Abstract
Bacterial microcompartments are subcellular compartments found in many prokaryotes; they consist of a protein shell that encapsulates enzymes that perform a variety of functions. The shell protects the cell from potentially toxic intermediates and co-localizes enzymes for higher efficiency. Accordingly, it is of considerable interest for biotechnological applications. We have previously structurally characterized an intact 40 nm shell comprised of three different types of proteins. One of those proteins, BMC-H, forms a cyclic hexamer; here we have engineered a synthetic protein that consists of a tandem duplication of BMC-H connected by a short linker. The synthetic protein forms cyclic trimers that self-assemble to form a smaller (25 nm) icosahedral shell with gaps at the pentamer positions. When co-expressed in vivo with the pentamer fused to an affinity tag we can purify complete icosahedral shells. This engineered shell protein constitutes a minimal shell system to study permeability; reducing symmetry from six- to three-fold will allow for finer control of the pore environment. We have determined a crystal structure of this shell to guide rational engineering of this microcompartment shell for biotechnological applications.
Keywords: bacterial microcompartment, synthetic biology, protein engineering
Introduction
Bacterial Microcompartments (BMCs) are organelles consisting of a selectively-permeable protein shell encapsulating a segment of a metabolic pathway. They are found across the Kingdom Bacteria where they play functionally diverse roles including CO2 fixation and the catabolism of a range of organic compounds1. Compartmentalization of enzymes contributes to catalytic efficiency, and prevents the leakage of substrate or toxic intermediates into the cytosol. Because they self-assemble, and can be programmed to contain heterologous cargo, synthetic BMCs 2–5 hold promise as metabolic modules that can be deployed for bioengineering in vivo or in vitro (e.g. cell-free metabolic engineering).
While the compartmentalized enzymes dictate the function, and hence the diversity of BMCs, the shells of all BMCs are architecturally similar, being composed of homohexamers, homotrimers (that resemble the hexamers in size and shape) and pentamers that assemble into an icosahedral shell. The basic unit of the hexagonal facet building blocks is the BMC domain, pfam00936. BMC-H proteins contain a single copy of this domain and assemble into cyclic hexamers. Loop residues from the six subunits converging at the cyclic symmetry axis dictate the size and charge of the resulting pores that serve as the conduits for substrates and products of the encapsulated enzymes. Hexamers formed by BMC-H proteins are the majority building block of shell facets in all characterized systems, and every BMC locus bioinformatically identified contains at least one BMC-H gene. BMC-T proteins are composed of a fusion of two pfam00936 domains, accordingly only three copies are needed to form a hexagonal shell protein comparable in size and shape to the hexamers. Gene and domain duplication and fusion is a well-accepted principle of protein evolution 6–9; BMC-T proteins likely originated from a duplication/fusion event of a BMC-H gene. While a duplication of a pfam00936 domain gene would likely result in increased expression, a fusion of the two copies of the pfam00936 domain into a single polypeptide ensures co-localization and interaction. Additionally, the reduced symmetry facilitates diversification of function. This is evident in the BMC-T proteins which have been structurally characterized: they exhibit pores that are gated 10–12, off the symmetry axis 13, 14 or that bind FeS clusters 15, 16. Furthermore, the hexagonal building blocks formed by BMC-T proteins have two types of edges distinct at the level of primary structure, and presumably confer different affinities for other shell proteins. This differential affinity could be useful for controlling the composition of the shell surface; indeed, there are BMCs with as many as eight BMC-H paralogs, and up to four BMC-T proteins. It is likely that the distinct edges on paralogous proteins play some role in defining the rules for shell self-assembly.
We have developed and structurally characterized a synthetic shell system based on the metabolosome encoded by Haliangium ochraceum (HO) 3. High yields of stable shells can be produced in vivo or in vitro by combining the BMC-H, a BMC-P and three types of BMC-T proteins encoded in the HO genome. The crystal structure of the resulting 40 nm shell showed that the BMC-H hexamer was the predominant component of the facets. While we have been able to construct similarly sized shells with a single BMC-T, BMC-P and BMC-H protein (the minimal HO shell) 17, the latter seems to be an absolute requirement for shell formation. Here we further minimized the shell system by constructing a synthetic BMC-T protein composed of two copies of the HO BMC-H domain. We show that the synthetic shell protein, BMC-H2, when co-expressed with pentamers, is capable of forming shells. We have determined the crystal structure of BMC-H2 and the crystal packing recapitulates shell assembly, forming 25 nm uncapped shells in the crystal. The BMC-H2 shell system constitutes a relatively simple generic building block that could be used to construct designed shells with a relatively highly tunable pore. More generally, our results demonstrate the composability of the BMC shell using the principles of protein evolution to inform design.
Results and Discussion
Design of an engineered BMC shell protein
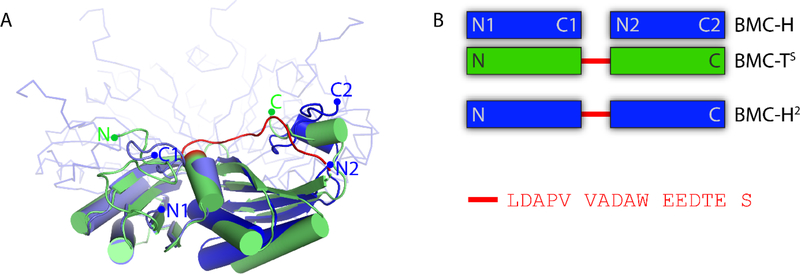
Pfam00936 domains are about 90–100 amino acids long and contain a structurally conserved α/β fold (Fig. 1A). The structures of HO-BMC-H and HO-BMC-TS have been previously determined 18,19; they have low sequence identity (25/28% identity over 97/89 aligned residues for BMC-TS domain 1 and 2, respectively). Structurally, however, the individual pfam00936 domains of BMC-TS each align well to BMC-H with low rmsd values (0.90/0.66 Å over 56/51 aligned Cα atoms for the N- and C-terminal domain respectively) (Fig. 1A). We have designed a synthetic BMC-TS consisting of two copies of the pfam00936 domain of HO-BMC-H connected by the natural linker sequence that connects the two domains of HO-BMC-TS protein (Fig. 1B) and named this fusion BMC-H2.
Figure 1. Design of a synthetic BMC-T protein: BMC-H2.
A: Structural alignment of BMC-TS (green cartoon) with BMC-H (blue cartoon for aligned chains and ribbon for the other chains). The linker connecting the two pfam00936 domains in BMC-TS is highlighted in red. B: Schematic comparison of the native and synthetic shell proteins with the primary structure of the interdomain linker.
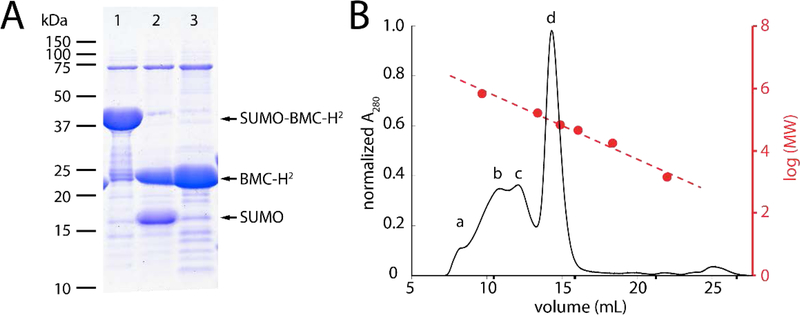
When recombinantly expressed in Escherichia coli, BMC-H2 (22 kDa) was mostly soluble (Fig. S1A). To further reduce aggregation, facilitate purification and increase yields, we employed a previously described strategy 20 of attaching a 6xHis-SUMO (Small Ubiquitin-like Modifier) solubility tag at the N-terminus (15 kDa) of BMC-H2. The fusion protein was highly expressed and could be cleaved by ULP protease and, following subtractive Ni-NTA chromatography, yielded pure BMC-H2 (Fig. 2A).
Figure 2. Purification and characterization of SUMO-BMC-H2.
A: Purification and cleavage of SUMO-BMC-H2 (37 kDa) into BMC-H2 (22 kDa) and His-SUMO (15 kDa). Lane 1: Ni-NTA elution; Lane 2: completed cleavage reaction and Lane 3: Ni-NTA flow-through containing pure BMC-H2. B: Size-exclusion chromatography elution profile of BMC-H2 corresponding to protein in lane 3 from (A). Protein molecular weight standards (red) were used to estimate the molecular weight of each species in the purified BMC-H2 sample; a:>670 kDa, b:356 kDa, c:188 kDa, d:60 kDa).
Size exclusion chromatography of BMC-H2 shows the predominant species has a molecular weight of ~60 kDa, consistent with the assembly of BMC-H2 trimers (Fig. 2B). However, despite being mostly soluble, several complexes with higher molecular weight are also present (Fig. 2B); all fractions consisted primarily of BMC-H2 (Fig. S1B).
Structural Characterization of BMC-H2
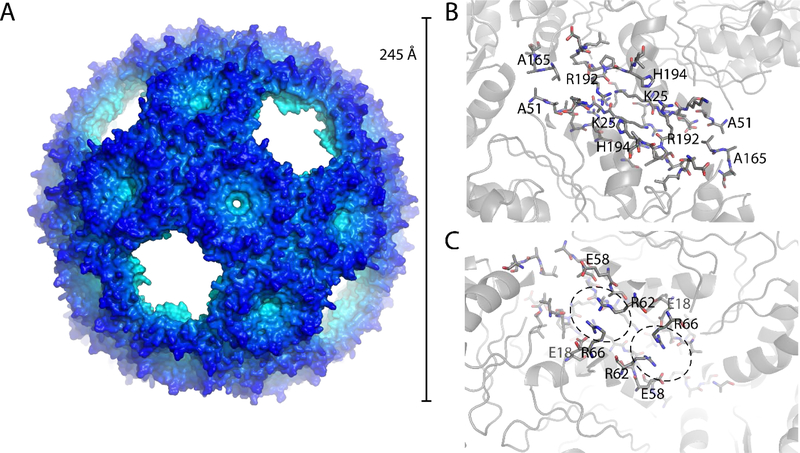
BMC-H2 was concentrated to 9 mg/ml and crystallized by vapor diffusion. BMC-H2 crystallized in space group P213 with 10 trimers in the asymmetric unit. The asymmetric unit comprises 1/3 of a large icosahedral shell consisting of 90 BMC-H2 chains (30 trimers) with a diameter of 245 Å (Fig. 3A) and a total mass of 1.96 MDa. Data collection and refinement statistics are summarized in Table 1. The density is of good quality considering the resolution (3.6 Å) and overall size of the particle (Fig. S4) and the model has good geometry (Table 1).
Figure 3. Structure of the BMC-H2 shell.
A: Surface representation of the crystal structure of the BMC-H2 T=4 icosahedral shell. B: View on the trimer-trimer interface from the outside with interacting residues shown as sticks. C: View from the inside, interacting arginine residues are highlighted as well as charge complementing glutamic acid residues.
Table 1.
Data collection and refinement statistics.
| BMC-H2 | |
|---|---|
| Data collection | |
| Resolution range (Å) | 49.6 – 3.59 (3.65 – 3.59) |
| Space group | P 21 3 |
| Unit cell dimensions (Å / °) | 325.5 325.5 325.590 90 90 |
| Total reflections | 5,385,926 (254,230) |
| Unique reflections | 133,312 (6,556) |
| Multiplicity | 40.4 (38.8) |
| Completeness (%) | 100.0 (99.9) |
| Mean I/sigma(I) | 8.7 (1.5) |
| R-merge | 0.58 (4.1) |
| R-meas | 0.60 (4.2) |
| CC1/2 | 0.938 (0.808) |
| Refinement | |
| Resolution range (Å) | 49.1 – 3.59 (3.72 – 3.59) |
| Number of reflections | 133,266 (13,205) |
| Number of reflections used for R-free | 1,676 (167) |
| R-work (%) | 17.1 (27.5) |
| R-free (%) | 20.9 (31.1) |
| Number of non-hydrogen atoms | 39,510 |
| macromolecules | 39,460 |
| ligands | 50 |
| solvent | 0 |
| Protein residues | 5,521 |
| RMS (bonds, Å) | 0.001 |
| RMS (angles, °) | 0.37 |
| Ramachandran favored (%) | 96.2 |
| Ramachandran allowed (%) | 3.8 |
| Ramachandran outliers (%) | 0 |
| Clashscore | 4.3 |
| Average B-factor (Å2) | 113 |
Statistics for the highest-resolution shell are shown in parentheses.
The positions where the pentameric subunits would be located are empty, resulting in a “wiffle ball” type shell. The linkers connecting the two BMC-H domains to form BMC-H2 were not visible in the density but the protein chain corresponding to BMC-H is fully ordered. Because the majority of the purified protein appears to be a trimeric species in size-exclusion chromatography (Fig. 2B) it is likely that the shells self-assemble over time under the crystallization conditions. It also appears that the inter-domain linker is cleaved, possibly due to a trace amount of protease in the sample (Fig. S2) which explains the absence of electron density for the linker.
The BMC-H2-trimers are arranged in T4 icosahedral symmetry with vacant vertex (pentamer) positions. The two trimers interact with each other at a 30° angle, the same as observed between two BMC-H hexamers adjacent to a BMC-P pentamer in the pseudo-T9 icosahedral shell composed of BMC-H, BMC-TS/D and BMC-P 21. The specific interactions are mostly identical to those observed for the T9 shell with only minor differences in side chain rotamers (Fig. S3). The position of the backbone of the BMC-H2 chain adjacent to the where the pentamer would be (residues 48–58) is also slightly different, possibly due to the lack of constraints by adjacent pentamer chains.
Distinctive features of the T4 icosahedral shell include the arginine stacking (R62:R66) on the interior surface (Fig. 3C). This interaction might stabilize the specific 30° angle between two BMC-H2 trimers; this type of parallel stacking interaction seems unusual since two positively charged residues are thought to repel each other. However, the energy gain from solvation combined with nearby negative charged residues (glutamic acid residues 18 and 58) can make this a stabilizing interaction 22. In the pore of each trimer there is a large spherical density that, considering its shape and intensity seems to be a sulfate ion (Fig. S4). There is no residual difference density after refining the model with a sulfate in this position with full occupancy. Sulfate has been previously observed in the center of the BMC-H pore, for example in the carboxysome protein CsoS1A 23.
It is not obvious why the linear fusion of two BMC-H domains would form a full shell particle instead of layers as observed before for the purified BMC-H protein 19.
The interfaces between the BMC-H2 are almost identical to the interactions between BMC-H observed in the HO shell, indicating that it might be possible to form similar shells from just BMC-H given the right conditions. While this might be possible for an engineered construct it is unlikely to be desired for assembling a native BMC in vivo, possibly the presence of other subunits and/or cargo will prevent formation of such small shells. This is also the second observation that icosahedral shells can be formed without a pentamer at the vertices, both highlighting the specificity of the BMC-H interactions with itself as well as raising the question whether the “pentamer capping” could have a role in native BMC assembly.
In vivo assembly of capped BMC-H2 shells
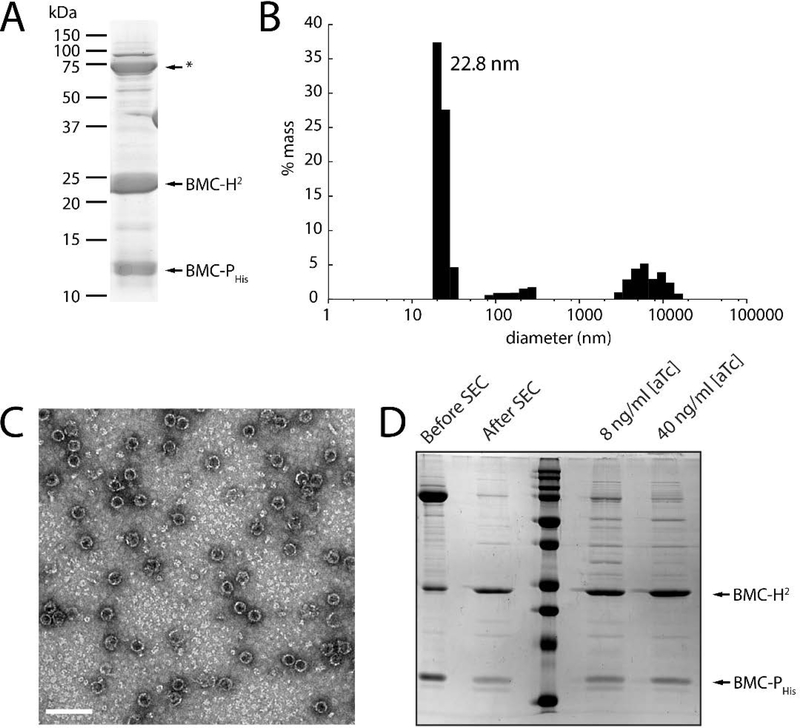
Our structural data suggest that shells composed of only BMC-H2, lacking vertex proteins can form. However, adding Strep-II-tagged pentamers to BMC-H2 samples did not yield any apparent complexes/shells when using the Strep-tag as an affinity handle (not shown). We tested whether it is possible to generate fully closed shells, containing pentamers at the vertices in vivo by co-expressing BMC-P with BMC-H2 proteins. BMC-P containing a C-terminal His tag (on a separate, compatible plasmid) was co-expressed with BMC-H2 and the complex was purified from cleared lysate using a HisTrap affinity column; in the absence of BMC-Phis, BMC-H2 does not appreciably bind to the HisTrap resin (Fig. S5). The shells in the eluate from the Ni column (Fig. 4A) were then pelleted using ultracentrifugation and further purified by anion exchange chromatography. This sample was then analyzed by Dynamic Light Scattering and the majority of the sample had a diameter of 22.8 nm (Fig. 4B). Shells with a diameter consistent with the crystal structure were observed by TEM (Fig. 4C), indicating that we are able to form and purify capped shells.
Figure 4. Characterization of in vivo assembled BMC-H2 shells in the presence of BMC-PHis.
A: SDS-PAGE analysis of concentrated HisTrap elution fractions. A common contaminant from the HisTrap is indicated by an asterisk. B: Size distribution of concentrated HisTrap elution fractions from (A) using Dynamic Light Scattering. C: TEM micrograph of the sample in (A). Diameter = 26 ± 2 nm (n=407). Scale bar = 100 nm. D: SDS-PAGE analysis showing the removal of excess pentamer with a size exclusion step (left two lanes) and samples purified from strains expressing BMC-Phis at two different induction levels.
To test whether the shells we generated are fully capped we removed excess BMC-Phis with an additional size exclusion step (Fig. 4D) and compared two different induction conditions of the pentamer; both samples show the same ratio between BMC-P and BMC-H2 (Fig. 4D). Densitometry analysis of the bands shows a ration of BMC-H2 to BMC-Phis of 3:1, which is consistent with the expected mass ratio for a fully capped shell. When we also co-expressed HO-BMC-TS we did not see a change in shell size as would be expected if BMC-H2 acted to replace the BMC-H (Fig. S6), indicating that the BMC-H2 shell formation is quicker and/or more stable than forming a full 40 nm diameter shell containing BMC-H2, BMC-P and BMC-TS.
Conclusions
We have designed an artificial BMC-T shell protein by connecting two BMC-H domains with a linker from a BMC-T protein. BMC-H2 expressed well and under conditions for used for crystallization formed shells lacking the twelve vertex pentamers with icosahedral T=4 symmetry. Co-expression with BMC-P led to apparently closed shells of the same dimensions (Fig. 4). In contrast to native BMC shell systems, BMC-H2-BMC-Phis shells constitute a very simple system that can be further customized for fundamental studies and bioengineering. Because they are composed of only one type of BMC-H protein, they provide a model system to characterize the influence of a single type of pore configuration on permeability. Moreover, as the pore in BMC-H2 is composed of residues from two distinct BMC domains, the three-fold symmetry permits constructing pores composed of two sets of residues, rather than six copies a single pore motif. This provides relatively finer control over or the potential for evolution of the duplicated pore and edge residues, of the hexagonal building block than is possible with a homohexameric construct.
While the BMC-H2 shell is smaller than synthetic shells 3, 21, 25 or natural BMCs formed in vivo, it is comparable in size to a smaller type of compartment composed of a single protein known as Encapsulin 26, 27. Both BMC-H2 and Encapsulins are composed of a single subunit and so are simple to express and potentially modify. Encapsulins form completely closed shells. In contrast, the wiffle ball type shell formed by BMC-H2 has 45 Å gaps at the vertices, a distinctive architectural feature that could enable of exchange larger substrates and products without the need to engineer the pores while still providing the scaffolding for co-localizing enzymes, not only on the interior but on the exterior shell surface as well.
The x-ray crystallographic coordinates and structure-factor files for BMC-H2 have been deposited in the Protein Data Bank (PDB) under the accession ID 6NER.
Materials and Methods
Cloning and Protein Expression:
The gene coding for BMC-H2 is composed of two copies of the coding sequence for Hoch_5815 (BMC-H) from Haliangium ochraceum, connected by the 16 amino acid linker (LDAPVVADAWEEDTES) that joins the two Pfam00936 domains in Hoch_5812 (BMC-TS); only the starting methionine residue from the second BMC-H sequence was omitted in the BMC-H2 construct (See Table S1 for a complete list of amino acid sequences used in this study). The gene was synthesized as a gBlock (IDT) and codon optimized for E. coli and inserted into a pET11 DNA vector using the NdeI and BamHI restriction sites (p CA154). Using Gibson assembly we introduced BMC-H2 into a pBbE2k vector containing a His-tagged Small Ubiquitin-like modifier (SUMO) to generate the SUMO-BMC-H2 plasmid (pCA181). The plasmids encoding BMC-H2 (pCA154) or SUMO-BMC-H2 (pCA181) were transformed into E. coli BL21 (DE3) for protein expression. E. coli containing pCA154 were grown in LB broth (Miller) with 100 mg L−1 of ampicillin at 37 °C while agitating (180 RPM). At an OD600 of 0.8, cultures were induced with 0.45 mM of isopropyl thio-β-D-thiogalactoside (IPTG) and grown overnight at 18°C. E. coli expressing pCA181 were grown in LB broth with 50 mg L−1 of kanamycin at 37 °C while agitating (180 RPM). At an OD600 of 0.8, cultures were induced with 50 ng /ml of anhydrotetracycline (aTc) and grown overnight at 18 °C.
For the in vivo production of shells with (pSM7) or without (pCA154) BMC-TS, E. coli BL21 (DE3) cells were co-transformed with a plasmid expressing a C-terminally His-tagged BMC-P protein (pARH252). pSM7 was made by first deleting the BMC-H gene from a pET11 DNA vector containing both BMC-H and BMC-TS (pARH329) using inverse PCR; BMC-H2 was subsequently added using Gibson assembly, resulting in a plasmid coding for BMC-H2 and BMC-TS. See Table S2 for a list of primers used in this study. E. coli harboring the plasmids pSM7 or pCA154 and pARH252 were grown as described above, and induced at an OD600 of 0.8 with 0.45 mM IPTG and 8 ng/ml of aTc.
Protein Purification of BMC-H2:
Cell pellets of E. coli expressing BMC-H2 (typically from 50 ml of culture) were resuspended in lysis buffer (50 mM Tris-HCl pH 7.8, 10 mM MgCl2, 100 mM NaCl) in the presence of lysozyme. Sonication was used to lyse the cells which were incubated for 15 min at room temperature in the presence of the solubilizing agent, Triton™ × 100. The solution was centrifuged at 22,000 ×g and the supernatant was collected, the pellet was resuspended in lysis buffer containing 1% Triton™ ×100 and incubated as before. The centrifugation was repeated and the pellet was resuspended in lysis buffer to remove excess Triton.
SUMO-BMC-H2 was purified as follows: cell pellets from E. coli (generally 2 L cultures) expressing SUMO-BMC-H2 were resuspended in 50 mM Tris-HCl pH 7.8, 300 mM NaCl (Buffer A) in the presence of DNase and a protease inhibitor cocktail (SIGMAFAST™) and lysed by two passes through a French Pressure Cell Press at 4 °C and 137 MPa. The crude lysate was pelleted by centrifugation at 40,000 ×g for 45 minutes to remove cellular debris and the supernatant was filtered through a 0.22 |am filter and applied to a Protino® Ni-IDA resin (Macherey-Nigel) gravity-column equilibrated in Buffer A. The column was washed with 10 column volumes (CV) of Buffer A, and the flow-through collected. Bound protein was eluted with 5 CV of 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 250 mM imidazole buffer. Fractions containing SUMO-BMC-H2 were concentrated with a 50 kDa molecular weight cutoff (MWCO) centrifugal concentrator (Amicon) and buffer exchanged into 25 mM Tris-HCl pH 7.8, 500 mM NaCl using a PD-10 column (GE Healthcare). Protein concentration was determined using bicinchoninic acid (BCA) kit (Sigma-Aldrich).
BMC-H2 was purified for crystallization by incubating the SUMOylated protein overnight in the presence of ULP-protease. The cleaved protein was applied to a Ni-NTA resin (GE Healthcare) gravity column, washed with 8 CV of 50 mM Tris-HCl pH 7.8, 150 mM NaCl and the flow-through containing BMC-H2 was concentrated with a 30 kDa MWCO centrifugal concentrator (Amicon). BMC-H2 was then buffer-exchanged into 10 mM Tris-HCl pH 8.0, 50 mM NaCl using a PD-10 column and concentrated to 9 mg/ml (measured by BCA). For size exclusion chromatography, purified BMC-H2 was loaded onto a gel filtration column (HiLoad 10/300 Superdex 200, GE Healthcare) equilibrated in 25 mM Tris-HCl pH 7.8, 500 mM NaCl.
Cells from 2 L of E. coli culture harboring the plasmids pSM7 or pCA154 and pARH252 (BMC-PHis) were resuspended in 50 mM Tris-HCl pH 7.8, 300 mM NaCl, 20 mM imidazole (binding buffer) in the presence of DNase and lysozyme (1 μl of 25 mg/ml per 1 ml of cell suspension). The cells were lysed with a French Press as described above. The crude lysate was centrifuged at 40,000 ×g for 45 min to remove cell debris and the supernatant collected and centrifuged once more at 30,000 ×g for 30 min. The supernatant was filtered using a 0.22 μm filter and loaded onto a HisTrap (GE Healthcare) column on an Akta Pure FPLC (GE Healthcare). The column was washed with 8 CV binding buffer and the intact shells and free BMC-PHis were eluted by addition of 50 mM Tris-HCl pH 7.8, 300 mM NaCl, and 500 mM imidazole. The purified shells were then concentrated over a 100 kDa MWCO centrifugal concentrator (Amicon). The retentate was collected and centrifuged in a Beckman-Coulter Optima™ L-90K ultracentrifuge with a Ti-70 rotor at 150,000 ×g for 4 hrs. The supernatant was removed and the pellet containing intact shells was resuspended in 2 ml of 50 mM Tris-HCl pH 7.8, 300 mM NaCl buffer. The shell fractions were cleaned by application to a gel filtration column (HiLoad 10/300 Superdex 200, GE Healthcare) equilibrated in 25 mM Tris-HCl pH 7.8, 500 mM NaCl to remove unincorporated BMC-Phis. Void volumefractions containing shells, as confirmed on SDS-PAGE, were pooled and concentrated using a 100 kDa MWCO centrifugal concentrator (Amicon).
Alternatively, for the comparison of BMC-Phis induction tests, cells containing the plasmids pCA154 and pARH252 were grown in the presence of 0.45 mM IPTG to induce BMC-H2 (pCA154) and either 8 ng/ml or 40 ng/ml aTc to induce BMC-Phis (pARH252) were lysed as described above and clarified by centrifugation at 45,000 ×g for 30 min at 4 °C. The soluble fractions were loaded onto 10 ml 25% sucrose cushions and centrifuged for 16 h at 182,000 ×g for 16 h at 4 °C using Beckman-Coulter OptimaTM L-90K ultracentrifuge with a Ti-70 rotor to remove unincorporated BMC-Phis. Pellets containing BMC-H2 + BMC-Phis shells were resuspended in 50 mM Tris-HCl pH 7.8, 300 mM NaCl, 20 mM imidazole (binding buffer), filtered using a 0.22 μm filter and loaded onto a HisTrap (GE Healthcare) column on an Akta Pure FPLC (GE Healthcare) and eluted as described above. Elution fractions from the HisTrap column were concentrated and buffer exchanged into 50 mM Tris-HCl pH 7.8, 300 mM NaCl using a 100 kDa MWCO centrifugal concentrator (Amicon) and loaded onto a gel filtration column (HiLoad 10/300 Superdex 200, GE Healthcare) equilibrated in 25 mM Tris-HCl pH 7.8, 500 mM NaCl. Shells eluted in the void volume and were collected and concentrated using a 100 kDa MWCO centrifugal concentrator (Amicon).
Crystallization and Structure Determination:
For crystallization, BMC-H2 was concentrated to 9 mg/ml in 10 mM Tris pH 8.0, 50mM NaCl. Conditions for vapor diffusion crystallization purified BMC-H2 were screened using an Art Robbins Crystal Phoenix robot, finding a candidate for crystallization and improving upon the initial condition in a 24-well, sitting-drop, HR3–160 Cryschem plate (Hampton Research). Rhombic bipyramidal crystals were observed in reservoir conditions of 0.1 M HEPES pH 7−8, 1−2% [w/v] tryptone, 12–16% [w/v] PEG 3,350 in a 4 μL drop consisting of 1 μL of reservoir condition and 3 μL of purified protein. Crystals were cryo-protected by the addition of PEG 3,350 to a final concentration of 35% [w/v], looped and then flash frozen in liquid nitrogen. Diffraction data were collected at ALS beam line 5.0.3, integrated with XDS 28 and scaled with SCALA (CCP4) 29. The BMC-H2 shell structure was solved by molecular replacement with phenix.phaser 30 using a search model consisting of one BMC-H hexamer (HO_5815, PDB ID 5DJB). The resulting solution was refined and rebuilt using phenix.refine 30 and COOT 31. Statistics for diffraction data collection, structure determination and refinement are summarized in Table 1.
Dynamic Light Scattering:
Concentrated and pooled eluates from the HisTrap that contained purified shells (in in 50 mM Tris-HCl pH 7.8 and 300 mM NaCl) were analyzed with a Dynapro Nanostar (Wyatt Technologies) with 20 acquisitions of 5 s each, at room temperature.
Transmission Electron Microscopy:
5 μL of the shell containing fractions were spotted onto a carbon-coated copper grid (Electron Microscopy Sciences, CF150-Cu) for 30 seconds, wicked dry using filter paper and negatively stained with 5 μL of 1% [w/v] uranyl acetate solution for 10 seconds. Samples were imaged using a JEOL 100CXII microscope at an accelerating voltage of 100 kV with a Gatan Orius SC200 CCD camera.
Supplementary Material
Acknowledgements
We thank C. Aussignargues for help with construct design and initial characterization. CAK and MS acknowledge support of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI114975–01. BF and SM were supported by the U.S. Department of Energy, Basic Energy Sciences, Contract DE-FG02–91ER20021. This work used resources of the Advanced Light Source which is supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. DOE under Contract No. DE-AC02–05CH11231.
Abbreviations
- BMC
Bacterial Microcompartment
- BMC-H/T/P
Bacterial microcompartment shell proteins: hexamer/trimer/pentamer
- HO
Haliangium ochraceum
- SUMO
Small Ubiquitin-like Modifier
- TEM
Transmission Electron Microscopy
References
- [1].Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, and Sutter M (2018) Bacterial microcompartments, Nature Reviews Microbiology 16, 277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Frank S, Lawrence AD, Prentice MB, and Warren MJ (2013) Bacterial microcompartments moving into a synthetic biological world, Journal of Biotechnology 163, 273–279. [DOI] [PubMed] [Google Scholar]
- [3].Lassila JK, Bernstein SL, Kinney JN, Axen SD, and Kerfeld CA (2014) Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function, J Mol Biol 426, 2217–2228. [DOI] [PubMed] [Google Scholar]
- [4].Liang M, Frank S, Lunsdorf H, Warren MJ, and Prentice MB (2017) Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E coli, Biotechnol J. 12. [DOI] [PubMed] [Google Scholar]
- [5].Parsons JB, Lawrence AD, McLean KJ, Munro AW, Rigby SE, and Warren MJ (2010) Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment, Plos One 5, e14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pasek S, Risler JL, and Brezellec P (2006) Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins, Bioinformatics 22, 1418–1423. [DOI] [PubMed] [Google Scholar]
- [7].Vogel C, Bashton M, Kerrison ND, Chothia C, and Teichmann SA (2004) Structure, function and evolution of multidomain proteins, Current Opinion in Structural Biology 14, 208–216. [DOI] [PubMed] [Google Scholar]
- [8].Kondrashov FA, and Kondrashov AS (2006) Role of selection in fixation of gene duplications, J Theor Biol 239, 141–151. [DOI] [PubMed] [Google Scholar]
- [9].Lees JG, Dawson NL, Sillitoe I, and Orengo CA (2016) Functional innovation from changes in protein domains and their combinations, Current Opinion in Structural Biology 38, 44–52. [DOI] [PubMed] [Google Scholar]
- [10].Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, and Kerfeld A (2009) Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport, J Mol Biol 392, 319–333. [DOI] [PubMed] [Google Scholar]
- [11].Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, and Kerfeld CA (2013) The Structure of CcmP, a Tandem Bacterial Microcompartment Domain Protein from the beta-Carboxysome, Forms a Subcompartment Within a Microcompartment, J Biol Chem 288, 16055–16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Takenoya M, Nikolakakis K, and Sagermann M (2010) Crystallographic Insights into the Pore Structures and Mechanisms of the EutL and EutM Shell Proteins of the Ethanolamine-Utilizing Microcompartment of Escherichia coli, Journal of Bacteriology 192, 6056–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Heldt D, Frank S, Seyedarabi A, Ladikis D, Parsons JB, Warren MJ, and Pickersgill RW (2009) Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri, Biochem J 423, 199–207. [DOI] [PubMed] [Google Scholar]
- [14].Sagermann M, Ohtaki A, and Nikolakakis K (2009) Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment, Proc Natl Acad Sci U S A 106, 8883–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pang A, Warren MJ, and Pickersgill RW (2011) Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site, Acta Crystallogr D Biol Crystallogr 67, 91–96. [DOI] [PubMed] [Google Scholar]
- [16].Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, and Yeates TO (2010) Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell, J Biol Chem 285, 37838–37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hagen A, Sutter M, Sloan N, and Kerfeld CA (2018) Programmed loading and rapid purification of engineered bacterial microcompartment shells, Nature communications 9, 2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Aussignargues C, Pandelia ME, Sutter M, Plegaria JS, Zarzycki J, Turmo A, Huang J, Ducat C, Hegg EL, Gibney BR, and Kerfeld CA (2016) Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster, J Am Chem Soc 138, 5262–5270. [DOI] [PubMed] [Google Scholar]
- [19].Sutter M, Faulkner M, Aussignargues C, Paasch BC, Barrett S, Kerfeld CA, and Liu LN (2016) Visualization of Bacterial Microcompartment Facet Assembly Using High-Speed Atomic Force Microscopy, Nano Lett 16, 1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hagen AR, Plegaria JS, Sloan N, Ferlez B, Aussignargues C, Burton R, and Kerfeld CA (2018) In Vitro Assembly of Diverse Bacterial Microcompartment Shell Architectures, Nano Lett 18, 7030–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sutter M, Greber B, Aussignargues C, and Kerfeld CA (2017) Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell, Science 356, 1293–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee D, Lee J, and Seok C (2013) What stabilizes close arginine pairing in proteins?, Phys Chem Chem Phys 15, 5844–5853. [DOI] [PubMed] [Google Scholar]
- [23].Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, and Yeates TO (2007) Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome, PLoS Biol 5, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, and Yeates TO (2005) Protein structures forming the shell of primitive bacterial organelles, Science 309, 936–938. [DOI] [PubMed] [Google Scholar]
- [25].Choudhary S, Quin MB, Sanders MA, Johnson ET, and Schmidt-Dannert C (2012) Engineered protein nano-compartments for targeted enzyme localization, Plos One 7, e33342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sutter M, Boehringer D, Gutmann S, Guenther S, Prangishvili D, Loessner MJ, Stetter KO, Weber-Ban E, and Ban N (2008) Structural basis of enzyme encapsulation into a bacterial nanocompartment, Nat Struct Mol Biol 15, 939–947. [DOI] [PubMed] [Google Scholar]
- [27].Nichols RJ, Cassidy-Amstutz C, Chaijarasphong T, and Savage DF (2017) Encapsulins: molecular biology of the shell, Crit Rev Biochem Mol Biol 52, 583–594. [DOI] [PubMed] [Google Scholar]
- [28].Kabsch W (2010) XDS, Acta Cryst D 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS (2011) Overview of the CCP4 suite and current developments, Acta Cryst D 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine, Acta Cryst D 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics, Acta Cryst D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.