Abstract
Metabolomics refers to study of metabolites in biospecimens such as blood serum, urine and tissues. NMR (Nuclear magnetic resonance spectroscopy) and UPLC-MS (mass spectrometry coupled to liquid chromatography) are most frequently employed to analyze the complex biological/clinical samples. NMR is a relatively insensitive tool compared to UPLC-MS, but offers straightforward quantification, identification and easy sample processing. One dimensional 1H NMR spectroscopy is inherently quantitative and can be readily used for metabolite quantification without individual metabolite standards. Two-dimensional spectroscopy is most commonly used for identification of metabolites but can also be used quantitively. Although NMR experiments are unbiased to the chemical nature of the analyte, it is crucial to adhere to proper metabolite extraction protocol for optimum results. Selection and implementation of appropriate NMR pulse programs is also equally important. Finally, employment of correct metabolite quantification strategy is also crucial. In this protocol, a step-by-step guidance of running NMR metabolomics experiment from typical biospecimens are presented. The protocol describes an optimized metabolite extraction protocol, followed by implementation of NMR experiments and quantification strategies using the so-called ‘targeted profiling’ technique. This approach relied on an underlying basis-set of metabolite spectra acquired under similar conditions. Finally, some strategies towards statistical analysis of the data is also presented. Overall, this protocol should serve as a guide for anyone who wishes to enter the world of NMR based metabolomics analysis.
Keywords: NMR, Metabolomics, Metabolites, Targeted profiling
INTRODUCTION
Success of NMR metabolomics studies depend on using optimized protocols for sample processing, selection of appropriate pulse programs and using the correct data analysis technique for quantification of metabolites. 1H NMR spectroscopy is frequently the method of choice owing to high natural abundance and sensitivity of the 1H nucleus. Other nuclei, such as 13C or 15N, may be used in special cases (1,2). The majority of NMR metabolomics studies use 1-dimensional 1H NMR for metabolic profiling purposes. Such experiments often suffer from complex overlapping signals from metabolites. This issue may be resolved either by using two-dimensional (2D) NMR experiments (3,4), or targeted profiling of metabolites using a library-based approach (5). 2D NMR is also the method of choice for elucidation of the chemical structure of unknown metabolites/small molecules (6).
The protocols described in this manuscript will be enough for establishing an NMR metabolomics pipeline. Extraction/sample processing for the most frequently encountered biospecimens (urine, blood serum/plasma/tissues) are described first. Then, typical NMR experiments (1D and 2D) are explained followed by the data analysis strategies. The commentary presents the critical parameters that need special attention. It is important to point out that metabolomics is still a growing field and several protocols are not standardized per se. Therefore, we will describe protocols that our laboratory successfully uses in a routine NMR metabolomics experiment (7–9), however, alternative protocols will be pointed out when necessary.
BASIC PROTOCOL 1
METABOLITE EXTRACTION FROM DIFFERENT BIOSPECIMEN
Several studies have tried to optimize metabolite extraction strategies for biospecimens (10–13). Extraction of small molecular metabolites from protein based biofluid/tissue matrix is crucial for NMR experiments since broad signals from large protein molecules can mask the signals of metabolites. Metabolites such as lactate, citrate and several amino acids are also known to form complexes with protein and thus remain partly “NMR invisible” (14,15) but may be quantified using specialized techniques (16).
Metabolite extraction from biofluids such as blood serum is straightforward and separates the proteins from the metabolites by using either organic solvents or ultrafiltration with 3kDa cutoff filters (17–19). Tissue/organ metabolite extraction require an additional step of tissue homogenization, preferably in the extraction solvent. In this section, a biphasic solvent extraction protocol adapted from the Bligh-Dyer method (20,21) will be described (22). An advantage of this method is that it also collects the lipid fraction from biospecimens that may be used for further lipidomics analysis, thereby providing complementary information (22).
Materials
Three sets of 2ml microtubes per sample. (Note: If using a tissue homogenizer (see below), use a safe-lock microtube, such as Eppendorf safe-lock tubes 2.0 ml, catalog no 022363352)
-
Pre-cooled (on ice) solvents – UPLC-MS grade methanol (Fisher Scientific product A456–4) and chloroform (Fisher Scientific product C298–1), Milli-Q® water.
Note: Treatment involving chloroform should be performed under fume hood and with proper personal protective equipments.
Micropipette and fresh pipette tips (do not need to be autoclaved).
-
Tissue homogenizer – electronic systems such as TissueLyzer II (Qiagen) is preferred for minimizing manual intervention/ analytical variation and increased throughput. Otherwise, mechanical homogenizers may also be used.
(Note: If using a tissue homogenizer system, pre-cool the blocks to −80°C.)
Pre-cooled (4°C) microcentrifuge (Eppendorf centrifuge 5430-R).
Vacuum concentrator (Eppendorf Vacufuge plus).
Sonicator bath (VWR model 97043–976).
Protocol steps
Note: The following protocol is valid for 50 μl serum/plasma or 50 mg soft tissue or 1×106 cells. For different amounts, the solvent volumes should be scaled up or down.
-
A. Serum or plasma: Place 50 μl serum or plasma in a microtube on ice.
B. Solid tissues: weigh out 50 mg tissue, snap freeze in liquid nitrogen till further processing. Place in ice just before sample processing.
C. Cells: Use cell pellet previously collected and stored at −80°C. (For collection of cell pellet see (23)).
Prepare 2:1 methanol/chloroform in a glass solvent bottle and place in ice.
Add 300 μl 2:1 methanol/chloroform to the serum/plasma/tissue/cell pellet.
Vortex the samples until a homogeneous suspension is formed.
If processing cell pellets, go to step 6, otherwise go to step 7.
-
Sonicate the cell pellet samples continuously (35 kHz, 240 Watts) for 15 minutes.
Note: make sure the caps of the tubes are secure.
Add 100 μl of chloroform and 100 μl water to the sample.
Vortex again to ensure a good mix.
If processing a tissue sample, go to step 10, otherwise go to step 11.
-
Use the tissue homogenizer system to homogenize the tissue.
Note: The time and speed of tissue homogenizer will depend upon the system. For TissueLyzer II – two rounds of 3 min, 25 Hz cycles provide satisfactory results.
-
Put the sample tubes in centrifuge and spin for 7 mins, 18787 RCF
Note: the RCF value is translated to 13300 RPM for Eppendorf centrifuge 5430R.
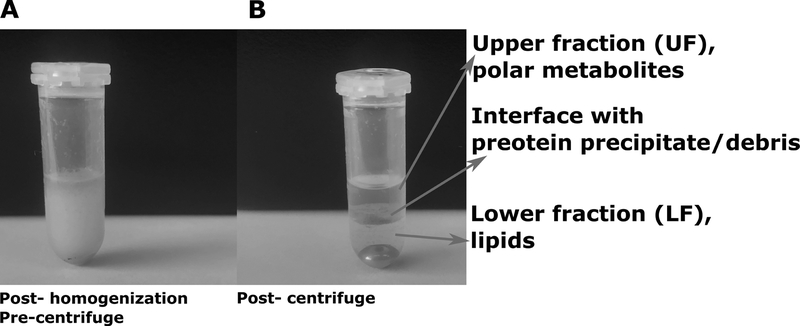
The sample should show three clearly separate layers – a clear upper fraction, clear lower fraction and a layer of protein/debris at the interface (Figure 1).
Carefully remove the upper fraction with an air displacement micropipette (c. 200 μl) and put in a new microtube (UF).
Allow the lower fraction and the solid protein/debris interface to settle, then pipette out (c. 150 μl) the lower fraction from the bottom of the tube (LF).
Use the vacuum concentrator to evaporate the UF to dryness.
Put the LF tubes in the fume hood and allow to dry overnight.
The UF will be used for NMR analysis. The LF may be stored in −20°C for further lipidomics analysis. UF may be stored in −80°C till further analysis.
Figure 1: Appearance of samples during extraction.
Before centrifugation the sample appears as a homogeneous colloidal suspension (A). After centrifugation, the sample separates into three layers – upper fraction of polar metabolites, interface with proteins and cellular/tissue debris and lower fraction comprising of lipids.
BASIC PROTOCOL 2
PREPARING DRIED UF FOR NMR ANALYSIS
The dried UF is reconstituted using a 0.1 M phosphate buffer that contains D2O and the internal standard for NMR experiments. The buffer pH is kept as close to the physiological pH as possible. Preparation of the sample for final NMR measurement is described below.
Materials
Sodium phosphate monobasic dihydrate (NaH2PO4.2H2O; MW 156.01).
Sodium phosphate dibasic (Na2HPO4; MW 141.96).
D2O (Cambridge Isotope Laboratories product DLM-4–100) and Milli-Q® water.
4,4-Dimethyl-4-silapentane-1-sulfonic acid (DSS, Cambridge Isotope Laboratories product DLM-32–1), alternatively the hexadeuterated version (DSS-D6, CIL product DLM-8206–1) may be used. Trimethylsilylpropionate (TSP-D4, CIL product DLM-48–1) is another widely used internal standard for aqueous samples, however it suffers from pH sensitivity and protein complex formation. Alternative NMR standards have been proposed to address some of the concerns with currently used internal standards (24).
pH meter.
Balance.
Hydrochloric acid (HCl) and sodium hydroxide (NaOH).
Vortexer
Microcentrifuge, as mentioned earlier.
NMR tubes (Bruker samplejet NMR tube rack with 96 3mm tubes - product ID Z112272, 5mm tubes - Z105684, 1.7 mm tubes – Z106463).
Protocol steps
Weigh out 6.2404 g monobasic sodium phosphate dihydrate and 8.5176 g dibasic sodium phosphate. Put in a clean, dry beaker.
Measure 100 ml D2O and add to the beaker.
Weigh out 98.17 mg DSS and add in the beaker.
Stir well to dissolve.
Add 900 ml Milli-Q® water.
Measure the pH, adjust using drops of HCl or NaOH as required. The pH of the buffer should be close to the physiological pH ~7.4.
Add 200 μl (for 3mm NMR tubes) or 600 μl (for 5 mm NMR tubes) buffer to the UF from step 13 of Basic Protocol 1.
This is the final sample for NMR analysis (referred to as sample from this point).
ALTERNATE PROTOCOL 1
PREPARING URINE SAMPLES FOR NMR ANALYSIS
Urine samples typically do not require extraction. A few alterations to the steps in Basic Protocols 1 and 2 should be sufficient for preparing urine samples for NMR analysis. The key changes are described below.
Protocol steps
Note: Urine samples should be collected in sterile tubes containing sodium azide (final concentration 0.05% wt/vol).
Collect 300 μl of urine sample and centrifuge for 10 mins in 13,300 rpm in 4°C.
From the supernatant, collect 180 μl in a microtube.
For preparation of buffer, scale up the protocol described in Basic Protocol 2 to 10 times with respect to DSS, monobasic sodium phosphate dihydrate and dibasic sodium phosphate concentration (final concentration 1M). Also, make sure that this buffer is made completely in D2O.
Add 20 μl of the buffer to the microtube containing urine sample (see step 2).
This is the final sample for NMR analysis (referred to as sample from this point).
BASIC PROTOCOL 3
NMR EXPERIMENTS
All NMR experiments described in this section use standard Bruker terms for pulse programs. Spectrometers from other vendors can certainly be used, although a consultation with the equipment vendor is advised prior to running experiments. In this section, special attention will be paid to acquiring 1-dimensional and 2-dimensional NMR data for samples extracted as described in the previous Protocols. However, it is to be noted that whole serum/plasma may also be analyzed using NMR spectroscopy. Briefly, T2 edited CPMG (Curr-Purcell-Meiboom-Gill, Bruker sequence cpmgpr1d) sequence (25,26) is used for suppression of broad signals from lipids and proteins in unextracted serum/plasma while observing the signals from small molecules. Alternately, the diffusion edited spectrum (Bruker sequence ledbpgppr2s1d) may be recorded (27) for profiling macromolecules such as lipids and lipo-protein signals. However, the blood serum/plasma NMR spectroscopy typically yields lower number of metabolite peaks compared to extracted samples. Furthermore, the internal standards (DSS/TSP) often form complexes with proteins leading to loss of quantitative information, although TSP is worse compared to DSS in this regard. For the purpose of this protocol, 1-dimensional version of NOESY (Bruker sequence noesygppr1d1h) will be described.
Materials
NMR spectrometer – Most current metabolomic studies are performed using spectrometers with field strength 500 MHz or higher with probes equipped with gradient coils. Spectrometers equipped with autosamplers are advised for high throughput studies in order to minimize human intervention.
NMR tubes – compatible with the autosampler configuration, refer to materials section of Basic Protocol 2 for details.
Pipettes and tips.
Optional - Gilson liquid handler for sample preparation.
Centrifuge and vortexer.
Protocol steps
Vortex the sample (refer to Basic Protocol 2, step 7 and Alternate Protocol 1, step 4) and centrifuge at 4°C at 13,300 rpm for 5 mins.
-
Use a pipette to carefully transfer the supernatant into the NMR tube
Note: recommended volumes – 40 μl, 200 μl and 600 μl for 1.7 mm, 3 mm and 5 mm NMR tubes.
Optionally, use Gilson liquid handler for step 2.
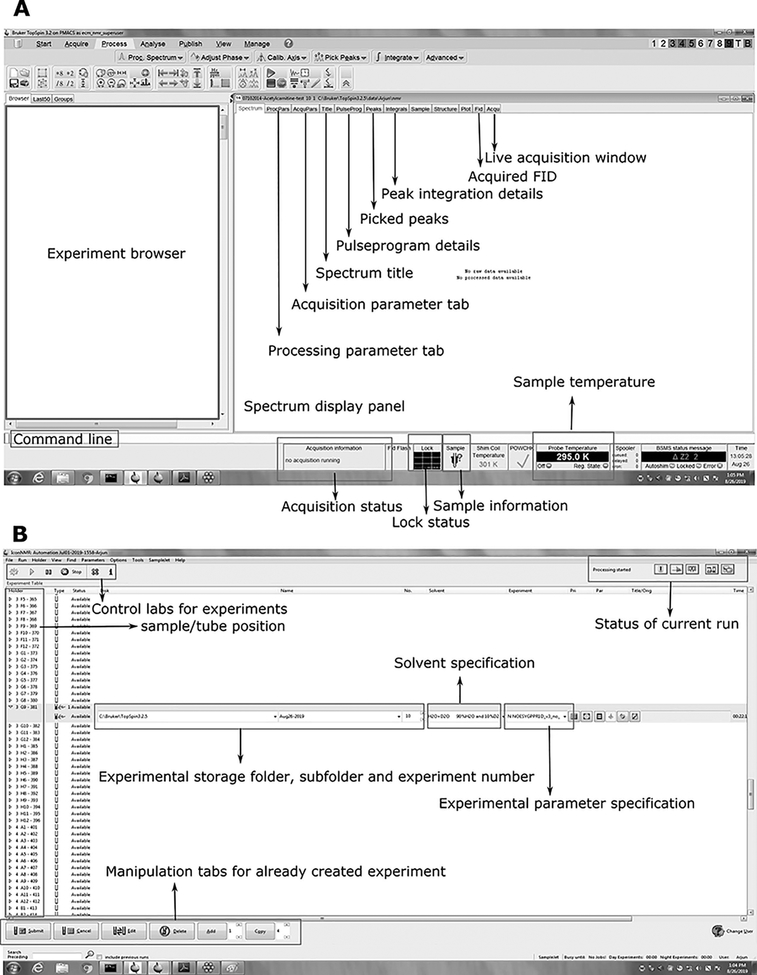
- Setting up NMR experiments – once all the samples are in NMR tubes, the samples are ready for data acquisition. In this section, acquisition of 1-dimensional 1H NMR and 2-dimensional 1H-1H homonuclear and 1H-13C heteronuclear experiments using Bruker automation module (iconnmr) is described. Refer to Figure 2 for the working space of Topspin and IconNMR software (Bruker Biospin).
- Make sure the magnet legs in the spectrometer are up.
- Open topspin and use the command line to open iconnmr (command: iconnmr).
- Switch on the probe temperature and set the experimental temperature. For plasma/serum extracts 298 K is used.
- Insert first sample in the probe and allow enough time to equilibrate (command: sx abc, where abc defines the sample position in the autosampler).
- Create the experiment – this may be done by reading a previously acquired experiment or reading the Bruker parameter files (for 1D - NOESYGPPR1D, 2D TOCSY – TOCSYESGP, 2D HSQC – HSQCETGPSISP etc. ).
- Lock the sample (command: lock h2o+d2o, Note: the solvent may change depending on the sample, in this protocol 90% H2O + 10% D2O is used as the solvent).
- Tune and match the probe (command: atma for automatic tune-match, atmm for assisted manual tune-match).
- Shim the sample: this step is crucial since shimming achieves homogenous magnetic field and helps in narrowing the linewidth of spectral peaks. Attaining good shim is highly important which may be achieved by 3-dimensional shimming (command: topshim 3d) on the first sample. Once a good shim is achieved, the shim file may be saved (command: wsh) and used for later samples, provided the samples have similar matrix. Even then, it is recommended to perform 1-dimensional topshim on every sample (this may be pre-programmed in iconnmr). (Note: steps f – h may be performed in iconnmr automation module). Please refer to the “Anticipated results” section for how to judge good shim is achieved.
- Set the offset (O1) – the offset serves two purposes. First, it defines the center of the spectrum and second, it specifies the position for solvent suppression when using solvent signal saturation (presaturation). For this purpose, a single pulse experiment (Bruker sequence zg/zg30) is acquired to obtain the signal from water and the center of the signal is defined as the offset. Copy the O1 and set it in the actual experiment created in e.
- Specify the spectral width (command: SW). In most cases, the water resonance will be around 5 ppm. Therefore, spectral width 14 – 16 ppm should be enough.
- Specify the other important experimental parameter (necessary Bruker commands are in the parentheses)–number of FID data points (TD) = 76k for blood serum/plasma, number of scans (NS) = 64 is sufficient for 200 μl starting material of blood serum/plasma. Pulse length (p1) = see l, relaxation delay (d1) = 1 – 5 sec (if a library based targeted profiling approach is followed, it is important to adhere to the total experimental time of the library spectra, therefore d1 may have to be modified accordingly), noesy mixing time (d8) = typically 0.1 sec.
- For acquiring 2-dimensional datasets some additional parameters are needed. Number of increments in the indirect dimension (TD2), spectral width in the indirect dimension (SW2) that depends on the nucleus observed (~200 ppm) for 13C, offset (O2P) in the indirect dimension and tocsy mixing time (d20) for TOCSY type experiments.
- Specify the proper power level needed for the experiment. Specifically for the 2D experiments, adhering to the manufacturer recommended power level is crucial. The probehead and solvent dependent parameters may be copied to the specific acquisition file using “getprosol” command for Bruker spectrometers. It is advisable to use adiabatic decoupling schemes for HSQC like experiments, particularly at high field strengths.
- Calibrate 90° pulse length at the specified power level (command: pulsecal).
- Set the receiver gain (command: rga). The receiver gain should preferably be kept constant across samples. If the variation in dilution is large across samples (e.g. urine), auto-receiver gain should be performed for each sample. (Note: The samples used in metabolomics experiments are typically highly aqueous in nature, therefore it is crucial to accurately suppress water (see i.). Otherwise, a receiver overflow warning might appear.)
- Start the spectral acquisition. (Note: if using iconnmr, the experiment list has to be submitted and start from the automation module. Otherwise “zg” may be used to start a specific experiment in topspin. Multiple experiments can be submitted using “multizg”).
Figure 2: Screenshot of typical NMR acquisition working area for a Topspin and IconNMR modules of Bruker spectrometer.
Topspin is the spectral acquisition and processing software coupled with the Bruker NMR spectrometer (A) while IconNMR (B) handles the automation module and is useful for running large batches of sample typically encountered in the metabolomics field.
BASIC PROTOCOL 4
SPECTRAL PROCESSING AND QUANTIFICATION OF METABOLITES
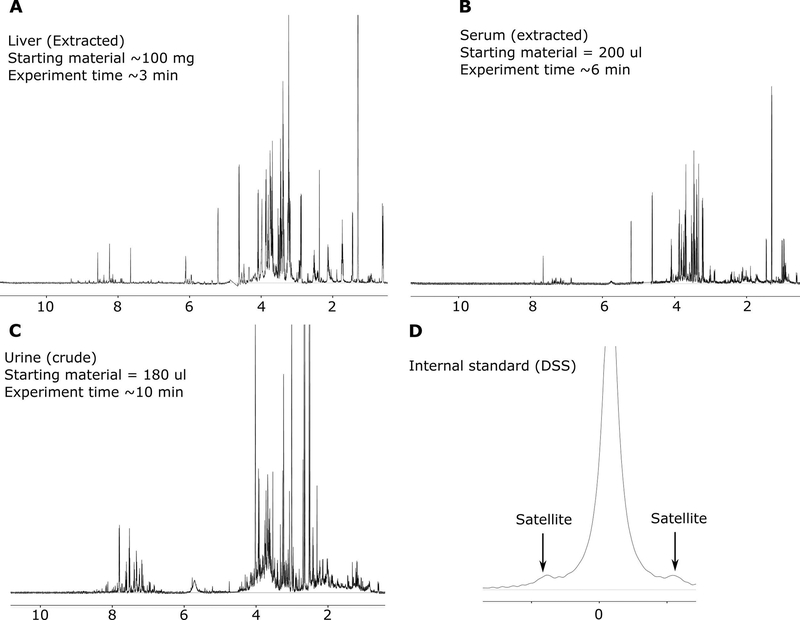
Typically, NMR data appears in the form of a time domain free induction decay (FID). In order to obtain a conventional frequency domain spectrum, Fourier transformation of the FID is performed. Bruker commands “xaup” and “xfb” may be used for the Fourier transformation of 1-dimensional and 2-dimensional NMR data, respectively. Note that further fine-tuning of phase and baseline correction (described below) may still be needed. For biofluid/tissue extracts, such operation leads to a heavily crowded spectrum from overlapping metabolite peaks, with the peak intensity directly proportional to the metabolite concentration (Figure 3). Spectral crowding also depends on the sample type. Typically, urine yields much more crowded spectrum than blood serum/plasma (Figure 3).
Figure 3: Representative NMR spectra from different biospecimens.
Typical spectra obtained from extracted liver (A), extracted serum (B) and crude urine (C) (buffered at pH ~ 7.0) are presented. Note that the spectral crowding depends on the nature of sample. Typically, urine and liver produces the most spectral crowding. The internal standard peak (DSS) (D) is investigated to judge the quality of the spectrum from linewidth and peak shape context. A narrow linewidth and good Lorentzian peak shape allows better quantification of metabolites. The DSS peak width should be < 1.5 Hz for usable spectrum and the Si-satellites should be visible around the CH3 peak (0 ppm).
Historically, quantification of metabolites from such spectrum are performed using three strategies – 1. Spectral binning: the spectrum is divided into small frequency window (typically 0.01 – 0.04 ppm) and each such window (bin) is integrated. While this approach is quick, it is prone to errors from inter sample variation in chemical properties such as pH and ionic strength and peak shifts as a result of variation in such properties (5,28). 2. Peak integration: individual peaks may be integrated manually and compared relative to the internal standard (DSS) to obtain the concentration. Such approach may be performed manually or automatically (using topspin after peak picking). This strategy, however, only works well for isolated peaks with a clearly defined baseline and fails for overlapped peaks which is often the case for metabolomics samples. 3. Targeted profiling: this approach uses a pre-recorded library of metabolite NMR spectra and uses the library spectrum of individual metabolite to fit the experimentally recorded NMR. While this approach is somewhat slow, it delivers both identification and quantification of individual metabolites, thereby utilizing the complete potential of NMR metabolomics. This approach is still considered a gold-standard for quantification studies (29).
In this section, spectral processing followed by targeted profiling will be described in detail. Specifically, targeted profiling using Chenomx NMR suite (Chenomx Inc. Edmonton, AB, Canada) is described. The basic principle behind the approach is explained elsewhere (Chang et al., 2008; Weljie et al., 2006) However, similar analysis may be performed using other proprietary software (AssureNMR from Bruker) and open source packages, such as BATMAN, with limitations (31).
Materials
Workstation with pre-loaded Chenomx NMR suite.
Protocol steps
- Processing the spectra -
- Open the Chenomx processor module and import the raw FID file.
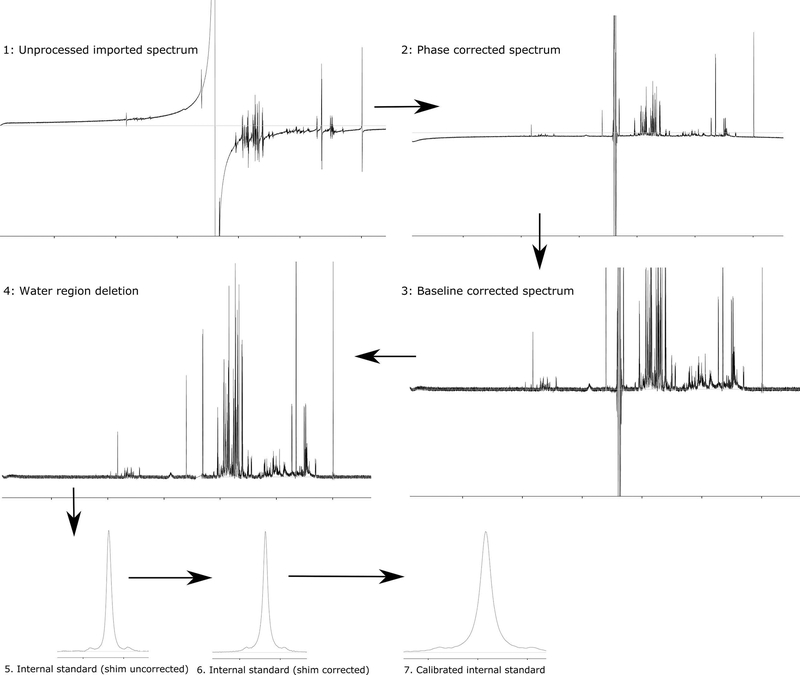
- Process the spectrum by putting in the necessary parameters – nature and concentration of the internal standard and zero filling parameter of the FID. If the zero fill is set to “automatic”, the software zero fill to the nearest power of two in order to double the number of data points acquired. Apply line broadening (typically 0.5 Hz). Optionally, one can select automatic phase, baseline and shim correction, delete the water region and calibrate internal standard.
- Phase correct the spectrum: Auto phase correction could serve as a starting point for refining the phase further. Iteratively perform zero order and first order phase correction till the right phase is achieved (Figure 4). (Tip: while performing a zero order phase correction, focus on the DSS/internal standard peak and try to achieve an in-phase peakshape, during a first order phase correction, focus on the water peak.)
- Baseline correction of the spectrum: There are two widely used strategies for baseline correction – linear and spline. For metabolomics samples, spline is more suitable. In order to do that, perform auto spline thereby generating several points for baseline correction. Closely inspect the spline curve and incorporate points as needed. (Note: baseline correction is crucial for binning type approaches. Tip: if right baseline correction is achieved the base of the peaks and the noise region should be at the same plane with zero intensity line. Refer to Figure 4.)
- Delete region: Deleting the water region (and the urea region for urine samples) using the processing tab (Figure 4).
- Shim correction: Shim correction is optional, but recommended. This step is performed to correct for any systematic distortions in the spectral peak as a result of variable shimming. The correction is performed using the isolated internal standard peak. Check the “use DSS/TSP satellite” box for optimum results (Figure 4).
- Calibrate internal standard (CSI): Calibrate the internal standard using the calibrate CSI mode. This step is crucial for further profiling and quantification. In most cases, this can be done automatically. However, this step should be performed after all processing steps (2 – 6) are finalized (Figure 4).
- Save the processed spectrum as a .cnx file and proceed to the next spectrum.
- At this stage, the saved .cnx files may be used for binning using the spectral binning module. The detail of binning procedure may be found in the user manual of the Chenomx Profiler software. Interested readers are also directed to literature that describes different binning strategies (32–36).(However, that will not be described in detail here.
- Targeted profiling of the processed spectra -
- Open the chenomx profiler module and import the first spectrum (.cnx file).
- The spectral profiling is performed using peak clusters. Thus, a good strategy is to start from one end of the spectrum and move to the other end.
- Activate the spectrum line, fit line and residual line from the left pane.
- Position the cursor on the most downfield peak (left side of the spectrum) and activate the compound list near that frequency range by right click and appropriate selection.
-
Check the peak pattern of each compound near the region and select the most appropriate compound (Figure 5).Note: the peak multiplicity, coupling constant and peak width should match with the experimental peak. Also, note the peak position. In most cases, the fitted peak should be within 0.02 ppm of the library peak.In case of overlapped cluster of peaks, identify other compounds and move the peak position until the shape of spectrum line and fitted line appear same. At this stage, move the peak heights to minimize the residual line. Contribution from each metabolite should be judged by peaks from other spectral region. Refer to Figure 5 for an example.In case of isolated peaks (i.e not within a multi-peak cluster), minimize the residual until it matched the spectral baseline. Then move to the next peak from the same metabolite and fit using similar strategy. If the next peak is within a multi-peak cluster, keep the peak height intact and come back later to the cluster. Refer to Figure 5 for example.
-
After finishing one cluster/peak, move on to the next cluster/peak and fit iteratively as before.Note: the compound library may be updated by adding new compounds using the compound builder module. This is useful for drug metabolites, exposome etc.
-
Typically, the number of fitted compounds from different biospecimens are as follows; plasma/serum extract – 40–50, liver – 70–80, urine – 70–80.Note: the number will vary depending on the starting material and spectral signal to noise).
- Certain compounds are difficult to fit using this approach because of their chemical nature and/or chemical shift. Notable among them are citrate and glucose (specifically, doublet near 4.6 ppm, which is subjected to artifacts from water suppression).
- After finishing profiling of the first spectrum, save the profile. This profile may be recalled as the base profile for processing a series of spectra from same biospecimen.
- After completion of the whole series, the data can be exported as a .csv file containing the concentration values of the fitted compounds.
Figure 4: Processing pipeline for NMR spectrum.
The Chenomx processing pipeline is described here. However, the general steps will remain the same for other software. Raw FID is imported, and Fourier transformed, followed by phase correction, baseline correction, region deletion, shim correction and internal standard calibration. Perfect baseline and phase correction are crucial. The base of the peak and noise should be centered around the zero line (white line in panel 4) if a perfect phase and baseline correction is achieved.
Figure 5: Targeted profiling of the NMR spectrum.
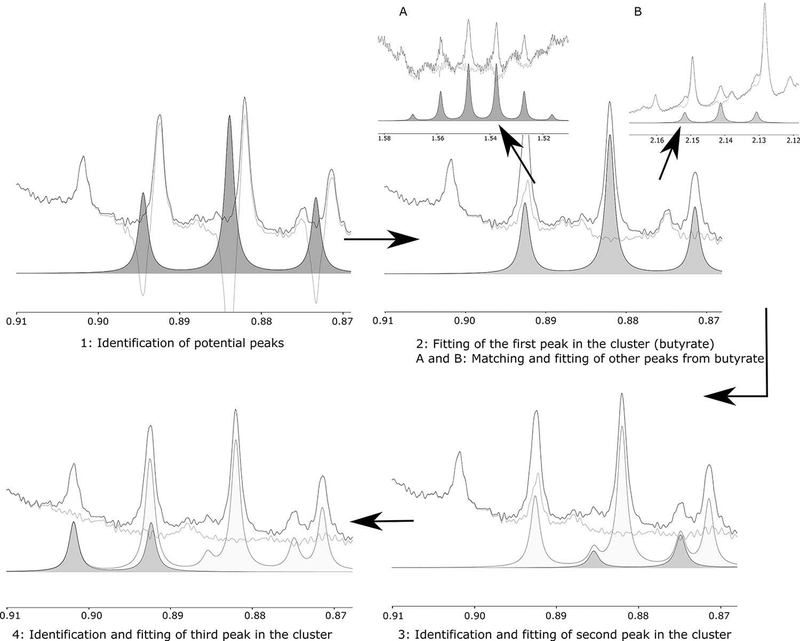
Targeted profiling of NMR peak clusters starts with identification of potential metabolite peaks based on the chemical shift. The most likely peak is profiled (2 - butyrate), followed by other peaks from the same compound (2A, 2B), potentially in different clusters. Then, other peaks in the same cluster are fitted (3 – valerate, 4 – isovalerate). If perfect profiling is achieved, the residual line (green) will become flat near the base of the peak.
BASIC PROTOCOL 5
STATISTICAL ANALYSIS OF THE DATA
Statistical analysis of metabolomics data can often be overwhelming because of large samples and multidimensional nature of the data. However, certain basic protocol may be followed to simplify the multivariate dataset. Typically, principal component analysis (PCA) (37) may be used to identify any hidden structure in the data and to identify outlier samples. PCA is, however, an unsupervised method of analysis and thus represents only the most dominant variation in the dataset. Partial least square - discriminant analysis (PLS-DA) (37) or orthogonal partial least square – discriminant analysis (OPLS-DA) (38) may be used for supervised data analysis in order to identify group specific differences (treated/control, wild type/mutant etc.). In this section, basic multivariate data analysis will be described. Proprietary software (Simca-P) as well as freely available R-packages (39,40) and servers (Metaboanalyst (41–43)) may be used for this purpose. It should be noted that, while this type of analysis strategy is predominant in the field, depending on data structure and specific questions, other statistical methods are often used.
Materials
Workstation with necessary software for statistical analysis. (Note: data processing and analysis steps are described in accordance with Simca-P software. However, the basic idea will be the same irrespective of the analysis platform)
Protocol steps
Import the compound concentration table in the software and identify the sample name column/row (NMR profile name of each sample) and variable name row/column (metabolite IDs).
Mean center the data and scale to unit variance.
- Perform the statistical analysis, preferably using the following pipeline.
- Begin by fitting a PCA model. This should result in components jointly describing most variation in the dataset. By definition, the components are mutually orthogonal. The explained variance of each component may be measured by R2X values.
- Analyze the PCA scores plot for obvious patterns and outlier samples. Also, analyze the loadings plot to identify the metabolites leading to the scores distribution.
- If any obvious outlier(s) is (are) detected (further from 95% confidence interval ellipse), exclude the sample and perform PCA again. This process should be iteratively performed until an outlier free clean data is obtained.
- Perform the supervised analysis, preferably OPLS-DA as that leads to better visualization of the variation compared to PLS-DA.
- Performing a cross validation method (44) (leave one out, 1/7 fold, etc.) during the supervised analysis, particularly in absence of an independently acquired test set, is highly recommended.
- The OPLS-DA model may be judged using CV-ANOVA p value and Q2, a parameter for cross-validation.Q2 measures the predictive ability of the model. Accordingly, Q2 = 1 indicates perfect predictability. Generally, Q2 ≥ 0.5, coupled to low CV-ANOVA p (< 0.05) indicates significantly predictive model.
- If a significant model is generated, the scores plot may be investigated for the patterns and clustering.
- The loadings plot and the VIP (variable importance on projection) values are investigated for significantly different metabolites. The VIP values associate a multivariate significance of the metabolites relevant to the sample clustering while the loadings are associated to directionality of the metabolites. Typically, VIP > 1.0 may be considered significant.
COMMENTARY
Background information
Metabolomics is defined as the study of cellular metabolites in cell, tissue, organ or organism of interest. Metabolomics is advantageous over other “omics” fields of study, such as transcriptomics and proteomics, as the metabolite concentration often reflect underlying biochemical activities and effects of environmental stressors (45). Metabolite identification and quantification are at the heart of metabolomics studies. Nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) coupled with liquid/gas chromatography are most frequently used for this purpose (46,47). These techniques are highly complementary to each other. For example, NMR offers unbiased and highly quantitative data with minimal sample processing. However, the technique is not suitable for less abundant metabolites (detection limit up to low micromolar). MS, on the other hand, is a much more sensitive technique. However, sample processing and quantification from MS experiments depend on the nature of the analyte. Ideally, combination of NMR and ultra performance liquid chromatography- tandem mass spectrometry (UPLC-MS) techniques is advised for obtaining maximum metabolome coverage, although it may be logistically challenging due to high equipment cost and requirement of specialized skills. Despite being a relatively insensitive technique, NMR spectroscopy is thought to have tremendous potential in the metabolomics field, largely due to recent technological and experimental advancements (4,48).
Critical parameters and troubleshooting
The most crucial limiting factor of NMR based metabolic profiling is the comparatively low sensitivity which is inherent to the technique itself and not related to complexity of the spectra. In fact, the targeted profiling approach presented here utilizes the full potential of 1H NMR spectroscopy and is able to recover detailed quantitative information from most crowded spectrum. Typically, millimolar to micromolar concentrations of analytes are needed. In case of limited amount of starting sample, NMR tubes with smaller inside diameter may be used to increase the effective concentration of the analyte by decreasing the reconstituting solvent volume (Basic protocol 2, step 8 and/or alternate protocol 1, step 5). For example, instead of reconstituting the sample in 600 μl buffer in 5 mm tube, the effective concentration of the analyte may be increased three-fold by using a 3 mm tube that needs 200 μl of reconstituting solvent.
Improper extraction can significantly impact the quality of the spectra. In some cases, residual peaks from protein/macromolecules can completely mask some of the less abundant metabolite peaks. Therefore, proper care should be taken during the extraction protocol.
Certain biospecimens are known to be affected by the chemical nature of the matrix such as pH and ionic strength. This is particularly relevant for urine samples and may be exacerbated by diseases that leads to acidity/alkalinity. This issue can be handled by proper buffering in most of the cases. However, ionic strength of the buffer may pose an issue during tuning and matching of the probe. Such effects may be attenuated using smaller NMR tube diameter.
It is advisable to keep the sample volume to at least the minimum prescribed value for the NMR tube diameter used in order to avoid issues with locking and shimming.
Perhaps the most important consideration for metabolomics by NMR is the high water content of the samples. Suppression of water signal by presaturation efficiently addresses the issue most of the time. However, if that fails, excitation sculpting (Bruker pulseprogram ZGESGP) offers a good alternative strategy for water suppression (49). In order to properly perform presaturation, it is important to ensure proper offset of the spectrum window, perhaps using a representative sample. Similarly, the representative sample may be used to calibrate the 90° pulse width as well.
In certain cases, sample dilution may vary widely (e.g. urine samples). Automatic receiver gain should be used in those cases to avoid receiver overflow and decay in FID, hence spectral, quality. Wide variation in sample dilution also poses some issues with statistical analysis. Specifically, some normalization strategies are warranted prior to data analysis. Total spectral normalization and probability quotient normalization are two of the most widely used normalization strategies for NMR metabolomics, however, several other normalization protocols are also employed (50). For a comparative analysis of different normalization protocols, see elsewhere (50–53).
Finally, proper care should be taken while processing the spectra. Distortion in the baseline due to improper baseline and/or phase correction may have unwanted effects in the data analysis, especially if a binning approach of data analysis is sought.
Anticipated results
Typical NMR spectra from different biospecimens are presented in Figure 3. In almost all the cases, complex overlapping patterns of metabolite peaks are observed. The water region (4.6 ppm – 4.8 ppm) displays a suppressed water artifact while the urea peak (in urine) is also suppressed heavily as a result of presaturation strategy. The spectral quality may be judged by the linewidth of the peaks. Typically, an isolated peak (internal standard in most cases) is chosen and the full width at half maxima (Bruker command: hwcal) is calculated. Peak width < 1.0 Hz is considered ideal, however < 1.5 is readily usable. If TSP/DSS is used as internal standard, the satellite humps (Figure 3D) of the methyl peak should be clearly visible when the target linewidth is achieved.
Time considerations
Sample extraction typically takes 3 – 4 hours of wet bench work. Further time is needed for drying down the solvents – typically 6–8 hours per batch.
Typical throughput of NMR spectroscopy is about 80 serum/plasma extracts per day (starting material 200 μl) or 100 tissue extracts (starting material 100 mg liver tissue) per day on a 600 or 700 MHz instrument without cryogenic probes.
Processing and profiling also requires considerable time. Typically, an experienced technician can process and profile 5–6 blood serum/plasma extract spectra per hour.(30)
Acknowledgement
This work was supported by NIH/NIA grant R21-AG-052905, NIH/NCI grant R21-CA-213234 and NIH/NCI grant P01-CA-165997–05. The authors acknowledge Ms. Dania Malik for help with generating the Figure 1.
References
- 1.Clendinen CS, Lee-McMullen B, Williams CM, Stupp GS, Vandenborne K, Hahn DA, et al. 13C NMR metabolomics: Applications at natural abundance. Anal Chem. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhinderwala F, Lonergan S, Woods J, Zhou C, Fey PD, Powers R. Expanding the Coverage of the Metabolome with Nitrogen-Based NMR. Anal Chem. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guennec A Le, Giraudeau P, Caldarelli S. Evaluation of fast 2D NMR for metabolomics. Anal Chem. 2014; [DOI] [PubMed] [Google Scholar]
- 4.Marchand J, Martineau E, Guitton Y, Dervilly-Pinel G, Giraudeau P. Multidimensional NMR approaches towards highly resolved, sensitive and high-throughput quantitative metabolomics. Curr. Opin. Biotechnol 2017. page 49–55. [DOI] [PubMed] [Google Scholar]
- 5.Weljie AM, Newton J, Mercier P, Carlson E, Slupsky CM. Targeted Profiling: Quantitative Analysis of 1 H NMR Metabolomics Data. Anal Chem [Internet]. 2006. [cited 2016 May 25];78:4430–42. Available from: http://pubs.acs.org/doi/abs/10.1021/ac060209g [DOI] [PubMed] [Google Scholar]
- 6.Dona AC, Kyriakides M, Scott F, Shepherd EA, Varshavi D, Veselkov K, et al. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Comput Struct Biotechnol. 2016;14:135–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab [Internet]. Elsevier; 2015 [cited 2016. May 25];22:1009–19. Available from: http://linkinghub.elsevier.com/retrieve/pii/S155041311500460X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sengupta A, Krishnaiah SY, Rhoades S, Growe J, Slaff B, Venkataraman A, et al. Deciphering the duality of clock and growth metabolism in a cell autonomous system using NMR profiling of the secretome. Metabolites. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehrman P, Sengupta A, Harders E, Ubeydullah E, Pack AI, Weljie A. Altered diurnal states in insomnia reflect peripheral hyperarousal and metabolic desynchrony: A preliminary study. Sleep. 2018;41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu R, Chou J, Hou S, Liu X, Yu J, Zhao X, et al. Evaluation of two-step liquid-liquid extraction protocol for untargeted metabolic profiling of serum samples to achieve broader metabolome coverage by UPLC-Q-TOF-MS. Anal Chim Acta. 2018; [DOI] [PubMed] [Google Scholar]
- 11.Masson P, Alves AC, Ebbels TMD, Nicholson JK, Want EJ. Optimization and evaluation of metabolite extraction protocols for untargeted metabolic profiling of liver samples by UPLC-MS. Anal Chem. 2010; [DOI] [PubMed] [Google Scholar]
- 12.Zukunft S, Prehn C, Röhring C, Möller G, Hrabě de Angelis M, Adamski J, et al. High-throughput extraction and quantification method for targeted metabolomics in murine tissues. Metabolomics. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wawrzyniak R, Kosnowska A, Macioszek S, Bartoszewski Rafałand Markuszewski M. New plasma preparation approach to enrich metabolome coverage in untargeted metabolomics: Plasma protein bound hydrophobic metabolite release with proteinase K. Sci Rep. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell JD, Brown JCC, Kubal G, Sadler PJ. NMR-invisible lactate in blood plasma. FEBS Lett. 1988; [DOI] [PubMed] [Google Scholar]
- 15.Nicholson JK, Gartland KPR. 1H NMR studies on protein binding of histidine, tyrosine and phenylalanine in blood plasma. NMR Biomed. 1989; [DOI] [PubMed] [Google Scholar]
- 16.Barrilero R, Ramírez N, Vallvé JC, Taverner D, Fuertes R, Amigó N, et al. Unravelling and Quantifying the “nMR-Invisible” Metabolites Interacting with Human Serum Albumin by Binding Competition and T2 Relaxation-Based Decomposition Analysis. J Proteome Res. 2017; [DOI] [PubMed] [Google Scholar]
- 17.Weljie AM, Dowlatabadi R, Miller BJ, Vogel HJ, Jirik FR. An inflammatory arthritis-associated metabolite biomarker pattern revealed by 1H NMR spectroscopy. J Proteome Res. 2007; [DOI] [PubMed] [Google Scholar]
- 18.Daykin CA, Foxall PJD, Connor SC, Lindon JC, Nicholson JK. The comparison of plasma deproteinization methods for the detection of low-molecular-weight metabolites by 1H nuclear magnetic resonance spectroscopy. Anal Biochem. 2002; [DOI] [PubMed] [Google Scholar]
- 19.Nagana Gowda GA, Raftery D. Quantitating metabolites in protein precipitated serum using NMR spectroscopy. Anal Chem. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tambellini N, Zaremberg V, Turner R, Weljie A. Evaluation of Extraction Protocols for Simultaneous Polar and Non-Polar Yeast Metabolite Analysis Using Multivariate Projection Methods. Metabolites. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bligh EG, Dyer WJ. Canadian Journal of Biochemistry and Physiology A RAPID METHOD OF TOTAL LIPID EXTRACTION AND PURIFICATION1. Can J Biochem Physiol. 1959; [DOI] [PubMed] [Google Scholar]
- 22.Malik DM, Rhoades S, Weljie AM. Extraction and Analysis of Pan-metabolome Polar Metabolites by Ultra Performance Liquid Chromatography–Tandem Mass Spectrometry (UPLC-MS/MS). Bio-protocol. 2018;8:e2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, et al. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metab. 2017;25. [DOI] [PubMed] [Google Scholar]
- 24.Nowick JS, Khakshoor O, Hashemzadeh M, Brower JO. DSA: A New Internal Standard for NMR Studies in Aqueous Solution. Org Lett. 2003; [DOI] [PubMed] [Google Scholar]
- 25.Nicholson JK, Foxall PJD, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR Spectroscopy of Human Blood Plasma. Anal Chem [Internet]. 1995 [cited 2016. June 2];67:793–811. Available from: http://pubs.acs.org/doi/abs/10.1021/ac00101a004 [DOI] [PubMed] [Google Scholar]
- 26.Ala-Korpela M 1H NMR spectroscopy of human blood plasma. Prog Nucl Magn Reson Spectrosc. 1995; [Google Scholar]
- 27.Tenori L, Oakman C, Morris PG, Gralka E, Turner N, Cappadona S, et al. Serum metabolomic profiles evaluated after surgery may identify patients with oestrogen receptor negative early breast cancer at increased risk of disease recurrence. Results from a retrospective study. Mol Oncol. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weljie AM, Newton J, Jirik FR, Vogel HJ. Evaluating low-intensity unknown signals in quantitative proton NMR mixture analysis. Anal Chem. 2008; [DOI] [PubMed] [Google Scholar]
- 29.Maulidiani Rudiyanto, Mediani A, Khatib A, Ismail A, Hamid M, et al. Application of BATMAN and BAYESIL for quantitative 1 H-NMR based metabolomics of urine: discriminant analysis of lean, obese, and obese-diabetic rats. Metabolomics. 2017; [Google Scholar]
- 30.CHANG D, WELJIE A, NEWTON J. LEVERAGING LATENT INFORMATION IN NMR SPECTRA FOR ROBUST PREDICTIVE MODELS. 2008. [PubMed]
- 31.Hao J, Astle W, De iorio M, Ebbels TMD. Batman-an R package for the automated quantification of metabolites from nuclear magnetic resonance spectra using a bayesian model. Bioinformatics. 2012; [DOI] [PubMed] [Google Scholar]
- 32.Sousa SAA, Magalhães A, Ferreira MMC. Optimized bucketing for NMR spectra: Three case studies. Chemom Intell Lab Syst. 2013; [Google Scholar]
- 33.Davis RA, Charlton AJ, Godward J, Jones SA, Harrison M, Wilson JC. Adaptive binning: An improved binning method for metabolomics data using the undecimated wavelet transform. Chemom Intell Lab Syst. 2007; [Google Scholar]
- 34.Anderson PE, Mahle DA, Doom TE, Reo NV, DelRaso NJ, Raymer ML. Dynamic adaptive binning: An improved quantification technique for NMR spectroscopic data. Metabolomics. 2011; [Google Scholar]
- 35.De Meyer T, Sinnaeve D, Van Gasse B, Tsiporkova E, Rietzschel ER, De Buyzere ML, et al. NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal Chem. 2008; [DOI] [PubMed] [Google Scholar]
- 36.Anderson PE, Reo NV, DelRaso NJ, Doom TE, Raymer ML. Gaussian binning: A new kernel-based method for processing NMR spectroscopic data for metabolomics. Metabolomics. 2008; [Google Scholar]
- 37.Madsen R, Lundstedt T, Trygg J. Chemometrics in metabolomics-A review in human disease diagnosis. Anal Chim Acta. 2010; [DOI] [PubMed] [Google Scholar]
- 38.Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemom [Internet]. John Wiley {&} Sons, Ltd.; 2006;20:341–51. Available from: http://doi.wiley.com/10.1002/cem.1006 [Google Scholar]
- 39.Costa C, Maraschin M, Rocha M. An R package for the integrated analysis of metabolomics and spectral data. Comput Methods Programs Biomed. 2016; [DOI] [PubMed] [Google Scholar]
- 40.Gaude E, Chignola F, Spiliotopoulos D, Spitaleri A, Ghitti M, M Garcia-Manteiga J, et al. muma, An R Package for Metabolomics Univariate and Multivariate Statistical Analysis. Curr Metabolomics. 2013; [Google Scholar]
- 41.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011; [DOI] [PubMed] [Google Scholar]
- 42.Xia J, Wishart DS. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr Protoc Bioinforma. 2011; [DOI] [PubMed] [Google Scholar]
- 43.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Triba M, Le Moyec L, Amathieu R, Goossens C, Bouchemal N, Nahon P, et al. PLS/OPLS models in metabolomics: the impact of permutation of dataset rows on the K-fold cross-validation quality parameters. Mol Biosyst. 2015;11:13–9. [DOI] [PubMed] [Google Scholar]
- 45.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: A Global Biochemical Approach to Drug Response and Disease. Annu Rev Pharmacol Toxicol. 2008; [DOI] [PubMed] [Google Scholar]
- 46.Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst. 2012. [DOI] [PubMed] [Google Scholar]
- 47.Lenz EM, Wilson ID. Analytical strategies in metabonomics. J. Proteome Res. 2007. [DOI] [PubMed] [Google Scholar]
- 48.Markley JL, Brüschweiler R, Edison AS, Eghbalnia HR, Powers R, Raftery D, et al. The future of NMR-based metabolomics. Curr. Opin. Biotechnol 2017. page 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross A, Schlotterbeck G, Dieterle F, Senn H. NMR Spectroscopy Techniques for Application to Metabonomics. Handb Metabonomics Metabolomics. 2007. [Google Scholar]
- 50.Kohl SM, Klein MS, Hochrein J, Oefner PJ, Spang R, Gronwald W. State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolomics. 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saccenti E Correlation Patterns in Experimental Data Are Affected by Normalization Procedures: Consequences for Data Analysis and Network Inference. J Proteome Res. 2017; [DOI] [PubMed] [Google Scholar]
- 52.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in1H NMR metabonomics. Anal Chem. 2006; [DOI] [PubMed] [Google Scholar]
- 53.Hochrein J, Zacharias HU, Taruttis F, Samol C, Engelmann JC, Spang R, et al. Data Normalization of 1H NMR Metabolite Fingerprinting Data Sets in the Presence of Unbalanced Metabolite Regulation. J Proteome Res. 2015; [DOI] [PubMed] [Google Scholar]