA group IB intron represents a basal branch of Microstromatales. Int. J. Syst. Evol. Microbiol. 59 914–920. 10.1099/ijs.0.003939-0 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Sugiyama J., Tokuoka K., Suh S.-O., Hirata A., Komagata K. (1991). Sympodiomycopsis: a new yeast-like anamorph genus with basidiomycetous nature from orchid nectar. Antonie Van Leeuwenhoek 59 95–108. 10.1007/bf00445653 [DOI] [PubMed] [Google Scholar]
- Takashima M., Nakase T. (1996). A phylogenetic study of the genus Tilletiopsis, Tilletiaria anomala and related taxa based on the small subunit ribosomal DNA sequences. J. Gen. Appl. Microbiol. 42 421–429. 10.2323/jgam.42.421 [DOI] [Google Scholar]
- Takashima M., Nakase T. (2001). Tilletiopsis derxii, Tilletiopsis oryzicola and Tilletiopsis penniseti, three new species of the ustilagionomycetous anamorphic genus Tilletiopsis isolated from leaves in Thailand. Antonie Van Leeuwenhoek 80 43–56. [DOI] [PubMed] [Google Scholar]
- Takashima M., Sugita T., Van B. H., Nakamura M., Endoh R., Ohkuma M. (2012). Taxonomic richness of yeasts in Japan within subtropical and cool temperate areas. PLoS One 7:e50784. 10.1371/journal.pone.0050784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G., Castresana J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56 564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Taylor J. W. (2011). One Fungus = One Name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus 2 113–120. 10.5598/imafungus.2011.02.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubaki K. (1952). Studies on the Sporobolomycetaceae in Japan: I. Tilletiopsis. Nagaoa 1 26–31. [Google Scholar]
- Turland N. J., Wiersema J. H., Barrie F. R., Greuter W., Hawksworth D. L., Herendeen P. S., et al. (eds) (2018). International Code of Nomenclature for Algae, Fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. Glashütten: Koeltz Botanical Books. [Google Scholar]
- Urquhart E. J., Punja Z. K. (2002). Hydrolytic enzymes and antifungal compounds produced by Tilletiopsis species, phyllosphere yeasts that are antagonists of powdery mildew fungi. Can. J. Microbiol. 48 219–229. 10.1139/w02-008 [DOI] [PubMed] [Google Scholar]
- Wang Q.-M., Begerow D., Groenewald M., Liu X. Z., Theelen B., Bai F. Y., et al. (2015). Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Stud. Mycol. 81 55–83. 10.1016/j.simyco.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber R. W. S., Zabel D. (2011). White haze and scarf skin, two little-known cosmetic defects of apples in northern Germany. Eur. J. Hortic Sci. 76 45–50. [Google Scholar]
- White T. J., Bruns T. D., Lee S. B., Taylor J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: a Guide to Methods and Applications, eds Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (Cambridge: Academic Press; ), 315–322. 10.1016/b978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yue Y., Yu H., Li R., Xing R., Liu S., Li P. (2015). Exploring the antibacterial and antifungal potential of jellyfish-associated marine fungi by cultivation-dependent approaches. PLoS One 10:e0144394. 10.1371/journal.pone.0144394 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be accessed from GenBank, provided in Tables 1, 2.