Abstract
OBJECTIVE:
Increasing evidence has indicated an association between gut microbiota in gastrointestinal cancer and clinical outcome. Herein, we aim to develop a prognosis-prediction tool based on an immune-lipid metabolism signature, tumor cell-associated immune microenvironment, and lipid metabolism proteins inferred from the function of gut microbiota.
METHODS:
16S gene ribosomal RNA sequencing was performed on 10 fecal samples obtained after tumor resection but before chemotherapy (EBVaGC = 4 and EBVnGC = 6). Least absolute shrinkage and selection operator (LASSO) Cox regression was applied to screening for highly accurate marker proteins. A compound score based on the fraction of screened markers was then constructed using a LASSO logistic regression model.
RESULTS:
The Tax4Fun analysis based on Kyoto Encyclopedia of Genes and Genomes data indicated differentially expressed tumor pathway between EBVnGC and EBVaGC. Using the LASSO logistic model, a compound score was established consisting of 14 types of immune microenvironment and lipid metabolism proteins. In the training set (378 patients), significant differences were found between high- and low-compound score groups in overall survival across and within subpopulations with an identical EBV. Multivariable analysis revealed that the compound score was an independent prognostic factor (hazard ratio, 2.26; 95% confidence interval = 2.28–3.36). The prognostic value ;of the compound score was also confirmed in the validation (162 patients) and entire (540 patients) sets.
DISCUSSION:
The proposed compound score is a promising signature for estimating overall survival in patients with gastric cancer having EBVaGCs or EBVnGCs.
INTRODUCTION
Treatment regimens based on the TNM staging have diverse outcomes (1–4). Recent studies suggest that the gut microbiota influences the pathogenesis and prognosis of digestive tract tumors (3). The consensus for gastric adenocarcinoma is that an EBV infection is most likely involved. Accordingly, understanding the impact of EBV infection on the survival outcomes may help clinicians predict patients' prognosis (5,6).
The gut microbiota affects the metabolism of the host. Changes in the composition of microbiota, meanwhile, may activate the host's immunity and participate in the process of protein secretion to act on the microenvironment, which interacts in a highly coordinated manner (3,7–9). However, their prognostic impact of the host and the markers of prognosis are still lacking. Therefore, enumerating the gut microbiota functional protein components according to their biologic function using bioinformatic analysis may be necessary for improving studies of the diverse biologic response in gastric adenocarcinoma and improve on its clinical management (10).
Collection of gut microbiologic specimens from patients with EBV-associated (EBVaGC) and EBV-negative (EBVnGC) gastric adenocarcinomas, 16S gene ribosomal RNA sequencing function prediction, and enrichment analysis are accepted methods for studying the effects of gut microbiota on host biologic activities. In the present study, gut microbiota its functional predictive expression was used to estimate the fractions of 21 biomolecules based on clinically annotated gastric cancer proteins expression profiles. Least absolute shrinkage and selection operator (LASSO) Cox regression analysis was used to screen 14 immune-lipid proteins, and then, to establish a compound score, LASSO logistic model was used to provide a statistically powerful means of predicting survival of patients with gastric adenocarcinoma.
METHODS
Study design and patient selection
Subjects were patients who underwent surgical resection of gastric adenocarcinoma at the Affiliated Hospital of Jiangnan University from December 2017 to May 2018. The resected adenocarcinoma tissues were formalin fixed and paraffin embedded. Within 72 hours of resection, the paraffin sections were analyzed with in situ hybridization (ISH) to determine Epstein-Barr virus-encoded RNA (EBER) and EBV latent membrane protein 1 (EBV-LMP1) and EBV nuclear antigen 1 (EBNA1) protein detection via immunohistochemistry (IHC). The patients were classified as EBVaGC and EBVnGC based on on their adenocarcinoma being EBV positive or EBV negative, respectively. The patients were followed up for 3 months and were contacted during the adjuvant before chemotherapy to collect their intestinal feces. This study was approved by the Ethics Review Board at the Affiliated Hospital of Jiangnan University.
Fecal sequencing and data analysis
The feces were collected from each subject in a sterile stool container, frozen immediately with liquid nitrogen, and stored at −80 °C. Because fecal samples differed in their collection dates, total bacterial DNA was extracted from the fecal samples within 1 month using the QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA) with minor adjustments to the manufacturer's protocol. The V3-V4 region of the 16S ribosomal RNA (rRNA) gene was amplified and sequenced on the Illumina MiSeq platform (Illumina, San Diego, CA) in multiple runs, pooling together all 10 samples using a 2 × 250 bp paired-end protocol, according to the manufacturer's instructions. Raw reads from the microbiota sequencing were analyzed using Pandaseq, processed through the QIIME (version 1.8.0), clustered into operational taxonomic units at 97% identity level, and taxonomically assigned via Ribosomal Database Project classifier against the Greengenes database (release 13.5; http://greengenes.secondgenome.com). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database was used to predict differences in bacterial biochemical pathways in de novo EBVaGCs and EBVnGCs. P value of <0.001 was considered statistically significant.
Collection of clinicopathologic variables and tissue samples
Formalin-fixed and paraffin-embedded tissue samples were obtained from 540 patients with gastric adenocarcinoma who underwent major surgery at the Affiliated Hospital of Jiangnan University between 2006 and 2011. For each case, 2 pathologists reviewed all original hematoxylin and eosin-stained sections to confirm the diagnosis of gastric adenocarcinoma. Tissue microarrays (TMAs) were spotted using a Quick-Ray manual TMA spotting (UNITMA, Seoul, Korea). Details on patients' gender, age, tumor size, TNM stage, and Lauren classification were obtained from medical records.
In situ hybridization
The EBER was detected with an EBER ISH kit (TIB Biotechnology, Xiamen, China). Briefly, 4-μm-thick slides were applied to the probe, denatured at 85 °C for 10 minutes, and then hybridized at 37 °C for 1 hour. The slides were first incubated with an antifluorescein monoclonal antibody and then with DAB as per the manufacturer's protocol. The slides were counterstained with nuclear fast red (11). Hematoxylin was used for negative controls.
Immunohistochemistry
Serial 4-μm-thick sections of fixed tissue were cut and placed onto glass slides, after which they underwent IHC staining with primary polyclonal antibodies targeting EBV-LMP1, EBNA1, ACOT1, ACOT4, ALCAM, BRAF, PDL1, B7H3, CD4, CD87, CXCL12, CXCL13, fatty acid desaturase (FADS) 2, FADS3, FADS6, hydroxyacyl-CoA dehydrogenase (HADHA), human leukocyte antigens (HLAA), human leukocyte antigens B (HLAB), human leukocyte antigens C (HLAC), PPT1, PPT2, TRAF1, and TRAF2 (all primary antibodies were purchased from Abcam, Hong Kong, China, and are listed in the Supplemental Digital Content 1 [see Table S1, http://links.lww.com/CTG/A87]). The sections were washed and incubated with an amplification agent and a polymerase. Nuclei were counterstained with hematoxylin. The expression of proteins was scored by 2 pathologists who were blinded and were unaware of all clinical parameters. Where serious discrepancies arose, a final score was determined by reassessment of the staining using a multihead microscope.
Random grouping method of study patients and the study population outcome
The patients were randomized 7:3 to a training or validation set as per the stratified randomization method. Patients were followed up long term by telephone, courier, mail, and in person. Overall survival was the primary endpoint. This was defined as the interval between the date of diagnosis and date of death from any cause. The 5-year survival rate was calculated as the proportion of patients surviving for 5 years after the diagnosis.
Statistical analyses
The differences in continuous and categorical variables between EBVaGC and EBVnGC groups were assessed using one-way analysis of variance and χ2 or Fisher exact tests, respectively. The most appropriate cutoff value for the expression of each protein was obtained by generating receiver operating characteristics (ROC) curves. The cutoff value was defined as that yielding the highest area under the curve (AUC), as a split line. Protein scores were thereafter dichotomized with the cutoff value. Correlations between the KEGG pathways differentially expressed in gut microbiota and IHC protein expression were analyzed by means of Pearson correlation test. Survival curves were constructed as per the Kaplan-Meier method and compared by means of the log-rank test. Hazard ratios (HRs) for univariable analyses were calculated using a univariable Cox proportional hazards regression model. A LASSO-penalized Cox regression model was used to select the most useful prognostic markers among the 14 proteins. A compound LASSO logistic model was then constructed based on the fraction of the selected proteins using logistic regression coefficients in the training set, and the optimal values of the penalty parameter λ were determined by 10-fold crossvalidations. The optimal cutoff values were evaluated based on the association between the 5-year survival rate and compound score. Only patients with complete clinical information were included in the multivariable survival analyses. The sensitivity and specificity of the survival prediction based on the compound score were depicted as a time-dependent ROC curve. A multivariable Cox regression model with the enter method was used to determine independent prognostic factors in 2 subgroups of EBV expression (see Figure S1, Supplemental Digital Content 2, http://links.lww.com/CTG/A88). All statistical tests were 2-sided. P value of <0.05 was considered statistically significant. Statistical analyses were conducted using R (https://www.R-project.org/), package “survminer” (https://CRAN.R-project.org/package=survminer), and IBM SPSS Stats (version 20.0; Chicago, IL). This study was conducted and reported in line with the The Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) statement for risk-prediction models (12).
RESULTS
Microbial composition in patients with EBVaGC and EBVnGC
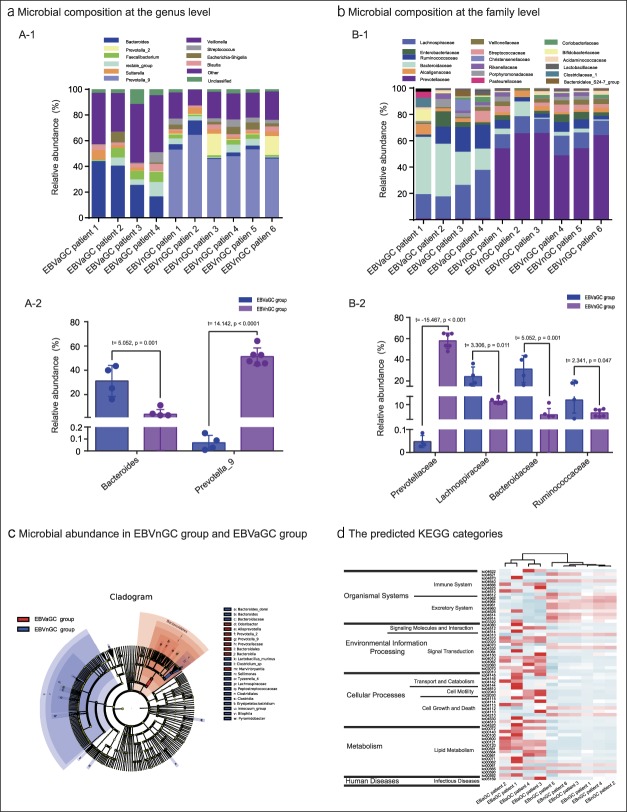
Ten patients diagnosed with gastric adenocarcinoma had fecal samples available for the study. Four of these patients were positive for EBV in their adenocarcinoma tissues. The microbial composition at the genus level (relative abundance > 1% in the 10 samples) is presented in Figure 1a-1. Bacteroides was the most abundant genus across all samples. The level of Bacteroides in EBVaGCs (31.271 ± 6.403) was significantly higher than that in EBVnGCs (3.825 ± 1.559) (t = 5.052, P = 0.001). The abundance of prevotella_9 in EBVaGCs (0.069 ± 0.031) was significantly less than that in EBVnGCs (51.321 ± 2.903) (t = 14.142, P < 0.0001), as shown in Figure 1a-2. Across all 10 fecal samples, species of the Blautia, Veillonella, and Sutterella genera accounted for 7.1%, 6.4%, and 8.2% of the overall microbiota, respectively.
Figure 1.
Analysis of microbial abundance in EBVnGC and EBVaGC. (a) Microbial composition at the genus level. (a-1) Comparisons of microbial abundance at the genus level. (a-2) Differences in the abundance of Bacteroides and Prevotella_9 between the EBVaGC and EBVnGC groups. (b) Microbial composition at the family level. (b-1) Comparisons of microbial abundance at the family level. (b-2) Differences in the abundance of Prevotellaceae and Prevotella_9 between the EBVaGC and EBVnGC groups. (c) LEfSe analysis of microbial abundance of patients' fecal microbial communities in EBVnGC and EBVaGC groups. (d) The predicted KEGG categories abundance of the expression in EBVaGC and EBVnGC groups. EBVaGC, EBV-associated gastric adenocarcinomas; EBVnGC, EBV-associated gastric adenocarcinomas; KEGG, Kyoto Encyclopedia of Genes and Genomes; LEfSe, linear discriminant analysis effect size.
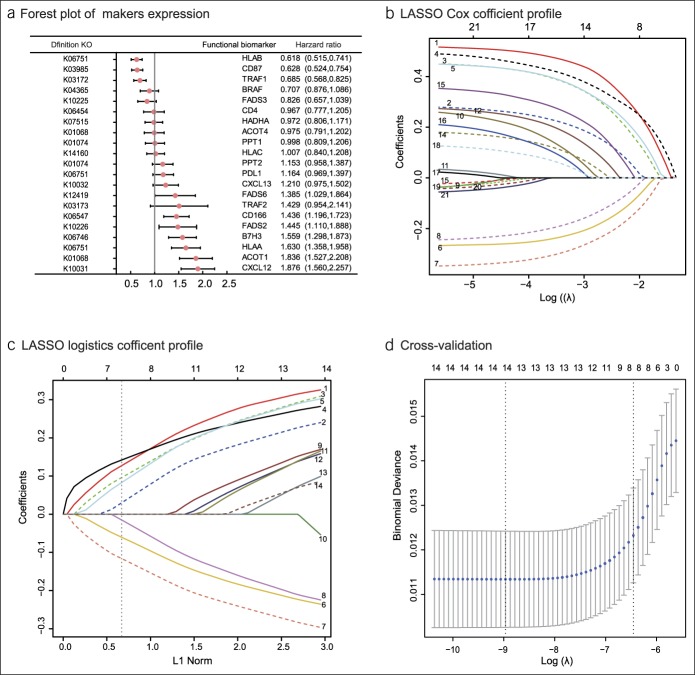
Figure 2.
Construction of the compound score model. (a) A forest plot showing associations between different proteins and overall survival in the training set. Unadjusted hazard ratios are shown with 95% confidence intervals. (b) LASSO Cox regression coefficient profiles of the fractions of 21 immune and lipid metabolism proteins in the entire set. (c) LASSO logistic regression coefficient profiles of the fractions of 14 functional proteins in the training set. 1, ACOT1; 2, CD166; 3, B7H3; 4, CXCL12; 5, HLAA; 6, CD87; 7, HLAB; 8, TRAF1; 9, CXCL13; 10, PDL1; 11, TRAF2; 12, FADS2; 13, FADS6; and 14, PPT2. The dotted line indicates the value chosen by 10-fold cross-test. (d) Ten-fold cross-test for tuning parameter selection in the LASSO model. The partial likelihood deviance is plotted against log (λ), where λ is the tuning parameter. Partial likelihood deviance values are shown, with error bars representing s.e. The dotted vertical lines are drawn at the optimal values by minimum criteria and 1 − s.e. criteria. LASSO, least absolute shrinkage and selection operator.
The microbiota at the family level is presented in Figure 1b. The average abundance was 0.1%. The abundance of 8 species was above average in all 10 samples, as shown in Figure 1b-1. The average abundance of Prevotellaceae was lower in patients with an EBVaGC (0.237 ± 0.132) compared with those with an EBVnGC (57.959 ± 7.312). The average abundance of Lachnospiraceae (24.194 ± 9.045), Bacteroidaceae (31.229 ± 12.805), and Ruminococcaceae (12.859 ± 8.020) in EBVaGC group was higher than that in EBVnGC group (Lachnospiraceae [12.056 ± 1.640], Bacteroidaceae [3.825 ± 3.819], and Ruminococcaceae [4.980 ± 2.217]) (Figure 1b-2). The linear discriminant analysis coupled with effect size measurements method were used to detect groups or species causing significant differences among the species difference levels, as shown in Figure 1c. The functional contributions of the bacteria in the EBVaGC and EBVnGC samples were predicted based on operational taxonomic units using the Tax4Fun package in R software. A total of 283 KEGG orthologues were found across all samples—most of them belonging to pathways governing metabolism, environmental information processing, cellular processes, organismal systems, and human diseases. The most abundant functional pathways are presented in Figure 1d.
Discrimination of the immune-lipid metabolism functional proteins and derivation of compound score
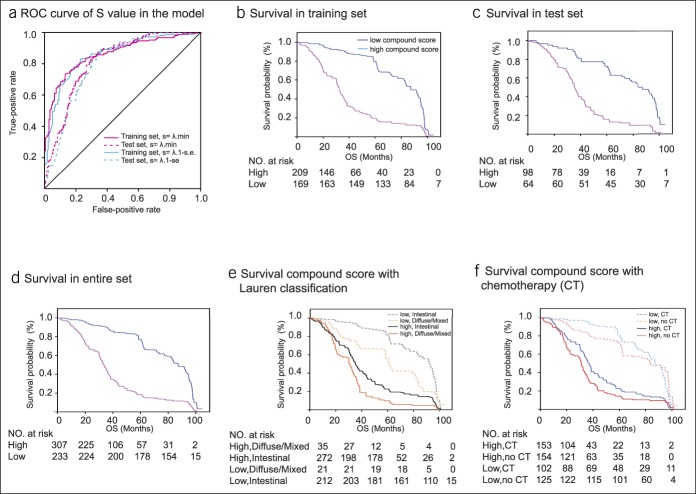
The difference in the expression of KEGG pathways between the EBVaGC and EBVnGCs groups was obtained from the intestinal bacterial group KEGG analysis. Correlations between the KEGG pathway in gut microbiota and IHC protein expression were analyzed using Pearson correlation test (see Table S1, Supplemental Digital Content 1, http://links.lww.com/CTG/A87 and Figure S2, Supplemental Digital Content 2, http://links.lww.com/CTG/A88). The survminer package was used to generate the optimal cutoff values for the fraction of each immune-lipid metabolism functional protein in the 540 patients with gastric adenocarcinoma (see Figure S3, Supplemental Digital Content 2, http://links.lww.com/CTG/A88). Presents a forest plot of the associations between each of the proteins and overall survival. Upregulation of FADS6, CD166, FADS2, B7H3, HLAA, ACOT1, and CXCL12, and downregulation of HLAB, CD87, and TRAF1 increased the risk of survival (P < 0.05) (Figure 2a). Thereafter, LASSO Cox regression selected 14 prognostic proteins involved in the immune response to tumors or in lipid metabolism or was a microenvironmental marker (Figure 2b). LASSO logistic regression analysis was used to build a compound score model in the training set of 378 patients (Figure 2c, d). The formula for the compound score is presented in Table S4 (see Supplemental Digital Content 1, http://links.lww.com/CTG/A87). In this formula, protein expression in the compound score fraction level was assigned as 0:1. This was defined as 0 if the fraction of one type of cell was less than the corresponding cutoff value or 1 if otherwise. To assess the accuracy of the compound score for the prognosis of patients with gastric adenocarcinoma, the ROC curve was used to judge the patient's five-year survival status. (Figure 2d). Patients in the training set were then assigned to a high- or low-compound score group using the cutoff value (0.016). Five-year survival rates were 79.2% and 19.6%, respectively, for the low- and high-compound score groups (HR = 2.93, 95% confidence interval [CI] = 2.36–3.71) (Figure 3a). The association between the compound score and overall survival was also significant when evaluated in the multivariable Cox regression model (HR = 2.89, 95% CI = 2.28–3.61) (Tables 1 and 2). The results of the univariable analyses of clinicopathologic variables are shown in Table S2 (see Supplemental Digital Content 1, http://links.lww.com/CTG/A87).
Figure 3.
Survival impact of the compound score. (a) Compound score measured by time-dependent ROC curves in the training and test sets. The area under the ROC curve was 0.845, 0.703, 0.842, and 0.697 for the compound score for the training set in minimum criteria, test set in minimum criteria, training set in 1-s.e., and test set in 1-s.e. (b-d) Kaplan-Meier curves for overall survival by compound score group in the training set (b), test set (c), and entire set (d). (e) For patients with Lauren classification in subgroups stratified compound score. HRs are shown with 95% CIs. P < 0.001 (log-rank test). (f) For patients with adjuvant CT in subgroups stratified compound score. HTs are shown with 95% CIs. P < 0.001 (log-rank test). CI, confidence interval; CT, chemotherapy; HR, hazard ratio; ROC, receiver–operating characteristic.
Table 1.
Baseline patient clinicopathologic characteristicsa
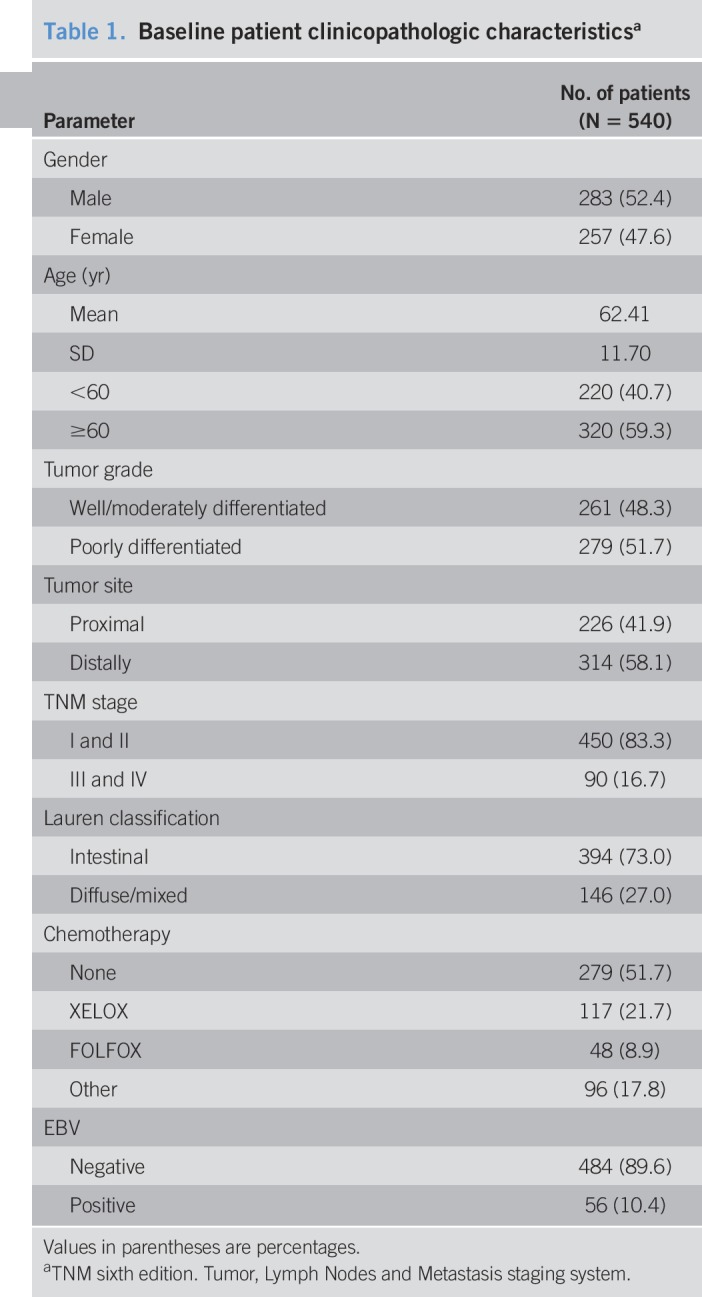
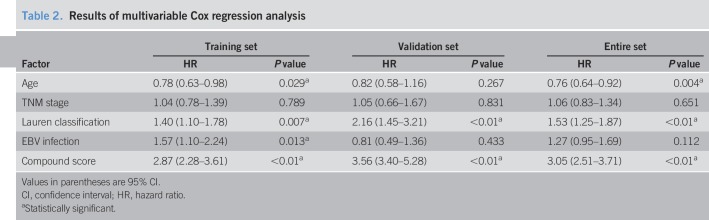
Table 2.
Results of multivariable Cox regression analysis
Validation of the compound score for predicting survival in the validation set and entire set
The compound score model underwent internal evaluation using the validation set and entire set. The compound score was standardized across the validation and entire sets. Consistent with the findings in the training set, patients in the high-compound score group had a significantly lower overall survival rate than those in the low-compound score group in both validation set (HR = 3.43, 95% CI = 2.36–4.97) (Figure 3b) and entire set (HR = 3.14, 95% CI = 2.60–3.81) (Figure 3c). The compound score model was also an independent prognostic factor when analyzed as a binary variable in multivariable analysis using both validation set (HR = 3.56, 95% CI = 3.40–5.28) and entire set (HR = 3.05, 95% CI = 2.51–3.71) (Table 2).
Compound score with Lauren classification and adjuvant chemotherapy
Stratification analysis was performed in the entire set of patients grouped by Lauren classification. The compound score differed among patients with different prognosis as per Lauren classification (Lauren intestinal type and diffuse or mixed type), although the result was significant for intestinal (P < 0.01) and diffuse/mixed (Figure 3e). Based on comparisons between the compound score and Lauren classification in the entire set, the ability of the compound score to predict survival was inferior to that of the Lauren classification for patients with diffuse/mixed. A similar tendency was also observed in both training and validation sets. The compound score identified patients with different prognoses in each chemotherapy variable as a binary (no chemotherapy [CT] and CT) subgroup. There was a survival advantage for patients who received chemotherapy in both low and high compound scores, regardless of the chemotherapy regimen (Figure 3f).
Compound score and the Epstein-Barr virus infection
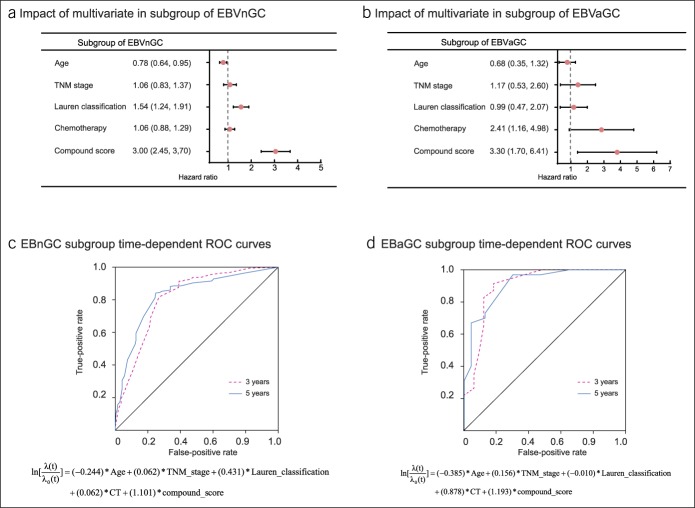
To confirm the compound score model and clinicopathologic variable values in different groups of EBV-infected patients, the same formula was applied to patients with an EBVaGC or EBVnGC. Detection of IHC protein detection of EBV-LMP1, EBNA1 and ISH detection of EBER. The difference in the compound score between the EBVaGC group and EBVnGC group was statistically significant (P = 0.0025) (see Table S5, Supplemental Digital Content 1, http://links.lww.com/CTG/A87 and Figure S4, Supplemental Digital Content 2, http://links.lww.com/CTG/A88). Figure 4a, b shows a forest plot of the associations between each of the clinicopathologic variables and overall survival (see Tables S6 and S7, Supplemental Digital Content 1, http://links.lww.com/CTG/A87). The prognostic accuracy of the multivariate Cox regression model (age, TNM stage, Lauren classification, chemotherapy, and compound score) assessed as a continuous variable was investigated in the entire set by plotting time-dependent ROC for overall survival at 3 and 5 years (Figure 4c, d). For EBVnGC, the Cox regression model had an area under the curve of 0.812 (95% CI = 0.773–0.850) at 3 years and 0.828 (95% CI = 0.790–0.866) at 5 years. For EBVaGC, the area under the curve was 0.901 (95% CI = 0.819–0.982) at 3 years and 0.910 (95% CI = 0.832–0.987) at 5 years.
Figure 4.
Model analysis of subgroups of EBVaGC and EBVaGC. (a) A forest plot showing the survival impact of age, TNM stage, Lauren classification, adjuvant chemotherapy, and compound score among patients in the EBVnGC subgroup. (b) A forest plot showing the survival impact of age, TNM stage, Lauren classification, adjuvant chemotherapy, and compound score among patients in the EBVaGC subgroup. HRs are shown with 95% CIs. (c) Cox multivariate regression model measured by time-dependent ROC curves in the subgroup of EBVnGC. The area under the ROC curve was 0.812 and 0.828 for the 3 and 5 years, respectively. (d) Cox multivariate regression model measured by time-dependent receiver–operating characteristic (ROC) curves in the subgroup of EBVaGC. The area under the ROC curve was 0.901 and 0.910 for the 3 and 5 years, respectively. CI, confidence interval; EBVaGC, EBV-associated gastric adenocarcinomas; EBVnGC, EBV-associated gastric adenocarcinomas; HR, hazard ratio; ROC, receiver–operating characteristic.
DISCUSSION
The present study found that the expression of KEGG functional pathways differed between EBVaGC and EBVnGC gut microbiota, and combined with tumor microenvironment and lipid metabolism biomolecules, this combination was predictive of the prognosis of gastric adenocarcinoma. Herein, we have proposed and validated a compound score to predict overall survival after the diagnosis of gastric adenocarcinoma. The compound score is based on the fractions of 8 immune proteins and metabolic proteins. The results show a clear separation of overall survival curves between patients with high and low compound scores. Furthermore, the compound score predicted survival in groups of patients with identical Lauren classification and EBV status, thereby suggesting that this model could have a prognostic value that complements EBV infection.
The role of gut microbiota in carcinogenesis has in the recent years attracted much attention (4,13) Several studies have reported that the microbiota affects chemotherapy resistance and the prognosis of patients with cancer. In the study of gastric cancer-related microorganisms, Helicobacter pylori is an important tumor-associated microbiota, which affects the occurrence of gastric cancer (14). However, with the development of tumor-associated gastrointestinal microbial research, the relationship between tumor progression and gut microbiota imbalance has become more important. Accordingly, the transplantation of flora is a suggested treatment for improving the prognosis, which has shown encouraging results in animal subjects (15,16). Owing to technical restrictions, however, these studies are limited to evidence from experimental animal models, and the impact of fecal microbiota transplantation on the human body may be more complicated (17). Moreover, the ability to standardize and repeat the measurement of transplanted flora and the predictability and regulation of biologic functions that are affected is also inherently difficult.
In contrast to previous studies, the candidate immune-lipid metabolism markers used to build the present compound score model were estimated based on 16S gene ribosomal RNA sequencing using the Tax4Fun analysis and linear discriminant analysis effect size (18–20). By applying LASSO Cox regression as a statistical method for screening protein variables to remove proteins with more accurate predictions and furthermore by using LASSO logistic regression models to establish the compound score model, the predictive accuracy could be improved significantly. It is worth mentioning that in the process of selecting the S value in the LASSO logistic model operation, we chose 1 standard error instead of the minimum S (see Table S4, Supplemental Digital Content 1, http://links.lww.com/CTG/A87), although both are commonly used alternatives (Figure 3a) (10,21). But in the case where the prediction effect is extremely close, we choose fewer variables in the model. The C-index suggests that the predictive ability of the compound score for survival was inferior to that of Lauren classification for patients receiving no adjuvant chemotherapy. Moreover, the predictive ability of the compound score for survival was inferior to that of EBVaGC. This could be attributed to the universally recognized poor prognosis associated with Lauren classification, EBV infection, and the multiple risk factors affecting prognosis. Therefore, the effect of the immune microenvironment and lipid metabolism proteins on the prognosis of patients with EBV infection is increased (22–24). Nonetheless, patients with an EBVaGC and who received no adjuvant chemotherapy are at an increased risk, thereby implying that the compound score and adjuvant chemotherapy could be used to reinforce the prognostic ability of EBVaGC (6,25). The value of the compound score was further demonstrated through internal evaluation on a validation set, which indicated good reproducibility.
We acknowledge several limitations. First, this study was based on gut microbiota sequencing data for clinical patients, and a minority of these were lost prematurely to follow-up. Moreover, our sample size is relatively small given the effect chemotherapy has on the intestinal flora. Second, the methodology for interpreting biomolecular proteins and the appropriate cutoff value needs to be standardized. Third, given the clinical importance of distinct tumor regions, it would be appropriate to apply the compound score to systematically evaluate gastric adenocarcinoma using TMAs. However, the protein expression assays used here were all derived from a core sample of adenocarcinoma tissue in IHC, making it impossible to use the protein expression results as a continuous variable when establishing the compound score model. Fourth, H. pylori plays a very important role in the progression of gastric inflammation and cancer, and it also affects the prognosis of patients with gastric cancer. In this study, we focus on the gut microbiota; the study of H. pylori and other stomach microbiota with the occurrence and prognosis of gastric cancer will be involved in our future research. Finally, the patients in our study were selected retrospectively, which has potential biases relating to unbalanced clinicopathologic parameters and heterogeneity of treatment that should not be overlooked. In the future, we plan to conduct a prospective study to verify this result.
CONFLICTS OF INTEREST
Guarantor of the article: Yong Mao, MD, PhD.
Specific author contributions: Study design, data acquisition, statistical analysis, interpretation, and manuscript drafting/revision. F.W. and Y.M.: study concept, design, study supervision, obtained funding, and critical revision of the manuscript for important intellectual content. J.W. and D.H.: study concept, design, and critical revision of the manuscript for important intellectual content. Y.Q., Y.L., and X.Q.: sample acquisition and pathologic diagnosis and provided facilities. Y.W., Y.J., X.J., and Z.Q.: study concept and critical revision of the manuscript for important intellectual content. Y.C. and X.G.: development of methodology and study supervision. All authors read and approved the manuscript.
Financial support: This work was supported by the National Natural Science Foundation of China (no. 81372375 to D.H. and no. 81502042 to X.G.), a grant from Fundamental Research Funds for the Central Universities funded by the Ministry of Education of China (no. JUSRP51710A to Y.C.), and a grant from the Natural Science Foundation Youth Project of Jiangsu Province (no. BK20140171 to X.G.).
Potential competing interests: The authors declare no conflicts of interest.
Study Highlights.
WHAT IS KNOWN
✓ Gut microbiota in patients with digestive tract tumors has a large immunologic and metabolic impact on tumor progression.
✓ Evidence is limited regarding whether a single biomarker can predict the prognosis in patients having EBVaGCs or EBVnGCs.
WHAT IS NEW HERE
✓ The gut microbiota in EBVaGCs and EBVnGCs is significantly different, which may affect the different biologic functions of patients.
✓ EBVaGC and EBVaGC patients differentially expressed in the gut microbiota affect the host biomarker protein construction model with accurate prediction results.
TRANSLATIONAL IMPACT
✓ The influence of gut microbiota and EBV has important significance in the evaluation of prognosis of patients with gastric cancer.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the collaborators of Carbohydrate Chemistry & Biotechnology of Ministry of Education in Jiangnan University. We thank all investigators at the Department of Pathology in the Affiliated Hospital of Jiangnan University for specimen collection and result recording. We acknowledge Dr. Jianbo Zhang for providing constructive comments.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A87 and http://links.lww.com/CTG/A88
REFERENCES
- 1.van Beek J, zur Hausen A, Klein Kranenbarg E, et al. EBV-positive gastric adenocarcinomas: A distinct clinicopathologic entity with a low frequency of lymph node involvement. J Clin Oncol 2004;22(4):664–70. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018;67:226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dejea CM, Fathi P, Craig JM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018;359(6375):592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogdill AP, Gaudreau PO, Arora R, et al. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol 2018;39(11):900–20. [DOI] [PubMed] [Google Scholar]
- 5.Song HJ, Srivastava A, Lee J. Host inflammatory response predicts survival of patients with Epstein-Barr virus–associated gastric carcinoma. Gastroenterology 2010;139(1):84–92.e2. [DOI] [PubMed] [Google Scholar]
- 6.Sohn BH, Hwang JE, Jang HJ, et al. Clinical significance of four molecular subtypes of gastric cancer identified by the cancer genome atlas Project. Clin Cancer Res 2017;23(15):4441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obata Y, Pachnis V. The effect of microbiota and the immune system on the development and organization of the enteric nervous system. Gastroenterology 2016:151:836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iida N, Dzutsev A, Stewart CA. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342(6161):967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horai R, Zárate-Bladés, Dillenburg-Pilla R, et al. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity 2015;43(2):343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneoka D, Saito E, Nakaoka S. New algorithm for constructing area-based index with geographical heterogeneities and variable selection: An application to gastric cancer screening. Scientific Rep 2016;6(1):26582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, Kim EK, Kim YH, et al. Epstein–Barr virus positivity, not mismatch repair-deficiency, is a favorable risk factor for lymph node metastasis in submucosa-invasive early gastric cancer. Gastric Cancer 2016;19(4):1041–51. [DOI] [PubMed] [Google Scholar]
- 12.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. Br J Surg 2015;102(3):148–58. [DOI] [PubMed] [Google Scholar]
- 13.Coker O, Dai Z, Nie Y, et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018;67:1024–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Re V, Orzes E, Canzonieri V, et al. Pepsinogens to distinguish patients with gastric intestinal metaplasia and Helicobacter pylori infection among populations at risk for gastric cancer. Clin Transl Gastroenterol 2016;7(7):e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosshart SP, Vassallo BG, Angeletti D, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 2017:171:1015–28.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juan-Juan G, Yang Z, Markus G, et al. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front Cell Infect Microbiol 2018;8:202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pamer EG. Fecal microbiota transplantation: Effectiveness, complexities, and lingering concerns. Mucosal Immunol, 2014, 7(2):210–4. [DOI] [PubMed] [Google Scholar]
- 18.Shoko I, Thomas W, Schmidt BL, et al. Piphillin: Improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One 2016;11(11):e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreno-Indias I, Sánchez-Alcoholado L, García-Fuentes E, et al. Insulin resistance is associated with specific gut microbiota in appendix samples from morbidly obese patients. Am J Transl Res 2016;8(12):5672–84. [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12(6):R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol 2016, 34(4):2157–64. [DOI] [PubMed] [Google Scholar]
- 22.Kim SY, Park C, Kim HJ, et al. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology 2015;148(1):137–47.e9. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Li J, Hao Y, et al. Differentiated tumor immune microenvironment of Epstein-Barr virus-associated and negative gastric cancer: Implication in prognosis and immunotherapy. Oncotarget 2017;8(40):67094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi E, Byeon SJ, Kim SH, et al. Implication of leptin-signaling proteins and Epstein-Barr virus in gastric carcinomas. PLoS One 2015;10(7):e0130839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moehler M, Delic M, Goepfert K, et al. Immunotherapy in gastrointestinal cancer: Recent results, current studies and future perspectives. Eur J Cancer 2016;59:160–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.