Abstract
Cellular senescence is an irreversible form of cell cycle arrest. Senescent cells have a unique gene expression profile that is frequently accompanied by senescence-associated heterochromatic foci (SAHFs). Protein kinase CK2 (CK2) downregulation can induce trimethylation of histone H3 Lys 9 (H3K9me3) and SAHFs formation by activating SUV39h1. Here, we present evidence that the PI3K-AKT-mTOR-reactive oxygen species-p53 pathway is necessary for CK2 downregulation-mediated H3K9me3 and SAHFs formation. CK2 downregulation promotes SUV39h1 stability by inhibiting its proteasomal degradation in a p53-dependent manner. Moreover, the dephosphorylation status of Ser 392 on p53, a possible CK2 target site, enhances the nuclear import and subsequent stabilization of SUV39h1 by inhibiting the interactions between p53, MDM2, and SUV39h1. Furthermore, p21Cip1/WAF1 is required for CK2 downregulation-mediated H3K9me3, and dephosphorylation of Ser 392 on p53 is important for efficient transcription of p21Cip1/WAF. Taken together, these results suggest that CK2 downregulation induces dephosphorylation of Ser 392 on p53, which subsequently increases the stability of SUV39h1 and the expression of p21Cip1/WAF1, leading to H3K9me3 and SAHFs formation.
Keywords: CK2, H3K9me3, p53, SAHFs, SUV39h1
INTRODUCTION
The p53 tumor suppressor protein is a latent transcription factor that increases genomic stability and protects against malignant transformations (Bieging et al., 2014). p53 is usually present at low levels in cells under normal conditions due to MDM2 (murine double minute 2, also used interchangeably to represent the human homolog)-mediated ubiquitination and proteasomal degradation (Wade et al., 2013). However, p53 becomes rapidly stabilized and activated in response to a variety of cellular stress conditions, such as DNA damage and oncogenic activation. Post-translational modifications of p53, including phosphorylation, methylation, acetylation, and sumoylation weaken the p53-MDM2 interaction, which causes the active p53 to accumulate of in the nucleus (Kruse and Gu, 2009). Depending on the amount of active p53, it can either induce apoptosis (by transactivation of Bax) or lead to cell cycle arrest (by transactivation of p21Cip1/WAF1) (Simabuco et al., 2018). The p53 pathway also plays an important role in the induction and maintenance of cellular senescence, which is an irreversible arrest of cell proliferation (Campisi, 2005; Roninson, 2003).
Senescence-associated heterochromatic foci (SAHFs), which result from the condensation of chromatin into characteristically punctate heterochromatic domains, repress genes encoding cell cycle progression-associated proteins (Funayama and Ishikawa, 2007; Kim, 2018; Narita et al., 2003). SAHFs contain several markers of transcriptionally silent heterochromatin, such as accumulation of trimethylated histone H3 Lys 9 (H3K9me3) and selective binding of heterochromatin proteins 1γ (HP1γ) to H3K9me3 (Kouzarides, 2007; Maison and Almouzni, 2004). The methylation status of H3K9 is modulated by H3K9 tri-methylase, SUV39h1, and di-methylases including G9a, GLP, and SETDB1 (Fritsch et al., 2010; Hublitz et al., 2009; Tachibana et al., 2005). The NAD+-dependent histone deacetylase, SIRT1, deacetylates lysine 266 on SUV39h1 and regulates SUV39h1 activity (Vaquero et al., 2007). Furthermore, it has been shown that MDM2 mediates the ubiquitination of lysine 87 within SUV39h1 to promote proteasomal degradation (Bosch-Presegue et al., 2011). However, it remains unclear how SUV39h1 activity and SAHFs formation are regulated during cellular senescence.
We previously showed that protein kinase CK2 (CK2) downregulation induces senescence-associated β-galactosidase activity and activation of the p53 and p21Cip1/WAF1 axis, suggesting that cellular senescence is mediated via the p53-p21Cip1/WAF1 pathway (Kang et al., 2009; Ryu et al., 2006). Additionally, we recently reported that CK2 downregulation induces H3K9me3 and SAHFs formation by increasing SUV39h1 and decreasing G9a (Park et al., 2018). In the present study, we investigated the role of p53 in CK2 downregulation-mediated SAHFs formation. This study indicates that CK2 downregulation inhibits SUV39h1 degradation in a p53-dependent manner and that this event promotes H3K9me3.
MATERIALS AND METHODS
Materials
Antibodies against CK2α, p53, p21Cip1/WAF1, and β-actin were obtained from Santa Cruz Biotechnology (USA). An antibody against phospho-p53 serine 392 (S392) was purchased from Cell Signaling Technology (USA), and antibodies against HP1γ, SUV39h1, H3K9me3, histone H3, and MDM2 were from Abcam (UK). Both 4′,6-diamidino-2-phenylindole (DAPI) and rhodamine-conjugated goat anti-rabbit IgG were purchased from Invitrogen (USA). The anti-hemagglutinin (HA) antibody was obtained from Roche (Switzerland) and TrueBlot reagent was from Rockland (USA). Pifithrin-α, nutlin-3, cycloheximide, N-Acetyl-L-cysteine (NAC), wortmannin, triciribine, and rapamycin were obtained from Sigma Chemical (USA).
Cell culture and DNA transfection
Human colon cancer HCT116 cells (p53 and p21 wild-type, p53−/−, and p21−/−) and breast cancer cells MCF-7 and MDA-MB-435 (p53 contains a Gly to Glu mutation at residue 266) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Both pcDNA3.1-p53 and pcDNA3.1-HA-CK2α were transfected into cells using Polyfect (Invitrogen) for 48 h. The siRNA for CK2α was 5′-UCAAGAUGACUACCAGCUGdTdT-3′. The siRNA for the negative control was 5′-GCUCAGAUCAAUACGGAGAdTdT-3′. siRNAs were transfected into cells using Lipofectamine (Thermo Fisher Scientific, USA) for 48 h.
Immunoblotting
Proteins were separated on 10% SDS-polyacrylamide gels and then transferred by electrophoresis to nitrocellulose membranes. The membranes were blocked with 5% (w/v) non-fat, dry, skim milk dissolved in TBST (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.05% Tween 20) for 2 h and then incubated with specific antibodies diluted in TBST with 1% (w/v) non-fat, dry, skim milk for 1 h. The membranes were washed three times in TBST and then analyzed with an ECL system (Amersham Biosciences, UK).
Immunofluorescence
Cells were seeded on eight-well micro-chamber slides (Nunc; Thermo Fisher Scientific) and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at 25°C. Then, cells were permeabilized in 0.25% Triton X-100 before blocking with 2% bovine serum albumin in PBS. Anti-H3K9me3 (1:100) and HP1γ (1:100) antibodies were added at 25°C for 1 h. The secondary antibodies were rhodamine-conjugated, goat anti-rabbit IgG (1:100). Next, DAPI was used to counterstain nuclei; fluorescence signals were detected using a Carl Zeiss Axioplan 2 microscope (Carl Zeiss, Germany). Fluorescence images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/).
Reverse transcription-PCR
Total RNA was extracted from cells. RNA was reverse-transcribed using gene-specific primers and reverse transcriptase (Takara Bio, Japan), and the resulting cDNAs were PCR-amplified. Primers used for the assays are listed in Supplementary Table S1. PCR products were resolved on a 1.5% agarose gel. Quantification of the reverse transcription-PCR bands was performed using densitometry. β-actin RNA levels were used to standardize the amounts of RNA in each sample.
Construction of p53 mutants
To replace p53 Ser 392 (S392) with glutamic acid or alanine, p53 cDNA was PCR-amplified using C-terminal mutagenic primers S392E (5′-CGAATTCTCAGTCTTCGTCAGGCCC TTCTGT-3′) or S392A (5′-CGAATTCTCAGTCTGCGTCAGGCCCTTCTGT-3′). The mutation sites are underlined. After digesting the PCR products with EcoRI and BamHI, the fragment was ligated into the EcoRI/BamHI sites of pcDNA3.1.
Immunoprecipitation
Cell lysates were pre-cleared with normal mouse or rabbit IgG and protein A Sepharose (Amersham Biosciences) for 1 h at 4°C. The supernatant was then incubated with anti-SUV39h1 or anti-p53 antibodies and protein A Sepharose by mixing for 12 h at 4°C. Then, the beads were collected by centrifugation and washed three times with PBS.
Isolation of nuclear and cytoplasmic extracts
Cytoplasmic and nuclear extracts were prepared using an NE-PER Nuclear Cytoplasmic Extraction Reagent kit (Pierce, USA) according to the manufacturer’s instructions.
Statistical analysis
Data were analyzed by one-way ANOVA with the IBM SPSS Statistics program ver. 25.0 (IBM, USA). The results were considered significant if the P value was < 0.05. Duncan’s multiple-range test was performed if significant differences between the groups were identified, using α = 0.05.
RESULTS
The PI3K-AKT-mTOR-ROS pathway is required for CK2 downregulation-mediated stimulation of SUV39h1 expression, H3K9me3, and SAHFs formation
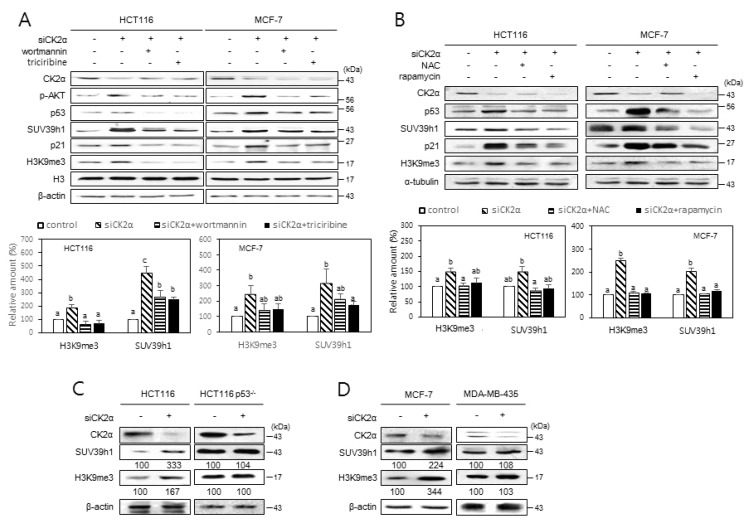
We previously showed that CK2 downregulation induces H3K9me3 by increasing SUV39h1 and decreasing G9a, GLP, and SETDB1 in both HCT116 and MCF-7 cells (Park et al., 2018). The PI3K-AKT-mTOR-reactive oxygen species (ROS) pathway is involved in CK2 downregulation-mediated senescence (Jeon et al., 2010; Park et al., 2013). To investigate the impact of the PI3K-AKT-mTOR-ROS pathway on SUV39h1 expression and H3K9me3, HCT116 and MCF-7 cells were treated with CK2α siRNA in the presence of the PI3K inhibitor wortmannin (100 nM), the AKT inhibitor triciribine (10 μM), the mTOR inhibitor rapamycin (100 nM), or the ROS scavenger NAC (5 mM). Consistent with our previous study (Park et al., 2018), CK2α knockdown resulted in increased levels of H3K9me3, SUV39h1, p-AKT, p53, and p21Cip1/WAF1 in these cells. However, treatment with wortmannin, triciribine, rapamycin, or NAC successfully suppressed the CK2 knockdown-mediated stimulation of SUV39h1 expression and H3K9me3 (Figs. 1A and 1B). We next examined the role of the PI3K-AKT-mTOR-ROS pathway in CK2 downregulation-mediated SAHFs formation. SAHFs formation was detected by staining with DAPI and by immunofluorescence using antibodies against H3K9me3 and HP1γ. Consistent with our previous study (Park et al., 2018), CK2α knockdown stimulated co-localization of HP1γ (green) and H3K9me3 (red) with DAPI foci (blue), suggesting that SAHFs formation is induced during CK2α downregulation-mediated senescence. However, treatment with wortmannin (100 nM), triciribine (10 μM), rapamycin (100 nM), or NAC (5 mM) interrupted the SAHFs formation induced by CK2α downregulation (Supplementary Fig. S1). Taken together, these results suggest that the PI3K-AKT-mTOR-ROS pathway is necessary for the CK2 downregulation-mediated induction of SUV39h1 expression, H3K9me3, and SAHF formation.
Fig. 1. Induction of H3K9me3 and SUV39h1 after CK2 downregulation requires PI3K-AKT-mTOR-ROS-p53 pathway.
(A and B) Cells were treated with CK2α siRNA for 48 h in the presence of 100 nM wortmannin or 10 μM triciribine (A) and 5 mM NAC or 100 nM rapamycin (B). Cell extracts were then visualized by immunoblotting (upper panels). Representative data from three independent experiments are shown. The graph shows the quantification of each protein relative to α-tubulin (A) or β-actin (B) (bottom panels). Values indicate mean ± SEM. Bars that do not share a common letter (a, b, or c) are significantly different between groups at P < 0.05. (C and D) Essential role of p53 in inducing H3K9me3 and SUV39h1 upon CK2 downregulation. Wild-type p53 cells (HCT116 and MCF-7) and mutant p53 cells (HCT116 p53−/− and MDA-MB-435) were treated with CK2α siRNA, and cell extracts were visualized by immunoblotting. The values below each band represent the mean fold differences (n = 3) in expression levels compared to the control, which was assigned a value of 100.
p53 is required for CK2 downregulation-mediated stimulation of SUV39h1 expression, H3K9me3, and SAHFs formation
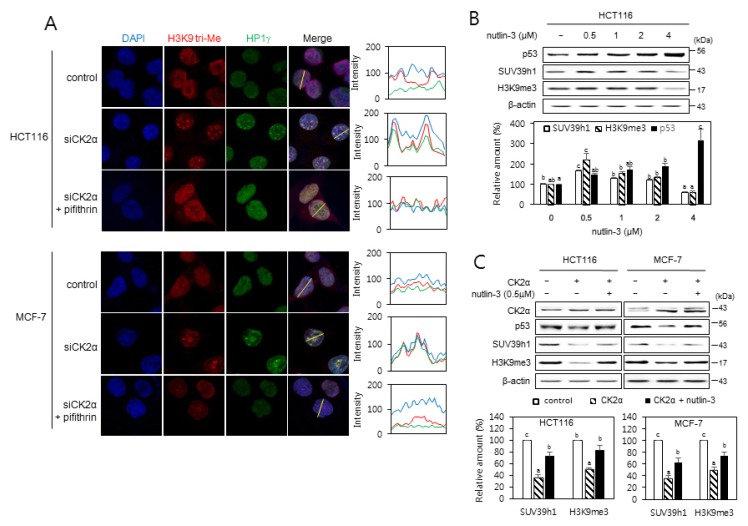
Because p53, a downstream regulator of ROS, is necessary for the senescence induced by CK2 silencing (Kang et al., 2009), we aimed to determine the role of p53 in the expression of SUV39h1 and H3K9me3 during CK2 downregulation-mediated senescence using HCT116 p53−/− cells and MDA-MB-435 cells (which contain a point mutation within p53). In contrast with cells containing wild-type p53, CK2α knockdown did not elevate SUV39h1 expression or H3K9me3 levels in either HCT116 p53−/− or MDA-MB-435 cells (Figs. 1C and 1D), suggesting that CK2 downregulation promotes SUV39h1 expression and H3K9me3 by activating the p53 pathway. We examined the role of p53 in CK2 downregulation-mediated SAHFs formation. Treatment with the p53 inhibitor pifithrin-α (30 μM) successfully interrupted SAHFs formation induced by CK2α downregulation (Fig. 2A). To determine the relationship between p53 levels and SUV39h1 expression, cells were treated with various concentrations of the MDM2 inhibitor nutlin-3, which inhibits the p53–MDM2 interaction and stabilizes p53. Expectedly, protein levels of p53 increased in a concentration-dependent manner, from 0.5 to 4 μM of nutlin-3. However, although the amounts of SUV39h1 and H3K9me3 increased at 0.5 μM of nutlin-3, they decreased at 1 to 4 μM of nutlin-3, also in a concentration-dependent manner (Fig. 2B). Treatment with 0.5 μM nutlin-3 successfully abrogated the reduction of SUV39h1 and H3K9me3 mediated by CK2α overexpression (Fig. 2C). Therefore, these data indicate that p53 is an upstream regulator of SUV39h1 expression and H3K9me3 in CK2 downregulation-mediated SAHFs formation.
Fig. 2. p53-dependent induction of SAHFs formation after CK2 downregulation.
(A) Confocal immunofluorescent images (×800 magnification) showing the co-localization of chromatin foci with H3K9me3 (red) and HP1γ (green) in cells treated with CK2α siRNA in the presence of the p53 inhibitor pifithrin-α (30 μM). DAPI staining (blue) was used to visualize DNA foci. Fluorescence intensity was quantified using ImageJ software (right panels). Arbitrary intensity values for H3K9me3 (red), HP1γ (green), or DAPI (blue) are shown relative to the reference line (white) used for the analysis. (B and C) The effect of nutlin-3 on the expression of p53, SUV39h1, and H3K9me3. (B) Cells were treated with the MDM2 inhibitor nutlin-3 (0.5 to 4 μM) for 48 h. (C) Cells were treated with pcDNA3.1-HA-CK2α in the absence or presence of nutlin-3 (0.5 μM) for 48 h. Cell lysates were visualized by immunoblotting (upper panels). The graph shows the quantification of each protein relative to β-actin (bottom panels). The values indicate mean ± SEM. Bars that do not share a common letter (a, b, or c) are significantly different between groups at P < 0.05.
CK2 downregulation promotes SUV39h1 protein stability
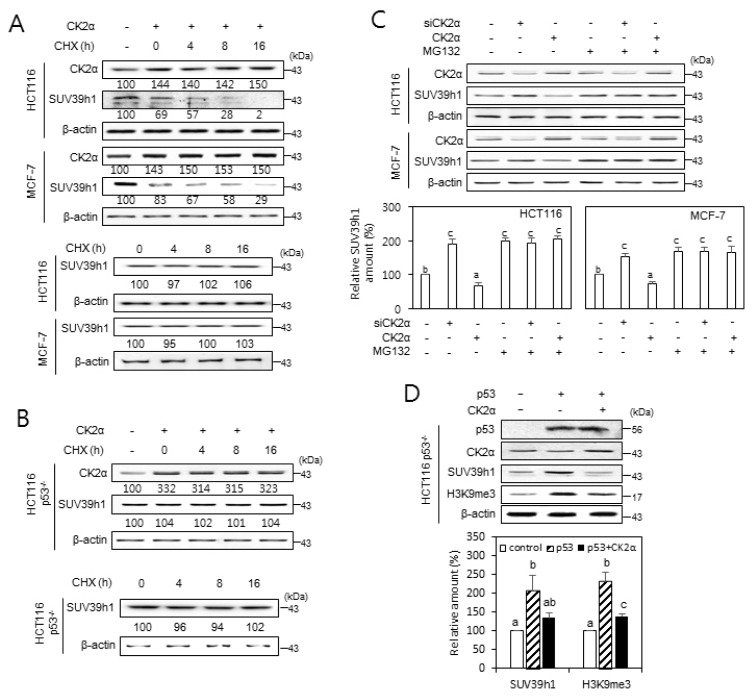
Our previous observation that the knockdown or overexpression of CK2α does not change the amount of SUV39h1 mRNA indicated that CK2 regulates the expression of SUV39h1 at a post-transcriptional level (Park et al., 2018). To investigate whether CK2 regulates SUV39h1 expression at translational or post-translational levels, cells transfected with pcDNA3.1-HA-CK2α were grown in the presence of the protein synthesis inhibitor cycloheximide (50 μg/ml). Protein levels of SUV39h1 decreased steadily in a time-dependent manner in cells overexpressing CK2α, suggesting that CK2 promotes the degradation of SUV39h1 protein. Without CK2α upregulation (control experiment), cycloheximide treatment did not decrease protein levels of SUV39h1 (Fig. 3A). Next, we investigated a possible role for p53 in the regulation of SUV39h1 stability using p53-null cells. Importantly, CK2α upregulation, even in the presence of cycloheximide, did not decrease the level of SUV39h1 protein in HCT116 p53−/− cells (Fig. 3B). To examine whether CK2-induced SUV39h1 destabilization is accompanied by proteasomal degradation, cells were treated with pcDNA3.1-HA-CK2α or CK2α siRNA in the presence of the proteasome inhibitor MG132 (10 μM). SUV39h1 protein levels were increased or decreased by CK2α downregulation or upregulation, respectively, when compared to levels in control cells. Furthermore, treatment with MG132 suppressed CK2 upregulation-mediated SUV39h1 reduction, suggesting that CK2 upregulation destabilizes SUV39h1 protein via proteasomal degradation (Fig. 3C). To confirm p53-dependency of CK2-mediated SUV39h1 destabilization, HCT116 p53−/− cells were transfected with pcDNA3.1-p53 and/or pcDNA3.1-HA-CK2α. Ectopic expression of wild-type p53 increased protein levels of SUV39h1 and H3K9me3 in p53-null cells, and additional overexpression of CK2α suppressed this event (Fig. 3D). Collectively, these observations suggest that CK2 downregulation stabilizes SUV39h1 protein by inhibiting its proteasomal degradation in a p53-dependent manner.
Fig. 3. The role of CK2 in regulating SUV39h1 expression.
(A and B) p53-dependent destabilization of SUV39h1 protein after CK2 upregulation. Wild-type p53 cells (HCT116 and MCF-7) (A) and p53-null cells (HCT116 p53−/−) (B) were treated with the protein synthesis inhibitor cycloheximide (CHX, 50 μg/ml) for the indicated times in the absence or presence of pcDNA3.1-HA-CK2α. The cell lysates were visualized by immunoblotting. The values below each band represent the mean fold differences (n = 3) in expression levels compared to the control, which was assigned a value of 100. (C) Cells were treated with CK2α siRNA or pcDNA3.1-HA-CK2α in the absence or presence of the proteasome inhibitor MG132 (10 μM) for 6 h. Cell lysates were visualized by immunoblotting (upper panel). The graph shows the quantification of each protein relative to β-actin (bottom panel). (D) CK2 overexpression compromises the p53-mediated induction of SUV39h1. Immunoblot analysis of cells treated with CK2α siRNA or pcDNA3.1-p53 (upper panel). The graph shows the quantification of SUV39h1 and H3K9me3 relative to β-actin (bottom panel). The values indicate mean ± SEM. Bars that do not share a common letter (a, b, or c) are significantly different between groups at P < 0.05.
The dephosphorylation status of S392 on p53 is associated with SUV39h1 stability and SAHFs formation
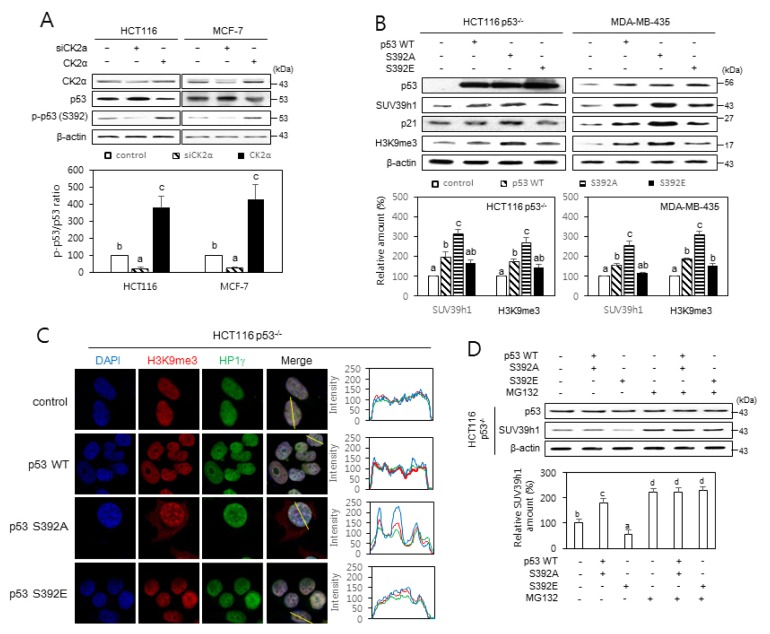
CK2 phosphorylates Ser 392 (S392) on p53 (Kim et al., 2004; Meek, 1999). Our immuno-blotting analysis using a phospho-p53 S392-specific antibody also revealed that the phosphorylation of S392 on p53 was decreased in CK2-downregulated cells and increased in CK2-upregulated cells (Fig. 4A). To investigate whether the phosphorylation status of S392 on p53 affected SUV39h1 destabilization, S392 was replaced with alanine or glutamic acid, resulting in p53S392A and p53S392E. Ectopic expression of p53S392A resulted in increased levels of SUV39h1 protein in p53-null or -mutant cells, when compared to those ectopically expressing wild-type p53 (Fig. 4B). Consistently, more SAHFs were detected in cells expressing p53S392A than in cells expressing either p53S392E or wild-type p53 (Fig. 4C). To examine whether the phosphorylation status of S392 on p53 regulates SUV39h1 destabilization by proteasomal degradation, p53-null cells ectopically expressing p53 variants were treated with MG132 (10 μM). When compared to wild-type p53 expression, p53S392A expression increased SUV39h1 protein levels, whereas p53S392E expression decreased them. Furthermore, treatment with MG132 suppressed p53S392E-mediated SUV39h1 reduction, suggesting that the phosphorylation status of S392 on p53 promotes SUV39h1 proteasomal degradation (Fig. 4D). Collectively, these results suggest that the dephosphorylation status of S392 on p53 is important for enhanced stability of SUV39h1, as well as SAHFs formation in CK2 downregulation-mediated senescence.
Fig. 4. The effect of p53 S392 phosphorylation on the stability of SUV39h1.
(A) CK2 phosphorylates S392 on p53. Immunoblot analysis of cells treated with CK2α siRNA or pcDNA3.1-p53 (upper panel). The graph shows the quantification of phospho-p53 S392 relative to p53 (bottom panel). The values indicate means ± SEM. Bars that do not share a common letter (a, b, or c) are significantly different between groups at P < 0.05. (B–D) Cells were treated with pcDNA3.1-p53 wild-type (WT) or -p53 mutants (S392A and S392E). (B) Induction of SUV39h1 and H3K9me3 by p53S392A relative to WT p53 or p53S392E. Protein extracts were visualized by immunoblotting (upper panels). The graph shows the quantification of SUV39h1 and H3K9me3 relative to β-actin (bottom panels). (C) Confocal immunofluorescent images (×800 magnification) of co-localization of chromatin foci with H3K9me3 (red) and HP1γ (green). DAPI staining (blue) was used to visualize DNA foci. The fluorescence intensity was quantified using ImageJ software (right panels). Arbitrary intensity values for H3K9me3 (red), HP1γ (green), and DAPI (blue) are shown relative to the reference line (white) used for the analysis. (D) Cells were treated with or without MG132 (10 μM) for 6 h. Cell lysates were visualized by immunoblotting (upper panel). The graph shows the quantification of each protein relative to β-actin (bottom panel). The values indicate means ± SEM. Bars that do not share a common letter (a, b, c, or d) are significantly different between groups at P < 0.05.
Phosphorylation of S392 on p53 enhances the interactions between p53, MDM2, and SUV39h1, as well as the nuclear export of SUV39h1
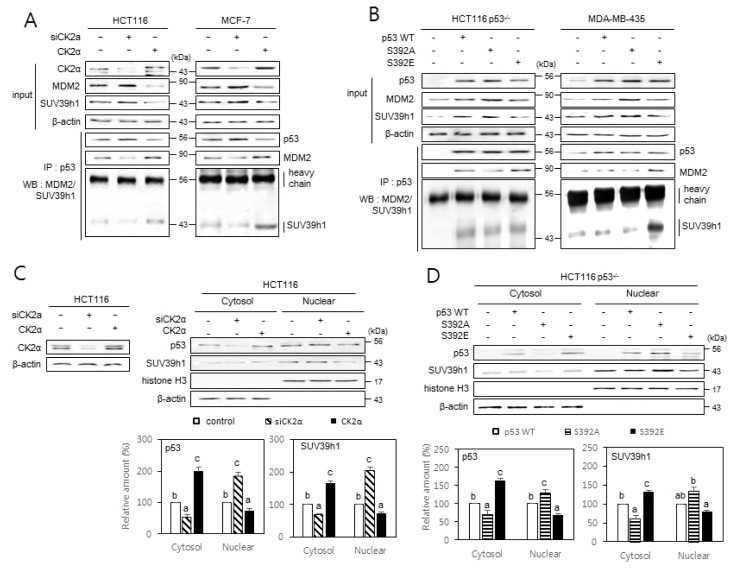
To determine whether CK2 regulates the interactions between p53, SUV39h1, and MDM2 (a ubiquitin ligase for SUV39h1) proteins, we carried out immunoprecipitation experiments. When compared to control cells, less SUV39h1 and MDM2 were co-precipitated with p53 in CK2α-downregulated cells, whereas increased amounts of SUV39h1 and MDM2 were co-precipitated with p53 in CK2α-upregulated cells (Fig. 5A). To investigate the effect of S392 phosphorylation on the interactions between p53, MDM2, and SUV39h1, p53 variants were immunoprecipitated from p53-null cells overexpressing either p53S392A or p53S392E. The results show that SUV39h1 and MDM2 preferentially co-precipitated with p53S392E, rather than with p53S392A (Fig. 5B). To determine whether CK2 regulates the nuclear localization of p53 and SUV39h1, we isolated the cytoplasm and nuclei from cells downregulating or upregulating CK2. More accumulation of p53 and SUV39h1 was detected in nuclear extracts than in cytosolic extracts of cells downregulating CK2α. Conversely, a greater accumulation of p53 and SUV39h1 was observed in cytosolic extracts than in nuclear extracts of cells overexpressing CK2α (Fig. 5C). Similarly, we examined the effect of S392 phosphorylation on the nuclear localization of p53 and SUV39h1. More accumulation of p53 and SUV39h1 was detected in nuclear extracts rather than in cytosolic extracts of HCT116 p53−/− cells overexpressing p53S392A. In contrast, greater accumulation of p53 and SUV39h1 was observed in cytosolic extracts rather than in nuclear extracts of HCT116 p53−/− cells overexpressing p53S392E (Fig. 5D). We next examined whether MDM2 protein levels were decreased in cells downregulating CK2 or overexpressing p53S392A. We found that MDM2 expression is stimulated in cells downregulating CK2 or overexpressing p53S392A when compared with levels in either control cells, wild-type p53-expressing cells, or p53S392E-expressing cells (Supplementary Fig. S2). Taken together, these results suggest that dephosphorylation of S392 on p53 reduces the nuclear export of SUV39h1 by inhibiting the interactions between p53, MDM2, and SUV39h1.
Fig. 5. The effect of p53 S392 phosphorylation on the interactions between p53, MDM2, and SUV39h1, and the nuclear localization of SUV39h1.
Cells were treated with CK2α siRNA or pcDNA3.1-CK2 (A and C) or with pcDNA3.1-p53 variants (B and D). (A and B) The inhibitory effect of CK2 downregulation on the interactions between p53, MDM2, and SUV39h1 (A) and the induction of p53, MDM2, and SUV39h1 interactions by p53S392E relative to wild-type (WT) p53 and p53S392A (B). Cell lysates were immunoprecipitated (IP) with anti-p53 antibodies followed by immunoblotting with anti-MDM2 and SUV39h1 antibodies. Cell lysate IP with IgG heavy chain served as a loading control. The same membranes were reprobed with anti-p53 antibodies and TrueBlot reagent, which eliminates the detection of the immunoprecipitated antibodies. Representative data from three independent experiments are shown. (C and D) The inhibitory effect of CK2 upregulation on the nuclear localization of p53 and SUV39h1 (C) and the induction of SUV39h1 nuclear localization by p53S392A relative to WT p53 and p53S392E (D). Cytoplasm and nuclei were isolated from p53-null cells overexpressing variants of p53 and were visualized by immunoblotting. β-actin (cytoplasmic marker) and histone H3 (nuclear marker) were quantified as loading controls (upper panels). The levels of CK2α protein in whole cells extracts are shown (left on the C panel). The graph shows the quantification of p53 and SUV39h1 relative to the subcellular markers (bottom panels). The values indicate mean ± SE. Bars that do not share a common letter (a, b, or c) are significantly different between groups at P < 0.05.
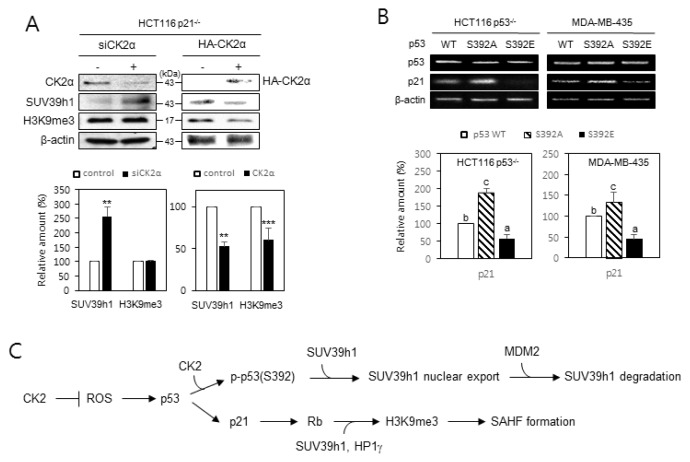
p21Cip1/WAF1 is required for CK2 downregulation-mediated H3K9me3, and dephosphorylation of S392 on p53 is important for efficient transcription of p21Cip1/WAF1
Since p21Cip1/WAF1, a downstream regulator of p53, is necessary for the senescence induced by CK2 silencing (Kang et al., 2009), we examined the role of p21Cip1/WAF1 in SUV39h1 expression and H3K9me3 during CK2 downregulation-mediated SAHFs formation. To investigate the impact of p21Cip1/WAF1 on CK2 downregulation-induced H3K9me3, we used HCT116 p21−/− cells. Knocking down CK2α elevated both SUV39h1 expression and H3K9me3 in HCT116 cells containing wild-type p21Cip1/WAF1 (Fig. 1C). In HCT116 p21−/− cells, however, knocking down CK2α increased the expression of SUV39h1, but H3K9me3 remained unchanged, suggesting that p21Cip1/WAF1 acts upstream of H3K9me3. Ectopic expression of CK2α reduced the amounts of both SUV39h1 and H3K9me3 in HCT116 p21−/− cells, suggesting that enhanced CK2 activity negatively regulates H3K9me3 by reducing SUV39h1 expression (Fig. 6A). We next examined whether the phosphorylation status of S392 on p53 affected mRNA levels of p21Cip1/WAF1. When compared to cells expressing wild-type p53, ectopic expression of p53S392A resulted in increased amounts of p21Cip1/WAF1 mRNA, whereas ectopic expression of p53S392E decreased the expression of p21Cip1/WAF1 (Fig. 6B). Collectively, these data indicate that CK2 regulates H3K9me3 in a p21Cip1/WAF1-dependent manner, and dephosphorylation of S392 on p53 is necessary for efficient transcription of p21Cip1/WAF.
Fig. 6. p21Cip1/WAF1 is required for CK2 downregulation-mediated H3K9me3, and dephosphorylation of S392 on p53 is important for efficient transcription of p21Cip1/WAF.
(A) p21Cip1/WAF1 is essential in CK2 downregulation-mediated induction of H3K9me3, but not for the induction of SUV39h1. Immunoblotting analysis was conducted on HCT116 p21−/− cells treated with CK2α siRNA or pcDNA3.1-HA-CK2α (upper panel). The graph shows the quantification of each protein relative to the levels of β-actin (bottom panel). The values indicate mean ± SEM. **P < 0.01, ***P < 0.001. (B) Induction of p21Cip1/WAF1 mRNA levels by p53S392A relative to wild-type (WT) p53 or p53S392E. RT-PCR analyses were performed using specific primers for p53 and p21Cip1/WAF1 (upper panel). The graph shows the quantification of p21Cip1/WAF1 mRNA levels relative to those of β-actin (bottom panel). The values indicate mean ± SE. Bars that do not share a common letter (a, b, or c) were significantly different between groups at P < 0.05. (C) A model possibly illustrating CK2 downregulation-induced SAHFs formation. CK2 downregulation induces not only p53 accumulation through increased ROS, but also dephosphorylation of S392 on p53. This process elevates H3K9me3 by impeding the interactions between p53, MDM2, and SUV39h1, which causes the nuclear import and subsequent stabilization of SUV39h1, and by enhancing p21Cip1/WAF1 expression, which results in Rb activation and subsequent recruitment of SUV39h1 and HP1γ.
DISCUSSION
CK2 downregulation induces H3K9me3 and SAHFs formation by activating the H3K9 trimethylase SUV39h1 and inactivating H3K9 dimethylases G9a and GLP (Park et al., 2018). Since CK2 downregulation induces premature senescence in human cells through the PI3K-AKT-mTOR-ROS-p53 pathway (Jeon et al., 2010; Kang et al., 2009; Park et al., 2013), we investigated the potential role of this pathway in CK2 downregulation-mediated expression of SUV39h1 and H3K9me3. Our present study demonstrates that the PI3K-AKT-mTOR-ROS pathway is associated with CK2 downregulation-induced SUV39h1 overexpression, H3K9me3, and SAHFs formation (Figs. 1A and 1B, Supplementary Fig. S1). In addition, this study demonstrates that p53 is also required for H3K9me3, SUV39h1 upregulation, and SAHFs formation in cells where CK2 is silenced (Figs. 1C, 1D, and 2A). Consistently, treatment with the p53 activator, nutlin-3 (0.5 μM), increased SUV39h1 protein level and H3K9me3 (Figs. 2B and 2C). Therefore, the present results collectively suggest that CK2 downregulation induces H3K9me3 and SAHFs formation through upregulation of p53. Moreover, we showed here that higher concentrations (1 to 4 μM) of nutlin-3 reduce SUV39h1 protein levels and H3K9me3, indicating that the effect of nutlin-3 resembles an inverted U-shaped dose–response relationship between p53 and SUV39h1 amounts in cells (Figs. 2B and 2C). This study is consistent with a previous report demonstrating that p53 upregulation induced by nutlin-3 (20 μM) leads to decreased SUV39h1 protein levels and H3K9me3 (Mungamuri et al., 2012). Based on these data, we propose that SUV39h1 expression is tightly regulated by p53 concentration; therefore, maintaining p53 levels within a narrow range is required for H3K9me3.
How does p53 regulate the amounts of SUV39h1 protein? Our recent study showed that CK2 does not affect SUV39h1mRNA levels (Park et al., 2018). The present study, which used the protein synthesis inhibitor cycloheximide and a proteasome inhibitor, shows that CK2 downregulation stabilizes SUV39h1 post-translationally by inhibiting proteasomal degradation in a p53-dependent manner (Fig. 3). In addition, the present study indicates that qualitative control of p53 is also associated with CK2 downregulation-induced H3K9me3, where p53 S392 is a possible CK2 target site (Kim et al., 2004; Meek, 1999) (Fig. 4A). Here, we demonstrate that ectopic expression of p53S392A accelerates the induction of SUV39h1, H3K9me3, and SAHFs formation, relative to expression of wild-type p53 (Figs. 4B and 4C). Furthermore, interactions between p53, MDM2, and SUV39h1, and the nuclear export of SUV39h1 were reduced by CK2 downregulation and ectopic expression of p53S392A, relative to CK2 upregulation and wild-type p53, respectively. Conversely, interactions between p53, MDM2, and SUV39h1, and the nuclear export of SUV39h1 were accelerated by CK2 upregulation and ectopic expression of p53S392E, when compared with control cells or wild-type p53 (Fig. 5). It has been previously reported that MDM2 ubiquitinates both SUV39h1 and p53 and causes their proteasomal degradation (Kruse and Gu, 2009; Mungamuri et al., 2016; Wade et al., 2013). p53 contains a nuclear localization signal in its C-terminus, whereas the nuclear export of p53 is mediated by MDM2 binding. In addition, SUV39h1 binds to the MDM2 acidic domain, possibly forming a p53-MDM2-SUV39h1 complex (Cross et al., 2011; Mungamuri et al., 2012). Because CK2 downregulation or ectopic expression of p53S392A reduces SUV39h1 ubiquitination and the interactions between p53, MDM2 and SUV39h1, we speculate that MDM2 decreases in cells downregulating CK2 or overexpressing p53S392A. However, the present study shows that MDM2 expression increases in cells downregulating CK2 or overexpressing p53S392A (Supplementary Fig. S2). This effect is more pertinent because MDM2 is upregulated by p53 (Bieging et al., 2014; Kruse and Gu, 2009; Wade et al., 2013). Therefore, the previous and present studies collectively suggest that CK2 downregulation-induced dephosphorylation of S392 on p53 promotes H3K9me3 by inhibiting interactions between p53, MDM2, and SUV39h1. This results in the nuclear import and subsequent stabilization of SUV39h1 (Fig. 6C).
p21Cip1/WAF1 is a key mediator of p53-dependent cell cycle arrest by inhibiting of cyclin-dependent kinases that phosphorylate Rb (Harper et al., 1993). Rb interacts with the heterochromatin protein HP1γ and associates with SUV39h1, which mediates H3K9me3 accumulation (Ait-Si-Ali et al., 2004; Nielsen et al., 2001). Our previous study showed that p21Cip1/WAF1 expression and hypophosphorylated forms of Rb increase during CK2 downregulation-induced senescence (Kang et al., 2009). In this study, we report not only that p21Cip1/WAF1 is involved in CK2 downregulation-mediated induction of H3K9me3, but also that it is not involved in the expression of SUV39h1 (Fig. 6A). In addition, ectopic expression of p53S392A accelerates the expression of p21Cip1/WAF1, relative to wild-type p53. Conversely, ectopic expression of p53S392E decreases the expression of p21Cip1/WAF1, relative to wild-type p53 (Fig. 6B). The previous and present studies, therefore, suggest that CK2 downregulation-mediated p53 S392 dephosphorylation status promotes SAHFs formation through the enhanced nuclear import of p53 and subsequent p21Cip1/WAF1 expression. This could result in Rb activation and subsequent recruitment of SUV39h1 and HP1γ (Fig. 6C).
In summary, CK2 downregulation stabilizes SUV39h1 by inhibiting ubiquitination in a p53-dependent manner. CK2 downregulation-mediated dephosphorylation of S392 on p53 represses the interactions between p53, MDM2, and SUV39h1, allowing the nuclear import and stabilization of SUV39h1. Since senescence acts as an important tumor suppression mechanism, elucidating the molecular mechanism by which p53 S392 phosphorylation status modulates SAHFs formation will extend our understanding of the role of p53 post-translational modifications in carcinogenesis and cancer prevention.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning (NRF-2015R1A2A2A01004593 and NRF-2019R1A2C1005219).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ait-Si-Ali S., Guasconi V., Fritsch L., Yahi H., Sekhri R., Naguibneva I., Robin P., Cabon F., Polesskaya A., Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegue L., Raurell-Vila H., Marazuela-Duque A., Kane-Goldsmith N., Valle A., Oliver J., Serrano L., Vaquero A. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol Cell. 2011;42:210–223. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Cross B., Chen L., Cheng Q., Li B., Yuan Z.M., Chen J. Inhibition of p53 DNA binding function by the MDM2 protein acidic domain. J Biol Chem. 2011;286:16018–16029. doi: 10.1074/jbc.M111.228981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch L., Robin P., Mathieu J.R., Souidi M., Hinaux H., Rougeulle C., Harel-Bellan A., Ameyar-Zazoua M., Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Funayama R., Ishikawa F. Cellular senescence and chromatin structure. Chromosoma. 2007;116:431–440. doi: 10.1007/s00412-007-0115-7. [DOI] [PubMed] [Google Scholar]
- Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-G. [DOI] [PubMed] [Google Scholar]
- Hublitz P., Albert M., Peters A.H. Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol. 2009;53:335–354. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- Jeon S.M., Lee S.J., Kwon T.K., Bae Y.S. NADPH oxidase is involved in protein kinase CKII down-regulation-mediated senescence through elevation of the level of reactive oxygen species in human colon cancer cells. FEBS Lett. 2010;584:3137–3142. doi: 10.1016/j.febslet.2010.05.054. [DOI] [PubMed] [Google Scholar]
- Kang J.Y., Kim J.J., Jang S.Y., Bae Y.S. The p53-p21(Cip1/WAF1) pathway is necessary for cellular senescence induced by the inhibition of protein kinase CKII in human colon cancer cells. Mol Cells. 2009;28:489–494. doi: 10.1007/s10059-009-0141-9. [DOI] [PubMed] [Google Scholar]
- Kim J.A. Cooperative instruction of signaling and metabolic pathways on the epigenetic landscape. Mol Cells. 2018;41:264–270. doi: 10.14348/molcells.2018.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.Y., Park B.J., Kim D.J., Kim W.H., Kim S., Oh K.S., Lim J.Y., Kim J., Park C., Park S.I. Modification of serine 392 is a critical event in the regulation of p53 nuclear export and stability. FEBS Lett. 2004;572:92–98. doi: 10.1016/j.febslet.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kruse J.P., Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- Meek D.W. Mechanisms of switching on p53: a role for covalent modification? Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- Mungamuri S.K., Benson E.K., Wang S., Gu W., Lee S.W., Aaronson S.A. p53-Mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol. 2012;19:478–484. doi: 10.1038/nsmb.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungamuri S.K., Qiao R.F., Yao S., Manfredi J.J., Gu W., Aaronson S.A. USP7 enforces heterochromatinization of p53 target promoters by protecting SUV39H1 from MDM2-mediated degradation. Cell Rep. 2016;14:2528–2537. doi: 10.1016/j.celrep.2016.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- Nielsen S.J., Schneider R., Bauer U.M., Bannister A.J., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Park J.H., Kim J.J., Bae Y.S. Involvement of PI3K-AKT-mTOR pathway in protein kinase CKII inhibition-mediated senescence in human colon cancer cells. Biochem Biophys Res Commun. 2013;433:420–425. doi: 10.1016/j.bbrc.2013.02.108. [DOI] [PubMed] [Google Scholar]
- Park J.W., Kim J.J., Bae Y.S. CK2 downregulation induces senescence-associated heterochromatic foci formation through activating SUV39h1 and inactivating G9a. Biochem Biophys Res Commun. 2018;505:67–73. doi: 10.1016/j.bbrc.2018.09.099. [DOI] [PubMed] [Google Scholar]
- Roninson I.B. Tumor cell senescence in cancer treatment. Cancer Res. 2003;63:2705–2715. [PubMed] [Google Scholar]
- Ryu S.W., Woo J.H., Kim Y.H., Lee Y.S., Park J.W., Bae Y.S. Downregulation of protein kinase CKII is associated with cellular senescence. FEBS Lett. 2006;580:988–994. doi: 10.1016/j.febslet.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Simabuco F.M., Morale M.G., Pavan I.C.B., Morelli A.P., Silva F.R., Tamura R.E. p53 and metabolism: from mechanism to therapeutics. Oncotarget. 2018;9:23780–23823. doi: 10.18632/oncotarget.25267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A., Scher M., Erdjument-Bromage H., Tempst P., Serrano L., Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Wade M., Li Y.C., Wahl G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.