Abstract
Oncogenic gain-of-function mutations are clinical biomarkers for most targeted therapies, as well as represent direct targets for drug treatment. Although loss-of-function mutations involving the tumor suppressor gene, STK11 (LKB1) are important in lung cancer progression, STK11 is not the direct target for anticancer agents. We attempted to identify cancer transcriptome signatures associated with STK11 loss-of-function mutations. Several new sensitive and specific gene expression markers (ENO3, TTC39C, LGALS3, and MAML2) were identified using two orthogonal measures, i.e., fold change and odds ratio analyses of transcriptome data from cell lines and tissue samples. Among the markers identified, the ENO3 gene over-expression was found to be the direct consequence of STK11 loss-of-function. Furthermore, the knockdown of ENO3 expression exhibited selective anticancer effect in STK11 mutant cells compared with STK11 wild type (or recovered) cells. These findings suggest that ENO3-based targeted therapy might be promising for patients with lung cancer harboring STK11 mutations.
Keywords: Enolase 3, lung adenocarcinoma, STK11 loss-of-function mutation
INTRODUCTION
Non-small-cell lung carcinoma (NSCLC) is a type of epithelial lung cancer, constituting ~80% to 90% of all lung cancers (Novello et al., 2016; Planchard et al., 2018). NSCLC is classified into three types: lung adenocarcinoma (LUAD), squamous cell carcinoma, and large cell carcinoma. LUAD accounts for approximately 30% of lung cancers (Gill et al., 2011). Serine/threonine kinase 11 (STK11), also known as liver kinase B1 (LKB1), is a major tumor suppressor gene in lung cancers. Particularly, STK11 is the most inactivated tumor suppressor gene in NSCLC (Carretero et al., 2004; Chen et al., 2016; Facchinetti et al., 2017). Although STK11 inactivation (i.e., loss-of-function mutation) occurs in ~30% of LUAD (Kim et al., 2016), its role in cancer progression has yet to be elucidated (Gao et al., 2010).
In previous studies, we found target genes and drugs with STK11 mutation-specific anticancer efficacy in lung cancer cell lines (He et al., 2014; Kim et al., 2016). The expression of phosphodiesterase-4D (PDE4D) was found to be upregulated in both lung cancer cell lines and tissue samples harboring STK11 homogeneous mutations. Knockdown of PDE4D gene expression and chemical inhibition of PDE4D protein enhanced the anticancer efficacy in STK11-mutant cell lines compared with other lung cancer cell lines (He et al., 2014). Drugs inhibiting ATPase Na+/K+ Transporting Subunit Alpha 1 (ATP1A1) also showed selective efficacy in STK11-mutant lung cancer cell lines (Kim et al., 2016). In the present study, we systematically analyzed the cancer transcriptome to further identify STK11-specific gene expression signatures in lung cancers. We compared the gene expression profile of STK11 loss-of-function cells (homozygous mutations) and STK11 wild type cells from the LUAD cell line panel and tissue samples. The expression of several genes exhibited significant sensitivity and specificity to the STK11 mutation. Particularly, the expression of Enolase 3 (ENO3) was altered by the loss or recovery of STK11 gene expression.
Enolase (ENO), also known as phosphopyruvate dehydratase, is a metalloenzyme that catalyzes the transformation of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP) during glycolysis (Ho et al., 2010). The three isoforms of ENOs in mammalian cells include: α or non-neuronal enolase (NNE), γ or neuron-specific enolase (NSE), and β or muscle-specific enolase (MSE) (Muller et al., 2012; Peshavaria and Day, 1991). They are cytoplasmic enzymes involved in glycolysis and gluconeogenesis (Lung et al., 2017). Specially, the expression of ENO1 was highlighted in small-cell lung carcinoma (SCLC) tissues, suggesting its association with cancer cell migration (Ho et al., 2010; Liu and Shih, 2007). The increased expression of ENO2 gene has also been reported in NSCLC (Ho et al., 2010; Isgrò et al., 2015). However, the role and function of ENO3 in cancer have yet to be elucidated. Here, we present the results of ENO3 regulation by the STK11 gene expression via STK11 mutation-specific transcriptome analysis. The therapeutic potential of ENO3 as the STK11 mutation-specific target was also investigated. This study extends our knowledge of STK11 mutation-driven cancer progress, and contributes to the development of new targeted therapies.
MATERIALS AND METHODS
Data acquisition
DNA microarray gene expression data of 47 LUAD cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012). mRNA expression levels obtained using Affymetrix U133 plus 2.0 arrays were represented as log2-transformed value after robust multichip average (RMA) normalization. The expression ranges are from 3 to 15 and the average expression is 7.5. Among 54,613 gene probes, the data available for the 15,080 gene probes in > 50% of selected LUAD cell lines were used in the present analysis. STK11 loss-of-function cell lines were defined by analyzing the accompanying exome sequencing data (Supplementary Table 1). For the analysis of STK11-dependent gene expression, we retrieved the DNA microarray data of two STK11 mutant, parental A549 cell lines and two STK11-recovered A549 cell lines (GSE6135) from Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (Clough and Barrett, 2016). Gene expression data were obtained using Agilent Homo sapiens 44k custom array and represented as log2 scale after normalization by locally weighted scatterplot smoothing (LOWESS). In addition, 230 patient tissue-based gene expression and exome sequencing data of LUAD were obtained from The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research Network et al., 2013). The RNA-seq expression level of each gene was represented with log2-transformed through upper quartile normalized RSEM (RNA-Seq by Expectation Maximization).
Sensitivity and specificity
To identify differentially expressed genes between STK11 mutant and wild-type cell lines, we used fold change and t-test P values. The fold change is defined as the difference between the averages of two compared groups. For statistical significance, we calculated P values from the t-statistic. To determine the selective association between gene expression and STK11 mutation, we also calculated the enrichment score using the odds ratio between the observed odds and the expected odds. The observed and the expected odds are defined as the ratio of cell lines with over threshold (fold change > 1) among STK11 mutant LUAD and among all LUAD cell lines, respectively. The statistical confidence of an odds ratio was calculated using Fisher’s exact test. Statistical analysis was performed using Perl (ver. 5.30.0; http://www.perl.org/).
Cell-based in vitro assays
A549, H23, H1993, and H322M cells were obtained from the National Institutes of Health, National Cancer Institute (NCI, USA). HCC-827 and H1975 cells were obtained from the American Type Culture Collection (ATCC, USA). The STK11-recovered cells from three parental cell lines (A549-STK11, H23-STK11, and H1993-STK11) were established in our laboratory (Kim et al., 2016). STK11 mutant and wild-type cell lines were cultured in RPMI 1640 (HyClone Laboratories, USA) supplemented with 10% fetal bovine serum (HyClone Laboratories) and 1% antibiotics (Thermo Fisher Scientific, USA) according to institutional laboratory safety guidelines. The STK11-recoverd cell lines were cultured in the same medium after supplementation with 1 μg/ml puromycin. The cells were maintained at 37°C in a 5% CO2 incubator and subcultured at 80% to 90% confluence. The media were changed every 2 to 3 days. A total of 1 to 3 × 105 cells per well were seeded on a 6-well culture plate for monolayer cell cultures for 3 days.
For siRNA transfection, we experimented with 10 nmol/L target siRNAs of NC (negative control), PLK1 (positive control), and ENO3 (Thermo Fisher Scientific, On-Target Plus SmartPool; GE Dharmacon, USA) (Song et al., 2017). Cells were seeded in a 96-well plate (Corning, USA) at an optimized density of 2,000 to 8,000 cells/well. After 72 h of seeding, the viability was measured using the CellTiter-Blue for Cell Viability Assay (Promega, USA). Simultaneously, cells were stained with DAPI and single cells were counted with Gen5TM software (BioTek Instruments, USA).
Quantitative real-time polymerase chain reaction analysis
Total RNA was extracted in 6-well plates using TRIzol. Synthesis of cDNA and polymerase chain reaction (PCR) amplification were carried out with the SuperScript One-step RT-PCR Platinum Taq Kit (Invitrogen, USA). The gene expression of ENO1, ENO2, and ENO3 was quantified via real-time PCR using specific primer sets targeting ENO1 (Hs00157360_m1, TaqMan), ENO2 (Hs00361415_m1, TagMan), and ENO3 (Hs01093275_m1, TaqMan), respectively, and the reference gene GAPDH (Hs02786624_g1, TagMan) (Ali et al., 2015; Liu et al., 2005). Quantitative real-time PCR (RT-qPCR) were carried out on an Applied Biosystems 7500 (Thermo Fisher Scientific) using a TaqMan real-time detection protocol. The delta cycle threshold (dCt) corresponds to the cycle threshold (Ct) values for ENO1, ENO2, and ENO3 RNA expression normalized to those for RNA expression of the internal control GAPDH. For intuitive interpretation, -dCt is used so as that the higher values indicate a higher normalized target gene expression.
RESULTS AND DISCUSSION
Analysis of STK11-associated gene expression in LUAD cell lines
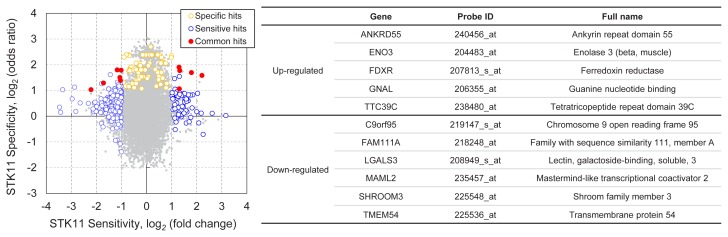
DNA microarray data of 47 LUAD cell lines were analyzed to identify the gene expression markers associated with STK11 mutations (Fig. 1). We compared the differences in gene expression between nine cell lines with STK11 loss-of-function mutations (Supplementary Table 1) and 38 STK11 wild-type cell lines based on fold change and odds ratio. The fold change reflects the sensitivity of the gene expression in STK11 mutant cells against STK11 wild-type cells, while the odds ratio quantifies the specificity (i.e., exclusiveness) of a gene expression in STK11-mutant cell lines compared with other cell lines (see Materials and Methods section for details). As a result, 222 gene probes (sensitive hits) were significantly (P < 0.01) over- or under-expressed (> 2-fold change) in STK11 mutation cells in LUAD, while the expression of 71 gene probes (specific hits) exhibited significantly (P < 0.01) exclusive expression (> 2-odds ratio). Since no general correlation was observed between sensitivity and specificity of gene expression, the selection of common hits based on these two measures provided unique gene expression signatures (or biomarkers) of STK11 mutant cancer cells. A total of 11 gene probes satisfied both sensitivity and specificity of the expression associated with STK11 loss-of-function mutations (Fig. 1). These 11 genes were identified from diverse functional classes and their functional or signaling aspects have never been reported in connection with the suppressive role of STK11 in the cancer progression.
Fig. 1. Analysis of gene expression associated with STK11 mutations.
A total of 15,080 gene probes were plotted to determine STK11 sensitive vs. STK11 specific expression in the CCLE dataset. The sensitivity (i.e., log2 fold-change) was determined based on the gene expression ratio in nine STK11-mutant cell lines compared with 38 STK11 wild-type cell lines. The specificity (i.e., log2 odds ratio) of a gene expression was based on the ratio of the observed odds in STK11-mutant cell lines over the expected odds in all cell lines. Orange circles represent the 71 STK11-specific gene probes with > 2 odds ratio (Fisher’s exact test, P < 0.01) in STK11-mutant cell lines. Blue circles represent the 222 STK11-sensitive gene probes with > 2 fold change (Student’s t-test, P < 0.01) in STK11-mutant cell lines. Filled red circles represent 11 common hits of specific (orange) and sensitive (blue) gene probes.
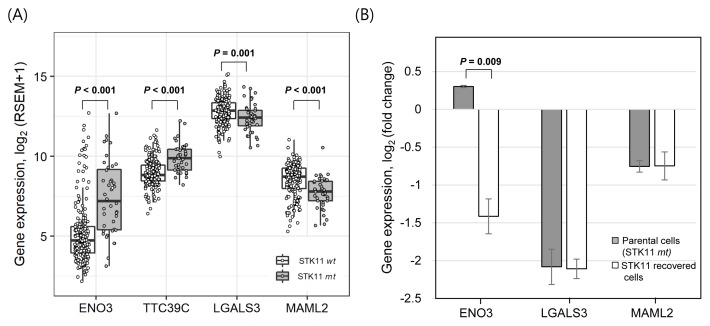
We further investigated the association of these 11 gene expression profiles associated with STK11 mutations using the RNA sequencing data of patient-derived tumor tissue samples. The gene expression was statistically compared between 40 STK11-mutant and 190 wild-type LUAD samples from the TCGA database (see Materials and Methods section). Among the 11 genes selected from the analysis of cell line data in Figure 1, the expression of ENO3 and TTC39C genes was found to be significantly up-regulated in STK11 mutant tissue samples (Fig. 2A). Down-regulation of LGALS3 and MAML2 genes in STK11-mutant cell lines was also confirmed in patient-derived LUAD samples. Particularly, the differential expression of the ENO3 gene in STK11-mutant and wild type samples was greater than 2-fold in tissue samples, displaying a minimal level of the ENO3 gene expression in most of the STK11 wild-type LUAD samples.
Fig. 2. STK11-specific expression of selected genes in LUAD tissue samples and STK11-recovered model cell line.
(A) Differential expression of genes plotted using LUAD tissue samples retrieved from TCGA. A total of 230 samples were classified into STK11 mutants (40 samples) and wild types (190 samples), and their gene expression (log2-transformed upper quartile values) was analyzed. Student’s t-test for LGALS3 and Wilcoxon rank-sum test for others were used to test statistical significance. (B) Differential gene expression was analyzed in STK11-mutant A549 cells and STK11-recovered A549 cells retrieved from GEO (GSE6135). Student’s t-test was used to test statistical significance. Error bars indicate SD.
STK11-dependent ENO3 gene expression
The gene expression of ENO3, TTC39C, LGALS3, and MAML2 was consistently up-or down-regulated in both STK11-mutant cell lines and tissue samples of LUAD. We thus investigated the changes in their gene expression based on the recovery of STK11 function in the STK11-mutant LUAD cell line (A549). DNA microarray data of STK11-mutant (loss-of-function) A549 and STK11-recovered A549 cell lines were retrieved from the GEO database (GSE6135). Since the gene expression data of TTC39C were not available in the dataset, we only analyzed the differential expression of ENO3, LGALS3, and MAML2 in STK11-mutant and STK11-recovered A549 cells (Fig. 2B). While the expression of LGALS3 and MAML2 was not changed by STK11 functional recovery, the ENO3 expression was greatly reduced in STK11-recovered A549 cells compared with STK11-mutant A549 cells suggesting that the ENO3 over-expression in both STK11-mutant LUAD cell lines and tissue samples was a direct the consequence of STK11 loss-of-function.
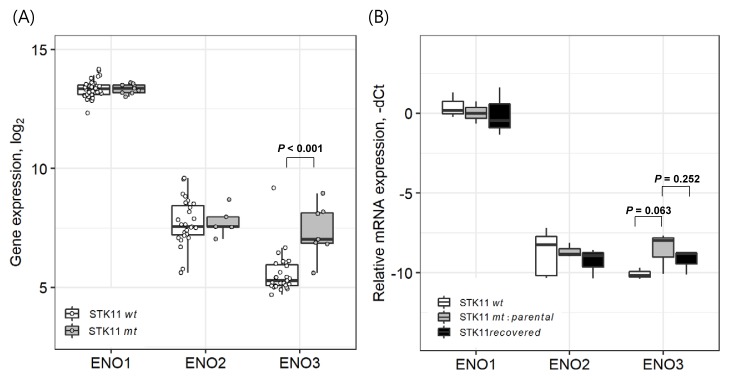
Although ENO1 and ENO2 have been widely studied in cancer context (Ho et al., 2010; Isgrò et al., 2015; Liu and Shih, 2007), the role of ENO3 in cancers has yet to be fully elucidated. In our analysis of CCLE dataset, the expression of ENO1 and ENO2 did not show any difference between STK11-mutant and -wild type cell lines (Fig. 3A). To further confirm the exclusive dependence of ENO3 overexpression on STK11 loss-of-function, we measured the mRNA levels of ENO1, ENO2, and ENO3 in diverse cell lines with/without STK11 function (Fig. 3B). As a result, only the expression of ENO3 exhibited exclusive sensitivity to STK11 functional status. In these qPCR experiments, cell lines harboring STK11 mutations (A549, H23, and H1993) displayed > 2-fold overexpression of the ENO3 mRNAs compared with STK11 wild-type cell lines (H322M, HCC827, and H1975). Recovery of STK11 function by the three STK11-mutant cell lines reduced the ENO3 gene expression, although the statistical significance was relatively weak (Fig. 3B). These qPCR validations strongly suggested that STK11 loss-of-function mutation directly induced the overexpression of ENO3 in LUAD cells.
Fig. 3. Expression of ENO gene family in DNA microarray data and qPCR experiment using LUAD cell lines.
(A) Comparison of ENO family gene expression (ENO1: 201231_s_at, ENO2: 201313_at, and ENO3: 204483_at) between nine STK11-mutant and 38 STK11 wild-type cell lines in the CCLE dataset. Wilcoxon rank-sum test for ENO3 and Student’s t-test for others were used to test statistical significance. (B) Experimental validation (qPCR) of ENO family gene expression in STK11 wide-type cell lines (H322M, HCC827, and H1975), STK11-mutant parental cell lines (A549, H23, and H1993) and STK11-restored cell lines. The expression level of GAPDH was observed to be varied within SD < 1 among 9 cell lines. Student’s t-test was used to test statistical significance. Error bars indicate SD.
Selective anticancer effect of ENO3 knockdown on STK11-mutant cancer cells
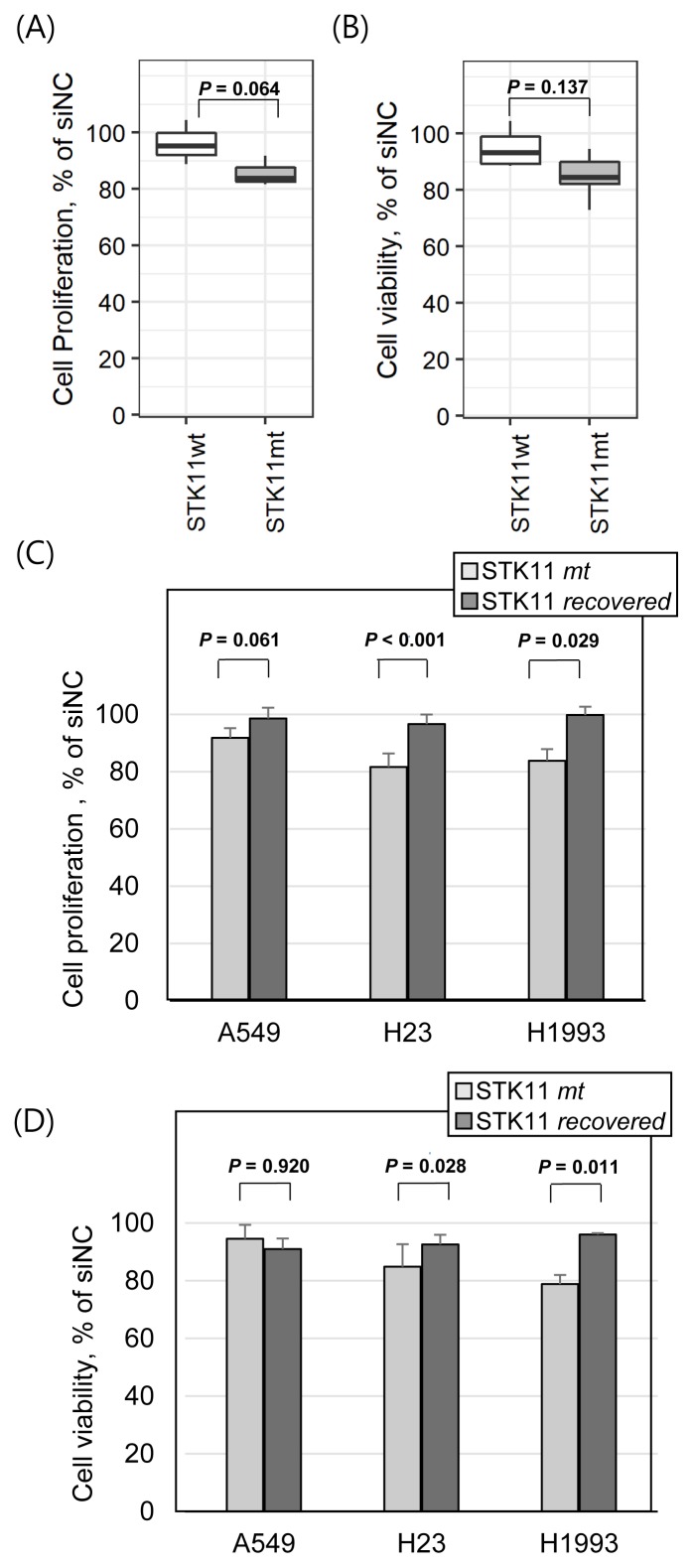
The observation of ENO3 overexpression under STK11 loss-of-function mutations implies that ENO3 might be a selective anticancer target in STK11-mutant cancer. We thus investigated the anticancer effect of ENO3 knockdown by siRNAs. In case of STK11 wild-type LUAD cell lines (H322M, HCC827, and H1975), the siENO3 treatment resulted in less than 5% inhibition of cell proliferation in 72 h in vitro assays (Fig. 4A). However, the anti-proliferative effect of siENO3 was increased on STK11-mutant cell lines (A549, H23, and H1993). The cell viability results were consistent with the cell proliferation assay (Fig. 4B). The viability of STK11-mutant cells was more sensitive to siENO3 treatment than STK11 wild-type cells. We further compared the efficacy of siENO3 in STK11-mutant cell lines compared with the corresponding cell lines with STK11 functional recovery (Figs. 4C and 4D). In both of cell proliferation and viability measures, STK11-recovered cells were less sensitive to siENO3 treatment than the parental mutant cells in H23 and H1993 cases. However, A549 cells showed inconsistent patterns between cell proliferation and viability assays, implying the existence of a cell line-specific effect of the STK11 recovery in cancer cells. Overall, these results in Figure 4 suggest that ENO3, induced by STK11 loss-of-function, might play a selective role in the development of targeted therapies against LUAD cell lines carrying STK11 mutations as an effective clinical biomarker.
Fig. 4. STK11 mutation-dependent anticancer efficacy of ENO3 gene knockdown.
Average variation in cell proliferation (A) and cell viability (B) of STK11 wild-type cell lines (H322M, HCC827, and H1975) and mutant cell lines (A549, H23, and H1993) by the ENO3 gene knockdown using siRNAs. Comparative changes in cell proliferation (C) and cell viability (D) in STK11-mutant parental cells and STK11 recovered cells by the ENO3 gene knockdown. Cell proliferation was measured based on the direct cell count in the well. Cell viability was measured using CellTiter Blue assay (see the Materials and Methods section for detail). All y-axes represent percentage of cell proliferation or cell viability divided by an average of negative control. Student’s one-tailed t-test was used to test statistical significance. Error bars indicate SD.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by grants from the National Research Foundation of Korea (KRF), including the Science Research Center Program (NRF-2016R1A5A1011974), and the Mid-career Researcher Program (NRF-2017R1A2B2007745 and NRF-2018R1A2B6009313), funded by the Korean government (MEST). This research was also financially supported by the Sookmyung Women’s University BK21 Plus Scholarship.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ali H., Du Z., Li X., Yang Q., Zhang Y.C., Wu M., Li Y., Zhang G. Identification of suitable reference genes for gene expression studies using quantitative polymerase chain reaction in lung cancer in vitro. Mol Med Rep. 2015;11:3767–3773. doi: 10.3892/mmr.2015.3159. [DOI] [PubMed] [Google Scholar]
- Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The Cancer Genome Atlas Pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero J., Medina P.P., Pio R., Montuenga L.M., Sanchez-Cespedes M. Novel and natural knockout lung cancer cell lines for the LKB1/STK11 tumor suppressor gene. Oncogene. 2004;23:4037–4040. doi: 10.1038/sj.onc.1207502. [DOI] [PubMed] [Google Scholar]
- Chen L., Engel B.E., Welsh E.A., Yoder S.J., Brantley S.G., Chen D.T., Beg A.A., Cao C., Kaye F.J., Haura E.B., et al. A sensitive NanoString-based assay to score STK11 (LKB1) pathway disruption in lung adenocarcinoma. J Thorac Oncol. 2016;11:838–849. doi: 10.1016/j.jtho.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough E., Barrett T. The gene expression omnibus database. In: Mathé E., Davis S., editors. Statistical Genomics. New York: Humana Press; 2016. pp. 93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti F., Bluthgen M.V., Tergemina-Clain G., Faivre L., Pignon J.P., Planchard D., Remon J., Soria J.C., Lacroix L., Besse B. LKB1/STK11 mutations in non-small cell lung cancer patients: descriptive analysis and prognostic value. Lung Cancer. 2017;112:62–68. doi: 10.1016/j.lungcan.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Gao Y., Xiao Q., Ma H., Li L., Liu J., Feng Y., Fang Z., Wu J., Han X., Zhang J., et al. LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proc Natl Acad Sci U S A. 2010;107:18892–18897. doi: 10.1073/pnas.1004952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R.K., Yang S.H., Meerzaman D., Mechanic L.E., Bowman E.D., Jeon H.S., Roy Chowdhuri S., Shakoori A., Dracheva T., Hong K.M., et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 2011;30:3784–3791. doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N., Kim N., Song M., Park C., Kim S., Park E.Y., Yim H.Y., Kim K., Park J.H., Kim K.I. Integrated analysis of transcriptomes of cancer cell lines and patient samples reveals STK11/LKB1-driven regulation of cAMP phosphodiesterase-4D. Mol Cancer Ther. 2014;13:2463–2473. doi: 10.1158/1535-7163.MCT-14-0297. [DOI] [PubMed] [Google Scholar]
- Ho J.A., Chang H.C., Shih N.Y., Wu L.C., Chang Y.F., Chen C.C., Chou C. Diagnostic detection of human lung cancer-associated antigen using a gold nanoparticle-based electrochemical immunosensor. Anal Chem. 2010;82:5944–5950. doi: 10.1021/ac1001959. [DOI] [PubMed] [Google Scholar]
- Isgrò M.A., Bottoni P., Scatena R. Neuron-specific enolase as a biomarker: biochemical and clinical aspects. In: Scatena R., editor. Advances in Cancer Biomarkers. Dordrecht, The Netherlands: Springer; 2015. pp. 125–143. [DOI] [PubMed] [Google Scholar]
- Kim N., Yim H.Y., He N., Lee C.J., Kim J.H., Choi J.S., Lee H.S., Kim S., Jeong E., Song M. Cardiac glycosides display selective efficacy for STK11 mutant lung cancer. Sci Rep. 2016;6:29721. doi: 10.1038/srep29721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D.W., Chen S.T., Liu H.P. Choice of endogenous control for gene expression in nonsmall cell lung cancer. Eur Respir J. 2005;26:1002–1008. doi: 10.1183/09031936.05.00050205. [DOI] [PubMed] [Google Scholar]
- Liu K.J., Shih N.Y. The role of enolase in tissue invasion and metastasis of pathogens and tumor cells. J Cancer Mol. 2007;3:45–48. [Google Scholar]
- Lung J., Chen K.L., Hung C.H., Chen C.C., Hung M.S., Lin Y.C., Wu C.Y., Lee K.D., Shih N.Y., Tsai Y.H. In silico-based identification of human α-enolase inhibitors to block cancer cell growth metabolically. Drug Des Devel Ther. 2017;11:3281–3290. doi: 10.2147/DDDT.S149214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F.L., Colla S., Aquilanti E., Manzo V.E., Genovese G., Lee J., Eisenson D., Narurkar R., Deng P., Nezi L. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novello S., Barlesi F., Califano R., Cufer T., Ekman S., Levra M.G., Kerr K., Popat S., Reck M., Senan S., et al. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27(Suppl 5):v1–v27. doi: 10.1093/annonc/mdw326. [DOI] [PubMed] [Google Scholar]
- Peshavaria M., Day I.N. Molecular structure of the human muscle-specific enolase gene (ENO3) Biochem J. 1991;275:427–433. doi: 10.1042/bj2750427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchard D., Popat S., Kerr K., Novello S., Smit E.F., Faivre-Finn C., Mok T.S., Reck M., Van Schil P.E., Hellmann M.D., et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(Suppl 4):iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- Song M., Lee H., Nam M.H., Jeong E., Kim S., Hong Y., Kim N., Yim H.Y., Yoo Y.J., Kim J.S., et al. Loss-of-function screens of druggable targetome against cancer stem-like cells. FASEB J. 2017;31:625–635. doi: 10.1096/fj.201600953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.