Abstract
Healthy tissues of the body express relatively low basal levels of interferons. However, following detection of microbial invasion by sentinel receptors, a cascade of events initiates leading to the transcriptional induction of interferon genes. Interferons are secreted and act primarily as paracrine cytokines to bind neighboring cell surface receptors. Binding to interferon receptors activates a signal pathway to the nucleus inducing a set of interferon-stimulated genes. The biological activity of these genes confers the unique antiviral and innate immune response of interferons. The rapid induction of interferons is critical to survival, and equally critical is the recovery from this defensive state. Either an aberrant response to infection or an inherited genetic disorder that leads to sustained or increased interferon levels can tip the balance towards pathogenesis.
Keywords: type I interferonopathy, autoinflammatory disease, inborn genetic mutations
1.1. Introduction
Interest in the clinical use of interferons (IFNs) was based initially on their unique antiviral activities. However, as their anti-angiogenic, immunoregulatory, and anti-tumor properties became obvious, their use expanded [1–5]. IFNs are classified into three types based on their recognition by distinct cell surface receptors. Most successful clinical therapeutic applications have centered on type I IFNs, notably IFN-α and IFN-β. Nevertheless, the beneficial clinical use of IFNs is not without adverse effects [6]. Adverse side effects can include fever, fatigue, myalgia, major depressive disorder, and autoimmunity. It is therefore not unexpected that a prolonged and sustained action of IFNs in response to infection or due to an inherited dysregulation of IFN homeostasis can promote pathogenic effects [7, 8].
1.2. Sentinel Receptors
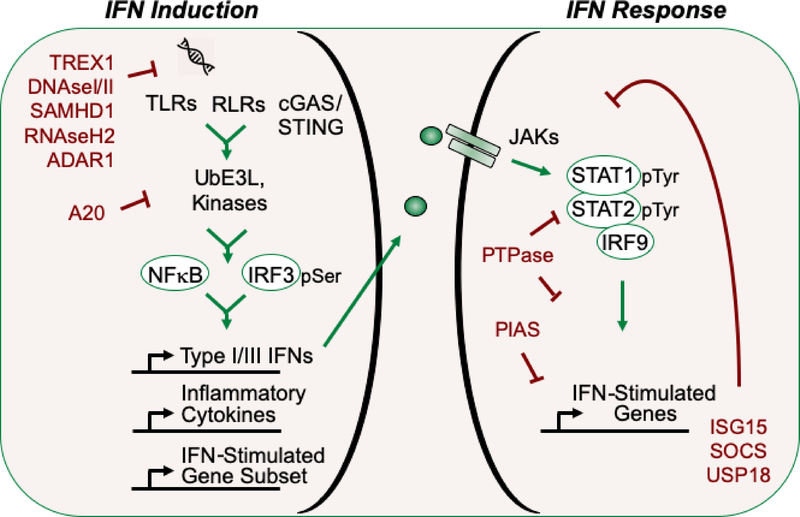
Survival depends on the ability of innate defenses to detect and respond rapidly to infection. Pattern recognition receptors (PRRs) are cellular sentinels that recognize pathogen associated molecular patterns (PAMPs) as foreign such as nucleic acids, glycoproteins, lipopolysaccharides, peptidoglycans, and flagellins [9]. The families of PRRs are diverse and include Toll-like receptors (TLRs), retinoic acid inducible gene I-like receptors (RLRs), nucleotide-binding oligomerization domain-like receptors (NLRs), Aim2-like receptors (ALRs), C-type lectin receptors (CLRs), and DNA sensors such as cyclic GMP-AMP synthase (cGAS) [10–14]. Activation of PPRs that bind nucleic acid ligands typically, although not exclusively, induce expression of the type I IFN genes. Recognition of PAMP nucleic acids by PRRs stimulates specific protein interactions, ubiquitination, and phosphorylation events that transmit danger signals to the nucleus leading to the production of IFNs (Fig.1). The induction of IFNs appears to have evolved in response to infection by viruses that possess nucleic acid genomes, whether RNA or DNA, single or double-stranded, positive or negative sense. However, since RNA and DNA structures are common to both pathogen and host, this defense strategy has the potential risk of recognizing self-ligands.
Figure 1: Conceptual Diagram of type I IFN Induction by Nucleic Acid Sensors and the cellular response to IFN.
Left) RNA and DNA sentinel receptors (TLRs, RLR, and cGAS) bind nucleic acids and mediate signals that stimulate ubiquitin E3 ligases (UbE3L) and kinases to activate latent transcription factors such as NFκB and IRF3. Type I IFN genes are induced and IFN proteins are secreted. Right) IFN binding to specific receptors activates latent JAKs and tyrosine phosphorylation of STAT1 and STAT2. Phosphorylation promotes dimerization and the specific DNA-binding ability of the STAT1/STAT2/IRF9 complex that induces expression of IFN-stimulated genes (ISGs). Some of the negative regulators of IFN induction and response are highlighted in red text.
1.2.1. Recognition of DNA by the cGAS/STING Pathway in the Cytoplasm
Considerable investigation has been devoted to understanding the specificity and the cellular localization of receptors that recognize foreign nucleic acid ligands. Cytoplasmic localization of foreign DNA can be distinguished from host since the mammalian DNA genome is sequestered within the nuclear envelope. The primary PRR that recognizes cytoplasmic DNA from virus or bacteria and leads to induction of IFN is the cyclic GMP-AMP synthase (cGAS) [15, 16]. cGAS activation is triggered following binding to cytoplasmic DNA to produce cyclic 2’3’-cyclic GMP-AMP (cGAMP). cGAMP functions as a small molecule second messenger and binds the adaptor, stimulator of interferon genes (STING), a transmembrane dimer associated with the endoplasmic reticulum [17–19]. Following binding to cGAMP (or cyclic dinucleotides produced by bacteria), STING relocalizes to the perinuclear Golgi area and recruits ubiquitin E3 ligases needed for activation of TANK-binding kinase 1 (TBK1). TBK1 phosphorylates STING which in turn recruits IFN regulator factor 3 (IRF3) for phosphorylation by TBK1 [20]. Phosphorylation of IRF3 promotes its dimerization, translocation to the nucleus, DNA binding, and induction of the IFN-β gene and a subset of IFN stimulated genes (ISGs)[21, 22]. The Iκ-B kinase (IKK) complex is also recruited and phosphorylates Iκ-B to release the NF-κB transcription factor that contributes to type I/III IFN gene induction as well as a number of other cytokine and chemokine genes [23].
1.2.2. Recognition of RNA by RLRs in the Cytoplasm
The induction of the type I IFN genes in response to dsRNA had long been recognized. Yet the molecular mechanism did not unfold until the discovery that members of the DEAD-box RNA helicase family were responsible for sensing foreign cytoplasmic RNA [24]. The retinoic acid-inducible gene I (RIG-I) and the melanoma differentiation-associated gene 5 (MDA5) helicases can bind RNA and undergo a conformational change revealing their caspase activation and recruitment domain (CARD) that is responsible for pathway activation [11, 13, 25]. The RLR CARD domain binds the CARD domain of the downstream signaling adaptor, mitochondrial antiviral-signaling protein (MAVS). This association promotes oligomerization, recruitment of ubiquitin E3 ligases, and activation of TBK1 and IKK complexes that phosphorylate MAVS [20]. IRF3 and Iκ-B are in turn recruited and phosphorylation leads to IRF3 and NF-κB activation and type I IFN gene induction. The response to infection by different viruses led to the findings that both RIG-I and MDA5 recognize dsRNA, and RIG-I recognizes 5’-triphosphate or 5’-diphosphate ends of RNA with significant secondary structure. Host mRNA, rRNA, and tRNA reside in the cytoplasm, but host RNA recognition is believed to be spared due to the 5’cap of many mRNAs, the 5’-monophosphate of tRNAs, the shielding of host RNA by specific binding proteins, and a low level of RNA degradation or editing.
1.2.3. DNA and RNA Recognition by TLRs in Endosomes
The Toll gene in Drosophila was the first PRR identified, shown to be essential for an antifungal defense response [26]. Since then, ten transmembrane Toll homologues (TLRs) have been described in humans that bind a variety of ligands [11, 27]. A subgroup, TLR3, TLR7, TLR8, and TLR9, are distinct for their cellular localization within endosomal membranes, and for their ability to recognize nucleic acids. TLR3 recognizes dsRNA, TLR7 and TLR8 recognize ssRNA, and TLR9 recognizes unmethylated CpG DNA. Since many viruses and bacteria gain entry into the cell by endocytosis, this TLR localization serves as a natural defense. In addition, TLR proteolytic processing in endolysosomes facilitates TLR activation. Ligand binding induces TLR oligomerization and association with cytoplasmic adaptor proteins. TLR3 binds TRIF (Toll/IL1 receptor-domain containing adapter inducing IFNβ), and TLR7/8/9 bind MyD88 (myeloid differentiation primary response 88). TRIF binding recruits ubiquitin E3 ligases leading to the activation of TBK1 and the IKK complex that result in nuclear translocation of IRF3 and NF-κB and transcriptional induction of type I IFN genes. MyD88 promotes ubiquitination that primarily activates NF-κB.
1.3. Interferon Production and Action
PRR-mediated activation of cytoplasmic IRF3 and NF-κB promotes their nuclear localization and cooperative induction of type I IFN genes, as well as other genes. Activated IRF3 can induce a subset of IFN stimulated genes (ISGs) prior to the action of IFNs [28], and NF-κB can induce type III IFNs and inflammatory cytokines and chemokines [29, 30]. IFNs must be secreted from cells to act by binding specific cell surface receptors that trigger a signal pathway to the nucleus, now referred to as a JAK-STAT pathway [2, 31]. Although type I and type III IFNs bind distinct receptors, both activate receptor-associated Janus kinases (JAK), JAK1 and Tyk2. These JAK tyrosine kinases phosphorylate a number of substrates in the cytoplasm including the signal transducers and activators of transcription (STATs), STAT1 and STAT2, that form a heterodimer via their phosphotyrosine and Src homology 2 (SH2) domains. STAT2 is continually associated with the IRF9 transcription factor [32], and therefore a trimeric complex forms, commonly known as ISGF3 (ISG factor 3). ISGF3 traffics to the nucleus, binds to genes containing the IFN stimulated response element (ISRE), and induces transcription of ISGs [33]. The ISGs include transcription factors such as IRF1 that elicit expression of a secondary set of response genes [34, 35]. ISGs confer both the beneficial and potentially detrimental effects of IFNs.
1.4. Putting on the Brakes
The induction of type I IFNs in response to foreign nucleic acids is critical for an acute anti-viral and inflammatory response. However, following this innate defense response, the PRR and IFN signal pathways need to be silenced to maintain homeostasis. Elimination of extraneous nucleic acids by nucleases, reversal of post-translational modifications, and proteasome degradation of signaling molecules are some mechanisms of pathway silencing. Much of our understanding of negative regulation derives from genetic engineering in murine models, and identification of genetic disorders in autoimmune and inflammatory human diseases.
1.4.1. Deubiquitination
The ubiquitin E3 ligases that catalyze K63-linked polyubiquitination are notable regulators of sentinel receptors and have been found to play a critical role in STING, RLR, and TLR signaling. Ubiquitination stimulates and recruits adaptors and kinases responsible for IRF3, IRF7, and NF-κB transcription factor activation. As might be expected, de-ubiquitination is a critical negative regulator [36, 37]. For example, the de-ubiquitinase activity of A20 (TNFAIP3) has been shown by murine gene knockout and biochemical means to be essential for termination of TLR signaling, for inhibition of RIG-I, and for suppression of IRF3 [38–41].
1.4.2. IFN Receptor
Multiple negative feedback mechanisms have been identified that regulate the type I IFN signal pathway. Two ISGs induced by type I IFN, the ubiquitin protease family member USP18 and ISG15, act in concert to shut down signaling at the IFN alpha receptor (IFNAR). USP18 is an isopeptidase that specifically deconjugates the ubiquitin-like ISG15 modification from proteins, but independent of its proteolytic activity, it binds to the IFNAR2 subunit, displaces JAK1, and silences signaling [42]. STAT2 has been described as an adaptor that mediates USP18 binding to IFNAR2 [43]. Intriguingly, human free ISG15 is needed to bind and stabilize the USP18 negative regulator, independent of ISG15 conjugation (ISGylation) [44].
1.4.3. Protein Tyrosine Phosphatases
A distinct aspect of STAT activation is their direct tyrosine phosphorylation which confers a gain of function and enables their specific DNA-binding ability [31]. It is not unexpected that dephosphorylation controls the longevity of STAT signaling, and this occurs both in the cytoplasm and the nucleus. STAT1 is distinct from other STATs in that it resides primarily in the cytoplasm in an unphosphorylated form and is transported into the nucleus following tyrosine phosphorylation [45–47]. Biochemical purification of a STAT1 phosphatase activity from nuclear extracts led to identification of the protein tyrosine phosphatase (PTP) PTPN2/TCPTP as the primary STAT1 nuclear phosphatase [48]. PTPN2 isoforms also reside in the cytoplasm and with PTPN11/SHP-2 have been shown to contribute to the dephosphorylation of STAT1 [49].
1.4.4. Protein Inhibitors of Activated STAT
Protein inhibitor of activated STAT1 (PIAS1) was identified in a yeast two-hybrid assay used to screen for STAT1-interacting proteins that might serve to regulate STAT1 function [50]. PIAS1 is a member of a family of proteins that have a SUMO (small ubiquitin-like modifier) E3 ligase activity [51]. Although sumoylation may not be involved, PIAS1 and PIASy are transcriptional repressors of STAT1. They primarily function by binding tyrosine phosphorylated STAT1 and blocking the DNA-binding ability of STAT1 at responsive gene promoters.
1.4.5. Suppressors of Cytokine Signaling
The family of suppressors of cytokine signaling (SOCS) are transcriptionally induced in response to cytokines and function as classical negative feedback regulators [52, 53]. SOCS1 was identified by three groups simultaneously either as an inhibitor of macrophage differentiation, a binding partner of JAK in a yeast two-hybrid assay, or a protein with similarity to STAT SH2 domain [54–56]. SOCS1 is transcriptionally induced in response to IFN and feeds back to reduce signals that originate at the receptor. SOCS proteins have a central SH2 domain and a conserved carboxyl terminal domain known as a SOCS box. The SH2 domain of SOCS proteins can bind to phosphorylated tyrosine residues on receptors or JAKs. The SOCS box contains elongin C and Cullin 5 binding sites which recruit the Rbx protein and forms an active E3 ubiquitin ligase able to modify targets by lysine 48 ubiquitination for proteasome-mediated degradation. SOCS1 has an additional amino terminal domain known as a kinase inhibitory domain (KIR). The KIR domain of SOCS1 is able to inhibit the catalytic activity of TYK2 and JAK1 associated with the IFNAR, and this appears to be its primary mechanism of IFN pathway silencing.
Specific and redundant mechanisms are in place to repress PRR signaling and the response to IFNs. More recently, an array of noncoding RNAs induced by IFNs have been found to negatively modulate signaling molecules and ISGs [57]. Rather than an on-off switch, the IFN response appears to be regulated as a rheostat.
1.5. Human Type I Interferonopathies
Early studies with complex autoimmune diseases such as systemic lupus erythematosus syndrome noted that patients expressed inappropriate high levels of type I IFNs in the absence of obvious infection. It wasn’t until 2011 that Yanick Crow proposed to conceptually unify this set of inherited autoimmune and inflammatory diseases with the terminology of type I interferonopathies [8]. Interferonopathies are now categorized as Mendelian disorders that produce high levels of IFNs either due to gain-of-function mutations in signaling molecules that stimulate the type I IFN response pathway, or loss-of-function mutations in negative regulators of the type I IFN system. Many of the mutations in type I interferonopathies are involved in recognition and response to self-nucleic acids. Although there is a diversity in clinical phenotypes, the unifying concept helps us to understand and potentially treat these diseases.
1.5.1. Systemic Lupus Erythematosus (SLE)
SLE is a chronic heterogeneous autoimmune disease with inflammatory manifestations in multiple organs and cumulative tissue damage [58]. Disease characteristics include nucleic acid autoantibodies and increased production of type I IFNs. An increased IFN-induced gene signature is a fundamental feature in peripheral blood leukocytes of patients with SLE [59, 60]. Much of our understanding of the effects of type I IFNs on immune cells is based on studies with microbial infections, but SLE patient symptoms persist in the absence of infection [61]. Type I IFNs are known to activate natural killer cells, differentiate dendritic cells, regulate macrophages, and shape T and B cell development [62–65]. These cells produce additional cytokines and chemokines that promote chronic inflammation and long-term detrimental effects. The detrimental effects include neutrophil recruitment and lysis forming neutrophil extracellular traps with networks of chromatin. One of the triggers of SLE is thought to result from inefficient apoptotic and immune clearance. A number of susceptibility loci have been identified and validated in murine models, and several genetic mutations identify the failure to clear extraneous nucleic acids.
DNase1 is a major nuclease in serum and secretions. A murine model of DNase1-deficiency was shown to produce classical symptoms of SLE [66]. Subsequently, loss-of-function mutations in DNASE1 and a homolog DNASE1L3 were found to be linked to a form of SLE [67, 68]. In addition to DNAse1 mutations, whole genome sequencing of samples from patients with an IFN gene signature and autoinflammatory disease identified causative mutations in DNASE2 [69]. The inability to degrade self-DNA leads to the activation of sentinel DNA sensors and the production and action of type I IFNs with chronic inflammation (Fig.1). Another nuclease deficiency was identified in SLE. An autosomal-dominant form of lupus was linked to aberrant DNA degradation with the identification of a missense mutation in TREX1 (three-prime repair exonuclease 1) [70]. Subsequently whole exome sequencing identified TREX1 mutations in a number of SLE patients [71]. TREX1 is an abundant intracellular DNase encoding a 3’−5’ repair exonuclease 1 and can be induced by IFN. TREX1 knockout mice die from overwhelming inflammation and myocarditis with a type I IFN gene signature, and specific depletion of TREX1 in dendritic cells is sufficient to cause systemic disease [72, 73].
1.5.2. Aicardi-Goutières Syndrome (AGS)
Aicardi and Goutières described a disease in children in 1984 with severe encephalopathy and immune cell infiltration in cerebrospinal fluid, and these children were found to have high levels of IFN-α in their cerebrospinal fluid and serum [74]. Many individuals with AGS do not survive childhood. In addition to neurological symptoms, painful skin lesions called chilblains often occur, which are also common in SLE. Similar to SLE, the inherited interferonopathies that lead to AGS result from aberrant activation of nucleic acid sentinel receptors.
Genetic loci were mapped in families that associated with AGS, and subsequently mutations in the TREX1 were identified at these loci [75]. Biochemical analyses were also performed with extracts of cell lines derived from AGS patients. Using poly(dA) as substrate, cells from AGS patients showed no detectable TREX1 activity. As with SLE, the inability to degrade self-DNA was linked to AGS, but not all patients had a TREX1 mutation. Screening for other mutations by SNP array and high-density genotyping of several families of AGS patients led to the finding of homozygous mutations in SAMHD1 (sterile alpha motif domain and histidine-aspartic domain-containing protein) [76]. SAMHD1 is a dNTP hydrolase that lowers intracellular dNTP pools and can stimulate exonuclease activity. As with SLE, a resultant increase in self-DNA with SAMHD1 deficiency can lead to STING activation and type I IFN production.
Additional genetic loci that track with AGS families identified defects in RNA degradation or RNA recognition to cause the inflammatory phenotype [77]. High-density genotyping of microsatellite markers identified mutations in the RNASEH2 enzyme complex. RNase H2 is an endonuclease that catalyzes RNA cleavage of RNA/DNA duplexes. It is a heterodimeric complex of three subunits, and mutations in any one of these subunits causes loss-of-function. Mutations in different AGS families were found in RNASEH2A, RNASEH2B, and RNASEH2C. More recently whole exome sequencing was used to evaluate Mendelian loci associated with AGS, and results identified a defective RNA-editing enzyme, ADAR1 (adenosine deaminase acting on dsRNA) [78]. ADAR1 is both continually expressed and can be induced by IFN [79]. It binds to dsRNA and deaminates adenosine C6 to produce inosine. Mutations in mice that produce a defective ADAR1 lead to embryonic death, however mice can be rescued with a concurrent deletion of the RLR RNA sensor, MDA5 [80]. This result provided evidence that an essential function of ADAR1 is the editing of self-RNA so that it is not recognized by the MDA5 RNA sensor to trigger inappropriate induction of type I IFNs. Supportive of this tenet was the discovery of gain-of-function mutations in the gene encoding MDA5 (IFIH1, IFN induced with helicase C domain 1) in AGS patients that were negative for mutations in TREX1, RNAseH2, SAMHD1, and ADAR1 [81]. Results indicate that activating mutations in MDA5 produce a protein that binds RNA more avidly leading to signal transmission and increased induction of type I IFNs.
1.5.3. STING-Associated Vasculopathy with Onset in Infancy (SAVI)/ Familial Chilbain Lupus (FCL)
Patients with a systemic inflammatory syndrome that includes cutaneous lupus erythematosus and pulmonary fibrosis are often characterized with SAVI or FCL. Symptoms can begin early in life and present with vasculopathy and severe skin lesions. The patients usually present with increased levels of type I IFN and ISGs. Whole genome sequencing identified unifying gain-of-function mutations in the gene encoding STING (TMEM173, transmembrane protein 173) [82–84]. Patient cells showed constitutive activity of the STAT1 transcription factor, localization of STING protein in the Golgi indicative of its activation, and enhanced STING dimerization in the absence of ligand. The continuous signaling of STING stimulates kinases and transcription factors that turn on type I IFN genes.
1.5.4. Singleton-Merten Syndrome (SMS)
SMS patients present with dental dysplasia, calcifications in a major arteries and heart valves, and osteoporosis. Although the clinical manifestations are distinct from other interferonopathies, patients display a chronic expression of type I IFN and IFN induced genes. Whole exome sequencing identified gain-of-function mutations in two RNA sensors. A specific mutation encoding the MDA5 gene, IFIH1, was found to cause classical SMS [85]. Introduction of the activated mutant MDA5 gene into tissue culture cells produced an increase in IFN-β expression in a dose dependent manner. Patients with a more atypical SMS who primarily manifested aortic calcification and skeletal abnormalities were found to harbor a different activating mutation in the gene encoding RIG-I, DDX58 (DExD/H-box helicase 58) [86]. Expression of the constitutive RIG-I mutant was shown to promote phosphorylation and activation of IRF3. Mice harboring the RIG-I gene mutation spontaneously developed lupus-like systemic autoimmune symptoms in the absence of viral infection.
1.5.5. Pseudo TORCH Syndrome (PTS)
The acronym TORCH (toxoplasmosis, other, rubella, cytomegalovirus, and herpes simplex) has been used to describe symptoms that resemble a viral infection. However PTS occurs in the absence of an infectious agent and is an autosomal recessive condition that frequently manifests with intracranial calcification and microcephaly [87]. The IFN gene signature of PTS patients led to identification of loss-of-function mutations in USP18 [88]. As described in section 1.4, USP18 is a negative regulator of the type I IFN receptor. In the absence of this negative regulation there is long-lasting IFN signaling. Cells from patients showed persistent STAT2 tyrosine phosphorylation and high levels of ISGs. Expression of defective USP18 in tissue culture cells led to an increase in protein ISGylation. ISG15 has multiple intracellular functions as a conjugated protein and as a free protein, and in addition, as a secreted form it was shown to promote IFN-γ production in T cells [89]. Patients identified with null alleles in ISG15 have an enhanced type I IFN signature, and a milder phenotype with respect to autoinflammatory disease [44]. The action of other ISGs to dampen the IFN response has been demonstrated in mice particularly in the context of microbial infection [90–92]. For example, the loss of IFIT2/ISG54 leads to increased inflammatory chemokine levels during Candida albicans infection [92]. The inflammatory response appears to reflect the ability of IFIT2 to inhibit NADPH oxidase and reactive oxygen species that induce chemokine production and immune cell recruitment. Future investigations are likely to identify other inborn errors associated with the type I IFN system that normally regulate inflammatory responses.
Summary
Elucidation of the mechanisms by which cells respond to nucleic acids with the induction and response to IFNs has advanced our understanding of inherited autoinflammatory diseases. The integration of a clinical phenotype with specific genetic mutations can be used to more accurately diagnose chronic inflammatory diseases. Autoinflammatory diseases may share some similar phenotypes and yet also have unique symptoms. Inherited genetic mutations may involve not one but multiple genes, some of which remain to be identified. A specific set of autoinflammatory diseases has found to have a unifying phenotype of an increased IFN gene signature, and they have been designated type I interferonopathies [93–95]. They include diseases mentioned above as well as those less understood such as systemic sclerosis, Sjögren’s syndrome, rheumatoid arthritis, and idiopathic myositis. Therapeutic interventions that block IFN signaling, such as the JAK inhibitors, are just beginning to be tested. Whether monogenic or multigenic, identifying type I interferonopathies may help to design effective clinical treatments for inherited autoinflammatory diseases.
Highlights.
Type I interferonopathies define a set of autoinflammatory diseases with an increased interferon gene signature
Mendelian inherited mutations are causally linked to type I interferonopathies
Continuous activation of sentinel nucleic acid sensors, or loss of interferon pathway silencing promotes type I interferonopathies
Acknowledgements
Thanks extend to Eleanor Fish for the invitation to contribute this review. Due to extensive literature, I apologize to those who were not cited. Support was provided by NIH RO1AI105114.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Friedman RM, Clinical uses of interferons. Br J Clin Pharmacol, 2008. 65(2): p. 158–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden EC, et al. , Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov, 2007. 6(12): p. 975–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunn GP, Koebel CM, and Schreiber RD, Interferons, immunity and cancer immunoediting. Nat Rev Immunol, 2006. 6(11): p. 836–48. [DOI] [PubMed] [Google Scholar]
- 4.Hervas-Stubbs S, et al. , Direct effects of type I interferons on cells of the immune system. Clin Cancer Res, 2011. 17(9): p. 2619–27. [DOI] [PubMed] [Google Scholar]
- 5.Platanias LC, Interferons and their antitumor properties. J Interferon Cytokine Res, 2013. 33(4): p. 143–4. [DOI] [PubMed] [Google Scholar]
- 6.Hauschild A, et al. , Practical guidelines for the management of interferon-alpha-2b side effects in patients receiving adjuvant treatment for melanoma: expert opinion. Cancer, 2008. 112(5): p. 982–94. [DOI] [PubMed] [Google Scholar]
- 7.Davidson S, Maini MK, and Wack A, Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res, 2015. 35(4): p. 252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crow YJ, Type I interferonopathies: a novel set of inborn errors of immunity. Ann N Y Acad Sci, 2011. 1238: p. 91–98. [DOI] [PubMed] [Google Scholar]
- 9.Janeway CA Jr. and Medzhitov R, Innate immune recognition. Annu Rev Immunol, 2002. 20: p. 197–216. [DOI] [PubMed] [Google Scholar]
- 10.Brubaker SW, et al. , Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol, 2015. 33: p. 257–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan X, et al. , Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu Rev Microbiol, 2018. 72: p. 447–478. [DOI] [PubMed] [Google Scholar]
- 12.Barrat FJ, Elkon KB, and Fitzgerald KA, Importance of Nucleic Acid Recognition in Inflammation and Autoimmunity. Annu Rev Med, 2016. 67: p. 323–36. [DOI] [PubMed] [Google Scholar]
- 13.Chow KT, Gale M Jr., and Loo YM, RIG-I and Other RNA Sensors in Antiviral Immunity. Annu Rev Immunol, 2018. 36: p. 667–694. [DOI] [PubMed] [Google Scholar]
- 14.Plato A, Hardison SE, and Brown GD, Pattern recognition receptors in antifungal immunity. Semin Immunopathol, 2015. 37(2): p. 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, et al. , Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science, 2013. 339(6121): p. 786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, et al. , Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science, 2013. 339(6121): p. 826–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa H and Barber GN, STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature, 2008. 455(7213): p. 674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun W, et al. , ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A, 2009. 106(21): p. 8653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong B, et al. , The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity, 2008. 29(4): p. 538–50. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, et al. , Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science, 2015. 347(6227): p. aaa2630. [DOI] [PubMed] [Google Scholar]
- 21.Kumar KP, et al. , Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol Cell Biol, 2000. 20(11): p. 4159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoneyama M, Suhara W, and Fujita T, Control of IRF-3 activation by phosphorylation. J Interferon Cytokine Res, 2002. 22(1): p. 73–6. [DOI] [PubMed] [Google Scholar]
- 23.Abe T and Barber GN, Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol, 2014. 88(10): p. 5328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoneyama M, et al. , The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol, 2004. 5(7): p. 730–7. [DOI] [PubMed] [Google Scholar]
- 25.Yoo JS, Kato H, and Fujita T, Sensing viral invasion by RIG-I like receptors. Curr Opin Microbiol, 2014. 20: p. 131–8. [DOI] [PubMed] [Google Scholar]
- 26.Lemaitre B, et al. , The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell, 1996. 86(6): p. 973–83. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T and Akira S, The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol, 2010. 11(5): p. 373–84. [DOI] [PubMed] [Google Scholar]
- 28.Daly C and Reich NC, Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J Biol Chem, 1995. 270(40): p. 23739–46. [DOI] [PubMed] [Google Scholar]
- 29.Brzostek-Racine S, et al. , The DNA damage response induces IFN. J Immunol, 2011. 187(10): p. 5336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onoguchi K, et al. , Viral infections activate types I and III interferon genes through a common mechanism. J Biol Chem, 2007. 282(10): p. 7576–81. [DOI] [PubMed] [Google Scholar]
- 31.Stark GR and Darnell JE Jr., The JAK-STAT pathway at twenty. Immunity, 2012. 36(4): p. 503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Moczygemba M, et al. , Distinct STAT structure promotes interaction of STAT2 with the p48 subunit of the interferon-alpha-stimulated transcription factor ISGF3. J Biol Chem, 1997. 272(32): p. 20070–6. [DOI] [PubMed] [Google Scholar]
- 33.Levy D, et al. , Transcriptional regulation of interferon-stimulated genes: a DNA response element and induced proteins that recognize it. Cold Spring Harb Symp Quant Biol, 1988. 53 Pt 2: p. 799–802. [DOI] [PubMed] [Google Scholar]
- 34.Hertzog P, Forster S, and Samarajiwa S, Systems biology of interferon responses. J Interferon Cytokine Res, 2011. 31(1): p. 5–11. [DOI] [PubMed] [Google Scholar]
- 35.Mostafavi S, et al. , Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell, 2016. 164(3): p. 564–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Qian C, and Cao X, Post-Translational Modification Control of Innate Immunity. Immunity, 2016. 45(1): p. 15–30. [DOI] [PubMed] [Google Scholar]
- 37.Arimoto KI, et al. , Negative regulation of type I IFN signaling. J Leukoc Biol, 2018. [DOI] [PubMed] [Google Scholar]
- 38.Boone DL, et al. , The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol, 2004. 5(10): p. 1052–60. [DOI] [PubMed] [Google Scholar]
- 39.Saitoh T, et al. , A20 is a negative regulator of IFN regulatory factor 3 signaling. J Immunol, 2005. 174(3): p. 1507–12. [DOI] [PubMed] [Google Scholar]
- 40.Catrysse L, et al. , A20 in inflammation and autoimmunity. Trends Immunol, 2014. 35(1): p. 22–31. [DOI] [PubMed] [Google Scholar]
- 41.Lin R, et al. , Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem, 2006. 281(4): p. 2095–103. [DOI] [PubMed] [Google Scholar]
- 42.Malakhova OA, et al. , Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev, 2003. 17(4): p. 455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arimoto KI, et al. , STAT2 is an essential adaptor in USP18-mediated suppression of type I interferon signaling. Nat Struct Mol Biol, 2017. 24(3): p. 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, et al. , Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature, 2015. 517(7532): p. 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McBride KM, McDonald C, and Reich NC, Nuclear export signal located within the DNA-binding domain of the STAT1transcription factor. EMBO J, 2000. 19(22): p. 6196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McBride KM, et al. , Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO J, 2002. 21(7): p. 1754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reich NC and Liu L, Tracking STAT nuclear traffic. Nat Rev Immunol, 2006. 6(8): p. 602–12. [DOI] [PubMed] [Google Scholar]
- 48.ten Hoeve J, et al. , Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol, 2002. 22(16): p. 5662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu TR, et al. , SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem, 2002. 277(49): p. 47572–80. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, et al. , Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A, 1998. 95(18): p. 10626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B and Shuai K, Targeting the PIAS1 SUMO ligase pathway to control inflammation. Trends Pharmacol Sci, 2008. 29(10): p. 505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linossi EM, et al. , Suppression of cytokine signaling: the SOCS perspective. Cytokine Growth Factor Rev, 2013. 24(3): p. 241–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimura A, et al. , Negative Regulation of Cytokine Signaling in Immunity. Cold Spring Harb Perspect Biol, 2018. 10(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naka T, et al. , Structure and function of a new STAT-induced STAT inhibitor. Nature, 1997. 387(6636): p. 924–9. [DOI] [PubMed] [Google Scholar]
- 55.Starr R, et al. , A family of cytokine-inducible inhibitors of signalling. Nature, 1997. 387(6636): p. 917–21. [DOI] [PubMed] [Google Scholar]
- 56.Endo TA, et al. , A new protein containing an SH2 domain that inhibits JAK kinases. Nature, 1997. 387(6636): p. 921–4. [DOI] [PubMed] [Google Scholar]
- 57.Forster SC, Tate MD, and Hertzog PJ, MicroRNA as Type I Interferon-Regulated Transcripts and Modulators of the Innate Immune Response. Front Immunol, 2015. 6: p. 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moulton VR, et al. , Pathogenesis of Human Systemic Lupus Erythematosus: A Cellular Perspective. Trends Mol Med, 2017. 23(7): p. 615–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirou KA, et al. , Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum, 2004. 50(12): p. 3958–67. [DOI] [PubMed] [Google Scholar]
- 60.Higgs BW, et al. , Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann Rheum Dis, 2011. 70(11): p. 2029–36. [DOI] [PubMed] [Google Scholar]
- 61.Trinchieri G, Type I interferon: friend or foe? J Exp Med, 2010. 207(10): p. 2053–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Biron CA, et al. , Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol, 1999. 17: p. 189–220. [DOI] [PubMed] [Google Scholar]
- 63.Luft T, et al. , Type I IFNs enhance the terminal differentiation of dendritic cells. J Immunol, 1998. 161(4): p. 1947–53. [PubMed] [Google Scholar]
- 64.Huber JP and Farrar JD, Regulation of effector and memory T-cell functions by type I interferon. Immunology, 2011. 132(4): p. 466–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bogdan C, Mattner J, and Schleicher U, The role of type I interferons in non-viral infections. Immunol Rev, 2004. 202: p. 33–48. [DOI] [PubMed] [Google Scholar]
- 66.Napirei M, et al. , Features of systemic lupus erythematosus in Dnase1-deficient mice. Nature Genetics, 2000. 25(2): p. 177–181. [DOI] [PubMed] [Google Scholar]
- 67.Yasutomo K, et al. , Mutation of DNASE1 in people with systemic lupus erythematosus. Nature Genetics, 2001. 28(4): p. 313–314. [DOI] [PubMed] [Google Scholar]
- 68.Al-Mayouf SM, et al. , Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nature Genetics, 2011. 43(12): p. 1186–1188. [DOI] [PubMed] [Google Scholar]
- 69.Rodero MP, et al. , Type I interferon-mediated autoinflammation due to DNase II deficiency. Nature Communications, 2017. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee-Kirsch MA, et al. , A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. Journal of Molecular Medicine-Jmm, 2007. 85(5): p. 531–537. [DOI] [PubMed] [Google Scholar]
- 71.Ellyard JI, et al. , Identification of a Pathogenic Variant in TREX1 in Early-Onset Cerebral Systemic Lupus Erythematosus by Whole-Exome Sequencing. Arthritis & Rheumatology, 2014. 66(12): p. 3382–3386. [DOI] [PubMed] [Google Scholar]
- 72.Stetson DB, et al. , Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell, 2008. 134(4): p. 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peschke K, et al. , Loss of Trex1 in Dendritic Cells Is Sufficient To Trigger Systemic Autoimmunity. Journal of Immunology, 2016. 197(6): p. 2157–2166. [DOI] [PubMed] [Google Scholar]
- 74.Stephenson JBP, Aicardi-Goutieres syndrome (AGS). European Journal of Paediatric Neurology, 2008. 12(5): p. 355–358. [DOI] [PubMed] [Google Scholar]
- 75.Crow YJ, et al. , Mutations in the gene encoding the 3 ‘−5 ‘ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature Genetics, 2006. 38(8): p. 917–920. [DOI] [PubMed] [Google Scholar]
- 76.Rice GI, et al. , Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature Genetics, 2009. 41(7): p. 829–U89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crow YJ, et al. , Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nature Genetics, 2006. 38(8): p. 910–916. [DOI] [PubMed] [Google Scholar]
- 78.Rice GI, et al. , Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nature Genetics, 2012. 44(11): p. 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samuel CE, Adenosine deaminase acting on RNA (ADAR1), a suppressor of double-stranded RNA-triggered innate immune responses. Journal of Biological Chemistry, 2019. 294(5): p. 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liddicoat BJ, et al. , RNA EDITING RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science, 2015. 349(6252): p. 1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rice GI, et al. , Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nature Genetics, 2014. 46(5): p. 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeremiah N, et al. , Inherited STING gain of function mutation in a familial inflammatory syndrome with lupus-like manifestations. Immunology, 2014. 143: p. 185–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Y, et al. , Activated STING in a Vascular and Pulmonary Syndrome. New England Journal of Medicine, 2014. 371(6): p. 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Konig N, et al. , Familial chilblain lupus due to a gain-of-function mutation in STING. Annals of the Rheumatic Diseases, 2017. 76(2): p. 468–472. [DOI] [PubMed] [Google Scholar]
- 85.Rutsch F, et al. , A Specific IFIH1 Gain-of-Function Mutation Causes Singleton-Merten Syndrome. American Journal of Human Genetics, 2015. 96(2): p. 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jang MA, et al. , Mutations in DDX58, which Encodes RIG-I, Cause Atypical Singleton-Merten Syndrome. American Journal of Human Genetics, 2015. 96(2): p. 266–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vivarelli R, et al. , Pseudo-TORCH syndrome or Baraitser-Reardon syndrome: diagnostic criteria. Brain & Development, 2001. 23(1): p. 18–23. [DOI] [PubMed] [Google Scholar]
- 88.Meuwissen MEC, et al. , Human USP18 deficiency underlies type 1 interferonopathy leading to severe pseudo-TOR CH syndrome. Journal of Experimental Medicine, 2016. 213(7): p. 1163–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Recht M, Borden EC, and Knight E Jr., A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J Immunol, 1991. 147(8): p. 2617–23. [PubMed] [Google Scholar]
- 90.Mankouri J, et al. , Optineurin negatively regulates the induction of IFNbeta in response to RNA virus infection. PLoS Pathog, 2010. 6(2): p. e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Das A, et al. , Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. J Virol, 2014. 88(6): p. 3103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stawowczyk M, et al. , Pathogenic Effects of IFIT2 and Interferon-beta during Fatal Systemic Candida albicans Infection. MBio, 2018. 9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uggenti C, Lepelley A, and Crow YJ, Self-Awareness: Nucleic Acid-Driven Inflammation and the Type I Interferonopathies. Annu Rev Immunol, 2019. 37: p. 247–267. [DOI] [PubMed] [Google Scholar]
- 94.Crow MK and Ronnblom L, Type I interferons in host defence and inflammatory diseases. Lupus Sci Med, 2019. 6(1): p. e000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee-Kirsch MA, The Type I Interferonopathies. Annu Rev Med, 2017. 68: p. 297–315. [DOI] [PubMed] [Google Scholar]