Abstract
Lung cancer is one of the most common causes of cancer death. Its poor prognosis can be attributed to the patients’ advanced or metastatic presentation at the time of diagnosis. To improve and accelerate the diagnosis, better therapeutic and diagnostic methods are constantly being sought. MicroRNAs (miRNAs) are short nucleotide sequences of single-stranded, non-coding RNA that function as critical post-transcriptional regulators of gene expression. They are identified not only intracellularly, but also in physiological and pathological body fluids. These molecules are responsible for the regulation of approximately 33% of human genes, either regulating the expression of both oncogenes and suppressor genes or acting directly as an oncogene or suppressor gene itself. MiRNAs can contribute to the formation of cancer. The high specificity and sensitivity of miRNAs have been demonstrated with various malignant diseases, and for this reason, they raise particular interest as new and perspective biomarkers of tumours. Our work summarises the available information from recent years regarding the possibility of using miRNAs as biomarkers in the diagnosis of neoplasms. In this review, we focused on malignant pleural effusions with an emphasis on non-small cell lung cancer (NSCLC).
Keywords: miRNA, lung cancer, pleural effusions, biomarker
Introduction
As per the Globocan 2018 database, 18.1 million new cases and 9.6 million cancer deaths worldwide were expected in 2018 [1]. In both sexes combined, the most commonly diagnosed cancer is lung cancer (11.6% of the total cases), followed by breast cancer (11.6%), prostate cancer (7.1%), and colorectal cancer (6.1%) [1–3]. When looking at mortality rates, lung cancer is the leading cause of cancer deaths (18.4% of the total cancer deaths), followed by colorectal cancer (9.2%), stomach cancer (8.2%), and liver cancer (8.2%) [4, 5].
Over the last two decades of identifying new mutations in oncogenes, gene suppressors, and genes responsible for genomic stability, the pathway of tumour formation could not be fully explained. In 1993, Ambros and Ruvkun correlated the amount of LIN-14 protein to 22-nucleotide long RNAs encoded by the LIN-4 gene, which are involved in the development of Caenorhabditis elegans. This discovery changed the perception of the genetic expression control system, indicating the possibility of silencing target mRNA with the use of these small RNA molecules [6, 7]. Numerous observations have provided evidence that short non-coding RNA sequences are involved in this complex process. Crucial elements of this regulatory mechanism are miRNAs. At the date of publication, 2588 mature miRNAs transcribed in the human genome have been identified [8]. The involvement of miRNAs has been described in the majority of life processes, both in physiological and pathological states [9], as in the presence of lung-associated malignant pleural effusions [10].
A single miRNA can modulate thousands of genes through the recognition of complementary sequences at the 3’ end of the UTR of the target mRNA [11]. Due to their discovery in various body fluids including blood, urine, synovial fluid, milk, saliva, and cerebrospinal fluid, as well as in fluids from bronchoalveolar lavage, the use of miRNA in diagnostics has significantly increased [12–14]. Depending on the genes that they influence, miRNAs can function either as “oncomirs”, activators of carcinogenic transformation, or as suppressor miRNA, which are inhibitors of oncogenes [15]. They can participate in various stages of cancer formation through their control over the expression of genes responsible for cellular events. For this reason, it has been suggested that miRNAs may be applied as biomarkers for the early diagnosis of cancer, or in monitoring its progression [16]. Their use provides many advantages. It has been shown that the specificity and sensitivity of miRNAs were higher than other biomarkers when used for screening colon, lung, and urinary bladder cancer, particularly in specific miRNA combinations [17]. Materials for miRNA studies can be obtained quickly and the analysis of miRNA profiles can be applied as a non-invasive method for cancer screening. Moreover, it is an inexpensive and rapid method, costing approximately 10 USD for raw materials and taking approximately 3.5 hours to obtain results [18]. These factors all indicate the utility of these molecules as biomarkers for tumour detection.
Diagnostic tests for the analysis of miRNA profiles in select cancers has recently been made available; however, such solutions have not yet been proposed for the analysis of pleural effusions.
Proper scientific investigations are beneficial, and more are required to facilitate the selection of potentially valuable biomarkers. Most of the studies published are focused on the circulating miRNA, for example the work of Switilik et al. [19]. In our review article we have attempted to collect and comprehensively characterise literature data concerning the use of miRNA as a biomarker in pleural effusion diagnostics.
MicroRNA – structure, synthesis, and functions
MicroRNAs are a group of single-stranded, noncoding, endogenous molecules composed of about 20 nucleotides. Genes encoding miRNA may constitute about 1–5% of all genes within human and animal genomes [20]. Genes coding miRNAs are localised not only in both the intron and exon sequences of protein-coding genes, but also in genomic regions that do not undergo the translation [21]. Often miRNAs are organised in groups that are subject to gene transcription as polycistronic units [22].
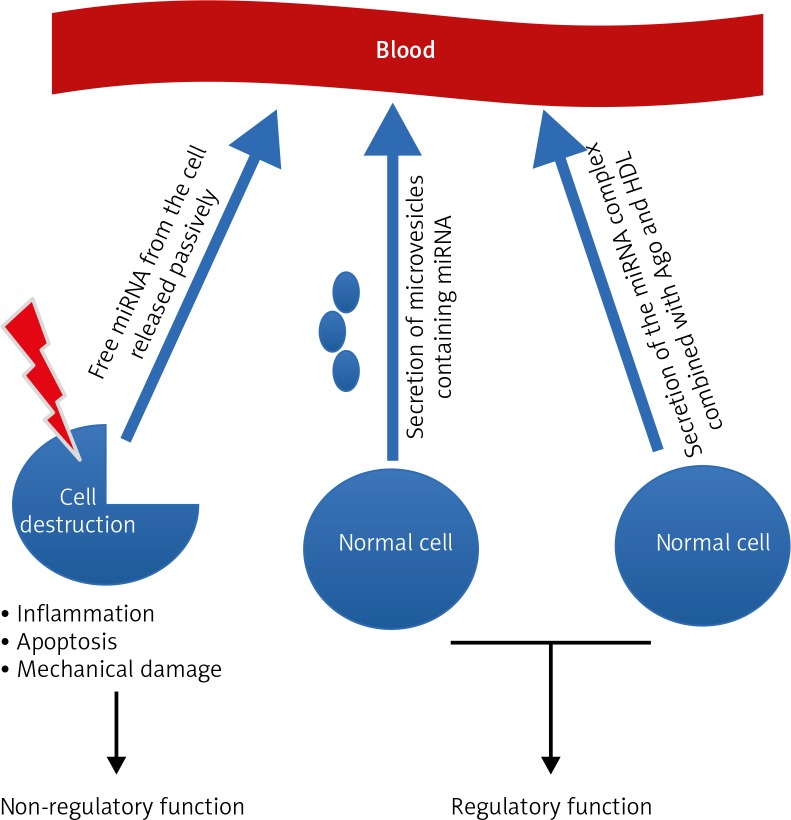
Many hypotheses attempt to explain the origin and properties of circulating miRNAs, and all are closely related without excluding each other. There are three leading theories (Fig. 1).
Fig. 1.
The three leading theories explaining the origin of circulating miRNAs
The first assumes that the occurrence of miRNA in the blood is a bystander effect of cell destruction and is due to the passive release of cellular miRNAs. This effect may occur as a result of damage to tissue and can thus be characteristic for particular stages of cell pathophysiology, including cancerogenesis, cell entry into inflammation, necrosis, apoptosis, or during the development of metastases. The second hypothesis accents the active process of miRNA release from the cell in a manner dependent on microvesicles (MV; exosomes). The third theory emphasises active and selective miRNA secretion in a free-form manner, independent of MVs, which is a consequence of the cell’s response to various stimuli [23]. This mechanism takes place owing to the formation of the miRNA complex together with proteins from the Argonaute family (Ago) and high-density lipoproteins (HDL). Microvesicles act as carriers of information between healthy cells and tumour cells alike. They can secrete exosomes that provoke an anti-tumour response from the host’s immune system, and tumour growth can be promoted by either inhibiting the immune response or by enhancing angiogenesis [24]. Exosomes from tumour cells contain tumour antigens, and these promote immunisation, followed by eradication of tumours by the activity of CD8+ and CD4+ T-lymphocytes [25]. This is accomplished intracellularly in tumour exosomes by transport of miRNA, which can act as a ligand of Toll-like receptors (TLR). It has been shown that miR-21/29a secreted from tumour cell exosomes can bind to TLR8 and lead to NF-kB activation and secretion of inflammatory cytokines such as TNF and IL-6 factors, which have a role in promoting metastasis.
There is increasing evidence suggesting that exosomes support oncogenesis by transporting oncoproteins, miRNAs, mRNAs, DNA, and immunosuppressive molecules responsible for promoting a phenotype that supports further tumour development. Ruiz-Martinez et al. have shown that miR-134 and miR-135b can regulate YKT6, which is a crucial exosome release control point. Approximately 30–80% of human genes are regulated by miRNAs [26–30]. Control of the silencing of genes can occur through direct degradation of mRNA or via inhibition of its translation [31], and in this way, a single molecule of miRNA can control the expression of hundreds of target genes simultaneously [32–35]. MicroRNAs participate in the regulation of cell division, cell-cycle control, cell differentiation, apoptosis, angiogenesis, and oncogenesis [36–42]. These molecules are capable of regulating cells of the immune system by influencing their production and differentiation, and by modulating the immune response during infection. In unicellular organisms, miRNA can also inhibit translation [43–53].
The first theory assumes that the occurrence of miRNA in the blood is a bystander effect of cell destruction and is due to the passive release of intracellular miRNAs. This effect may occur as a result of damage to tissue and thus is characteristic for particular stages of cancerogenesis, cell entry into inflammation, necrosis or apoptosis, or during the development of metastases. The second hypothesis accents the active process of miRNA release from the cell in a manner dependent on microvesicles. The third theory emphasises active and selective miRNA secretion in a free-form manner, independent of MVs, which is a consequence of the cell’s response to various stimuli.
The role of miRNA in the pathogenesis of cancer
It has been proven that miRNAs are not only able to regulate the expression of oncogenes and suppressors but can also function directly as an oncogene or as a suppressor [54]. The oncogenic effect of miRNAs is provided through the silencing of expression, or the deletion of mRNA, of oncogenes inhibitors, as well as the silencing of suppressor gene amplification or the up-regulation of their expression in neoplastic cells. The lack of miRNA expression results in severe cellular abnormalities, leading to tumorigenesis of the lungs [25]. These abnormalities may be due to the fact that almost 50% of the miRNA genes are localised within regions of the human genome prone to innate damage, known as fragile sites. Mutations in these areas are often associated with oncogenesis. This indicates a strong association between abnormal expression of miRNA and the development and progression of cancer [55]. Fragile sites of the genome are areas on chromosomes that are susceptible to frequent mutations, such as loss of genetic material or fragment rearrangement, abnormalities that are often observed in cancer cells. Expression of genes coding for miRNA located near these fragile sites may be altered pathologically. Examples of such molecules include miRNA-15a and miRNA-16/1, which are coded by genes localised in a fragile region on chromosome 13 known as area 14.2. Because of frequent gene deletions in this region, there is reduced expression or even a complete lack of these miRNAs observed in many patients with prostate cancer, chronic lymphocytic leukaemia (CLL), B-cell lymphoma, mantle cell lymphoma (MCL), and multiple myeloma (MM) [56].
MiRNAs in the pathogenesis and progression of non-small cell lung carcinoma
Many studies support the potential role of miRNAs as biomarkers in non-small cell lung cancer (NSCLC). Increasing evidence indicate the association between altered miRNA expression and tumour progression and survival. Patients with NSCLC who were found to have a down-regulation of let-7 expression have been shown to have lower postoperative survival and worse prognosis [57].
This decrease in levels of let-7c expression was reported to be significantly associated with cancer metastasis, venous invasion, advanced TNM stages, and poor survival in patients with NSCLC [58]. It was also shown that miRNA-520b plays a role as a tumour suppressor gene in the development of NSCLC. Its downregulated expression in lung cancer tissue was negatively correlated with tumour development, lymph node metastasis, and TNM staging [59].
The pathophysiology of malignant pleural effusion
A malignant pleural effusion (MPE) is an indicator of the high degree of histological malignancy of a tumour, especially concerning lung adenocarcinomas [60]. The annual incidence of lung cancer worldwide is approximately 1.8 million individuals [61]. Most patients present with advanced-stage disease, for which the treatment remains palliative. Pleural effusions can occur in many types of lung cancer. The presence of MPE in patients diagnosed with lung cancer is an unfavourable prognostic factor. The exact pathomechanism of the formation of the MPE has not been fully elucidated. One explanation includes an increase in capillary permeability, where an increase in the volume of a pleural effusion occurs when the fluid production exceeds its removal ability. Angiogenesis and growth in lymph production accompanying this proliferative process along with the formation of metastases promotes the development of effusion in the pleural cavity [60]. It is therefore imperative to recognise the aetiology of a patient’s pleural effusion using biomarkers with high sensitivity and specificity, such as miRNAs, which would allow earlier detection and treatment of patients with suspicious lesions.
Prognostic and diagnostic biomarkers in malignant pleural effusions
For the assessment of fluid accumulation, the localisation of solid tumours within the pleural cavity, and differentiating pleural thickening that could be suggestive of metastatic cancer, it is chest X-ray and computed tomography that are routinely performed [62]. In routine cytological testing of MPE, the presence of cancer cells in only confirmed in 60% of cases. Such detection is still insufficient for making clinical decisions, and usually the diagnosis is made after the use of more-invasive techniques such as thoracoscopy or thoracotomy [63]. It was found that for the determination of MPE, carcinoembryonic antigen (CEA) showed good specificity (94%) but a substantially low sensitivity (54%) [64]. In this situation, it would be beneficial to search for an additional marker or group of markers, allowing for a much earlier preliminary diagnosis. It was proposed that miRNAs can be used as such a factor in the differential diagnosis of malignant and non-malignant pleural effusions. Recent studies have shown several miRNAs present in pleural effusions, characterised by different levels of their expression. Wang et al. linked the high expression of miRNA-100 and a low miRNA-93, miRNA-134, miRNA-151, and miRNA-345 with poor survival rate [65]. Other studies have shown that miRNA-134, miRNA-185, and miRNA-22 were present at significantly lower levels in patients with malignant PE as compared to those with diagnoses of non-malignant PE (p < 0.001). Evaluation of the profile of miRNAs combined with the CEA antigen measurement provided a test with a sensitivity of 91.9% and a specificity of 92.5% [66]. Research has shown that MPE-associated adenocarcinoma of the lung expressed lower levels of miRNA-198 than in the case of non-malignant effusions [67]. Assessment of the expression of miR-198, CEA, and CYFRA 21/1 allowed for a test with a sensitivity of 89.2% and specificity of 85.0% [68]. The findings of this investigation encourage further efforts into the research of identifying additional biomarkers for the early detection of lung cancer, and in confirming the above conjectures.
Expression of miRNAs in malignant pleural effusions
The use of miRNAs as diagnostic biomarkers at this time is associated with certain limitations. For example, it has been proven that a single miRNA may regulate the transcription of more than one mRNA, while on the other hand, one specific mRNA may be controlled by several miRNAs, making it difficult to determine a target molecule [69, 70]. There are also studies implicating the same marker in entirely different diseases, for instance miRNA-134 in oesophageal cancer [71], as well as malignant pleural effusions associated with lung adenocarcinoma [10]. Laboratory investigations focusing on malignant pleural effusions are thus a major diagnostic challenge. An attempt was made to differentiate miRNA expression profiles in three groups of PE-positive patients. In this study, PE samples were selected according to Light’s criteria [72, 73]. Effusions that contained adenocarcinoma cells or pleural biopsy specimens were defined as APE; effusions with acid-fast bacilli and Mycobacterium tuberculosis were determined as TPE; and undiagnosed samples negative for cancer according to histological or cytological criteria were classified as controls (NPE). In this study, nine miRNAs (miRNA-200c-3p, miRNA-200b-3p, miRNA-200a-3p, miRNA-429 and miRNA-141-3p, miRNA-205-5p, miRNA-483-5p, miRNA-375, and miRNA-203a-3p) were the most frequently observed in APE, when compared with TPE or NPE. The indicated profile seems promising as a means by which to accurately differentiate APE from the other disease states. TPE and NPE, however, had a similar miRNA expression profile. These two groups differed from each other only in three miRNAs (miRNA-148a-3p, miRNA-451a, and miRNA-150-5p). Even so it still holds promise as a way to distinguish TPE from NPE [74].
The role of miRNA in cellular pathways
Currently, there is increasing interest in the role of miRNA as a modulator of the signal pathway associated with epidermal growth factor receptor (EGFR) [75]. Huang et al. found that approximately 80% of exosomes isolated from NSCLC expressed the EGFR receptor on their membrane, as compared to 2% in the case of chronic pneumonia [76]. Within the exosomes from the biopsy of lung tumours, 12 miRNAs were overexpressed: miRNA-17, miRNA-21, miR-106, miR-146, miR-155, miR-191, miR-192, miRNA-203, miRNA-205, miRNA-210, miRNA-212, and miRNA-214 [77]. Rabinowits et al. concluded that based on the differences in expression of miRNAs in this panel, there is potential for a lung cancer diagnosis. These sequences allowed the authors to differentiate between malignant and healthy lung tissue. The authors confirmed that the above-mentioned 12 miRNAs showed increased expression in NSCLC, and they proved its associations with circulating exosomes. Moreover, investigators found a correlation between relevant miRNAs and the resistance of tumours to biological drugs, which were based on the action of anti-EGFR antibodies. In summary, the discovery of the role of miRNA in cellular pathways has allowed for new targets for cancer therapy [78].
The pathological and molecular diagnosis of mesothelioma
Another fatal neoplasm presenting with a broad spectrum of nonspecific symptoms is mesothelioma. This primary pleural neoplasm has a very high level of malignancy and constitutes a serious diagnostic challenge. Patients with mesothelioma have a poorer prognosis and a reduced quality of life at the time of diagnosis. In this context, an analysis of miRNA patterns alongside classical cytology seems to be an interesting novel diagnostic method for mesothelioma-related pleural effusions. There is a proven association between altered profiles of miRNA and the presence of mesothelioma. Evaluation of the miRNA profile enables the investigation of discrepancies between the various types of human lung tumours. Gee et al. discovered that the expression level of miRNA-200 in mesothelioma-related pleural effusion is significantly lower than in the pleural effusion caused by lung adenocarcinoma [79]. Matsumoto et al. linked the decreased level of miRNA-17-5p, miRNA-30c, and miRNA-31 with a positive result for sarcomatoid carcinoma [80, 81]. Pass et al. suggested that miRNA-29c-5p can be used as an independent prognostic factor for establishing the disease progression, as well as post-surgical time of survival. The elevated level of miRNA-29c-5p is related to a more favourable prognosis [82]. A signature comprising the let-7c/5p and miR-151a/5p profile was indicated as a tool for predicting the time of survival of mesothelioma patients, and elevated levels of these miRNAs were linked with a less favourable prognosis [83]. Truini et al. found that down-regulation of the miRNA-99a/let-7/miRNA-125b, the miRNA cluster, predicts a poor survival rate in unresected mesothelioma [84]. In another study, it was suggested that a combination of six miRNAs (miRNA-21-5p, miRNA-23a-3p, miRNA-30e-5p, miRNA-221-3p, miRNA-222-3p, and miRNA-31-5p) provides approximately 90% accuracy for survival prediction of patients after surgically-resected mesothelioma [85].
Expression of miRNAs in pleural effusion of varying aetiologies
The diagnostic utility of specific microRNAs has been reported in differentiating between patients with malignant pleural mesothelioma and those with benign asbestos-related pleural effusion. A significant upregulation of multiple miRNAs (including miRNA-484, miRNA-320, let-7a, and miRNA-125a-5p) was discovered in malignant pleural mesothelioma compared to benign asbestos-related pleural effusion, suggesting a potential diagnostic and therapeutic role [86]. The role of specific miRNA, particularly miRNA-93, has been elucidated in the regulation of endothelial cell migration, proliferation, angiogenesis, and cell cycle, playing an important part in the occurrence and development of malignant pleural effusions. Its high expression is associated with a good prognosis, due to its inhibition of pleural effusion by regulating angiogenesis and lymphangiogenesis [87]. Furthermore, the differing expression levels of miRNAs, such as miRNA-182 and miRNA-210 expression, are seen in malignant pleural effusions associated with lung adenocarcinoma compared to benign pleural effusion, providing a useful role in the minimally invasive screening for diagnostic evaluations of pleural effusions [88].
MiRNAs for the diagnosis and management of malignant mesothelioma
Malignant mesothelioma develops from the mesothelial cells cushioning the serosal cavities [89, 90]. The incidence of mesothelioma is rising dramatically worldwide, and it is expected to reach maximum levels in the next 10 years [89, 90]. Long-term inhalation of asbestos fibres is the typical agent involved in the genesis of this neoplasm, and the pleura is the most affected structure [90]. Patients with mesothelioma usually present with recurrent pleural effusions. Fine needle biopsy and cytological examination of the exfoliated cells is the first line of the diagnostic process, and for this reason, miRNAs can be applied for diagnostic purposes, both in cell lines and histologic samples [91]. Cappellesso et al. reported that mainly miRNA-126 was down-regulated, whereas miRNA-19a, miRNA-19b, miRNA-21, miRNA-25, and miRNA-126 were over-expressed in malignant mesothelioma cell lines versus the mesothelial cell line [92, 93]. The distinction of mesothelioma from reactive mesothelial cells (RMCs) may also be challenging, even when immunocytochemistry or fluorescence in situ hybridisation is made [92, 93]. Investigators recognised the profiling of pleural effusions using miRNA-21 and miRNA-126 as a promising diagnostic tool that could be helpful for cytopathologists in the distinction of mesothelioma from RMCs [93]. The microRNA profile was also used for discriminating between mesothelioma and lung adenocarcinoma, with results showing that miRNA-130 was significantly higher in mesotheliomas when compared with lung adenocarcinomas. The use of miR-130a demonstrated a sensitivity of 77%, a specificity of 67%, a positive predictive value of 69%, a negative predictive value of 75%, and an accuracy of 72% in identifying mesotheliomas [93]. Another group of scientists discovered the differences in expression of miRNA between malignant pleural mesothelioma (MPM) and benign asbestos-related pleural effusion (BAPE) samples. Eleven miRNAs were significantly upregulated in MPM compared to BAPE and included miRNA-484, miRNA-320, let-7a, miRNA-744, miRNA-20a, miRNA-193b, let-7d miRNA-125a-5p, miRNA-92a, miRNA-155, and miRNA-152 [86].
Several studies support the role of miRNAs in the aetiology of mesothelioma, suggesting their possible use as diagnostic and prognostic markers of the disease [94]. MiRNA-16 was found to be directly related to mesothelioma patient prognosis. Its increased expression level has been shown to correlate with higher patient survival, possibly due to its role in cell cycle control and carcinogenesis [95]. Other than its role as a biomarker in disease prognosis, miRNA may also aid in discriminating between patients with malignant pleural mesothelioma and subjects with past asbestos exposure [96].
Clinical implications and future perspectives
The monitoring of mesothelioma on the basis of the evaluation of miRNA expression in the accompanying pleural effusions appears to be a promising approach. We suspect that the use of various miRNA profiles, particularly specific to the regulation of oncogenes, will soon be used routinely for fast and more precise diagnosis of patients with suspected lung neoplasms. Santoni-Rugiu et al. have highlighted that an Israeli company is already showing promising diagnostic methods from an in vivo study of tissue-specific miRNA signatures to differentiate malignant pleural mesothelioma from lung adenocarcinoma, based on differentially expressed miRNAs, with high sensitivity and specificity [97]. Santoni-Rugiu et al. have further elucidated miRNA-223 as a promising biomarker for detecting malignant pleural mesothelioma. When compared with non-malignant controls, malignant pleural mesothelioma expressed a diminished level on miRNA-223. This was modulated by the c-Jun N-terminal kinase signalling pathway, which inhibits the proliferation and motility of malignant pleural mesothelioma cell lines by down-regulating gene expression of their microtubule regulator [97]. As public interest continues to grow regarding the leading cause of cancer deaths when looking at mortality rates, i.e. lung cancer, research into miRNAs will only continue to expand with it. This is already being shown by the growing number of yearly entries in the public miRNA-registry, known as miRBase [8], which now offers more potential biomarkers to be tested by lung cancer researchers than ever before.
Conclusions
Currently, screening tests serving to detect primary or metastatic tumour cells in pleural effusions are labour-intensive and time-consuming. Such tests generate not only high utilisation costs but also a high index of false positive or negative results. This, in consequence, incurs additional costs for the lengthy continuation of the diagnostic workup. This review article aims to explore the significance of miRNAs and their promising new potential for utilisation as diagnostic biomarkers for the differential diagnosis of pleural effusions. We particularly emphasise the possibilities of using these molecules for the workup of the problematic diagnosis of mesothelioma, and for metastatic pleural diseases. The diagnostic approach discussed in our article can complement the existing panels of tumour markers and classical imaging techniques. Research on the prevention and early diagnosis of cancer has focused on the identification of biomarkers that can help in the detection of tumours using non-invasive diagnostic methods. The addition of miRNAs can currently increase the specificity and sensitivity of NSCLC detection. The specific signature of miRNA expression in malignant pleural effusions can further be utilised in the prognostication of NSCLC patient survivability. We have worked under the assumption that the pleural effusion environment, in contradiction to blood-circulating miRNA, is less likely to undergo fluctuations resulting from peripheral processes such as pregnancy, heart infarction, sepsis, or viral infection. Due to the promising capabilities offered by MPE-derived miRNAs, we necessitate that further research studies are required. We speculate that the possibilities of an early and low-cost detection of NSCLC- or mesothelioma-derived pleural effusions with the application of miRNA profiles will provide a greater choice in the personalisation of anticancer treatment along with more objective follow-up for those patients undergoing treatment. The assessment of miRNA occurrence in pleural effusions is highly attractive from the scientific point of view because it allows for the investigation of the oncogenic processes concerning the different elements presented in the cancer microenvironment. Such investigations enhance not only the scientific knowledge surrounding tumour biology, but also its direct and indirect implications.
Acknowledgements
The authors thank Jadzia Tin-Tsen Chou and Marcin Lenkowski for their help in article revision.
The study was supported by Student Scientific Society of the Poznan University of Medical Sciences grant No. 502-05-01127188-07727.
Footnotes
The authors declare no conflict of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Rawla P. Epidemiology of Prostate Cancer. World J Oncol. 2019;10:63–89. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Lancet GLOBOCAN 2018: counting the toll of cancer. Lancet. 2018;392:985. doi: 10.1016/S0140-6736(18)32252-9. [DOI] [PubMed] [Google Scholar]
- 4.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Gastroenterology Rev. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Gastroenterology Rev. 2018;14:26–38. doi: 10.5114/pg.2018.80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 8.miRBase. 06.2018.
- 9.Turchinovich A, Samatov TR, Tonevitsky AG, et al. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin YM, Yun J, Lee OJ, et al. Diagnostic Value of Circulating Extracellular miR-134, miR-185, and miR-22 Levels in Lung Adenocarcinoma-Associated Malignant Pleural Effusion. Cancer Res Treat. 2014;46:178–185. doi: 10.4143/crt.2014.46.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116(2 Suppl):S89–92. doi: 10.1016/s0092-8674(04)00035-2. [DOI] [PubMed] [Google Scholar]
- 17.Xiao JF, Yong X, Fan YH, Lü MH, Yang SM, Hu CJ. microRNA detection in feces, sputum, pleural effusion and urine: Novel tools for cancer screening. Oncol Rep. 2013;30:535–544. doi: 10.3892/or.2013.2525. [DOI] [PubMed] [Google Scholar]
- 18.Yamada Y, Enokida H, Kojima S, et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 19.Switlik WZ, Szemraj J. Circulating miRNAs as non-invasive biomarkers for non-small cell lung cancer diagnosis, prognosis and prediction of treatment response. Postepy Hig Med Dosw (Online) 2017;71:649–662. doi: 10.5604/01.3001.0010.3845. [DOI] [PubMed] [Google Scholar]
- 20.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2004;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Isik M, Korswagen HC, Berezikov E. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence. 2010;1:5. doi: 10.1186/1758-907X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truscott M, Islam A, Frolov MV. Novel regulation and functional interaction of polycistronic miRNAs. RNA. 2016;22:129–138. doi: 10.1261/rna.053264.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao W, Liu L, Lu X, Shu Y. Circulating MicroRNAs: Possible Prediction Biomarkers for Personalized Therapy of Non-Small-Cell Lung Carcinoma. Clin Lung Cancer. 2011;12:14–17. doi: 10.3816/CLC.2011.n.001. [DOI] [PubMed] [Google Scholar]
- 25.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 26.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Grenda A, Budzyński M, Filip A. Biogenesis of microRNAs and their role in the development and course of selected hematologic disorders. Postepy Hig Med Dosw. 2013;67:174–185. doi: 10.5604/17322693.1038361. [DOI] [PubMed] [Google Scholar]
- 32.Oliveto S, Mancino M, Manfrini N. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8:45–56. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mack GS. MicroRNA gets down to business. Nat Biotechnol. 2007;25:631–638. doi: 10.1038/nbt0607-631. [DOI] [PubMed] [Google Scholar]
- 36.Qi J, Yu JY, Shcherbata HR, et al. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otto T, Candido SV, Pilarz MS, et al. Cell cycle-targeting microRNAs promote differentiation by enforcing cell-cycle exit. Proc Natl Acad Sci U S A. 2017;114:10660–10665. doi: 10.1073/pnas.1702914114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 41.Bueno MJ, Perez de Castro I, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7:3143–3148. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- 42.Foshay KM, Gallicano GI. Small RNAs, big potential: the role of MicroRNAs in stem cell function. Curr Stem Cell Res Ther. 2007;2:264–271. doi: 10.2174/157488807782793781. [DOI] [PubMed] [Google Scholar]
- 43.Baltimore D, Boldyn MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 44.Carissimi C, Fulci V, Macino G. MicroRNAs: Novel regulators of immunity. Autoimmun Rev. 2009;8:520–524. doi: 10.1016/j.autrev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. MiR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 47.Johnnidis JB, Harris MH, Wheeler RT, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1130. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 48.Lindsay MA. MicroRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Linsley PS, Schelter J, Burchard J, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tili E, Michaille IJ, Cimino S, et al. Modulation of miR-155 and miR-125b levels following lipopolysac-charide/TNF-alpha stimulation and their possible role in regulation the response to endotoxin shock. J Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 52.Williams AE. Functional aspect of animal microRNAs. Cell Mol Life Sci. 2008;65:545–562. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao C, Srinivasan L, Calado DP, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majorek K, Krzyżosiak W. Role of microRNA in pathogenesis, diagnostics and therapy of cancer. Współcz Onkol. 2006;10:359–366. [Google Scholar]
- 55.Calin GA, Sevignani C, Dumitru CD, et al. Human genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calin G, Dumitru C, Shimizu M, et al. Frequent deletions and downregulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 58.Zhao B, Han H, Chen J, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Yu S. Role of miR-520b in non-small cell lung cancer. Exp Ther Med. 2018;16:3987–3995. doi: 10.3892/etm.2018.6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Igaz I, Igaz P. Possible role for microRNAs as inter-species mediators of epigenetic information in disease pathogenesis: Is the non-coding dark matter of the genome responsible for epigenetic interindividual or interspecies communication? Med Hypotheses. 2015;84:150–154. doi: 10.1016/j.mehy.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 61.World Health Organization . GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012. International Agency for Research on Cancer. [Google Scholar]
- 62.See KC, Lee P. Advances in the diagnosis of pleural disease in lung cancer. Ther Adv Respir Dis. 2011;5:409–418. doi: 10.1177/1753465811408637. [DOI] [PubMed] [Google Scholar]
- 63.Antonangelo L, Sales RK, AP Corá AP, Acencio MMP, Teixeira LR, Vargas FS. Pleural fluid tumour markers in malignant pleural effusion with inconclusive cytologic results. Curr Oncol. 2015;22:e336–e341. doi: 10.3747/co.22.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu P, Huang G, Chen Y, Zhu C, Yuan J, Sheng S. Diagnostic utility of pleural fluid arcinoembryonic antigen and CYFRA 21-1 in patients with pleural effusion: a systematic review and meta-analysis. J Clin Lab Anal. 2007;21:398–405. doi: 10.1002/jcla.20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang T, Lv M, Shen S, et al. Cell-Free MicroRNA Expression Profiles in Malignant Effusion Associated with Patient Survival in Non-Small Cell Lung Cancer. PLoS One. 2012;7:e43268. doi: 10.1371/journal.pone.0043268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shin YM, Yun J, Lee OJ, et al. Diagnostic Value of Circulating Extracellular miR-134, miR-185, and miR-22 Levels in Lung Adenocarcinoma-Associated Malignant Pleural Effusion. Cancer Res Treat. 2014;46:178–185. doi: 10.4143/crt.2014.46.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Han HS, Jo YN, Lee JY, Choi SY, Jeong Y, Yun J, Lee OJ. Identification of suitable reference genes for the relative quantification of microRNAs in pleural effusion. Oncol Lett. 2014;8:1889–1895. doi: 10.3892/ol.2014.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han HS, Yun J, Lim SN, et al. Downregulation of cell-free miR-198 as a diagnostic biomarker for lung adenocarcinoma-associated malignant pleural effusion. Int J Cancer. 2013;133:645–652. doi: 10.1002/ijc.28054. [DOI] [PubMed] [Google Scholar]
- 69.Puerta-Gil P, Garcia-Baquero R, Jia AY, et al. miR-143, miR-222, and miR-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. Am J Pathol. 2012;180:1808–1815. doi: 10.1016/j.ajpath.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 70.Janakiram NB, Rao CV. Molecular markers and targets for colorectal cancer prevention. Acta Pharmacol Sin. 2008;29:1–20. doi: 10.1111/j.1745-7254.2008.00742.x. [DOI] [PubMed] [Google Scholar]
- 71.Klimczak-Bitner AA, Kordek R, Bitner J, Musiał J, Szemraj J. Expression of MMP9, SERPINE1 and miR-134 as prognostic factors in esophageal cancer. Oncol Lett. 2016;12:4133–4138. doi: 10.3892/ol.2016.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Light RW. Clinical practice. Pleural effusion. N Engl J Med. 2002;346:1971–1977. doi: 10.1056/NEJMcp010731. [DOI] [PubMed] [Google Scholar]
- 73.Pastré J, Roussel S, Israël Biet D, Sanchez O. Pleural effusion: diagnosis and management. Rev Med Interne. 2015;36:248–255. doi: 10.1016/j.revmed.2014.10.360. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y, Xu YM, Zou YQ, et al. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Medicine (Baltimore) 2017;96:e8361. doi: 10.1097/MD.0000000000008361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han F, He J, Li F, Yang J, Wei J, Cho WC, Liu X. Emerging Roles of MicroRNAs in EGFR-Targeted Therapies for Lung Cancer. Biomed Res Int. 2015;2015:672759. doi: 10.1155/2015/672759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang SH, Li Y, Zhang J, et al. Epidermal growth factor receptorcontaining exosomes induce tumor-specific regulatory T cells. Cancer Invest. 2013;31:330–335. doi: 10.3109/07357907.2013.789905. [DOI] [PubMed] [Google Scholar]
- 77.Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: The new ambrosia of researchers. Biochim Biophys Acta. 2014;1846:539–546. doi: 10.1016/j.bbcan.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 78.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 79.Gee GV, Koestler DC, Christensen BC, et al. Downregulated microRNAs in the differential diagnosis of malignant pleural mesothelioma. Int J Cancer. 2010;127:2859–2869. doi: 10.1002/ijc.25285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Busacca S, Germano S, De Cecco L, et al. MicroRNA signature of malignant meosthlelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol. 2010;42:312–319. doi: 10.1165/rcmb.2009-0060OC. [DOI] [PubMed] [Google Scholar]
- 81.Matsumoto S, Nabeshima K, Hamasaki M, Shibuta T, Umemura T. Upregulation of microRNA-31 associates with a poor prognosis of malignant pleural mesothelioma with sarcomatoid component. Med Oncol. 2014;31:303. doi: 10.1007/s12032-014-0303-2. [DOI] [PubMed] [Google Scholar]
- 82.Pass HI, Goparaju C, Ivanov S, et al. hsa-miR-29c* is linked to the prognosis of malignant pleural mesothelioma. Cancer Res. 2010;70:1916–1924. doi: 10.1158/0008-5472.CAN-09-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.De Santi C, Melaiu O, Bonotti A, et al. Deregulation of miRNAs in malignant pleural mesothelioma is associated with prognosis and suggests an alteration of cell metabolism. Sci Rep. 2017;7:3140. doi: 10.1038/s41598-017-02694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Truini A, Coco S, Nadal E, et al. Downregulation of miR-99a/let-7c/miR-125b miRNA cluster predicts clinical outcome in patients with unresected malignant pleural mesothelioma. Oncotarget. 2017;8:68627–68640. doi: 10.18632/oncotarget.19800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Birnie KA, Prele CM, Thompson PJ, Badrian B, Mutsaers SE. Targeting microRNA to improve diagnostic and therapeutic approaches for malignant mesothelioma. Oncotarget. 2017;8:78193–78207. doi: 10.18632/oncotarget.20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ak G, Tomaszek SC, Kosari F, et al. MicroRNA and mRNA features of malignant pleural mesothelioma and benign asbestos-related pleural effusion. Biomed Res Int. 2015;2015:635748. doi: 10.1155/2015/635748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qian Q, Sun W, Zhu W, et al. The role of microRNA-93 regulating angiopoietin2 in the formation of malignant pleural effusion. Cancer Med. 2017;6:1036–1048. doi: 10.1002/cam4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tamiya H, Mitani A, Saito A, et al. Exosomal MicroRNA Expression Profiling in Patients with Lung Adenocarcinoma-associated Malignant Pleural Effusion. Anticancer Res. 2018;38:6707–6714. doi: 10.21873/anticanres.13039. [DOI] [PubMed] [Google Scholar]
- 89.Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med. 2005;353:1591–1603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- 90.Travis WD, Brambilla E, Burke AP, et al. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. Lyon, France: IARC Press; 2015. [DOI] [PubMed] [Google Scholar]
- 91.Ikeda K, Tate G, Suzuki T, Kitamura T, Mitsuya T. IMP3/L523S, a novel immunocytochemical marker that distinguishes benign and malignant cells: the expression profiles of IMP3/L523S in effusion cytology. Hum Pathol. 2010;41:745–750. doi: 10.1016/j.humpath.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 92.Cappellesso R, Galasso M, Nicole L, Dabrilli P, Volinia S, Fassina A. miR-130A as a Diagnostic Marker to Differentiate Malignant Mesothelioma From Lung Adenocarcinoma in Pleural Effusion Cytology. Cancer Cytopathol. 2017;125:635–643. doi: 10.1002/cncy.21869. [DOI] [PubMed] [Google Scholar]
- 93.Cappellesso R, Nicolè L, Caroccia B, Guzzardo V, Ventura L, Fassan M, Fassina A. Young Investigator Challenge: MicroRNA-21/MicroRNA-126. Profiling as a Novel Tool for the Diagnosis of Malignant Mesothelioma in Pleural Effusion Cytology. Cancer Cytopathol. 2016;124:28–36. doi: 10.1002/cncy.21646. [DOI] [PubMed] [Google Scholar]
- 94.Lo Russo G, Tessari A, Capece M, Galli G, de Braud F, Garassino MC, Palmieri D. MicroRNAs for the Diagnosis and Management of Malignant Pleural Mesothelioma: A Literature Review. Front Oncol. 2018;8:650. doi: 10.3389/fonc.2018.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mozzoni P, Ampollini L, Goldoni M, et al. MicroRNA Expression in Malignant Pleural Mesothelioma and Asbestosis: A Pilot Study. Dis Markers. 2017;2017:9645940. doi: 10.1155/2017/9645940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cavalleri T, Angelici L, Favero C, et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS One. 2017;12:e0176680. doi: 10.1371/journal.pone.0176680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santoni-Rugiu E, Andersen M, Grauslund M. MicroRNAs as potential biomarkers in malignant pleural mesothelioma. Curr Biomark Find. 2016;6:1–21. [Google Scholar]