Abstract
When endoplasmic reticulum (ER) functions are perturbed, the ER induces several signaling pathways called unfolded protein response to reestablish ER homeostasis through three ER transmembrane proteins: inositol-requiring enzyme 1 (IRE1), PKR-like ER kinase (PERK), and activating transcription factor 6 (ATF6). Although it is important to measure the activity of ATF6 that can indicate the status of the ER, no specific cell-based reporter assay is currently available. Here, we report a new cell-based method for monitoring ER stress based on the cleavage of ATF6α by sequential actions of proteases at the Golgi apparatus during ER stress. A new expressing vector was constructed by using fusion gene of GAL4 DNA binding domain (GAL4DBD) and activation domain derived from herpes simplex virus VP16 protein (VP16AD) followed by a human ATF6α N-terminal deletion variant. During ER stress, the GAL4DBD-VP16AD(GV)-hATF6α deletion variant was cleaved to liberate active transcription activator encompassing GV-hATF6α fragment which could translocate into the nucleus. The translocated GV-hATF6α fragment strongly induced the expression of firefly luciferase in HeLa Luciferase Reporter cell line containing a stably integrated 5X GAL4 site-luciferase gene. The established double stable reporter cell line HLR-GV-hATF6α(333) represents an innovative tool to investigate regulated intramembrane proteolysis of ATF6α. It can substitute active pATF6(N) binding motif-based reporter cell lines.
Keywords: activating transcription factor 6, ER stress, GAL4 binding site, luciferase, regulated intramembrane proteolysis, reporter cell line, unfolded protein response
INTRODUCTION
The endoplasmic reticulum (ER) is a specialized cell organelle that plays important roles in the biosynthesis of many membrane proteins and secreted proteins, their posttranslational modifications (assembly, glycosylation, and disulfide bond), calcium storage and gated release, and production of lipids and sterols. When ER functions are perturbed, the ER induces several signaling pathways called unfolded protein response (UPR) to reestablish ER homeostasis (Back and Kaufman, 2012; Ron and Walter, 2007; Wang and Kaufman, 2016).
In UPR, three ER transmembrane proteins, inositol-requiring enzyme 1 (IRE1), PKR-like ER kinase (PERK, EIF2AK3), and activating transcription factor 6 (ATF6), serve as proximal transducers of UPR pathways (Hetz et al., 2011; Ron and Walter, 2007; Rutkowski and Kaufman, 2007; Wek and Cavener, 2007). IRE1 has ER-stress regulated kinase and endonuclease activities that can initiate unconventional splicing of mRNA encoding basic leucine zipper transcription factor X-box binding protein 1 (XBP1) (Hetz and Glimcher, 2009). IRE1-mediated unconventional cytoplasmic splicing removes a 26-nucleotide intron from unspliced Xbp1 mRNA to induce a translational frameshift, producing a fusion protein encoded from two evolutionarily conserved open reading frames (Ron and Walter, 2007). The fusion protein, XBP1s, acts as a potent transcription factor that can activate the expression of UPR target genes involved in protein folding and export from the ER, export and degradation of misfolded proteins, and lipid biosynthesis to resolve ER stress (Back and Kaufman, 2012; Ron and Walter, 2007). Therefore, measurement or observation of the extent of Xbp1 mRNA splicing is a good tool for monitoring IRE1 activation under ER stress condition. Several studies (Back et al., 2006; Iwawaki et al., 2004; Spiotto et al., 2010) have developed reporter systems for monitoring IRE1-mediated Xbp1 mRNA splicing, a partial sequence of Xbp1, including 26-nt ER stress-specific intron fused to the gene encoding firefly luciferase (FLuc) or green fluorescence protein (GFP). During stress, the spliced indicator mRNA is translated into an XBP1ΔC-Fluc or GFP fusion protein that can be detected by its luminescence or fluorescence.
Activated PERK can phosphorylate serine 51 on the α-subunit of heterotrimeric eukaryotic translation initiation factor 2 (eIF2) (Proud, 2005). Phosphorylation of eIF2α in eIF2 complex results in global attenuation of general mRNA translation while simultaneously enhancing the translation of selected mRNAs (such as activating transcription factor 4 [Atf4] mRNA) that contain short upstream open reading frames in their 5′ untranslated regions (5′UTRs) (Pakos-Zebrucka et al., 2016; Wek et al., 2006). Therefore, under ER stress condition, measurement of Atf4 5′UTR-dependet translational activity is a useful tool for monitoring the status of eIF2α phosphorylation. A reporter construct consisting of translational fusions of the Atf4 5′UTR and the luciferase coding region has been reported (Harding et al., 2000). Recently, a transgenic mouse model for monitoring translational activation of ATF4 in response to cellular stress has been developed (Iwawaki et al., 2017). However, in mammalian cells, there are three more eIF2α kinases (general control nonderepressible 2 [GCN2], protein kinase RNA-activated [PKR], and heme-regulated inhibitor kinase [HRI]) that can be activated by distinct stress stimuli (Pakos-Zebrucka et al., 2016; Wek et al., 2006), suggesting that Atf4 5′UTR-mediated translation enhancement does not always represent activation of PERK-dependent eIF2α phosphorylation pathway.
ATF6 is the third proximal transducer of UPR pathways that is encoded by two related genes, Atf6α and Atf6β (Yoshida et al., 1998). The C-terminal of ATF6 protrudes into the ER lumen whereas its N-terminal faces the cytosol. The N-terminal cytosolic part encompasses a transactivation domain and a basic leucine zipper region followed by a 20-amino acid transmembrane domain (Haze et al., 1999). Under normal conditions, the ER luminal domain of ATF6 maintains ATF6 in an inactive state by binding to BiP, a major molecular chaperone in the ER. Upon ER stress, BiP dissociation from the ER luminal domain of ATF6 exposes Golgi localization signals, evoking ATF6 translocation to the Golgi apparatus (Shen et al., 2002). In addition, ER stress can reduce the extent of intra- and inter-molecular disulfide bridges formed between the two conserved cysteine residues in the luminal domain (Nadanaka et al., 2007). These two molecular mechanisms can induce translocated ATF6 proteins to be cleaved by sequential actions of site 1 protease (S1P) and site 2 protease (S2P) at the Golgi apparatus (Shen and Prywes, 2004; Ye et al., 2000), resulting in liberation of a soluble and active transcription factor designated pATF6(N) (Yoshida et al., 2000; 2001). Thus, activation of ATF6 is characterized as one regulated intramembrane proteolysis (RIP). Liberated pATF6(N) transcription factor can enter the nucleus and bind to ER stress-response elements (ERSEs) of genes that encode functions in ER protein folding, endoplasmic reticulum-associated degradation (ERAD), protein secretion, and ER biogenesis (Back and Kaufman, 2012; Yoshida, 2007). Therefore, measurement of ATF6 activation can be another tool for monitoring ER stress. The most commonly utilized assay systems for monitoring ATF6α activation are luciferase reporter constructs driven by tandem copies of either a UPR element (UPRE) (Lee et al., 2002; Wang et al., 2000) or by BiP promoter which contains three ERSE sites (Gallagher et al., 2016; Yoshida et al., 1998). However, both UPRE and ERSE element sequences of these reporter constructs are a consensus DNA binding motif for other transcription factors such as spliced XBP1 or activated ATF6β that can be generated during ER stress (Bommiasamy et al., 2009; Lee et al., 2003; Yamamoto et al., 2007). Therefore, under ER stress, UPRE or BiP promoter-driven luciferase expression can occur by binding of not only activated ATF6α and β but also spliced XBP1 in cell-based assays using these reporter constructs, suggesting that such promoter-based reporter assay will not solely represent activation of ATF6α during ET stress. On the other hand, previous studies (Chen et al., 2002; Wang et al., 2000) have reported that full length ATF6α fused to GAL4 DNA binding domain (GAL4DBD) could activate 5X GAL4 site-luciferase reporter under ER stress. However, it was difficult to detect GAL4DBD-ATF6α protein in immunoblots using SV40 promoter (Wang et al., 2000). In addition, the expression of GAL4DBD-ATF6α protein should be regulated at low level to prevent activation of reporter gene in uninduced cells (Chen et al., 2002). As such, the development of a reporter cell line for monitoring activation of ATF6α under ER stress conditions has not been reported yet.
Thus, the objective of this study was to develop a luciferase reporter cell line that could monitor activation of ATF6α under ER stress conditions. We constructed a new expressing vector using fusion gene of GAL4DBD and activation domain derived from herpes simplex virus VP16 protein (VP16AD) followed by a human ATF6α N-terminal deletion variant that could generate a cytosolic and active transcription activator (encompassing GAL4DBD-VP16AD and cytosolic part of hATF6α) by S1P and S2P-mediated ATF6α cleavage under ER stress conditions. The GAL4DBD-VP16AD(GV)-hATF6α deletion variant expressing construct was stably integrated into HeLa Luciferase Reporter (HLR) cell line containing a stably integrated 5X GAL4 site-luciferase gene. The expressed GAL4DBD-VP16AD(GV)-hATF6α deletion variant was efficiently cleaved in the established double stable reporter cell line in response to ER stress inducers. The active transcription factor encompassing GAL4DBD-VP16AD and cytosolic part of hATF6α can translocate into the nucleus and strongly induce the expression of firefly luciferase gene in the HLR cell line.
MATERIALS AND METHODS
Construction of pCMV-GAL4DBD-VP16AD-hATF6α full or deletion variants
pCMV-GAL4DBD-VP16AD-hATF6α full or deletion variants were generated by inserting coding sequences that could express a fusion protein of the GAL4 DNA binding domain (GAL4DBD, amino acids 1–147) and the VP16AD (amino acids 413–490) followed by human ATF6α full length or deletion variants into pcDNA 3.1(+) vector (Invitrogen, USA). The coding sequence fragment of GAL4DBD-VP16AD was amplified from pM3-VP16 vector (Clontech Laboratories, USA) by polymerase chain reaction (PCR) with the following primers: 5′-GGGTTTGCTAGCTTCCTGAAAGATGAAG-3′ and 5′-GGGTTTGATATCCCCACCGTACTCGTCAATTC-3′. cDNAs of human ATF6α full length or deletion variants were obtained from pCGNATF6α full length (Zhu et al., 1997) generously provided by Dr. Ron Prywes (Columbia University, USA) by PCR with the following pairs. Forward primers included hATF6α (full, aa 1–670), 5′-GGGTTTGATATCATGGGGGAGCCGGCTGGGGTTG-3′; hATF6α (aa 93–670), 5′-GGGTTTGATATCCTTCCAGCCTCCTCAAGTTAT-3′; hATF6α (aa 307–670), 5′-GGGTTTGATATCCTAAGGAGACAGCAACGTATGAT-3′; hATF6α (aa 333–670), 5′-GGGTTTGATATCGGGTTAGAGGCGAGATTAAAGGC-3′; hATF6α (aa 362–670), 5′-GGGTTTGATATCGTTGTGTCAGAGAACCAGAG-3′; and hATF6α (aa 374–670): 5′-GGGTTTGATATCCTAAGGAGACAGCAACGTAT-3′. Common reverse primer was 5′-GGGTTTGCGGCCGCCTAAGCGTAATCTGGAACATCGTATGGGTATTGTAATG ACTCAGGGATGG-3′.
To construct pCMV-GAL4DBD-VP16AD-hATF6α full or deletion variants, pCMV-GAL4DBD-VP16AD was generated at first. GAL4DBD-VP16AD PCR fragment digested with NheI and EcoRV restriction enzymes was inserted into pcDNA 3.1(+) vector (Invitrogen) digested with the same restriction endonucleases. Next, pCMV-GAL4DBD-VP16AD-hATF6α full or deletion variants were constructed by inserting hATF6α full or deletion variants’ PCR fragments digested with EcoRV and NotI into pCMV-GAL4DBD-VP16AD plasmid treated with the same restriction enzymes. Clones were confirmed by sequencing.
Cell culture and cell lines
HLR cells (Agilent Technologies, USA) containing a stably integrated 5X GAL4 binding site of luciferase reporter gene were grown in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, USA), 1% glutamax (Invitrogen), 1% penicillin G/streptomycin (Invitrogen), and 0.1 mg/ml of Hygromycin B (Invitrogen).
To generate stably transformed HLR cells expressing GAL4DBD-VP16AD-hATF6αΔN(aa 333–670) under the control of CMV enhancer-promoter, HLR cells were transfected with SspI-digested pCMV-GAL4DBD-VP16AD-hATF6αΔN(aa 333–670) vector. Transformants were selected with both hygromycin B (0.1 mg/ml) and G418 (0.5 mg/ml) antibiotics. Antibiotics-resistant stable cell clones were screened for the activity of firefly luciferase after treatment with dithiothreitol (DTT, 2 mM) as an ER stress inducer as described below. Among several positive clones identified, one clone (#3) that showed the highest firefly luciferase activity after DTT treatment was chosen. Thus, in subsequent experiments, we used a clone named HLR-GV-hATF6α(333) expressing GAL4DBD-VP16AD(GV)-hATF6αΔN(aa 333–670).
Dual luciferase assay and firefly luciferase assay
HLR cells were plated onto 48-well culture dishes the day before transfection. Expressing plasmids (0.5 μg of pcDNA3.1(+), pCMV-GAL4DBD-VP16AD-hATF6α full or deletion variants) and reference plasmid (0.01 μg of pRL-CMV; Promega, USA) were transfected into HLR cells using Fugene 6 (Promega) for 24 h. These cells were then further incubated with fresh medium with DMSO or ER stress inducers (1 μg tunicamycin [Tm] or 5 nM thapsigargin [Tg]) for 12 h. For DTT treatment, transfected cells were exposed to 2 mM DTT for 2 h and then incubated with DTT-free fresh medium for 10 h. At 36 h post transfection, cells were washed with phosphate-buffered saline (PBS) three times, harvested, and stored at −80°C for dual luciferase assay. Dual-Luciferase® assay (Promega) was carried out as described previously (Back et al., 2006). Firefly luciferase activity was normalized against Renilla luciferase activity reflecting transfection efficiency. All transfections were performed at least three times to obtain mean ± SD.
For firefly luciferase assay in double stable cell lines expressing GV- hATF6αΔN(aa 333–670), cells were plated onto 48-well culture dishes the day before treatment. These cells were treated with different amounts of ER stress inducers (Tm or Tg) for 12 h or specified amounts of ER stress inducers (1 μg Tm or 5 nM Tg) for indicated times. For DTT treatment, cells were exposed to different amounts of DTT or specified amount (2 mM DTT) for 2 h and then incubated with DTT-free fresh medium for indicated times. After treatment, cells were washed with PBS three times, harvested, and stored at −80°C for firefly luciferase assay. Luciferase assay (Promega) was carried out according to the manufacturer’s instructions. Firefly luciferase activities were normalized to protein contents (relative light units per microgram of protein).
Immunoblot analysis
HLR-GV-hATF6α(333) cells, but not HLR cells, were treated with 2 mM DTT for 2 h and then incubated with DTT-free fresh medium for indicated times. HLR-GV-hATF6α(333) cells were treated with or without 1 μg/ml Tm or 5 nM Tg for 12 h, respectively. For DTT treatment, cells were exposed to 2 mM DTT for 2 h and then incubated with DTT-free fresh medium for 10 h. Cell lysates were prepared from HLR cells or ER stress inducer-treated HLR-GV-hATF6α(333) cells using EzRIPA lysis kit (20 mM HEPES pH 7.5, 150 mM NaCl, 1% IGEPAL CA-630, 0.1% SDS, 0.5% sodium deoxycholate) including 1× protease inhibitors (aprotinin, pepstatin A, and leupeptin) and 1× phosphatase inhibitors (sodium fluoride, sodium vanadate, and sodium glycerophosphate) as specified by the manufacturer (ATTO, USA). Cell lysates were centrifuged at 13,000g for 15 min. Cellular proteins (70 μg) were resolved on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes. Immunoblot analyses were performed as described previously (Back et al., 2006) using anti-GAL4 DNA-BD monoclonal antibody (Clontech Laboratories) and anti-β-actin monoclonal antibody (Santa Cruz Biotechnology, USA).
Subcellular fractionation
To obtain nuclear and cytosolic fractions from HLR or HLR-GV-hATF6α(333) cells treated with or without three ER stress inducers, cell pellets were resuspended in 350 μl 1× hypotonic buffer (10 mM HEPES pH 7.4, 10 mM KCl, 0.1 mM EDTA, 0.5% NP-40, 1 mM DTT, protease inhibitor cocktail, and phosphatase inhibitor cocktail) by passing cell suspension through 20-gauge needle 15 to 20 times. Homogenates were incubated on ice for 40 min. During incubation, homogenates were vortexed for 20 seconds at the highest setting every 10 min. Samples were then centrifuged at 15,700g at 4°C for 15 min. Supernatants were kept as cytoplasmic fractions at −80°C. Cell pellets were resuspended in 80 μl nuclear extraction buffer (20 mM HEPES, 400 mM NaCl, 1 mM EDTA, 1 mM DTT, protease inhibitor cocktail, and phosphatase inhibitor cocktail), sonicated, and stored as nuclear fractions at −80°C until analysis. Cellular proteins of nuclear and cytosolic fractions were resolved on SDS-polyacrylamide gels and transferred to PVDF membranes. Immunoblot analyses were performed using anti-GAL4 DNA-BD monoclonal antibody (Clontech Laboratories), anti-α-tubulin monoclonal antibody (Sigma-Aldrich, USA), and anti-Histone H3 antibody (Abcam, USA).
Fluorescence microscopy analysis
For fluorescence microscopy, HLR-GV-hATF6α(333) cells were grown on 1% gelatin-coated coverslips. The cells were exposed to 2 mM DTT for 2 h and then incubated with DTT-free fresh medium for 4 h. Next, cells were washed with PBS three times and then fixed with 4% (w/v) paraformaldehyde (Sigma-Aldrich) at room temperature for 15 min. After washing with PBS three times, cells on coverslips were permeabilized in 0.1% Triton X-100 at room temperature for 2 min and washed with PBS three times. For GAL4DBD immunostaining, samples were blocked in a blocking solution (PBS containing 3% bovine serum albumin) for 1 h at room temperature and then incubated with anti-GAL4 antibody (Santa Cruz Biotechnology) at 4°C overnight. After washing with PBS three times, samples were treated with Hoechst 33342 at room temperature for 2 min and washed again with PBS three times. They were then incubated with rhodamine (tetramethyl rhodamine isothiocyanate [TRITC])-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, USA) at room temperature for 1 h. Finally, these coverslips were washed three times with PBS, placed on glass slides, and sealed with transparent nail polish. Images were captured with a cooled charge-coupled device camera and a confocal laser scanning microscope (FLUOVIEW FV10i; Olympus, Japan).
RESULTS AND DISCUSSION
Determination of the minimal domain of ATF6α required for RIP reporter system
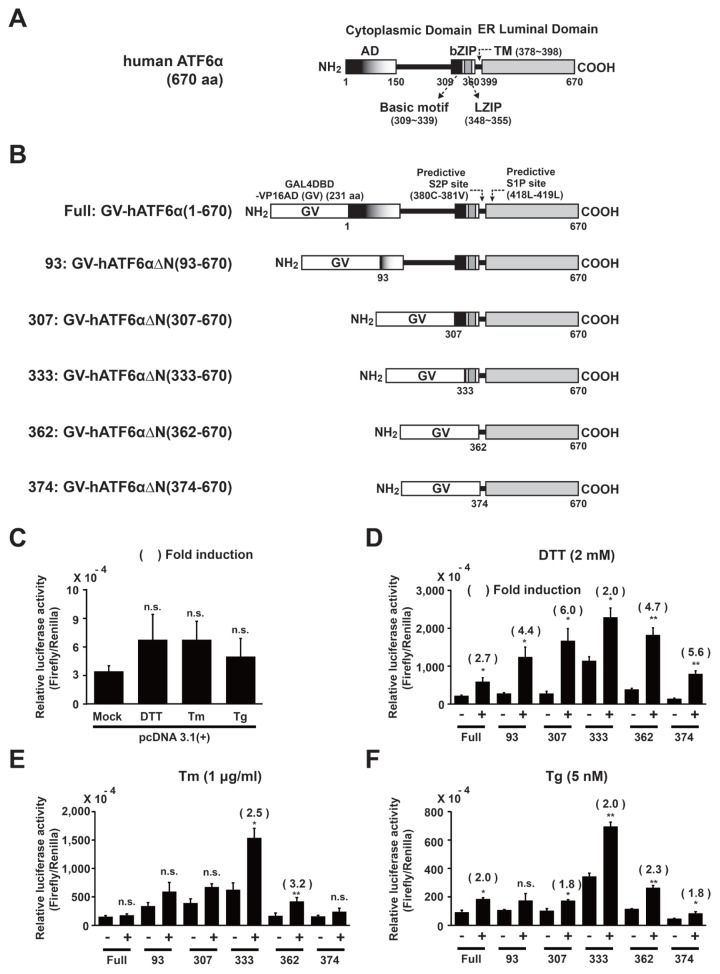
We determined the C-terminal minimal domain required by the ATF6α N-terminal deletion variant fused to a trans-activator protein (GAL4DBD-VP16AD, GV) consisting of the GAL4DBD and the VP16AD. Toward this end, we constructed six vectors expressing a fusion trans-activator protein (GV) followed by full-length or various N-terminal-deleted variants of human ATF6α (GV-hATF6α or GV-hATF6αΔNs; Figs. 1A and 1B). Resulting vectors were co-transfected into HLR cells, which carries a stably integrated firefly luciferase reporter gene, with Renilla luciferase-expressing vector pRL-CMV to normalize the transfection efficiency. The reporter gene consisted of five direct repeats of yeast GAL4 binding site that controls the expression of firefly luciferase gene. Under ER stress, if the expressed fusion protein is cleaved to release a specific fragment containing GAL4DBD-VP16AD(GV) for nuclear translocation, the nuclear trans-activator fragment binds to the 5X GAL4-binding site and induces the expression of firefly luciferase gene. Therefore, the firefly luciferase activity represents the direct cleavage efficiency of GV-ATF6αΔN in transfected cells.
Fig. 1. Selection of a GV-ATF6α fusion variant to measure regulated intermembrane proteolysis of ATF6α in HLR cell line during ER stress.
(A) Predicted domain structure of human ATF6α full length. (B) Schematic diagrams of fusion proteins expressed from pCMV-GAL4DBD-VP16AD(GV)-ATF6α full or deletion variants as described in Materials and Methods section. (C–F) HLR cells were transfected with each expressing plasmid, including pcDNA 3.1(+) and pRL-CMV as internal controls. DTT (2 mM), Tm (1 μg/ml), or Tg (5 nM) was used for treatment for 12 h. Firefly luciferase and Renilla luciferase activities were measured as described in Materials and Methods section. Results are given as absolute values of firefly luciferase normalized against Renilla luciferase activities in each cell lysate. The number of parenthesis indicates average fold induction relative to the untreated group. Data are expressed as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 (untreated group vs each treated group); n.s., not significant.
Through Dual-Luciferase® Reporter (DLRTM) assay, the ratio of firefly luciferase to Renilla luciferase activity in each different construct including an empty vector was calculated and fold induction values were compared for conditions with or without treatment with three different ER stress inducers (DTT, Tm, and Tg), respectively (Figs. 1C–1F). In empty vector-transfected HLR cells, no significant changes were observed in fold induction following treatment with three different ER stress inducers and the numeric values of luciferase activity were significantly lower (at least 10-fold) compared with HLR cells transfected with Full: GV-ATF6α(1–670) or all GV-ATF6αΔN deletion variants regardless of treatment (Fig. 1C vs Figs. 1D–1F). Therefore, the results indicate that the expression of the firefly luciferase reporter gene depends on the availability of a nuclear trans-activator protein (GAL4DBD-VP16AD). However, the transient overexpression of Full: GV-ATF6α(1–670) or all GV-ATF6αΔN deletion variants in HLR cells significantly increased luciferase activities compared with empty vector-transfected HLR cells without treatment (Fig. 1C vs Figs. 1D–1F). The findings suggest undesirable expression of the firefly luciferase reporter gene due to transactivation of a GAL4DBD-VP16AD-containing trans-activator fragment generated by constitutively activated proteolysis of overexpressed fusion proteins. The change was more significant in the 333: GV-hATF6α(333–670) expressing cells. Similarly, the N-terminal active 50-kDa pATF6(N) transcription factor fragment was detected in transient full-length ATF6α-overexpressing cells (Haze et al., 1999; Yoshida et al., 1998), suggesting that the fragment was produced via constitutively activated proteolysis of full-length ATF6α(1–670) overexpressed in transfected cells. Thus, in our reporter system, transient overexpression of Full: GV-ATF6α(1–670) or GV-ATF6αΔN deletion variants might cause dysregulation of regulated cleavage. Nevertheless, upon treatments with ER stress inducers, average fold inductions of firefly luciferase activities from most constructs were significantly increased up to 6-fold at different levels (Figs. 1D–1F), suggesting that ER stress could induce S1P and S2P-mediated cleavage of each GV-ATF6α fusion variant at the Golgi apparatus to generate active trans-activator proteins. In DTT (Fig. 1D) or Tg (Fig. 1F)-treated HLR cells, average fold inductions from Full: GV-hATF6α(1–670), 93: GV-hATF6αΔN(93–670) and 307: GV-hATF6αΔN(307–670) deletion variants were significantly increased. However, the average fold induction in the 333: GV-hATF6αΔN(333–670)-expressing HLR cells carrying a deletion (333: GV-hATF6αΔN(333–670)) extending until the basic motif of bZIP domain was only 2-fold (DTT and Tg) or 2.5-fold (Tm) because of increased luciferase activity in the absence of ER stress (Figs. 1D–1F). Nevertheless, the highest relative luciferase activities were observed in 333: GV-hATF6αΔN(333–670) expressing HLR cells during ER stress (Figs. 1D–1F). Further deletions (362 and 374 constructs) close to transmembrane domain (aa 378–398) gradually decreased relative luciferase activities under ER stress conditions (Figs. 1D–1F).
In Tm-treated HLR cells, no significant fold inductions were detected following the expression of Full: GV-hATF6α(1–670), 93: GV-hATF6αΔN(93–670), 307: GV-hATF6αΔN(307–670) and 374: GV-hATF6αΔN(374–670) deletion variants. Therefore, the four constructs were not selected for further studies. Next, among the remaining two constructs, the 333: GV-hATF6αΔN(333–670) deletion variant expressing HLR cells always showed higher luciferase activities than the 362: GV-hATF6αΔN(362–670) expressing HLR cells. Nevertheless, the former had a smaller fold induction than the latter because of strong basal firefly luciferase activity without treatments. Based on the constitutive proteolysis observed in overexpressed GAL4DBD-ATF6α (Chen et al., 2002) or full-length ATF6 (Haze et al., 1999; Yoshida et al., 1998), the highly basal firefly luciferase activity was induced by such proteolysis of 333: GV-hATF6αΔN(333–670) overexpressed in transfected cells. However, if the deletion variant is expressed at a lower level when 333: GV-hATF6αΔN(333–670)-expressing HLR stable cell lines are generated, the constitutively activated proteolysis of 333: GV-hATF6αΔN(333–670) may be decreased and the basal firefly luciferase activity may be lowered, resulting in higher fold induction under ER stress. Based on this rationale, the 333: GV-hATF6αΔN(333–670)-expressing construct was selected to generate double stable luciferase reporter cell lines to monitor RIP of ATF6α.
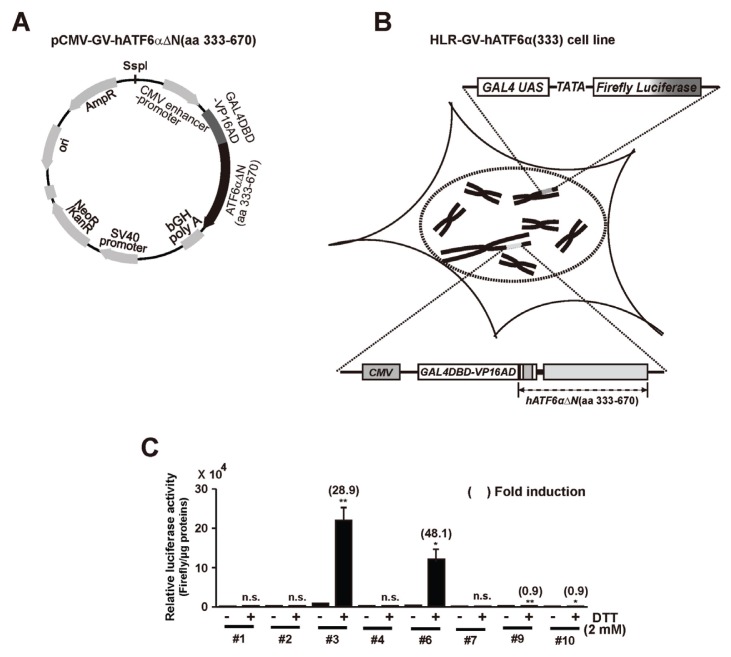
Generation of a double stable reporter cell line (HLR-GV-hATF6α(333) cell line) for monitoring RIP of ATF6α
To generate a double stable reporter cell line expressing firefly luciferase by regulated proteolysis of GV-hATF6αΔN(aa 333–670) chimeric protein, HLR cells were transfected with Ssp1-digested DNA fragment of pCMV-GV-hATF6αΔN(aa 333–670) vector encoding a fusion trans-activator protein (GAL4DBD-VP16AD) followed by N-terminal deleted variant (aa 333–670) of hATF6α (Figs. 2A and 2B). These transfected cells were selected using medium containing both G418 for GV-hATF6αΔN(aa 333–670) expressing cassette (Fig. 2A) and hygromycin B for firefly luciferase expressing cassette. After generation of double stable cell lines, the ability of the reporter cell line to monitor intramembrane proteolysis-dependent ATF6α activation was evaluated based on increased firefly luciferase activity under DDT treatment (Fig. 2C). Of 10 G418 and hygromycin B resistant clones, two clones showed ~30 folds (clone #3) or ~50 folds (clone #6) induction in firefly luciferase activity upon treatment with DTT, respectively. Because clone #3 showed the highest relative luciferase activity, this clone was chosen for further characterization and named HLR-GV-hATF6α(333) cell line.
Fig. 2. Isolation of a GV-hATF6αΔN(333–670) chimeric protein expressing HLR cell line.
(A) Graphical representation of the pCMV-GV-hATF6αΔN(aa 333–670) plasmid described in Materials and Methods section. (B) Graphical representation of a double-stable HLR cell line having both pGAL4USA-Luc plasmid and pCMV-GV-hATF6αΔN(aa 333–670) plasmid stably integrated into the genome. (C) Both hygromycin B (0.1 mg/ml) and G418 (0.5 mg/ml)-resistant stable cell clones were isolated. These clones were treated with DTT (2 mM) as described in Materials and Methods section. Firefly luciferase activities in cell lysates were measured. Results are given as absolute values of firefly luciferase activity to 1 μg of proteins in each cell lysate. The number in parenthesis indicates average fold induction relative to the untreated group. Data are expressed as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 (untreated group vs each treated group); n.s., not significant.
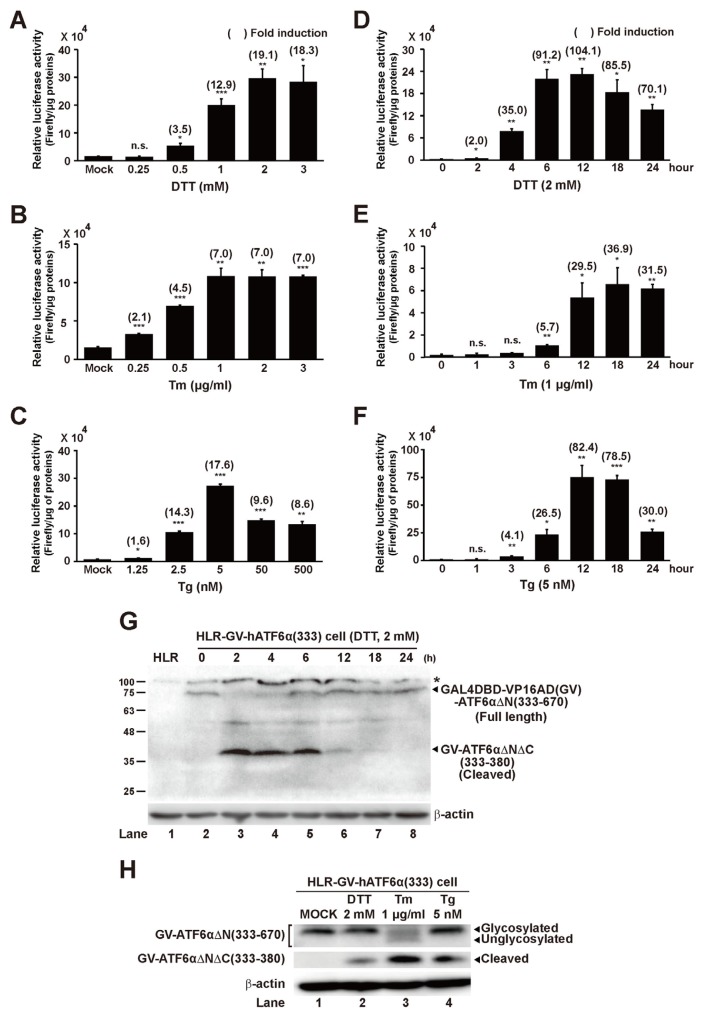
Characterization of a new RIP reporter system (HLR-GV-hATF6α(333) cell line) for monitoring regulated proteolysis of ATF6α
To check the ability of HLR-GV-hATF6α(333) reporter cell line in response to ER stress, luciferase activities of HLR-GV-hATF6α(333) cells were measured after treatment with DDT, Tm, and Tg at different concentrations or times. Concentrations that showed the maximal luciferase activities were found to be 2 mM for DTT, 1 mM for Tm, and 5 nM for Tg (Figs. 3A–3C). Higher concentrations did not further increase luciferase activity. Next, the best treatment time of each ER stress inducer was determined at these concentrations selected based on results from Figures 3A–3C. Significant increase of luciferase activity was detected after treatment with DDT for 2 h. The maximal luciferase activity was detected after treatment with DDT for 12 h, Tm for 18 h, or Tg for 12 h (Figs. 3D–3F). However, the maximal activity did not sustain after further incubation. Next, to confirm that HLR-GV-hATF6α(333) cell line expressed GV-hATF6αΔN(aa 333–670) fusion protein and ER stress could induce its proteolysis to generate GV-hATF6αΔNΔC(aa 333–380) fragment, Western blot analysis was performed using GAL4 antibody and each ER stress inducer-treated cell lysate (Figs. 3G and 3H). HLR-GV-hATF6α(333) cells constitutively expressed ~75 kDa GV-hATF6αΔN(aa 333–670) protein while the original HLR cells did not (Fig. 3G; lanes 1 and 2). However, DDT treatment immediately made the full length GV-hATF6αΔN(aa 333–670) disappear till 4 h after treatment (lanes 3 and 4). Simultaneously, ~40 kDa small cleaved products supposed to be GV-hATF6αΔNΔC(aa 333–380) fragments were detected till 18 h (lanes 3 to 6). As described for DTT treatment experiments in Materials and Methods section, DDT-free media replaced DDT-containing media after 2 h of treatment. Therefore, it was believed that DDT effect diminished and cells recovered from ER stress as time went by, resulting in reduction of GV-hATF6αΔNΔC(aa 333–380) fragments and appearance of full length GV-hATF6αΔN(aa 333–670) proteins (Fig. 3G; lanes 5–8). Moreover, RIP of GV-hATF6αΔN(aa 333–670) was also observed in Tm or Tg-treated cell lysate (Fig. 3H). Importantly, GV-hATF6αΔNΔC(aa 333–380) fragments generated by RIP were detected in only ER stress inducer-treated cell lysates, but not in mock-treated cell lysate (Figs. 3G; lane 2 and 3H; lane 1), indicating that GV-hATF6αΔN(aa 333–670) expressing HLR-GV-hATF6α(333) cell line could be utilized as a specific and novel reporter cell line for measuring RIP of ATF6α.
Fig. 3. Concentration and time-dependent effects of ER stress inducer treatments to HLR-GV-hATF6α(333) cell line and detection of GV-hATF6αΔN(aa 333–670) cleavage on ER stress-induced HLR-GV-hATF6α(333) cells.
(A–C) HLR-GV-hATF6α(333) cells were treated with DTT, Tm, or Tg at indicated concentrations for 12 h. (D–F) HLR-GV-hATF6α(3 33) cells were treated with DTT (2 mM), Tm (1 μg/ml), or Tg (5 nM) at indicated times. Firefly luciferase activities in cell lysates were measured. Results are given as absolute value of firefly luciferase activity to 1 μg of proteins in each cell lysate. The number in parenthesis indicates average fold induction relative to the untreated group. Data are expressed as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (untreated group vs each treated group); n.s., not significant. (G) HLR-GV-hATF6α(333) cells were treated with DTT (2 mM) for 2 h and then incubated with DTT-free medium for the indicated times. At indicated time points, cells were harvested and total cellular extracts were prepared and then subjected to Western blot analysis with anti-Gal4 and anti-β-actin antibodies. The asterisk (*) indicates non-specific bands in Western blot. (H) HLR-GV-hATF6α(333) cells were treated with DTT (2 mM), Tm (1 μg/ml), or Tg (5 nM) for 12 h as described in Materials and Methods section. Cells were harvested and total cellular extracts were prepared and then subjected to Western blot analysis.
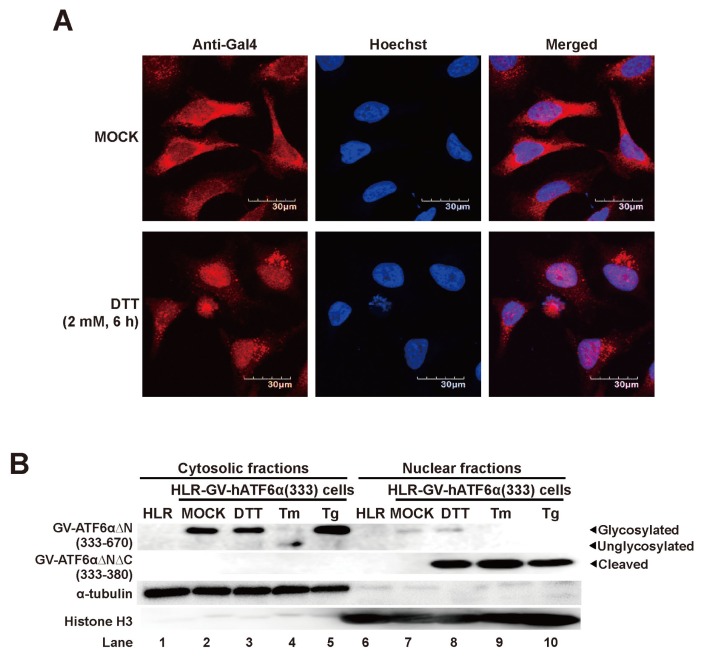
ER stress-mediated nuclear translocation of cleaved fragment of GV-hATF6αΔN(aa 333–670) in HLR-GV-hATF6α(333) cell line
As described our reporter system in the introduction part, the GV-hATF6αΔNΔC(aa 333–380) fragment working as a transcription factor should translocate into the nucleus to induce expression of firefly luciferase gene in HLR-GV-hATF6α(333) cell line during ER stress. To assess nuclear localization of the chimeric fragment, immunofluorescence analysis was performed using GAL4 antibody and DTT-treated cells (Fig. 4A). Without DTT treatment, majority of anti-GAL4 reactive proteins were localized in the cytosol (Fig. 4A; upper panels). However, with DTT treatment, most anti-GAL4 reactive proteins were observed in the nucleus of HLR-GV-hATF6α(333) cells (Fig. 4A; lower panels), suggesting that the GAL4DBD-containing cleavage fragment of GV-hATF6αΔN(aa 333–670) believed to be GAL4DBD-VP16AD(GV)-ATF6αΔNΔC(aa 333–380) fragment was generated after DTT treatment. It then translocated into the nucleus. Next, we further confirmed nuclear localization of GV-ATF6αΔNΔC(aa 333–380) fragment by subcellular fractionation. Cytosolic and nuclear fractions were extracted from original HLR cells and ER stress inducer-treated HLR-GV-hATF6α(333) cells. Western blot analysis of “house-keeper” (HK) proteins (α-tubulin and histone H3 for cytosolic and nuclear fractions, respectively) validated the purity of fractionated samples (Fig. 4B). In Western blot analysis of fractionated samples of HLR cells, there was no anti-GAL4 specific band in either fraction (lanes 1 and 6). Before ER stress inducer treatment, the GV-ATF6αΔNΔC(aa 333–380) fragment was virtually undetectable in the nuclear fraction of HLR-GV-hATF6α(333) cells (lane 7). Instead, full length GV-hATF6αΔN(aa 333–670) proteins were only detected in the cytosolic fraction of HLR-GV-hATF6α(333) cells (lane 2). However, GV-ATF6αΔNΔC(aa 333–380) fragments were clearly observed in nuclear factions of ER stress inducer (DTT, Tm, or Tg)-treated HLR-GV-hATF6α(333) cells (lanes 8–10), indicating that HLR-GV-hATF6α(333) cell lines could produce nuclear localized anti-GAL4 reactive protein as a transcription factor to induce expression of firefly luciferase gene under ER stress conditions.
Fig. 4. Subcellular localization of GV-hATF6αΔN(333–670) chimeric protein during ER stress.
(A) HLR-GV-hATF6α(333) cells were stained with Hoechst 33342 (blue fluorescence) and anti-Gal4 antibody (red fluorescence) after DTT treatment for 6 h and analyzed by confocal fluorescence microscopy. Scale bars = 30 μm. (B) HLR-GV-hATF6α(333) cells were treated with DTT (2 mM), Tm (1 μg/ml), or Tg (5 nM) for 12 h. Cytosolic and nuclear fractions extracted from HLR-GV-hATF6α(333) cells treated as indicated were subjected to Western blot analysis using anti-Gal4, anti-α-tubulin, and anti-histone H3 antibodies.
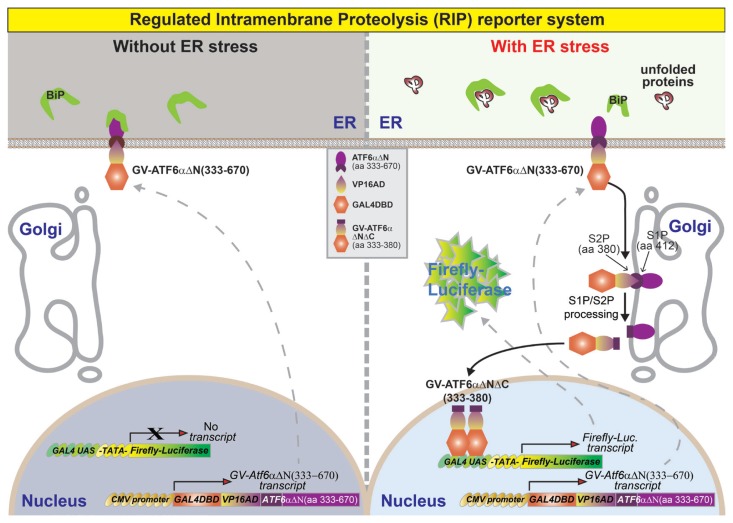
Summary of a reporter system monitoring RIP of transmembrane bZIP transcription factor ATF6α
In this report, we developed a specific, rapid, and convenient monitoring system for the in vivo activation of ATF6α that may also be used to determine RIP of other transmembrane bZIP transcription factors such as ATF6β and CREBH (Bailey and O’Hare, 2007). Therefore, we modified the HLR cell line carrying a stably integrated firefly luciferase reporter gene consisting of five direct repeats of yeast GAL4-binding site that controls the expression of firefly luciferase gene (Fig. 5). We introduced an expression cassette containing the gene of a fusion trans-activator protein (GAL4DBD-VP16AD-hATF6αΔN) into this cell line. The gene for the fusion trans-activator protein (GAL4DBD-VP16AD-hATF6αΔN(aa 333–670)) consists of the GAL4DBD and the VP16AD followed by N-terminal deletion variant (aa 333–670) of ATF6α. Thus, the final double stable cell line contains both the luciferase expression cassette and the fusion trans-activator expression cassette. Without ER stress, the ATF6αΔN-fused trans-activator protein (GAL4DB-VP16AD(GV)-ATF6αΔN(aa 333–670)) is localized to ER membrane by transmembrane and ER-luminal domains of ATF6α (Fig. 5). Therefore, no significant expression of the firefly luciferase gene is detected (Figs. 2C and 3A–3F). However, when cells were treated with ER stress inducers such as DTT, Tm, and Tg to activate ATF6α, the ER-localized GV-ATF6αΔN protein is translocated to the Golgi apparatus for sequential cleavage by S1P at aa 412, and by S2P at aa 380 (Ye et al., 2000) to release the cytoplasmic GV-ATF6αΔNΔC fragment (Figs. 4 and 5). The GV-ATF6αΔNΔC fragment is translocated to the nucleus to induce the transcription of firefly luciferase from the reporter cassette in HLR cells (Figs. 4 and 5). Thus, the firefly luciferase activity represents the cleavage efficiency of GAL4DBD-VP16AD-ATF6αΔN directly and endogenous ATF6α proteins indirectly in the double stable cell line.
Fig. 5. Schematic diagram of the reporter system for monitoring RIP of ATF6α.
The RIP reporter cell line carries two stably integrated expression constructs. The first construct is a firefly luciferase gene controlled by a synthetic promoter comprising five direct repeats of yeast GAL4-binding site (GAL4 Upstream Activating Sequences [GAL4 UAS]) and TATA box. The second expression cassette is composed of a fusion gene encoding GAL4DBD and the VP16AD followed by the N-terminal deletion variant of ATF6α (ATF6αΔN(aa 333–670)). In the absence of ER stress, the localization of GAL4DBD-VP16AD(GV)-ATF6αΔN(aa 333–670) protein is restricted at the ER membrane; therefore, there is no significant expression of the firefly luciferase gene because of the absence of active transcription factors. However, under ER stress, the GV-ATF6αΔN(aa 333–670) is translocated to the Golgi apparatus for cleavage by S1P and S2P. The cytosolic N-terminal fragment GV-ATF6αΔNΔC(aa 333–380) is translocated into the nucleus to bind with the GAL4-binding site (GAL4 UAS) as an active transcription factor and induces the expression of firefly luciferase.
Compared with other known reporter systems, the RIP reporter system is unique. It is distinct from other UPRE or ERSE element-based reporter systems (Gallagher et al., 2016; Lee et al., 2002; Wang et al., 2000; Yoshida et al., 1998) in that the transcription of luciferase is not induced by endogenous transcription factors (such as XBP1s or activated ATF6β) generated upon ATF6α activation, but is solely dependent on the translocation of GAL4DBD-VP16AD-ATF6αΔN to the Golgi followed by the cleavage of transmembrane domain under specific conditions. Therefore, our RIP reporter system is not only a unique tool to monitor RIP of ATF6α, but also represents a substitute for other nonspecific reporter systems that use an UPRE- or ERSE-based luciferase reporter (Gallagher et al., 2016; Lee et al., 2002; Wang et al., 2000; Yoshida et al., 1998). Furthermore, compared with GAL4DBD full-length ATF6α fusion protein reported by Ron Prywes’s group before (Chen et al., 2002; Wang et al., 2000), our GV-ATF6αΔN(aa 333–670) does not carry most functional domains of ATF6α N-terminal active transcription factor fragment (pATF6(N)). The active pATF6(N) fragment generated from full-length ATF6α interacts with other transcription factors (such as serum response factor [SRF] or spliced XBP1) (Yamamoto et al., 2007; Zhu et al., 1997). The pATF6(N)/SRF or pATF6(N)/XBP1s heterodimer complexes cooperatively lead to induce expression of several serum- or ER stress-dependent genes, respectively. Hence, the active GAL4DBD-pATF6(N) fragment generated from GAL4DBD-full length ATF6α can also form a heterodimer complex, which may affect the expression of various endogenous target genes. Consequently, a reporter cell line additionally expressing the active GAL4DBD-pATF6(N) fragment may show different cellular response compared to a normal cell line, and in turn the results of the reporter cell line may be distorted by exogenous and active GAL4DBD-pATF6(N) fragment. However, our GV-ATF6αΔN(aa 333–670) may not show the binding ability or transcriptional activity, which affect the expression of endogenous serum- or ER stress-dependent genes. Thus, we believe that our RIP reporter system represents a specific and reliable experimental tool to monitor RIP of ATF6α.
Therefore, this reporter system can be used to screen for genes, small-molecules, and protein-based drugs that can activate or inhibit RIP of ATF6α. Moreover, for other known transmembrane bZIP transcription factors including Luman/CREB3, OASIS/CREB3L1, BBF2H7/CREB3L2, CREB-H/CREB3L3, CREB4/AIbZIP/CREB3L4, and CREBL1/ATF6β (Bailey and O’Hare, 2007), if the optimal DNA fragment for cleavage of GAL4DBD-VP16AD(GV)-bZIP transcription factor is determined as shown in the present study, the resulting plasmid can be used to develop a new reporter system for monitoring RIP of the transmembrane bZIP transcription factor in the luciferase reporter cell line which contains a stably integrated 5X GAL4 site-luciferase gene.
ACKNOWLEDGMENTS
We thank Mi-Jeong Kim and Kyung-Ju Ahn at University of Ulsan for technical assistance with data preparation. This research was supported by a grant (2017R1D1A1B03035248 to J.Y.M.) of the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education. This research was also supported by the Priority Research Centers Program (2014R1A6A1030318 to S.H.B.) and the Bio & Medical Technology Development Program (2017M3A9G7072745 to S.H.B.) of the NRF funded by the Korean government. Portions of this work were supported by National Institutes of Health (NIH) grants (CA198103, DK113171 and AG062190 to R.J.K.).
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Back S.H., Kaufman R.J. Endoplasmic reticulum stress and type 2 diabetes. Annu Rev Biochem. 2012;81:767–793. doi: 10.1146/annurev-biochem-072909-095555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S.H., Lee K., Vink E., Kaufman R.J. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J Biol Chem. 2006;281:18691–18706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- Bailey D., O’Hare P. Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid Redox Signal. 2007;9:2305–2321. doi: 10.1089/ars.2007.1796. [DOI] [PubMed] [Google Scholar]
- Bommiasamy H., Back S.H., Fagone P., Lee K., Meshinchi S., Vink E., Sriburi R., Frank M., Jackowski S., Kaufman R.J., et al. ATF6alpha induces XBP1-independent expansion of the endoplasmic reticulum. J Cell Sci. 2009;122:1626–1636. doi: 10.1242/jcs.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Shen J., Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045–13052. doi: 10.1074/jbc.M110636200. [DOI] [PubMed] [Google Scholar]
- Gallagher C.M., Garri C., Cain E.L., Ang K.K., Wilson C.G., Chen S., Hearn B.R., Jaishankar P., Aranda-Diaz A., Arkin M.R., et al. Ceapins are a new class of unfolded protein response inhibitors, selectively targeting the ATF6alpha branch. Elife. 2016;5:e11878. doi: 10.7554/eLife.11878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Glimcher L.H. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–561. doi: 10.1016/j.molcel.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Martinon F., Rodriguez D., Glimcher L.H. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- Iwawaki T., Akai R., Kohno K., Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- Iwawaki T., Akai R., Toyoshima T., Takeda N., Ishikawa T.O., Yamamura K.I. Transgenic mouse model for imaging of ATF4 translational activation-related cellular stress responses in vivo. Sci Rep. 2017;7:46230. doi: 10.1038/srep46230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., Glimcher L.H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Tirasophon W., Shen X., Michalak M., Prywes R., Okada T., Yoshida H., Mori K., Kaufman R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452–466. doi: 10.1101/gad.964702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadanaka S., Okada T., Yoshida H., Mori K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol Cell Biol. 2007;27:1027–1043. doi: 10.1128/MCB.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C.G. eIF2 and the control of cell physiology. Semin Cell Dev Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rutkowski D.T., Kaufman R.J. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- Shen J., Chen X., Hendershot L., Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/S1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- Shen J., Prywes R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J Biol Chem. 2004;279:43046–43051. doi: 10.1074/jbc.M408466200. [DOI] [PubMed] [Google Scholar]
- Spiotto M.T., Banh A., Papandreou I., Cao H., Galvez M.G., Gurtner G.C., Denko N.C., Le Q.T., Koong A.C. Imaging the unfolded protein response in primary tumors reveals microenvironments with metabolic variations that predict tumor growth. Cancer Res. 2010;70:78–88. doi: 10.1158/0008-5472.CAN-09-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Kaufman R.J. Protein misfolding in the ER as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R.J., Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Wek R.C., Cavener D.R. Translational control and the unfolded protein response. Antioxid Redox Signal. 2007;9:2357–2371. doi: 10.1089/ars.2007.1764. [DOI] [PubMed] [Google Scholar]
- Wek R.C., Jiang H.Y., Anthony T.G. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST0340007. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Ye J., Rawson R.B., Komuro R., Chen X., Dave U.P., Prywes R., Brown M.S., Goldstein J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Haze K., Yanagi H., Yura T., Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/MCB.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Okada T., Haze K., Yanagi H., Yura T., Negishi M., Mori K. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6alpha and 6beta that activates the mammalian unfolded protein response. Mol Cell Biol. 2001;21:1239–1248. doi: 10.1128/MCB.21.4.1239-1248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Johansen F.E., Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/MCB.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]