Abstract
INTRODUCTION:
The etiology of acute liver failure (ALF) remains an important prognostic factor. The Acute Liver Failure Study Group recently reported that 150 of 2,718 adult patients with ALF (5.5%) had an indeterminate etiology. Our aim was to use whole exome sequencing to identify genetic variants associated with phenotypic, biochemical, and histologic features among patients with indeterminate ALF.
METHODS:
This effort has defined a cohort of well-pedigreed patients with indeterminate ALF; DNA samples extracted from whole blood samples were obtained from 26 respective patients with indeterminate ALF. These samples were kept at the Acute Liver Failure Study Group repository at the NIDDK, Bethesda. Whole exome sequencing and bioinformatics analysis were performed at the Mayo Clinic Center of Individualized Medicine in Rochester, MN.
RESULTS:
Of the 26 patients, 8 survived spontaneously, 6 died, and 12 underwent a liver transplantation; all those transplanted were alive at 21 days after enrollment in the study. Twenty-two of the 26 patients presented as ALF. We found 12 variants associated with 11 genes. The most common variant was rs4940595 in the SERPINB11 gene which was found in 23 of the 26 patients. This variant had a stop codon; no reports of disorders have been associated with this variant. The next most commonly found variant was rs1135840 in the CYP2D6 gene; this mutation is a missense_variant and has been reported to be associated with hepatotoxicity of antituberculous therapy. None of our patients were receiving this therapy. We also found a significant asymmetric distribution of rs1800754 of the CYP2D7 gene and rs1135840 of the CYP2D6 gene between patients who survived spontaneously (75%) and those who died or underwent liver transplantation (30.5% and 25%, respectively).
DISCUSSION:
We found 12 variants of 11 genes significantly associated with ALF among adults with indeterminate etiology. We also found a significant asymmetric distribution of 2 variants belonging to the CYP2D7 and CYP2D6 genes, respectively, between those who survived spontaneously and those who died or underwent liver transplantation. The 2 most common variants, rs4940595 and rs1135840, of the SERPINB11 and CYP2D6 genes, respectively, found in our patients with ALF have been described as potentially important in the adaptive response combating the emergence of infectious diseases and associated with hepatotoxicity of antituberculous therapy, respectively. Our findings need to be expanded to include more patients with indeterminate ALF as well as viral, drug toxicity, and autoimmune etiologies to determine whether our findings are associated with the specific etiology, indeterminate, or with the overall ALF syndrome itself.
INTRODUCTION
The etiology of acute liver failure (ALF) remains an important prognostic factor. The Acute Liver Failure Study Group (ALFSG) initiated a registry in January 1998 to characterize the etiologies, clinical features, and outcomes of this condition among adults in the United States and Canada. In a recent publication, Ganger et al. (1) from the ALFSG reported that 150 of 2,718 patients (5.5%) had an indeterminate etiology.
Among children with liver failure, the indeterminate etiology is more commonly found and estimated to affect around 50% of the cases. Vilarinho et al. (2) studied 3 children with whole exome sequencing (WES) to investigate the possibility of an atypical presentation of inherited metabolic liver diseases. One patient with the clinical diagnosis of Wilson disease was found to have a recessive MPV17 mutation; a second patient presented with neonatal fatal liver failure and was found to have a homozygous splice site mutation in the SERAC1 gene. Last, a third patient who presented with progressive cholestasis and liver failure was found to have a NOTCH2 gene mutation. A more recent publication reported deep sequencing among 12 children with ALF and tissue evidence of impaired energy metabolism (3); the authors customized a next-generation sequencing panel for 26 genes associated with mitochondrial and fatty acid oxidation defects. Five of the 12 patients had variants involving the genes ACAD9, POLG, POLG2, DGUOK, and RRM2B.
There have been no similar reports among adults with ALF. The ALFSG, National Institutes of Health-sponsored consortium which has been operating since 1998, has not only reported seminal publications regarding this condition but also collected serum and DNA samples from patients with ALF enrolled in this study. At the same time, the ALFSG has evaluated each patient enrolled and assigned an etiology by the implementation of a Causality Adjudication Committee (1). This effort has defined a cohort of well-pedigreed patients with indeterminate ALF from whom DNA samples extracted from whole blood samples have been collected. This collection provided a unique opportunity to perform WES.
Our aim was to use WES to identify genetic variants associated with phenotypic, biochemical, and histologic features among patients with indeterminate ALF. This diagnosis was established by strict criteria defined by the Causality Adjudication Committee of the ALFSG (1).
METHODS
Between 1998 and 2015, 2,670 patients with acute liver injury (ALI) and ALF have been enrolled in the ALFSG registry wherein detailed data and daily biosamples are collected over 7 days. All study patients met criteria for ALF or ALI as defined previously; to ensure patients with ALI had severe liver dysfunction, we have previously defined ALI as international normalized ratio ≥ 2.0 (1,4). Patients with chronic liver disease or cirrhosis were excluded, with the exception of certain patients with chronic hepatitis B or Wilson disease presenting with rapid onset of severe illness in the absence of a history of known liver disease. For each enrolled subject, detailed demographic, clinical, laboratory, radiologic, and outcomes data were recorded. Etiologic diagnoses were made by the principal investigator at each study site using standard criteria, based on the history and clinical presentation, laboratory, radiographic information, and liver biopsy results, when available. When a thorough investigation failed to disclose a cause, the etiology was designated indeterminate. In some instances, serologic data to complete the exclusion of other etiologies were not available. In particular, patients with indeterminate etiology were reviewed in detail by the Causality Committee composed of senior hepatologists for confirmation that most if not all diagnostic testing was complete and a specific etiology remained unknown. For each subject, written informed consent was obtained from next of kin because of the presence of hepatic encephalopathy, with approval consent revisited with the patient after recovery. Consent was provided by the patients if they exhibited no evidence of hepatic encephalopathy (ALI) and by the patients' legal next of kin if they had any signs of encephalopathy (ALF). Institutional Review Boards of each participating institution have approved the ALFSG registry. A total of 150 patients were deemed to have indeterminate ALF by the ALFSG Causality Adjudication Committee (1); only 26 of these well-pedigreed 150 patients had DNA samples that had been extracted from whole blood samples and kept at the ALFSG repository at the NIDDK, Bethesda. Although the enrollment of patients and their respective collection of clinical data began in 1998, we only began to collect serum DNA in 2005. In many instances, relatives allowed data collection at enrollment but were wary to provide permission for serum DNA collection. WES and bioinformatics analysis were performed at the Mayo Clinic Center of Individualized Medicine in Rochester, MN.
Paired-end libraries were prepared using 1.0 μg of genomic DNA following the manufacturer's protocol (Agilent) using the Agilent Bravo liquid handler. The concentration and size distribution of the completed libraries were determined using an Agilent Bioanalyzer DNA 1000 chip (Santa Clara, CA) and Qubit fluorometry (Invitrogen, Carlsbad, CA). Whole exon capture was carried out using 750 ng of the prepped library following the protocol for Agilent's SureSelect Human All Exon v5 + UTRs 75 MB kit. The purified capture products are then amplified using the SureSelect Post-Capture Indexing forward and Index polymerase chain reaction reverse primers (Agilent) for 12 cycles. The concentration and size distribution of the completed captured libraries were determined on Qubit (Invitrogen) and Agilent Bioanalyzer DNA 1000 chip.
Libraries were sequenced at an average coverage of ∼80X following Illumina's standard protocol using the Illumina cBot and HiSeq 3,000/4,000 PE Cluster Kit. The flow cells were sequenced as 150 × 2 paired-end reads on an Illumina HiSeq 4,000 using HiSeq 3,000/4,000 sequencing kit and HCS v3.3.52 collection software. Base-calling is performed using Illumina's RTA version 2.7.3. All samples undergoing whole genome sequencing were also run on a sample ID validation genotyping panel. An aliquot of each sample is taken before the library preparation (above), and sample ID validation genotype data are deposited into a database. Once sequencing data are available, the genotype data are compared with the sequencing data to confirm identity. Once confirmed, data are released to the investigator.
Genome_GPS v5 (formerly named as TREAT (5)) was used as a comprehensive secondary analysis pipeline for DNA sequencing data at Mayo Clinic. FASTQ files were aligned to the hg38 reference genome using bwa-mem (VN:V0.7.10) using default options. Realignment was performed using GATK (VN:3.4–46) (6). Multisample variant calling was performed using the GATK (VN: 3.4–46) Haplotype Caller, and variants were filtered using variant recalibration Variant Quality Score Recalibrator for both single nucleotide variants and insertions and deletions. Identified variants were annotated using BioR (7) framework with functional features, impact prediction, and clinical significance using Clinical Annotation of Variants, ClinVar, Human Gene Mutation Database, Mayo Biobank, and Exome Aggregation Consortium population frequencies. Copy number variations were detected using Pattern Copy number variation (8) (Figure 1).
Figure 1.
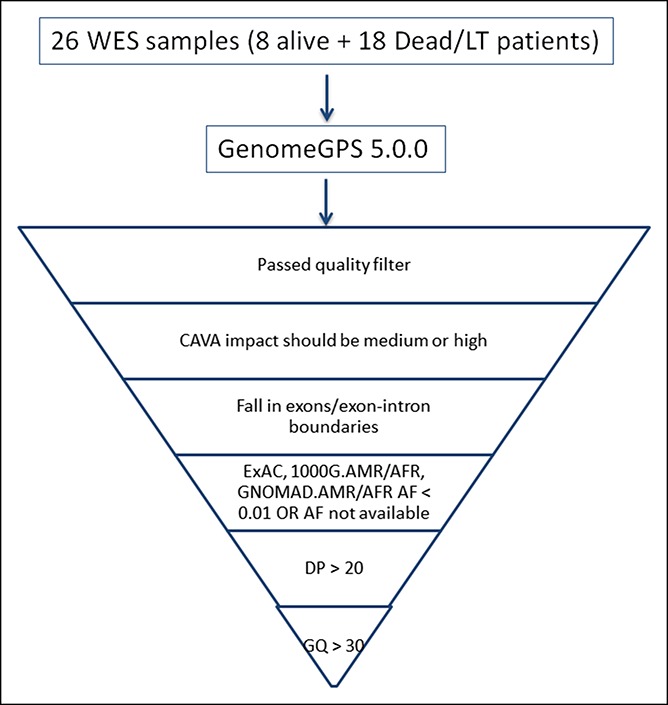
Functional filters applied in 26 DNA samples from 26 patients with indeterminate ALF. AF, allele frequency; ALF, acute liver failure; CAVA, clinical annotation of variants; DP, filtered depth of coverage for each sample; GQ, quality of the assigned genotype; LT, liver transplantation; WES, whole exome sequencing.
This project was approved by the Mayo Clinic Institutional Review Committee.
RESULTS
The 26 patients were enrolled in the ALFSG study between 1998 and 2013. Of these 26 patients, 22 presented as ALF, 3 presented as ALI and converted to ALF, and 1 presented as ALI. There were 13 women and 13 men; median age was 37.5 years (range, 22–87 years). Their self-declared ethnicity distribution was 5 African Americans, 1 Hispanic, 1 Asian, and 19 whites. Of the 26 patients, 8 survived spontaneously, 6 died, and 12 underwent a liver transplantation; all those transplanted were alive at 21 days after enrollment in the study. Twenty patients were alive at 21 days from study enrollment; of these 20 patients, 8 were spontaneous survivors and 12 underwent deceased donor liver transplantation. Six patients were dead at 21 days from enrollment. Median age among the 20 patients who survived was 35 years (range, 22–87 years), and median age among those who died was 43 years (range, 23–47 years). The median age of patients who underwent liver transplantation and were alive was 31.5 years (range, 18–52 years). All patients who were transplanted were alive at 21 days from enrollment.
The 26 patients presented with a classic pattern of serum liver biochemistries (aspartate aminotransferase, alanine aminotransferase, and alkaline phosphatase) to that of hepatocellular injury, with elevation of aspartate aminotransferase/alanine aminotransferase and normal or minimal elevation of serum alkaline phosphatase. Liver biopsies or explants examination showed massive or sub-massive hepatic necrosis with necroinflammatory changes. None of our patients showed evidence of fatty deposits in hepatocytes suggestive of underlying fatty liver disorders.
There were 12 variants of 11 genes significantly associated with ALF in the 26 patients (Table 1). The variant rs4940595 in the SERPINB11 gene was found in 23 of the 26 patients with ALF; this variant has a stop_gained codon (c.268 G > Tp.Glu90X). There have not been any publications associating this variant with any specific disorder.
Table 1.
Twelve variants from 11 genes significantly associated with indeterminate ALF among 26 patients with ALF
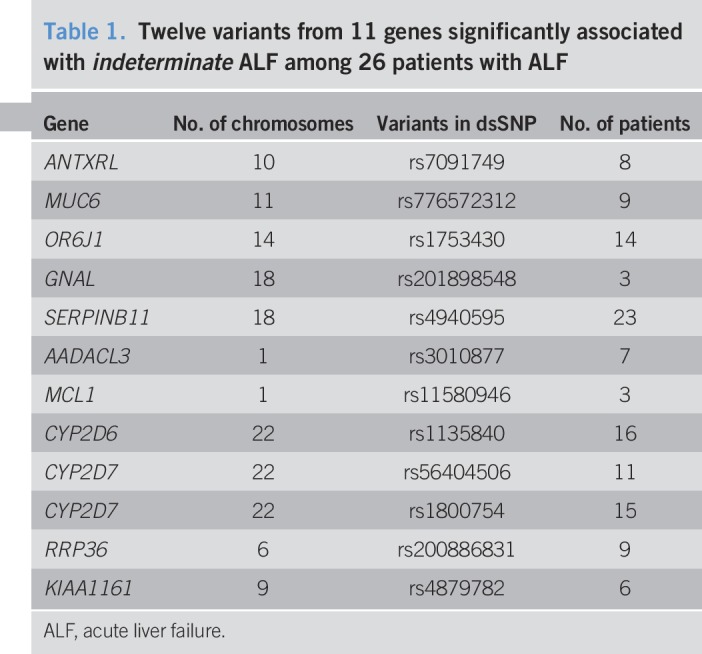
The next commonly found variant was rs1135840 in the CYP2D6 gene; the mutation is a missense_variant ((c.1457 G > C_p.Ser488Thr). This variant has been reported in association with hepatotoxicity of antituberculous drug (9); however, none of our patients gave a history of antituberculous therapy before their clinical presentation.
The analysis of the distribution of these 12 variants among the 8 patients who survived spontaneously vs those who died or underwent liver transplantation showed that moderate impact variants in drug metabolism associated gene CYP2D7, rs1800754, and its paralog CYP2D6, rs1135840, were found in 75% of the patients who survived spontaneously and in 30.5%–25%, respectively, of the patients who died or were transplanted. This asymmetric variant distribution was significant, P < 0.05, by exact test module in Rvtests genetic association package.
All variants listed in Table 1 have had no disorders associated with them in the National Center for Biotechnology Information databases, with the exception of rs1135840, a variant of CYP2D6 which has been associated with hepatotoxicity of antituberculous drugs (9).
DISCUSSION
We have postulated in the past that the etiology is an important prognostic factor of ALF (4). Patients with ALF indeterminate etiology pose a challenge in determining the underlying pathophysiology that lead to this serious clinical presentation. Our WES data show that 12 unique variants of 11 genes are significantly associated with ALF indeterminate etiology in the 26 patients studied. These variants have not been reported to be associated with any disorder so far and have not been described before in any of the databases of the National Center for Biotechnology Information, with the exception of rs1135840 of the CYP2D6 gene which has been reported to be associated with hepatotoxicity of antituberculous drugs (9). Our observation of an asymmetric distribution of 2 variants, rs1135840 and rs1800754, of the CYP2D6 and CYP2D7 genes, respectively, is noteworthy because they seem to have a survival protective effect; however, drawing conclusions on the significance of the group-specific variant distribution is limited by the cohort size. It would be statistically sound to determine whether the above-mentioned variant distributions hold true on a larger cohort.
The most commonly found variant, rs4940595, of the SERPINB11 gene has not been reported to be associated with any specific disorder. This gene has been described as having a potentially important role in the adaptive immune response combating the emergence of infectious diseases in recent human evolution (10). However, this variant has been found in 73% of the participants of the Mayo Biobank, who had no specific disorder, as a requisite to be part of this biobank.
Although our findings are the first reported in adult patients with indeterminate ALF, our conclusions are limited by the small size of our sample. We intend to expand our analysis with the enrollment of more patients with this condition in the future; we will also need to perform WES analysis of adult patients with ALF associated with viral, drug-induced, toxins, and autoimmune etiologies, and confirm our findings and determine whether these variants are associated with a specific etiology, indeterminate, or with the ALF syndrome itself.
CONFLICTS OF INTEREST
Guarantor of the article: Jorge Rakela, MD.
Specific author contributions: Develop the concept of the study, obtain clinical samples from the Acute Failure Study Group, obtain funding, coordinated the effort of the various contributors of the study, and wrote the manuscript.
Financial support: Grant 18-0011260 from the Mayo Clinic Center for Individualized Clinic. Acute Liver Failure Study Group (ALFSG) supported by NIDDK Grant U-01-058369.
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ Acute liver failure is a syndrome accompanied by a high fatality rate depending on its etiology; this can range from 40% to 80%. The most common etiologies are hepatitis viruses, autoimmune hepatitis, metabolic liver disorders such as Wilson disease, and drug-induced liver injury, among which acetaminophen toxicity is the most common. There are 5.5% of patients who have indeterminate ALF. The Acute Liver Failure Study Group, consortium funded by the NIH since 1998, has collected DNA samples of patients enrolled in the study. We were able to access 26 DNA samples from patients with indeterminate ALF and perform WES to evaluate gene variants that were significantly associated with indeterminate ALF and provide an insight into the diagnosis and possible pathogenic mechanisms.
WHAT IS NEW HERE
✓ We found 12 variants from 11 genes that were significantly associated with indeterminate ALF. In particular, rs4940595, a variant fromSERPINB11gene, was present in 23 of the 26 patients studied. We also found an asymmetric distribution of 2 variants belonging to theCYP2D7 andCYP2D6 genes which were more commonly present among patients who survived spontaneously.
TRANSLATIONAL IMPACT
✓ This is the first communication describing WES findings among 26 adult patients with indeterminate ALF. We found 12 variants of 11 genes significantly associated with indeterminate ALF.
ACKNOWLEDGEMENTS
Acute Liver Failure Study Group: Members and institutions participating in the Acute Liver Failure Study Group 1998-2013 were as follows: W.M. Lee, MD (Principal Investigator); Anne M. Larson1, MD, Iris Liou, MD, University of Washington, Seattle, Washington; Timothy Davern2, MD, Oren Fix1, MD, University of California, San Francisco; Timothy Davern2, MD, California Pacific Medical Center, San Francisco, California, Michael Schilsky3, MD, Mount Sinai School of Medicine, New York, New York Timothy McCashland, MD, University of Nebraska, Omaha, Nebraska; J. Eileen Hay, MBBS, Mayo Clinic, Rochester, Minnesota; Natalie Murray, MD, Baylor University Medical Center, Dallas, Texas; A. Obaid S. Shaikh, MD, University of Pittsburgh, Pittsburgh, Pennsylvania; Andres Blei, MD, (deceased), Daniel Ganger, MD, Northwestern University, Chicago, Illinois; Atif Zaman, MD, University of Oregon, Portland, Oregon; Steven H.B. Han, MD, University of California, Los Angeles, California; Robert Fontana, MD, University of Michigan, Ann Arbor, Michigan; Brendan McGuire, MD, University of Alabama, Birmingham, Alabama; Raymond T. Chung, MD, Massachusetts General Hospital, Boston, Massachusetts; Alastair Smith, MB, ChB, Duke University Medical Center, Durham, North Carolina; Robert Brown, MD, Cornell/Columbia University, New York, New York; Jeffrey Crippin, MD, Washington University, St Louis, Missouri; Edwyn Harrison, Mayo Clinic, Scottsdale, Arizona (closed); Adrian Reuben, MBBS, FRCP, FACG, David Koch, MD, MSR, Medical University of South Carolina, Charleston, South Carolina; Santiago Munoz, MD, Albert Einstein Medical Center, Philadelphia, Pennsylvania; Rajender Reddy, MD, University of Pennsylvania, Philadelphia, Pennsylvania; R. Todd Stravitz, MD, Virginia Commonwealth University, Richmond, Virginia; Lorenzo Rossaro, MD, University of California Davis, Sacramento, California Closed); Raj Satyanarayana, MD, Mayo Clinic, Jacksonville, Florida; Tarek Hassanein, MD, University of California, San Diego, California; James Hanje MD, The Ohio State University, Columbus Ohio; Jody Olson MD, University of Kansas Medical Center, Kansas City, Kansas; Ram Subramanian MD, Emory University Medical Center, Atlanta Georgia; Michael Schilsky MD, Yale University School of Medicine, New Haven Connecticut; Constantine Karvellas MD, University of Alberta, Edmonton, Canada; Eugene Schiff MD., University of Miami Florida.
REFERENCES
- 1.Ganger DR, Rule J, Rakela J, et al. Acute liver failure of indeterminate etiology: A comprehensive systematic approach by an expert committee to establish causality. Am J Gastroenterol 2018;113:1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vilarinho S, Choi M, Jain D, et al. Individual exome analysis in diagnosis and management of paediatric liver failure of indeterminate aetiology. J Hepatol 2014;61:1056–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valencia CA, Wang X, Wang J, et al. Deep sequencing reveals novel genetic variants in children with acute liver failure and tissue evidence of impaired energy metabolism. PLoS One 2016;11:e0156738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostapowicz G, Fontana RJ, Schiødt FV, et al. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med 2002;137:947–54. [DOI] [PubMed] [Google Scholar]
- 5.Asmann YW, Middha S, Hossain A, et al. TREAT: A bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics 2012;28:277–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kocher JP, Quest DJ, Duffy P, et al. The biological reference repository (BioR): A rapid and flexible system for genomics annotation. Bioinformatics 2014;30:1920–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Evans JM, Bhagwate AV, et al. PatternCNV: A versatile tool for detecting copy number changes from exome sequencing data. Bioinformatics 2014;30:2678–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu X, Zhang M, Bai H, et al. Antituberculosis drug-induced adverse events in the liver, kidneys, and blood: Clinical profiles and pharmacogenetic predictors. Clin Pharmacol Ther 2018;104:326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seixas S, Ivanova N, Ferreira Z, et al. Loss and gain of function in SERPINB11: An example of a gene under selection on standing variation, with implications for host-pathogen interactions. PLoS One 2012;7:e32518. [DOI] [PMC free article] [PubMed] [Google Scholar]


