Abstract
Ultraviolet light (UV)-induced cellular response has been studied by numerous investigators for many years. Long noncoding RNAs (lncRNAs) are emerging as new regulators of diverse cellular process; however, little is known about the role of lncRNAs in the cellular response to UV treatment. Here, we demonstrate that levels of lncRNA-HOTTIP significantly increases after UV stimulation and regulates the UV-mediated cellular response to UV through the coordinate activation of its neighboring gene Hoxa13 in GC-1 cells (spermatogonia germ cell line). UV-induced, G2/M-phase arrest and early apoptosis can be regulated by lncRNA-HOTTIP and Hoxa13. Furthermore, lncRNA-HOTTIP can up-regulate γ-H2AX and p53 expression via Hoxa13 in UV-irradiated GC-1 cells. In addition, p53 has the ability to regulate the expression of both lncRNA-HOTTIP and Hoxa13 in vitro and in vivo. Our results provide new data regarding the role lncRNAs play in the UV response in spermatogenic cells.
Keywords: DNA damage, HOTTIP, Hoxa13, spermatogenic cell, ultraviolet light
INTRODUCTION
Solar ultraviolet light (UV) irradiation is the most prominent and ubiquitous physical carcinogen in our natural environment and UV-mediated DNA damage plays a critical role in the development of skin cancer (Armstrong and Kricker, 2001; de Gruijl, 1999; de Gruijl et al., 2001). DNA damage induced by UV irradiation initiates cellular recovery mechanisms, including the activation of signaling pathways that respond to DNA damage, cell cycle arrest and apoptosis (Gentile et al., 2003). UV irradiation can regulate testicular germ cell fate through DNA damage (Koberle et al., 2008). Recent studies have indicated that the mechanism for the cellular response to UV involves noncoding RNAs (Degueurce et al., 2016; Li et al., 2012; Williamson et al., 2017). UV stimulation promotes the transcription of the long noncoding RNA (lncRNA), ASCC3, which inhibits the expression of the ASCC3 protein encoded by the same gene, and plays important role in the cellular response induced by UV light (Williamson et al., 2017). Further, the expression of intermediate-size (70–500 nucleotides) ncRNAs have been shown to be altered post UV irradiation in Caenorhabditis elegans (Li et al., 2012). In addition, knockdown of miR-21-3p can inhibit UV-induced skin inflammation (Degueurce et al., 2016).
Throughout eukaryotic genomes, lncRNAs are pervasively transcribed and range in size from 200 bp to over 100 kb (Brosnan and Voinnet, 2009; Chen and Carmichael, 2010; Kapranov et al., 2007). Depending on their proximity to protein-coding genes, lncRNAs may be classified as antisense, intronic, bidirectional or intergenic lncRNAs (Rinn and Chang, 2012). Mounting evidence have recently suggested that lncRNAs are important molecules with roles in a in a diverse set of cellular processes, including growth, cell cycle, differentiation and apoptosis, tumor suppression and promotion, and the stress response (Liu et al., 2012; Tripathi et al., 2013; Zhou et al., 2007). Furthermore, lncRNAs can regulate the expression of neighboring protein-coding genes at the level of chromatin remodelling, and transcriptional and posttranscriptional processing (Mercer et al., 2009). Although lncRNAs have been investigated extensively, very little is known about the function of lncRNAs with respect to the testicular germ cell response to UV stimulation.
The lncRNA-HOTTIP (HOXA transcript at the distal tip) is spliced and polyadenylated transcript located at the 5′ tip of the Hoxa13 gene. The lncRNA-HOTTIP coordinates the activation of several HOXA genes through binding to and driving MLL1/WDR5 occupancy and H3K4 trimethylation of the HOXA gene promoter in human primary fibroblasts (Burgess, 2011; Wang et al., 2011). In addition, HOTTIP regulates human cartilage development and destruction by modulating integrin-α1 transcription via Hoxa13, and the HOTTIP transcript could be a potent predictive biomarker for osteoarthritis (Kim et al., 2013). In hepatocellular carcinoma patients, HOTTIP and Hoxa13 expression are associated with disease progression and can predict patient outcome (Quagliata et al., 2013). Furthermore, the expression levels of HOTTIP RNA are up-regulated in both osteoarthritis chondrocytes and hepatocellular carcinoma specimens (Kim et al., 2013; Quagliata et al., 2013), suggesting that HOTTIP is involved in multiple types of conditions in which the misregulation of cellular functioning occurrs. HOTTIP can be used as aprognostic biomarker for early-stages of human non–small-cell lung cancer and is correlated with a number of mRNAs and miRNAs signatures (Navarro et al., 2019). Moreover, HOTTIP enhances insulin secretion and regulates cell proliferation and the cell cycle by modifying the MEK/ERK cascade in islet-β cells (Xu et al., 2018). It is not clear; however, whether HOTTIP participates in the cellular response to UV-induced DNA damage in germ cells.
In the current study, we have elucidated that expression of lncRNA-HOTTIP and Hoxa13 in mouse tissues and cells, and HOTTIP-Hoxa13 expression is involved in the response to UV-mediated DNA damage in the spermatogonia germ cell line GC-1. Moreover, we have found that HOTTIP-Hoxa13 plays a major role in UV-induced cell cycle arrest and apoptosis via regulating γ-H2AX and p53 expression. Collectively, this study uncovers new insights into the function of HOTTIP-Hoxa13 in response to UV damage in spermatogenic cells.
MATERIALS AND METHODS
Animals
C57BL6/J mice were originally acquired from the Laboratory Animal Center, Bengbu Medical College (Bengbu, China), and housed at room temperature (23 ± 2°C) under of 14 h light and 10 h dark. The mice had free access to water and food. This study received ethical approval from the Ethical Committee for Bengbu Medical College (approval No. 2016004).
Plasmids and siRNAs
The mouse Hoxa13 expression vector was constructed by cloning the mouse Hoxa13 cDNA into the pcDNA3.1(+) vector at the BamH I and EcoR I restriction sites according to a protocol outlined previously (Liang et al., 2013). Primer sequences have been listed in Supplementary Table S1. The construct generated was verified by sequencing. The siRNAs for mouse lncRNA-HOTTIP (si-HOTTIP) and siRNAs for negative control (si-NC) were obtained from Shanghai GenePharma Company Limited (Shanghai, China).
Cell culture and treatments
NIH3T3, STO, GC-1, GC-2 and C18-4 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Life Technologies, USA) or DMEM/F12 (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) and 1% penicillin and streptomycin (100 U/ml penicillin and 100 μg/ml streptomycin; Life Technologies) and incubated at 37°C and 5% CO2. Transfection of GC-1 cells were carried out with Lipofectamine 2000 reagent (Invitrogen, USA) following the manufacturer’s protocol and cells were transfected with 100 nM si-HOTTIP or si-NC. The DNA plasmids, si-HOTTIP and si-NC were diluted in Opti-MEM I reduced serum medium (Life Technologies).
For UV irradiation, the cultured medium of transfected cells was discarded and cells were rinsed with phosphate-buffered saline (PBS) (van der Wees et al., 2003). GC-1 cells were irradiated at 0, 5 or 10 J/m2 using UV Crosslinker with a peak emission at 254 nm (UVP CL-1000; Analytic Jena US LLC, USA). Immediately after UV treatment, the cells were cultured at 5% CO2 and 37°C in fresh medium for 24 h and then collected to study cellular responses to UV.
RNA isolation and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from either cultured cells or mouse tissues with Trizol (Invitrogen) (Ma et al., 2013). RT-qPCR assays were used to analyze lncRNA-HOTTIP and Hoxa13 mRNA expression according to protocols outlined previously (Bustin et al., 2009). Briefly, RNA was reverse transcribed into cDNA by using the PrimeScript RT reagent kit (TaKaRa, Japan) and real-time PCR was executed on ABI Step One System (Applied Biosystems, USA) using the SYBR Premix Ex Taq II kit (TaKaRa) according to the manufacturer’s instructions. The relative expression levels of lncRNA-HOTTIP and Hoxa13 mRNA were normalized using endogenous β-actin mRNA. Primers for β-actin, HOTTIP, and Hoxa13 have been listed in Supplementary Table S2.
Cell proliferation and flow cytometric analysis (FACS)
The proliferation of GC-1 cells was measured using a CCK-8 kit (Dojindo Laboratories, Japan) according the manufacturer’s protocols. After 24 h of transfection with si-NC/si-HOTTIP and another 24 h after UV treatment, the GC-1 cells were cultured in fresh medium with 10% CCK-8 reagent for 2 h. Subsequently, absorbance at 450 nm for GC-1 cells were measured using a Cytation 3 imaging reader (BioTek, USA).
Cell apoptosis and cell cycle were analyzed using a FACScalibur Flow Cytometer (BD Biosciences, USA). Data analysis was carried out using WinMDI software. To assess cell apoptosis, cells were collected 48-h post treatment, stained with the Annexin V-FITC Apoptosis Detection Kit (Beyotime Biotechnology, China) and analyzed by FACS. To assess cell cycle, cells were collected 48-h post treatment, washed with PBS and fixed overnight in 70% ethanol. Cells were then harvested, washed with PBS, incubated at 37°C for 10 minutes with PBS, 100 μg/ml RNase A (Sigma-Aldrich, USA) and 25 μg/ml propidium iodide (PI) (Sigma-Aldrich), and analyzed by FACS.
Western blotting
GC-1 cells were lysed in RIPA buffer (Millipore, USA) with 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml CompleteTM EDTA free protease inhibitor cocktail (Roche, USA) and phosphatase inhibitors (5 mM sodium orthovanadate). Protein lysates were loaded on to SDS-PAGE gels, transferred to nitrocellulose membranes (Amersham Biosciences, Germany), immunoblotted with antibodies and visualized using enhanced chemiluminescence substrate (Thermo Fisher Scientific, USA). Protein levels of p53, p21 and γ-H2AX were normalized to GAPDH. The primary antibodies in this study were the following: anti-GAPDH (Cell Signaling Technology, USA), anti-p53 (Santa Cruz, USA), anti-γ-H2AX (Abcam, USA) and anti-p21 (Santa Cruz).
Statistical analysis
Experiments were repeated at least three times and performed in triplicate. Data have been shown as mean ± SEM. Means for groups were analyzed using the Student’s t-test through IBM SPSS Statistics 21.0 (IBM, USA). P values of less than 0.05 was considered statistically significant.
RESULTS
Expression of lncRNA-HOTTIP and Hoxa13 in response to UV exposure
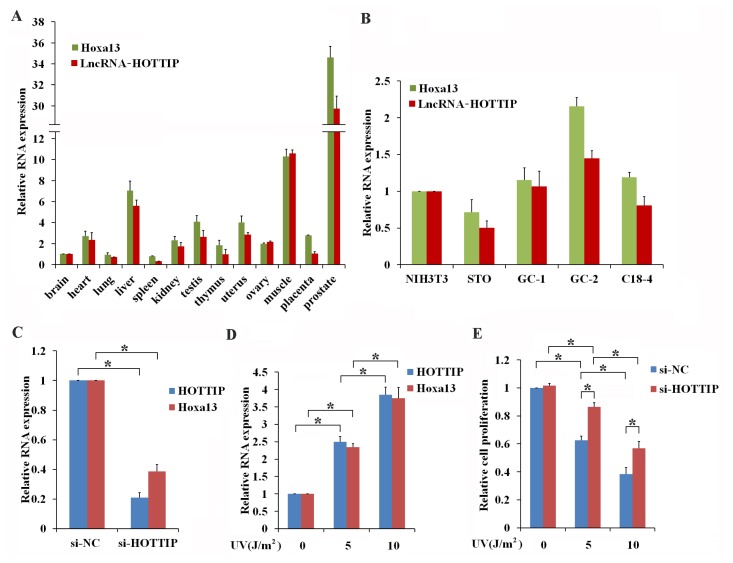
According to the UCSC Genome Browser (http://genome.ucsc.edu/), lncRNA-HOTTIP is located at the 5′ tip of the Hoxa13 gene on chromosome 6qB3. RT-qPCR results have shown that HOTTIP and Hoxa13 are co-expressed in multiple types of mouse tissues (Fig. 1A) and cell lines (Fig. 1B), indicating that HOTTIP is coordinately expressed with Hoxa13 both in vivo and in vitro. Expression of lncRNA-HOTTIP is reported to be involved in cancer progression associated with UV exposure (Jiang and Bikle, 2014; Li et al., 2015). These results suggest that HOTTIP may play critical roles in the male reproductive system, so we chose the spermatogonia germ cell line GC-1 to analyze the potential roles of HOTTIP in vitro. Knockdown of HOTTIP significantly inhibited the level of Hoxa13 mRNA (Fig. 1C) and overexpression of HOTTIP significantly enhanced the level of Hoxa13 mRNA in GC-1 cells (Supplementary Fig. S1), which was in agreement with the relationship of coordinated expression for HOTTIP and Hoxa13. To determine whether HOTTIP and Hoxa13 participate in UV-induced germ cell activity. Three intensities of UV irradiation (0, 5, and 10 J/m2) were used to stimulate GC-1 cells. As shown in Figure 1D, results revealed that stimulation with both 5 J/m2 and 10 J/m2 significantly increases relative levels of both HOTTIP and Hoxa13 mRNA when compared to cells that have not been irradiated with UV light. Furthermore, irradiation with both 5 J/m2 and 10 J/m2 of UV light inhibited the proliferation of GC-1 cells (Fig. 1E). Knockdown of HOTTIP attenuated the repressive effects of irradiation of cells with both 5 J/m2 and 10 J/m2 UV light on the proliferation of GC-1 cells (Fig. 1E). These data not only indicated that lncRNA-HOTTIP mediates the effects of UVvstimulation in GC-1 cells, but also that lncRNA-HOTTIP and Hoxa13 may cooperate to regulate the UV-induced, cellular response in the reproductive system.
Fig. 1. UV induces the coordinated expression of lncRNA-HOTTIP and Hoxa13.
(A and B) The coordinated expression of lncRNA-HOTTIP and Hoxa13. RT-qPCR was used to examine expression of both HOTTIP and Hoxa13 in mouse tissues (A) and cell lines (B). (C) LncRNA-HOTTIP promotes the expression of Hoxa13. The efficiency of the knockdown of HOTTIP and the Hoxa13 expression was confirmed using RT-qPCR to measure transcript levels in GC-1 cells after they had been transfected with si-NC or si-HOTTIP for 24 h. (D) UV induces the coordinated expression of lncRNA-HOTTIP and Hoxa13. GC-1 cells were treated with 0, 5, and 10 J/m2 UV. Expression levels of genes were then measured using RT-qPCR 24-h post treatment. (E) Knockdown of HOTTIP attenuates the inhibitory effects of UV treatment on GC-1 cell proliferation. GC-1 cells were transfected with si-NC or si-HOTTIP and were treated with 0, 5, and 10 J/m2 UV 24-h post transfection. Proliferation of GC-1 cells was measured by using the CCK-8 method 24-h post UV exposure. Data shown are the mean ± SEM of three independent experiments, each of which were performed in triplicate. *P < 0.05.
LncRNA-HOTTIP mediates the G2/M phase arrest and apoptosis in UV-irradiated GC-1 cells
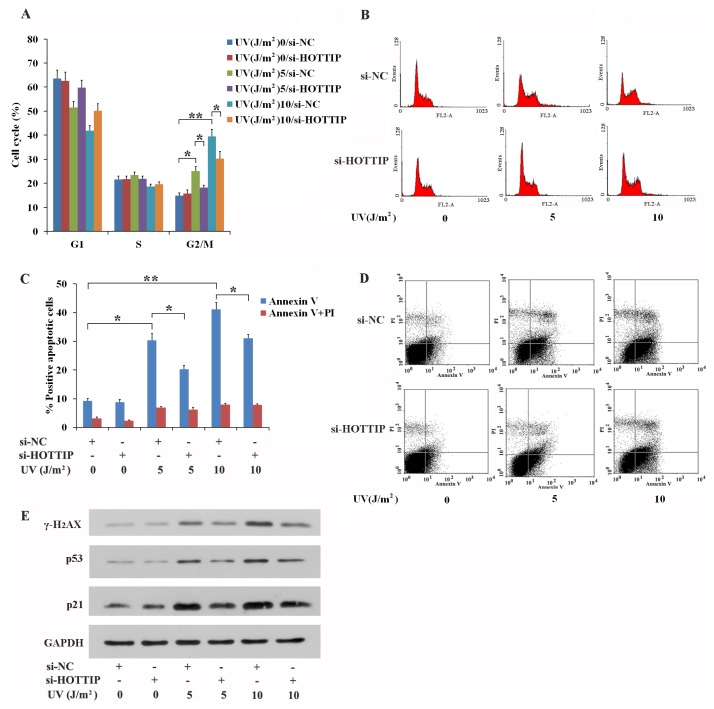
We next explored how lncRNA-HOTTIP regulates proliferation post UV irradiation in GC-1 cells. The effects of stimulation with 0, 5, 10 J/m2 UV on cell-cycle progression was determined using FACS analysis. These experiments revealed that stimulation with 5 J/m2 and 10 J/m2 UV can inhibit GC-1 from exiting G2/M phase relative to those that were not irradiated with UV (Figs. 2A and 2B). Knockdown of HOTTIP expression partially reversed the effects of UV, reducing the level of G2/M phase arrest in GC-1 cells stimulated with either 5 J/m2 or 10 J/m2 UV (Figs. 2A and 2B). Furthermore, stimulation with either 5 J/m2 or 10 J/m2 UV causes a significant increase in the proportion of cells that undergo early apoptosis compared with compared with those not subjected to UV treatment (Figs. 2C and 2D). Knockdown of HOTTIP partially reversed the occurrence of early apoptosis in GC-1 cells irradiated with either 5 J/m2 or 10 J/m2 UV (Figs. 2C and 2D).
Fig. 2. UV treatment leads to G2/M phase arrest and apoptosis through lncRNA-HOTTIP in GC-1 cells.
(A–D) Knockdown of HOTTIP partially rescues the level of G2/M phase arrest (A and B) and early apoptosis (C and D) in UV-stimulated GC-1 cells. GC-1 cells were transfected with si-NC or si-HOTTIP, and were treated using 0, 5, and 10 J/m2 UV 24-h post transfection. 24 h subsequently, cell cycle (A and B) and apoptosis (C and D) of GC-1 cells were detected by using FACS. Representative photographs of cell cycle and apoptosis were shown in panels (B) and (D), respectively. (E) HOTTIP inhibited the expression of proteins that regulate UV-induced cell cycle arrest, apoptosis and DNA damage repair in GC-1 cells. GC-1 cells were transfected with si-NC or si-HOTTIP, and were treated using 0, 5, and 10 J/m2 UV 24-h post transfection. Protein abundance was analyzed using western blotting. (A and C) Data shown are the mean ± SEM of three independent experiments each performed in triplicate. *P < 0.05, **P < 0.01.
To determine whether levels of the cell cycle and apoptosis-related proteins, p53 and p21, and the UV-induced, DNA-damage-repair protein, γ-H2AX, were affected, relative expression of the proteins were assessed by western blot. As shown in Figure 2E, irradiation with 5 J/m2 and 10 J/m2 UV promoted expression of both p53 and p21, proving a mechanistic explanation of why GC-1 cells accumulated in G2/M phase and underwent early apoptosis post UV exposure. Stimulation with 5 J/m2 and 10 J/m2 UV also enhanced γ-H2AX expression, which suggests that DNA damage occurs in UV-irradiated GC-1 cells (Fig. 2E). Knockdown of HOTTIP can reverse the effects stimulation with 5 J/m2 and 10 J/m2 UV with respect to p53, p21 and γ-H2AX levels (Fig. 2E). Furthermore, lncRNA-HOTTIP can promote γ-H2AX transcription in UV-induced GC-1 cells with DNA damage (Supplementary Fig. S2). These results suggest that HOTTIP may have the ability to inhibit UV-induced cell-cycle arrest, apoptosis and DNA damage repair. Induction of G2/M cell cycle arrest by lncRNA-HOTTIP indicates that HOTTIP acts as a pro-apoptotic activator and then causes DNA damage in UV-stimulated GC-1 cells.
Hoxa13 functions downstream from lncRNA-HOTTIP in UV-treated GC-1 cells
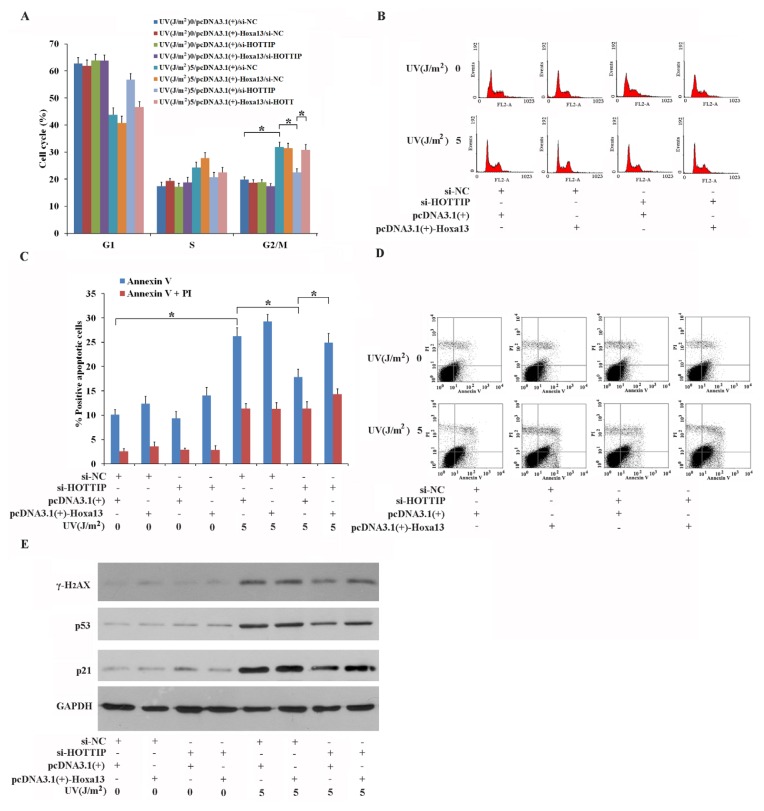
Previous studies have reported that Hoxa13 is involved in cell proliferation, the cell cycle and apoptosis both in vivo and in vitro (Morgan et al., 2003; Qu et al., 2017; Shi et al., 2018). These reports were able to show that expression of lncRNA-HOTTIP and Hoxa13 are coordinated, so we examined whether Hoxa13 expression affected HOTTIP-mediated UV responses in GC-1 cells. Results here demonstrated that overexpression of Hoxa13 reverses si-HOTTIP-mediated repression of G2/M phase arrest (Figs. 3A and 3B) and early apoptosis (Figs. 3C and 3D) in GC-1 cells irradiated with 5 J/m2 UV. Furthermore, overexpression of Hoxa13 can block si-HOTTIP-mediated inhibition of p53, p21 and γ-H2AX accumulation in GC-1 cells irradiated with 5 J/m2 UV (Fig. 3E). Overexpression of Hoxa13 has no significant effect on the cell cycle, apoptosis and the expression of proteins related to those processes without UV treatment, indicating that the high expression level of Hoxa13 does not likely alter GC-1 cell activity under normal cellular conditions. Taken together, these experiments suggest that a novel signaling pathway consisting of the coordinated expression of HOTTIP-Hoxa13 is involved in the response to UV irradiation in GC-1 cells in which expression of key regulatory proteins and corresponding cellular mechanisms are altered.
Fig. 3. Hoxa13 mediates the function of lncRNA-HOTTIP in UV-stimulated GC-1 cells.
(A–D) Overexpression of Hoxa13 blocks the inhibitory effects of si-HOTTIP on G2/M phase arrest (A and B) and early apoptosis (C and D). GC-1 cells were transfected with either si-NC or si-HOTTIP and pcDNA3.1(+) or pcDNA3.1(+)-Hoxa13, and were treated with 0 and 5 J/m2 UV 24-h post transfection. 24-h post UV irradiation, cell cycle progression (A and B) and apoptosis (C and D) of GC-1 cells were assessed using FACS. Representative photographs of cell cycle progression and apoptosis were shown (B) and (D), respectively. (E) Hoxa13-mediates the promotion of expression of p53, p21 and γ-H2AX proteins, which are reduced in si-HOTTIP post UV exposure in GC-1 cells. GC-1 cells were transfected with si-NC or si-HOTTIP, and were irradiated with and 5 J/m2 UV 24-h post transfection. Isolated proteins was analyzed using western blotting. (A and C) Data shown are the mean ± SEM of three independent experiments each performed in triplicate. *P < 0.05.
p53 regulates lncRNA-HOTTIP expression in vitro and in vivo
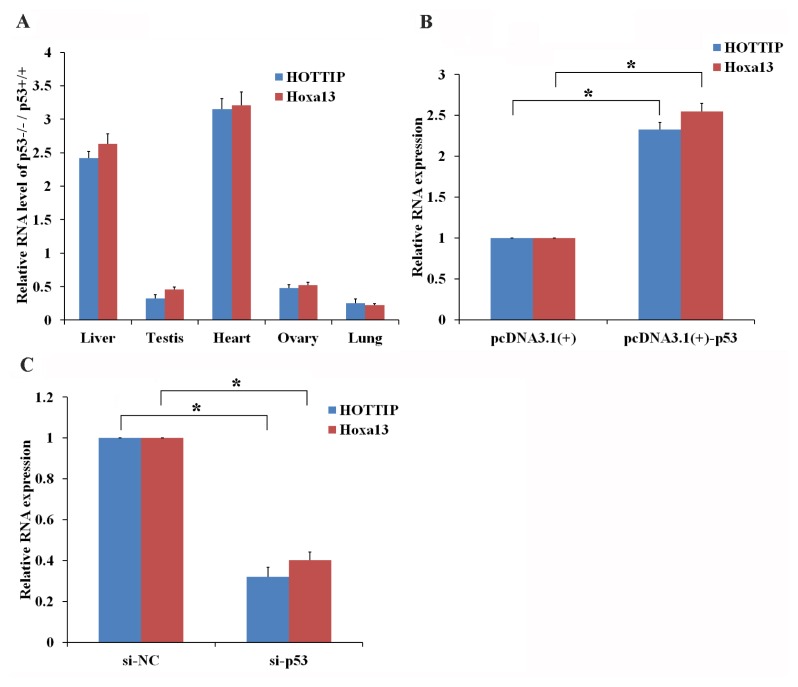
The mutual regulation of p53 and lncRNA has already been shown to vital roles in physiological and pathological processes such as development and cancer (Joo et al., 2019; Voce et al., 2019). In this research, we revealed that expression of lncRNA-HOTTIP and Hoxa13 was lower in p53 −/− mouse testis, ovaries and lung cells compared with the same type of cells from p53 +/+ mice (Fig. 4A). Furthermore, overexpression of p53 in GC-1 cells significantly increased the expression level of both lncRNA-HOTTIP and Hoxa13 (Fig. 4B), while knockdown of p53 in GC-1 cells significantly decreased the expression of lncRNA-HOTTIP and Hoxa13 (Fig. 4C). These data identified a p53-HOTTIP-hoxa13 signaling pathway that participates in the cellular response to UV. Further, expression of lncRNA-HOTTIP and Hoxa13 was higher in p53 −/− mouse liver and heart cells compared with liver and heart cells from a p53 +/+ mouse (Fig. 4A), these findings suggest that p53 regulates HOTTIP expression in a tissue-specific manner. In the future, further study will be conducted both in vitro and in vivo to elucidate the molecular mechanism employed by p53 in the regulation of lncRNA-HOTTIP in GC-1 cells post UV irradiation.
Fig. 4. p53 enhances the expression of lncRNA-HOTTIP in vitro and in vivo.
Expression of HOTTIP and Hox13 was measured in mouse tissues with and without p53 in vivo have been shown in (A). Total RNA was isolated from p53 +/+ mouse tissues and p53 −/− mouse tissues, and then RT-qPCR was used to measure HOTTIP and Hoxa13. (B and C) p53 promotes the expression of HOTTIP and Hoxa13 in vitro. GC-1 cells were transfected with pcDNA3.1(+)/pcDNA3.1(+)-Hoxa13 (B) or si-NC/si-p53 (C). Isolated RNA was analyzed using RT-qPCR after 24-h post transfection. (A–C) Data are presented as the mean ± SEM of three independent experiments, each performed in triplicate. *P < 0.05.
DISCUSSION
The majority of the animal genomes encode a large number of noncoding RNAs, which can be broadly divided into two groups comprised of small ncRNAs (18–200 bp) and long ncRNAs (200 bp to > 100 kb) (Bohacek and Mansuy, 2015; Prasanth and Spector, 2007). In addition to well-studied small ncRNAs (e.g., microRNAs, piwi-interacting RNAs and small interfering RNAs), lncRNAs have recently been identified as important molecules that are responsible for the regulation of diverse cellular processes including dosage compensation and genomic imprinting (Militti et al., 2014; Park et al., 2017). Furthermore, the coordinated expression of lncRNAs with proximal or distal genes plays an important role in physiological and pathological processes in lower and higher organisms (Carmona et al., 2018; Li et al., 2017; Tan et al., 2019; Zhao et al., 2018). For example, the expression of the β-catenin-coordinated lncRNA, MALAT1, and downstream signaling factors, affects the function of hepatocyte growth factors and promotes differentiation of bone marrow mesenchymal stem cells into hepatocytes (Tan et al., 2019). The lncRNA, Jpx, regulates expression of the lncRNA, Xist, in a dose-dependent manner using both trans and cis methods, which were confirmed using transgenic mouse models (Carmona et al., 2018). The lncRNA, MACC1-AS1, is an antisense version of the transcriptional regulator, MACC1, which regulates the mesenchymal to epithelial transition (MET). MACC1 expression increases under conditions of metabolic stimulation and facilitates metabolic plasticity through enhancing MACC1 expression via mRNA stabilization (Zhao et al., 2018). The signaling pathway including lncRNA linc1281, miRNA let-7 and DNA methylation promote the acute extinction of naive pluripotency, which is required for the rapid transition from the pre-implantation epiblast stage to gastrulation in rodents (Li et al., 2017). In this research, we demonstrated that lncRNA-HOTTIP is coordinated with expression of Hoxa13 both in vivo and in vitro. The synergistic expression of lncRNA-HOTTIP-Hoxa13 mediates the effects of UV stimulation on spermatogonia germ cell GC-1 cells. These data imply that lncRNA-HOTTIP and Hoxa13 may cooperate to regulate the reproductive system in physiological and pathological processes. We also revealed that UV induces the expression of lncRNA-HOTAIR/HOTTIP and inhibits the expression of lncRNA-p21/MD-1 in GC-1 cells (Supplementary Fig. S3). The transcriptome-wide examination of lncRNAs induced by UV using RNA-seq will be studied in future. In addition, UV can also promote the expression of lncRNA-HOTTIP in skin TE353-3K cells (Supplementary Fig. S4). Further research investigating UV-induced HOTTIP in TE353-3K cells will be need to elucidate the role of the lncRNA in skin cells.
Many studies have focused on the functions of lncRNAs with respect to cellular proliferation, cell cycle and apoptosis and have revealed both positive and negative effects of lncRNAs on the cellular functioning (Lei et al., 2018; Li et al., 2018b; Wang et al., 2018a; Zeng et al., 2017; Zhang et al., 2018). Low expression of lncRNA NBAT-1 is associated with the occurrence of lung cancer and overexpression of lncRNA NBAT-1 suppress cellular proliferation, the cell cycle and stimulate apoptosis in A549 cells (Lei et al., 2018). LncRNA AF113014 restrains proliferation of the hepatocllular carcinoma cell line both in vitro and in vivo through acting as the sponge for miR-20a, which directly targets Egr2 (Zeng et al., 2017). LncRNA MIR31HG can enhance cellular proliferation and cell cycle progression, and block apoptosis in vitro and in vivo by regulating HIF1A and p21 in head and neck squamous cell carcinoma (Wang et al., 2018a). LncRNA CACS11 affects the gastric cancer cell cycle signaling pathway consisting of miR-340-5p-CDK1 and then activates cellular proliferation, migration and invasion (Zhang et al., 2018). The lncRNA TP73AS1 is considered an ovarian cancer oncogenic lncRNA, which promotes proliferation of ovarian cancer by modifying the cell cycle and apoptosis signaling (Li et al., 2018b). In the present study, we demonstrated that lncRNA-HOTTIP has no significant effects on the proliferation, cycle and apoptosis of GC-1 cells under physiological conditions. However, GC-1 cells produce large amounts of lncRNA-HOTTIP which can regulate the GC-1 cellular proliferation, cycle and apoptosis post UV irradiation. These results demonstrated that the roles of some lncRNAs may be limited to the response to stress or to regulating cellular activities under conditions of abnormal cellular functioning such as cancer.
UV is a vital external stimulus that can regulate cellular activities in various biological systems through the modification of multiple signaling pathways (Ascer et al., 2015; Farrell et al., 2018; Kaneko et al., 2015; Matsunuma et al., 2016; Siegenthaler et al., 2018). UV irradiation facilitates the phosphorylation of histone acetyltransferases and affects CRL4 ubiquitylation and, subsequently, inhibits cellular proliferation (Matsunuma et al., 2016). Over-activation of CDC42 is induced by UV stimulation and results in G2/M phase arrest and suppresses proliferation and survival in Hela cells (Ascer et al., 2015). The deficiency of chromatin remodeling protein Brahma in keratinocytes enhances UV-induced carcinogenesis through promoting the cell cycle progression and promotes the accumulation of DNA mutations (Farrell et al., 2018). BV-2 microglial cells treated with lipopolysaccharide can resist UV-stimulated apoptosis via cell cycle arrest in G1 through the upregulation of expression of p21 and GADD45α (Kaneko et al., 2015). The transcription factor nuclear factor erythroid 2-related factor 3 (Nrf3) activates UV-induced apoptosis in keratinocytes through the inhibition of cell-cell and cell-matrix adhesion (Siegenthaler et al., 2018). This study has revealed that UV treatment significantly promotes lncRNA-HOTTIP expression, which coordinates the activation of Hoxa13, a neighboring gene, in GC-1 cells. In addition, UV-induced G2/M phase arrest and early apoptosis are regulated by HOTTIP and Hoxa13. Signaling through lncRNA-HOTTIP-Hoxa13 regulates the expression of p53, p21 and γ-H2AX in UV irradiated GC-1 cells. Taken together, these results have shown that a novel lncRNA-related signaling cascade participates in regulating the response to UV irradiation in reproductive cells.
A growing number of studies have indicated that lncRNA-HOTTIP contributes to a variety of human conditions and are key candidates as biomarkers of disease (Li et al., 2018a; Su et al., 2019; Sun and Liu, 2018; Wang et al., 2018b; Zhuang et al., 2019). Upregulation of lncRNA-HOTTIP in acute myeloid leukemia-M5 patients significantly activated cellular proliferation and cell cycle progression through the modification miR-608-DDA1 (Zhuang et al., 2019). In renal carcinoma cells, increased expression of lncRNA-HOTTIP promotes cellular proliferation, migration and invasion and inhibits cellular autophagy via the PI3K-Akt-Atg13 signaling pathway (Su et al., 2019). In addition, the lncRNA-HOTTIP levels were decreased in an ischemic stroke mouse model, while overexpression of lncRNA-HOTTIP weakened oxygen-glucose-deprivation-induced neuronal injury. The lncRNA affected glycolytic metabolism by sequestering miR-143, which directly targeted HK-2 expression (Wang et al., 2018b). Expression of lncRNA-HOTTIP was remarkably increased in diabetic retinopathy rat and mouse models, which aggravate diabetic retinal microangiopathy via p38-MAPK signaling (Sun and Liu, 2018). The reduced level of expression of lncRNA-HOTTIP in preeclampsia patients has been associated with maternal blood pressure and urinary protein levels. Overexpression of lncRNA-HOTTIP, however has been shown to promote trophoblast cellular proliferation and cell cycle progression through suppressing Rho family GTPase 3 (Li et al., 2018a). This research demonstrated that the expression of lncRNA-HOTTIP and Hoxa13 was significantly decreased in the testis of p53 −/− versus wild type mouse models. Overexpression of p53 in GC-1 cells significantly promoted the expression of lncRNA-HOTTIP and Hoxa13 in vitro. These results suggest that the mutual regulation of p53 and HOTTIP may occur in the cellular response to UV induction in GC-1 cells. To enhance our understanding of the detailed regulatory signaling pathway involving p53 and lncRNA-HOTTIP in GC-1 cells further research is required.
This research showed a novel lncRNA-HOTTIP-Hoxa13 signaling pathway in the regulation of the UV-induced GC-1 cellular response. UV irradiation causes spermatogonia germ cell line, GC-1, cells to arrest in the G2/M phase and undergo early apoptosis. This occurs, at least partiall0y, through the synergistic co-expression of HOTTIP and Hoxa13, along with increased expression of downstream signaling factors including p53, p21 and γ-H2AX (Fig. 5). Further analysis of the roles of lncRNA-HOTTIP in vivo may provide the information necessary for strengthening treatments for diseases caused by UV irradiation.
Fig. 5. Model for describing the lncRNA-HOTTIP-Hoxa13 molecular signaling cascade in UV-treated GC-1 cells.
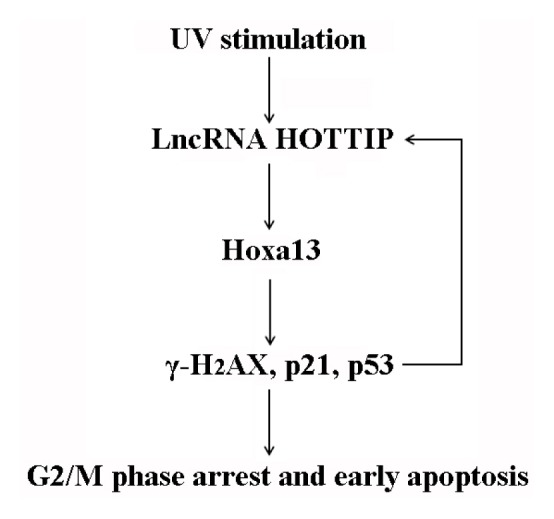
UV promotes G2/M phase arrest and early apoptosis in the spermatogonia germ cell line GC-1. This occurs, at least partially, through the coordinated induction of lncRNA-HOTTIP and Hoxa13 expression.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the following grants: National Natural Science Foundation of China (81801516), Key project of Natural Science Foundation of Anhui Provincial Department of Education (P.R. China) (KJ2018A0992).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Armstrong B.K., Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B. 2001;63:8–18. doi: 10.1016/S1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- Ascer L.G., Magalhaes Y.T., Espinha G., Osaki J.H., Souza R.C., Forti F.L. CDC42 Gtpase activation affects hela cell DNA repair and proliferation following UV radiation-induced genotoxic stress. J Cell Biochem. 2015;116:2086–2097. doi: 10.1002/jcb.25166. [DOI] [PubMed] [Google Scholar]
- Bohacek J., Mansuy I.M. Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nat Rev Genet. 2015;16:641–652. doi: 10.1038/nrg3964. [DOI] [PubMed] [Google Scholar]
- Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Burgess D.J. Non-coding RNA: HOTTIP goes the distance. Nat Rev Genet. 2011;12:300. doi: 10.1038/nrg2992. [DOI] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carmona S., Lin B., Chou T., Arroyo K., Sun S. LncRNA Jpx induces Xist expression in mice using both trans and cis mechanisms. PLoS Genet. 2018;14:e1007378. doi: 10.1371/journal.pgen.1007378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.L., Carmichael G.G. Decoding the function of nuclear long non-coding RNAs. Curr Opin Cell Biol. 2010;22:357–364. doi: 10.1016/j.ceb.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gruijl F.R. Skin cancer and solar UV radiation. Eur J Cancer. 1999;35:2003–2009. doi: 10.1016/S0959-8049(99)00283-X. [DOI] [PubMed] [Google Scholar]
- de Gruijl F.R., van Kranen H.J., Mullenders L.H. UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J Photochem Photobiol B. 2001;63:19–27. doi: 10.1016/S1011-1344(01)00199-3. [DOI] [PubMed] [Google Scholar]
- Degueurce G., D’Errico I., Pich C., Ibberson M., Schutz F., Montagner A., Sgandurra M., Mury L., Jafari P., Boda A., et al. Identification of a novel PPARbeta/delta/miR-21-3p axis in UV-induced skin inflammation. EMBO Mol Med. 2016;8:919–936. doi: 10.15252/emmm.201505384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell A.W., Halliday G.M., Lyons J.G. Brahma deficiency in keratinocytes promotes UV carcinogenesis by accelerating the escape from cell cycle arrest and the formation of DNA photolesions. J Dermatol Sci. 2018;92:254–263. doi: 10.1016/j.jdermsci.2018.11.006. [DOI] [PubMed] [Google Scholar]
- Gentile M., Latonen L., Laiho M. Cell cycle arrest and apoptosis provoked by UV radiation-induced DNA damage are transcriptionally highly divergent responses. Nucleic Acids Res. 2003;31:4779–4790. doi: 10.1093/nar/gkg675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.J., Bikle D.D. LncRNA profiling reveals new mechanism for VDR protection against skin cancer formation. J Steroid Biochem. 2014;144(Pt A):87–90. doi: 10.1016/j.jsbmb.2013.11.018. [DOI] [PubMed] [Google Scholar]
- Joo M.S., Shin S.B., Kim E.J., Koo J.H., Yim H., Kim S.G. Nrf2-lncRNA controls cell fate by modulating p53-dependent Nrf2 activation as an miRNA sponge for Plk2 and p21(cip1) FASEB J. 2019;33:7953–7969. doi: 10.1096/fj.201802744R. [DOI] [PubMed] [Google Scholar]
- Kaneko Y.S., Ota A., Nakashima A., Nagasaki H., Kodani Y., Mori K., Nagatsu T. Lipopolysaccharide treatment arrests the cell cycle of BV-2 microglial cells in G(1) phase and protects them from UV light-induced apoptosis. J Neural Transm. 2015;122:187–199. doi: 10.1007/s00702-014-1256-5. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kim D., Song J., Han J., Kim Y., Chun C.H., Jin E.J. Two non-coding RNAs, MicroRNA-101 and HOTTIP contribute cartilage integrity by epigenetic and homeotic regulation of integrin-alpha1. Cell Signal. 2013;25:2878–2887. doi: 10.1016/j.cellsig.2013.08.034. [DOI] [PubMed] [Google Scholar]
- Koberle B., Roginskaya V., Zima K.S., Masters J.R., Wood R.D. Elevation of XPA protein level in testis tumor cells without increasing resistance to cisplatin or UV radiation. Mol Carcinogen. 2008;47:580–586. doi: 10.1002/mc.20418. [DOI] [PubMed] [Google Scholar]
- Lei T., Lv Z.Y., Fu J.F., Wang Z., Fan Z., Wang Y. LncRNA NBAT-1 is down-regulated in lung cancer and influences cell proliferation, apoptosis and cell cycle. Eur Rev Med Pharmaco. 2018;22:1958–1962. doi: 10.26355/eurrev_201804_14721. [DOI] [PubMed] [Google Scholar]
- Li A., Wei G., Wang Y., Zhou Y., Zhang X.E., Bi L., Chen R. Identification of intermediate-size non-coding RNAs involved in the UV-induced DNA damage response in C. elegans PLoS One. 2012;7:e48066. doi: 10.1371/journal.pone.0048066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang X., Hong S.L., Chen Y., Ren G.H. Long non-coding HOTTIP regulates preeclampsia by inhibiting RND3. Eur Rev Med Pharmaco. 2018a;22:3277–3285. doi: 10.26355/eurrev_201806_15146. [DOI] [PubMed] [Google Scholar]
- Li M.A., Amaral P.P., Cheung P., Bergmann J.H., Kinoshita M., Kalkan T., Ralser M., Robson S., von Meyenn F., Paramor M., et al. A lncRNA fine tunes the dynamics of a cell state transition involving Lin28, let-7 and de novo DNA methylation. eLife. 2017;18:e23468. doi: 10.7554/eLife.23468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang X., Mao L., Zhao S., Wei H. LncRNA TP73AS1 predicts poor prognosis and promotes cell proliferation in ovarian cancer via cell cycle and apoptosis regulation. Mol Med Rep. 2018b;18:516–522. doi: 10.3892/mmr.2018.8951. [DOI] [PubMed] [Google Scholar]
- Li Z., Zhao X., Zhou Y., Liu Y., Zhou Q., Ye H., Wang Y., Zeng J., Song Y., Gao W., et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M., Yao G., Yin M., Lu M., Tian H., Liu L., Lian J., Huang X., Sun F. Transcriptional cooperation between p53 and NF-kappaB p65 regulates microRNA-224 transcription in mouse ovarian granulosa cells. Mol Cell Endocrinol. 2013;370:119–129. doi: 10.1016/j.mce.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Liu X., Li D., Zhang W., Guo M., Zhan Q. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 2012;31:4415–4427. doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Hu Y.W., Zhao Z.L., Zheng L., Qiu Y.R., Huang J.L., Wu X.J., Mao X.R., Yang J., Zhao J.Y., et al. Anti-inflammatory effects of propofol are mediated by apolipoprotein M in a hepatocyte nuclear factor-1alpha-dependent manner. Arch Biochem Biophys. 2013;533:1–10. doi: 10.1016/j.abb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Matsunuma R., Niida H., Ohhata T., Kitagawa K., Sakai S., Uchida C., Shiotani B., Matsumoto M., Nakayama K.I., Ogura H., et al. UV damage-induced phosphorylation of HBO1 triggers CRL4DDB2-mediated degradation to regulate cell proliferation. Mol Cell Biol. 2016;36:394–406. doi: 10.1128/MCB.00809-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T.R., Dinger M.E., Mattick J.S. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Militti C., Maenner S., Becker P.B., Gebauer F. UNR facilitates the interaction of MLE with the lncRNA roX2 during Drosophila dosage compensation. Nat Commun. 2014;5:4762. doi: 10.1038/ncomms5762. [DOI] [PubMed] [Google Scholar]
- Morgan E.A., Nguyen S.B., Scott V., Stadler H.S. Loss of Bmp7 and Fgf8 signaling in Hoxa13-mutant mice causes hypospadia. Development. 2003;130:3095–3109. doi: 10.1242/dev.00530. [DOI] [PubMed] [Google Scholar]
- Navarro A., Moises J., Santasusagna S., Marrades R.M., Vinolas N., Castellano J.J., Canals J., Munoz C., Ramirez J., Molins L., et al. Clinical significance of long non-coding RNA HOTTIP in early-stage non-small-cell lung cancer. BMC Pulm Med. 2019;19:55. doi: 10.1186/s12890-019-0816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K.S., Mitra A., Rahat B., Kim K., Pfeifer K. Loss of imprinting mutations define both distinct and overlapping roles for misexpression of IGF2 and of H19 lncRNA. Nucleic Acids Res. 2017;45:12766–12779. doi: 10.1093/nar/gkx896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K.V., Spector D.L. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Gene Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Qu L.P., Zhong Y.M., Zheng Z., Zhao R.X. CDH17 is a downstream effector of HOXA13 in modulating the Wnt/beta-catenin signaling pathway in gastric cancer. Eur Rev Med Pharmaco. 2017;21:1234–1241. [PubMed] [Google Scholar]
- Quagliata L., Matter M., Piscuoglio S., Makowska Z., Heim M., Tornillo L., Cillo C., Terracciano L. Hoxa13 and hottip expression levels predict patients’ survival and metastasis formation in hepatocellular carcinoma. J Hepatol. 2013;58:S39–S40. doi: 10.1016/S0168-8278(13)60092-6. [DOI] [Google Scholar]
- Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q., Shen L., Dong B., Fu H., Kang X., Dai L., Yang Y., Yan W., Chen K.N. Downregulation of HOXA13 sensitizes human esophageal squamous cell carcinoma to chemotherapy. Thorac Cancer. 2018;9:836–846. doi: 10.1111/1759-7714.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler B., Defila C., Muzumdar S., Beer H.D., Meyer M., Tanner S., Bloch W., Blank V., Schafer M., Werner S. Nrf3 promotes UV-induced keratinocyte apoptosis through suppression of cell adhesion. Cell Death Differ. 2018;25:1749–1765. doi: 10.1038/s41418-018-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Lu J., Chen X., Liang C., Luo P., Qin C., Zhang J. Long non-coding RNA HOTTIP affects renal cell carcinoma progression by regulating autophagy via the PI3K/Akt/Atg13 signaling pathway. J Cancer Res Clin. 2019;145:573–588. doi: 10.1007/s00432-018-2808-0. [DOI] [PubMed] [Google Scholar]
- Sun Y., Liu Y.X. LncRNA HOTTIP improves diabetic retinopathy by regulating the p38-MAPK pathway. Eur Rev Med Pharmaco. 2018;22:2941–2948. doi: 10.26355/eurrev_201805_15048. [DOI] [PubMed] [Google Scholar]
- Tan Y.F., Tang L., OuYang W.X., Jiang T., Zhang H., Li S.J. beta-catenin-coordinated lncRNA MALAT1 up-regulation of ZEB-1 could enhance the telomerase activity in HGF-mediated differentiation of bone marrow mesenchymal stem cells into hepatocytes. Pathol Res Pract. 2019;215:546–554. doi: 10.1016/j.prp.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Tripathi V., Shen Z., Chakraborty A., Giri S., Freier S.M., Wu X., Zhang Y., Gorospe M., Prasanth S.G., Lal A., et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wees C.G., Vreeswijk M.P., Persoon M., van der Laarse A., van Zeeland A.A., Mullenders L.H. Deficient global genome repair of UV-induced cyclobutane pyrimidine dimers in terminally differentiated myocytes and proliferating fibroblasts from the rat heart. DNA Repair. 2003;2:1297–1308. doi: 10.1016/j.dnarep.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Voce D.J., Bernal G.M., Wu L., Crawley C.D., Zhang W., Mansour N.M., Cahill K.E., Szymura S.J., Uppal A., Raleigh D.R., et al. Temozolomide treatment induces lncRNA MALAT1 in an NF-kappaB and p53 codependent manner in glioblastoma. Cancer Res. 2019;79:2536–2548. doi: 10.1158/0008-5472.CAN-18-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Ma Z., Feng L., Yang Y., Tan C., Shi Q., Lian M., He S., Ma H., Fang J. LncRNA MIR31HG targets HIF1A and P21 to facilitate head and neck cancer cell proliferation and tumorigenesis by promoting cell-cycle progression. Mol Cancer. 2018a;17:162. doi: 10.1186/s12943-018-0916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li G., Zhao L., Lv J. Long noncoding RNA HOTTIP alleviates oxygen-glucose deprivation-induced neuronal injury via modulating miR-143/hexokinase 2 pathway. J Cell Biochem. 2018b;119:10107–10117. doi: 10.1002/jcb.27348. [DOI] [PubMed] [Google Scholar]
- Williamson L., Saponaro M., Boeing S., East P., Mitter R., Kantidakis T., Kelly G.P., Lobley A., Walker J., Spencer-Dene B., et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell. 2017;168:843–855.e13. doi: 10.1016/j.cell.2017.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Tian J., Li Q.Y. Downregulation of HOTTIP regulates insulin secretion and cell cycle in islet beta cells via inhibiting MEK/ERK pathway. Eur Rev Med Pharmaco. 2018;22:4962–4968. doi: 10.26355/eurrev_201808_15636. [DOI] [PubMed] [Google Scholar]
- Zeng T., Wang D., Chen J., Tian Y., Cai X., Peng H., Zhu L., Huang A., Tang H. LncRNA-AF113014 promotes the expression of Egr2 by interaction with miR-20a to inhibit proliferation of hepatocellular carcinoma cells. PLoS One. 2017;12:e0177843. doi: 10.1371/journal.pone.0177843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kang W., Lu X., Ma S., Dong L., Zou B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17:1886–1900. doi: 10.1080/15384101.2018.1502574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao Y., Liu Y., Lin L., Huang Q., He W., Zhang S., Dong S., Wen Z., Rao J., Liao W., et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17:69. doi: 10.1186/s12943-018-0820-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R., Ansell P.J., Zhao J., Weng C., Klibanski A. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- Zhuang M.F., Li L.J., Ma J.B. LncRNA HOTTIP promotes proliferation and cell cycle progression of acute myeloid leukemia cells. Eur Rev Med Pharmaco. 2019;23:2908–2915. doi: 10.26355/eurrev_201904_17569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.