Abstract
Post-translational modifications of histone proteins play a pivotal role in DNA packaging and regulation of genome functions. Histone acetyltransferase 1 (Hat1) proteins are conserved enzymes that modify histones by acetylating lysine residues. Hat1 is implicated in chromatin assembly and DNA repair but its role in cell functions is not clearly elucidated. We report the generation and characterization of a Hat1 loss-of-function mutant in Drosophila. Hat1 mutants are viable and fertile with a mild sub-lethal phenotype showing that Hat1 is not essential in fruit flies. Lack of Hat1 results in the near complete loss of histone H4 lysine (K) 5 and K12 acetylation in embryos, indicating that Hat1 is the main acetyltransferase specific for these marks in this developmental stage. We found that Hat1 function and the presence of these acetyl marks are not required for the nuclear transport of histone H4 as histone variant His4r retained its nuclear localization both in Hat1 mutants and in His4r-K5R-K12R double point mutants. RNA-seq analysis of embryos indicate that in Hat1 mutants over 2000 genes are dysregulated and the observed transcriptional changes imply a delay in the developmental program of gene expression.
Subject terms: Histone post-translational modifications, Gene expression, Transcriptomics
Introduction
The hereditary material of eukaryotes can be found in the nucleus in the form of chromatin: a complex of DNA, RNA and protein molecules. The basic building units of chromatin are the nucleosomes, protein octamers containing two copies of each of the histone proteins H2A, H2B, H3 and H41,2. Histones can undergo several post-translational modifications (PTMs), including acetylation, methylation or phosphorylation that might influence transcription and other chromatin templated processes3. Histone acetylation that occurs on lysine (K) residues is positively associated with transcription and might exert its effect by charge neutralization and by providing binding surfaces to regulatory proteins4. Although histone acetylation mostly occurs in nucleosomal context, nascent pre-nucleosomal histones can also be acetylated4–6. These acetyl marks are then shortly removed after replication-coupled chromatin assembly7,8. Newly synthesized H4 histones are diacetylated on residues K5/K12 in several diverse organisms including Tetrahymena, Drosophila, chicken, mouse and humans6,9,10. As H4 acetyl-K5/K12 pre-deposition marks are rapidly removed after chromatin assembly8,10 it seems feasible that the K5/K12 diacetyl mark might play important roles in nuclear transport of histones and/or chromatin assembly/maturation. However, experimental results only partially supported these hypotheses11–13.
The well conserved H4 K5/K12 diacetylation mark is generated by cytoplasmic histone acetyltransferase 1 (HAT1) enzymes. HAT1 was first described in S. cerevisiae as an acetyltransferase showing activity towards the H4K12 residues14. Loss-of function mutation of Hat1, however, did not cause growth defects or any other apparent phenotypes14. HAT1 forms a catalytically active holoenzyme with a second protein, HAT2 that shows homology to mammalian Rbap46 and Rbap48 proteins15. HAT2 facilitates the binding of HAT1 to the histone H4 tail and dramatically increases its acetyltransferase activity15. Similarly, in human cells HAT1 can be found in a heterodimeric holoenzyme with p46/RBBP7 (Retinoblastoma-binding protein 7), a protein highly homologous to yeast HAT216. In contrast to yeast cells, Hat1 is indispensable in mice since it is essential for proper development. Hat1 null pups that are born show lung and cranial developmental defects and die shortly after birth10. In cultured murine cells Hat1 is required for the acetylation of nascent histones, illustrated by a decrease in the acetylation levels of H4K5 and H4K12 in Hat1 null cells. Surprisingly, similar change was also observed in the case of H3K9, H3K18 and H3K27 that are incorporated during replication. Loss of Hat1 also leads to decreased proliferation rate, decreased genome stability and sensitivity to a wide range of DNA damaging agents in cultured murine cells10.
Previous studies suggested that in Drosophila melanogaster Hat1 might have functions in formation of centromeric chromatin. In flies Hat1 is most abundant in the cytoplasm, but it can also be detected in nuclear and chromatin fractions17. A small portion of Hat1 is associated with the centromeric histone H3 variant Centromeric Protein-A (CENP-A) both in the cytoplasm and in the nucleus17. Furthermore, Hat1 was found to be enriched in CENP-A containing chromatin, and in addition to euchromatic sites it was also localized to centromeres during interphase but not during mitosis18.
Here we report generation of Hat1 loss-of-function mutants in Drosophila and the phenotypic and molecular analysis of fruit flies lacking Hat1 activity. We show that although Hat1 is the major H4K5 and H4K12 specific acetyltransferase in embryos and its loss results in altered transcript levels of more than 2000 genes it is not essential for viability, fertility or nuclear localization of histone H4 or His4r. Loss of Hat1 does not affect inspected adult phenotypes such as daily activity, sleep pattern, or heat-stress tolerance but mitigates proteopathic stress induced neurodegeneration in a Huntigton’s disease model.
Results
Generation of Hat1 mutants
To be able to analyze the functions of Hat1 in vivo we generated loss of function Hat1 mutants. Several transposon insertion alleles of Hat1 were reported previously but none of these affect the coding region of the gene, therefore cannot be considered null alleles. We chose one of these alleles, P{EPgy2}Hat1EY21697 to generate Hat1 deletion mutants by P element jump-out. P{EPgy2}Hat1EY21697 is inserted in the first intron of Hat1 at genome position 3R:5,789,135 (Fig. 1A). The P{EPgy2} element carries Scer/UAS binding sites in correct orientation to overexpress Hat1 if a GAL4 driver is present. To remobilize the element we crossed P{EPgy2}Hat1EY21697 females to males carrying a third chromosomal Δ2-3 transposase source and selected flies that lost the transposon by screening for loss of the dominant mini-white marker in the second generation. Revertant lines were genotyped by PCR using primers Hat1.F and Hat1.R that straddle the insertion point of P{EPgy2}Hat1EY21697 and the coding sequence of Hat1 and produce a 2007 bp amplicon on wild-type template. Since Hat1 is located in a gene rich genomic region and partially overlaps with a neighboring gene we designed the primers to enable the identification of deletions affecting only Hat1: Hat1.F is situated in the intergenic region between Hat1 and its upstream neighbor, Rpn5, while Hat1.R is located at the end of the coding sequence of Hat1 juxtaposed to the 3′ UTR of the partially overlapping gene, kat-60L1. We tested 226 individual revertants and found 5 which gave a shorter PCR product than the wild-type template. We selected revertant line Hat1Δ57 that gave the shortest PCR amplicon of ~600 bp, therefore having the largest deletion of ~1400 bp, for further analysis (Fig. 1B). We used Sanger sequencing to determine the exact breakpoints of the Hat1Δ57 deletion (Fig. 1A). In Hat1Δ57 a 1409 bp deletion (3 R:5787709-5789127) removes the downstream Hat1-RA promoter, all splice sites and 80% of the coding region (326/405 codons) including the acetyltransferase domain rendering Hat1Δ57 a catalytically null allele. We also selected a precise revertant, Hat1rev16, that did not cause a deletion in the gene (Fig. 1B) to use as control.
Figure 1.

Generation of Hat1 deletion mutant. (A) The genomic region of Hat1 on chromosome 3R is shown under a kilobase scale. On the Hat1 transcript models black bars represent coding exons, empty bars show untranslated exonic regions, introns are represented by lines, arrows indicate transcriptional start sites. Neighboring genes are shown with grey bars. Red arrowhead marks the position of the P{EPgy2}Hat1EY21697 element used for mutagenesis, the breakpoints of the Hat1Δ57 deletion are shown by round brackets. Position of PCR primers are marked by blue arrows. (B) Agarose gel electrophoresis of PCR products amplified from wild-type and Hat1rev16 genomic DNA template with Hat1.F and Hat1.R primers show ~2 kbp bands. PCR on Hat1Δ57 template produce a fragment of approximately 600 bp while there is no PCR product from reactions with P{EPgy2}Hat1EY21697 template (having the 10908 bp long transposon inserted) or without template (NTC).
Hat1 is essential for H4K5 and H4K12 acetylation but not required for nuclear localization of H4
Hat1 enzymes were demonstrated to possess H4 specific acetyltransferase activities in several organisms including yeast, mice and humans. Therefore, we aimed to determine whether Drosophila Hat1 also has H4 specific acetyltransferase activity. We collected 0–6 hours old w1118 (wild-type control), Hat1Δ57 and da-GAL4/Hat1EY21697 (Hat1 overexpressing, Supplementary Fig. S1) embryos for immunoblot analysis. Using acetyl-H4K5 (Fig. 2A,D, Supplementary Fig. S2) and acetyl-H4K12 (Fig. 2B,E, Supplementary Fig. S3) specific antibodies we found that the level of these PTMs diminished significantly in Hat1Δ57 mutants to 1.8% (P = 0.0079, Kruskal-Wallis Test followed by Wilcoxon Rank Sum Test, n = 5) and to 3.95% (P = 0.025, ANOVA with Tukey HSD Test, n = 3), respectively, to that of the wild-type control. These results suggest not only that Hat1 is capable of acetylating H4K5 and H4K12 residues but also that it is the main H4K5 and H4K12 specific acetyltransferase of Drosophila in this developmental stage. Surprisingly, however, overexpression of Hat1 did not result in increased acetylation of either residues (Fig. 2A,B,D,E) suggesting that Hat1 is not a rate limiting component in this catalytic process. As a control experiment, we also analyzed the acetylation level of the H4K8 residue by immunoblotting Hat1Δ57 mutants and found no significant change in the level of this PTM in Hat1Δ57 mutants (Fig. 2C,F, Supplementary Fig. S4). To rule out the possibility that the observed reduction in H4K5 and H4K12 acetylation levels is not due to loss of Hat1 but is caused by background mutations we performed immunoblots on extracts of heterozygotes of Hat1Δ57 and two other Hat1 imprecise excision mutants and found that acetyl-H4K5 and acetyl-H4K12 levels were severely diminished in these heterozygotes (P < 0.01 in both cases, Kruskal-Wallis Test, n ≥ 3, Supplementary Fig. S5). It was previously shown that in murine cells steady state and replication dependent acetylation levels of specific lysine residues of histone H3 are reduced in the absence of Hat110. Therefore, we also tested the acetylation state of H3K18 (Fig. 2G,I, Supplementary Fig. S6) and H3K23 (Fig. 2H,J, Supplementary Fig. S7) residues in Hat1Δ57 and control embryos by immunoblotting. We found that the level of acetyl-H3K18 was significantly lower in Hat1Δ57 mutants than in wild-type control (40.6% of control, P = 0.028, Welch’s t-test, n = 3) while the level of acetyl-H3K23 did not change.
Figure 2.
Hat1 acetylates H4K5 and H4K12 residues in embryos. Immunoblot analysis of embryo lysates with antibodies against acetylated histones show that in Hat1Δ57 mutants the level of acetylated H4K5 (A,D) and H4K12 (B,E) residues is significantly reduced while overexpression of Hat1 does not affect H4K5ac and H4K12ac levels. Acetylation of H4K8 is not altered in Hat1Δ57 mutants (C,F). The level of H3K18 acetylation is lower in Hat1Δ57 embryos than in controls (G,I), while there is no change in the abundance of acetyl-H3K23 (H,J). Bar graphs show average signal intensities of acetylated histone bands normalized to signal intensities of H3 loading controls, whiskers indicate standard deviation. Asterisks mark significant differences compared to wild-type control, *P < 0.05, **P < 0.01.
As the cytoplasmic acetylation of H4 and His4r histones precedes their nuclear transport we investigated whether H4K5 and H4K12 acetylation by Hat1 is required for nuclear localization of histone H4. For these tests we generated transgenic Drosophila strains carrying an inducible His4r transgene with a C-terminal FLAG tag (UAS-His4r) or its mutated derivative in which K5 and K12 lysines were changed to arginine (UAS-His4r-K5R-K12R) mimicking unmodified lysine that is not (and cannot be) acetylated. The amino acid sequence of the protein encoded by the His4r gene is identical to the sequence of canonical H4, thus the proteins produced from these transgenes correspond to the histones encoded by both His4r and H4. To visualize the localization of His4r in the presence or absence of Hat1 we performed immunostaining of w; hs-GAL4/+; Hat1Δ57 (staining control, Fig. 3D,H,L), w; hs-GAL4/+; UAS-His4r (Fig. 3A,E,I), and w; hs-GAL4/+; UAS-His4r Hat1Δ57/Hat1Δ57 (Fig. 3B,F,J) larval tissues with anti-FLAG antibody. We found that His4r protein localized to the nucleus both in larvae wild-type for Hat1 and in homozygous Hat1Δ57 mutants. Having found that Hat1 is not required for the nuclear localization of His4r we investigated whether acetylation of Hat1 target lysines, H4K5 and H4K12, influences the localization of His4r by immunostaining w; hs-GAL4/+; UAS-His4r-K5R-K12R larvae with anti-FLAG antibody (Fig. 3C,G,K). We found that His4r-K5R-K12R that cannot be acetylated at the mutated positions has a nuclear localization similar to wild-type His4r, suggesting that these PTMs do not have a crucial role in the nuclear transport of His4r and H4 histones.
Figure 3.
Hat1 dependent H4K5 and H4K12 acetylation is not required for nuclear localization of His4r. His4r protein localizes to the cell nucleus as shown by anti-FLAG immunostaining of polytene larval salivary gland cells expressing transgenic His4r with a C-terminal FLAG tag (A,I), nuclei are visualized by DAPI staining (E). Nuclear localization of His4r is unaltered in homozygous Hat1Δ57 larvae lacking functional Hat1 (B,F,J). Mutated His4r proteins in which Hat1 target lysines were changed to arginines (His4r-K5R-K12R) are also correctly localized to the nucleus (C,G,K). In the negative control having the driver element but lacking a His4r-FLAG transgene there is no specific staining with anti-FLAG antibody (D,H,L).
Drosophila Hat1 is not required for viability or fertility
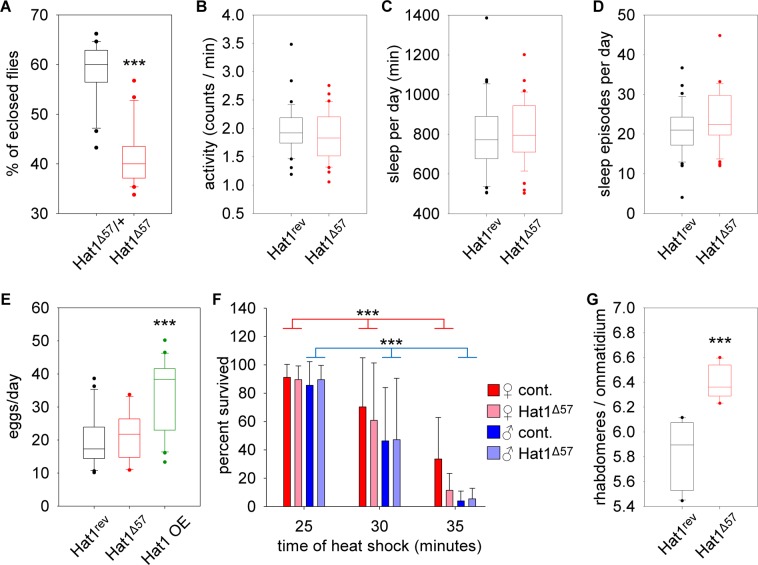
All recovered revertant lines, including Hat1Δ57, were homozygous viable and fertile proving that despite its fundamental role of H4 acetylation in early embryos Hat1 is not an essential gene. To determine whether loss of Hat1 has subtler developmental or adult effects we set up several experiments. First, we made genetic crosses to determine whether loss of Hat1 influences viability. We crossed Hat1Δ57 homozygous females to heterozygous males and by recording the number of homozygous and heterozygous F1 offspring per vials we found that the percentage of Hat1Δ57 homozygotes among the progeny was significantly lower (41.26 ± 5.92%, P = 4.77 * 10−5, Wilcoxon Signed Ranks Test) than that of heterozygotes (58.76 ± 5.92%) (Fig. 4A). Thus, we concluded that loss of Hat1 results in a sub-lethal phenotype.
Figure 4.
Phenotype analysis of Hat1 mutants. (A) Hat1Δ57 Drosophila have reduced viability. The graph shows the percentages per vials of Hat1Δ57/+ heterozygous and Hat1Δ57 homozygous offspring from crosses of homozygous females and heterozygous males. (B) Loss of Hat1 in Hat1Δ57 mutants does not affect the daily motor activity, (C) total sleep time and (D) the number of sleep episodes per day. (E) Hat1Δ57 does not affect, while overexpression of Hat1 increases female fecundity. The plots show the average number of eggs per female laid daily in vials containing Hat1rev16, Hat1Δ57, or da-GAL4/P{EPgy2}Hat1EY21697 (Hat1 OE) females mated with w males. (F) Loss of Hat1 does not affect heat-stress tolerance. The graph shows the percentage of 7-day-old Hat1rev16 (control) or Hat1Δ57 male or female flies that survived after a 25, 30 or 35 minutes heat-shock at 42 °C. (G) Heterozygous reduction of Hat1 ameliorates mutant Huntingtin induced neurodegeneration. The graph shows the average number of rhabdomeres per ommatidium in the eyes of flies overexpressing a Httex1p-Q120 transgene and heterozygous for Hat1rev16 (control) or Hat1Δ57. On boxplots the bottom and the top of boxes indicate 25th and 75th percentiles, the line within the box shows the median value, whiskers represent 10th and 90th percentiles, dots mark outliers. Bar graphs show the averages, whiskers represent standard deviation. Asterisks mark significant differences compared to control, ***P < 0.001.
As Hat1 is not essential during development we aimed to characterize its potential effects on adult functions. It was reported previously that RNAi mediated Hat1 silencing resulted in circadian phase delay19. Therefore, we investigated whether loss of Hat1 affects the daily activity of flies since altered circadian activity may reflect disturbed circadian regulation or in more general terms dysfunction of the nervous and/or neuro-muscular system. We used a Drosophila Activity Monitor (TriKinetics Inc) to measure daily motor activity and sleep patterns of individual flies for five days under 12 hr light and 12 hr dark (12:12 LD) cycles. In our hands loss of Hat1 had no statistically significant effect on either the activity index (1.979 ± 0.424 vs 1.852 ± 0.426 counts/minute in Hat1rev16 vs Hat1Δ57, respectively, Fig. 4B), the total amount of daily sleep (794.4 ± 185.8 vs 817.9 ± 162.7 minutes/day in Hat1rev16 vs Hat1Δ57, respectively, Fig. 4C) or the number of daily sleep episodes (20.97 ± 6.27 vs 23.74 ± 7.06 in Hat1rev16 vs Hat1Δ57, respectively, Fig. 4D) of flies.
Next, as gene expression level of Hat1 is highest in ovaries and early embryos20, we investigated whether Hat1 affects female fecundity. We compared the average number of eggs laid daily by Hat1rev16, Hat1Δ57, and da-GAL4/Hat1EY21697 females, the latter of which overexpressing Hat1, and found significant differences (P = 3.66 × 10−4, Kruskal-Wallis Test) among these groups (Fig. 4E). Surprisingly, however, the lack of Hat1 did not affect fecundity, the number of eggs laid per day by Hat1Δ57 homozygous females (21.83 ± 7.49) was similar to the amount seen in the Hat1rev16 controls (19.69 ± 7.81). On the other hand, females that over-expressed Hat1 laid significantly more eggs per day (33.27 ± 11.37, P = 1.55 × 10−4, Wilcoxon Rank Sum Test) than control females. Thus, our data suggest that although Hat1 per se is not required for female fertility the biochemical functions it performs are rate limiting in egg production.
Next, we aimed to determine whether Hat1 plays a role in response to stress conditions in adults. First, we analyzed heat-stress tolerance of Hat1 mutants. We heat-shocked 7-day-old Hat1rev16 and Hat1Δ57 male and female Drosophila at 40 °C for 25, 30 or 35 minutes and recorded the number of survivors after a 12-hour recovery period (Fig. 4F). Under these conditions the number of survivors significantly decreased with longer treatment times both in males and females (P < 0.0001 in both genders, Two-way ANOVA), however, we found no significant difference between the number of Hat1rev16 and Hat1Δ57 survivors in either gender.
We also tested another cellular stress condition, mutant Huntingtin induced proteopathy. Specific gain-of-function mutations of the human huntingtin (HTT) gene are responsible for Huntington’s disease (HD), a devastating neurodegenerative disorder21. In the mutant HTT allele a CAG triplet repeat is expanded resulting in an elongated polyglutamine region of the Huntingtin protein, which is primarily responsible for proteopathy in HD. We and others had previously shown that histone acetylation is perturbed in HD, however, the potential role of Hat1 in HD pathogenesis was not tested. We analyzed the effect of reduced Hat1 in mutant Huntingtin induced neurodegeneration in the retina by the pseudopupil assay comparing the number of visible rhabdomeres, light gathering structures of photoreceptor neurons, per ommatidium. In wild-type flies each ommatidum contains 7 visible rhabdomeres. In the eyes of elav-GAL4/w; HTTex1p-Q120/+; Hat1rev16/+ flies expressing mutant HTT in the nervous system the average number of rhabdomeres per ommatidium was reduced to 5.83 ± 0.26, while in elav-GAL4/w; HTTex1p-Q120/+; Hat1Δ57/+ flies neurodegeneration was significantly (P = 4.35 × 10−5, Welch’s t-test) ameliorated with 6.39 ± 0.13 rhabdomeres per ommatidium (Fig. 4G).
Transcription in the absence of Hat1
Several lines of evidence indicate that acetylation of nucleosomal histones positively correlates with gene expression4. Based on our data showing that Hat1 is responsible for the bulk of H4K5 and H4K12 acetylation in embryos we raised the question whether Hat1 is required for the proper regulation of gene expression. To investigate whether loss of Hat1 results in transcriptional dysregulation we compared gene expression levels of 6–12 hours old Hat1Δ57 mutant and w1118 (wild-type) control embryos by RNA-sequencing. In this developmental stage embryonic gene expression is already activated22.
In the embryo derived samples 13518 of the 17661 annotated transcriptional units could be detected on the RNA level, 8471 of these could be statistically tested for differential gene expression. We found that the transcript level of 2137 (25.2% of statistically tested, 12.1% total) genes changed significantly (false discovery rate q < 0.05). Nearly two thirds (1397 genes, 65.4%) of these genes were up-regulated, while those making up the remaining one third (740 genes, 34.6%) were down-regulated (Fig. 5A). Hat1 itself was in the set of down-regulated genes with a Fragments Per Kilobase of transcript per Million mapped reads (FPKM) value of 1.5 in Hat1Δ57 samples compared to 46 in control samples, hereby validating the method and the samples. Thus, the majority of observed gene expression changes was in the opposite direction of what would have been expected as a result of reduced histone acetylation (i.e. reduced gene expression). This suggests that either a large portion of the observed effects are indirect or the acetylation by Hat1 is inhibitory to gene expression, possibly by augmenting chromatin formation.
Figure 5.
Transcriptomic changes in Hat1 mutants. (A) RNA-seq analysis of 6-12 hours old embryos showed that 7.9% of annotated genes were significantly (q < 0.05) up-regulated while 4.2% were down-regulated. The graph shows the number of genes that were not expressed (black), were expressed but not tested statistically (grey), did not show change in expression (yellow), were up-regulated (green) or down-regulated (red). (B) Transcriptional profile of genes up-regulated in 6-12 hours old Hat1Δ57 embryos during embryonic development based on modENCODE data. (C) Transcriptional profile of genes down-regulated in 6-12 hours old Hat1Δ5 embryos during embryonic development. Charts (D,E) show the ten specialized Gene Onthology Biological Process terms (from the bottom of the hierarchy) with the most significant false discovery rates (FDR) that showed enrichment among the 154 genes up-regulated and the 216 genes down-regulated in Hat1Δ57 embryos, respectively.
To investigate whether loss of Hat1 can lead to the activation/derepression of inactive/silenced genes we compared the set of genes significantly up-regulated in Hat1Δ57 embryos to a set of genes not expressed in embryos (FPKM = 0) retrieved from Drosophila developmental modENCODE high-throughput RNA-seq dataset that contains embryonic transcriptional levels of annotated Drosophila genes with two hours resolution20. We found that out of the 3370 genes not expressed in embryos based on modENCODE data none was up-regulated in Hat1Δ57. Furthermore, in our RNA-seq dataset there were only 21 genes (0.98%) that suffered significant transcriptional changes in Hat1Δ57 and had a very low (FPKM < 3) expression level in the control samples. These data together suggest that loss of Hat1 alone is not sufficient for the activation or de-repression of inactive genes.
Next, we retrieved the embryonic transcriptional profile of genes showing significant expression changes in Hat1Δ57 embryos from the modENCODE developmental RNA-seq dataset20. We found that genes that were up-regulated or down-regulated in Hat1Δ57 embryos showed markedly different embryonic expression profiles (Fig. 5B,C). The expression levels of upregulated genes are higher in early embryos (with highest median expression level in 2–4 hours old embryos) and decrease as embryonic development progresses (Fig. 5B). As opposed to this, the expression levels of down-regulated genes are lower in early embryos (with lowest median expression level in 2–4 hours old embryos) and their expression levels increase during embryonic development (Fig. 5C). The characteristic transcriptional profiles of the two sets of dysregulated genes suggested that the majority of gene expression changes in Hat1Δ57 embryos could be explained by a delay in the transcriptional program that would cause the analyzed 6–12 hours old embryos having transcriptomic profiles similar to younger embryos. We selected those genes whose altered transcriptional levels in Hat1Δ57 embryos could not be explained by developmental delay by searching for up-regulated genes that have lower or equal expression levels between hours 0–6 than between hours 6–12 in wild-type embryos, and for down-regulated ones that have higher or equal expression levels between hours 0–6 than between hours 6–12. Altered transcript levels of 154 out of the 1397 up-regulated genes and 216 out of the 740 down-regulated genes could not be explained by developmental delay.
We used these subsets of dysregulated genes to identify the biological processes affected by loss of Hat1 by Gene Onthology (GO) term analysis. Overrepresentation test of biological process terms revealed that in the up-regulated gene set terms associated with development, translation and regulation of biosynthetic processes were enriched (Fig. 5D), while among the down-regulated genes biological process terms associated with cellular metabolism dominated (Fig. 5E). Thus, genes up- or down-regulated in Hat1 mutant embryos are involved in distinct, well defined functions.
Discussion
In this study we presented the phenotypic and molecular effects of a loss-of-function mutant of Drosophila Hat1 for the first time. Although the deletion allele we generated does not remove the whole gene it removes ~80% of the coding sequence including the catalytic acetyltransferase domain rendering the mutant a functional null. By immunoblot analysis of Hat1 mutants we confirmed that Drosophila Hat1, similarly to its orthologues, has histone H4K5 and H4K12 lysine specific acetyltransferase activities. However, Hat1 is not the only acetyltransferase in Drosophila showing this specificity. Previously it was reported that lack of the Ada2a subunit of the GCN5 acetyltransferase containing ATAC complex leads to reduced H4K5 and H4K12 acetylation in wandering L3 larvae23. Notwithstanding, our data shows that Hat1 is the dominant acetyltransferase with this substrate specificity in embryos as the level of acetylated H4K5 and H4K12 residues dropped to less than 4% of that of wild-type in Hat1 mutant embryos. Importantly, this enzymatic activity seems to be directed to specific lysine residues as the acetylation state of another H4 N-terminal lysine, H4K8, was not affected by loss of Hat1. We also analyzed the acetylation levels of specific histone H3 residues as previously it was reported, that both steady state acetyl-H3K18 levels and H3K18 acetylation on histones deposited during replication was reduced in Hat1 mutant murine cells, while the level of H3K23 acetylation was not sensitive for the presence of Hat110. Our results corroborate this data, we found that in Hat1 mutant embryos H3K18 acetylation levels were significantly decreased compared to wild-type controls while there was no change in H3K23 acetylation. Thus, beside acetylation of histone H4 Hat1 also influences that of histone H3. However, as Hat1 has no H3 specific catalytic activity15,17 this is most likely an indirect effect.
The histone H4 K5/K12 diacetyl mark was proposed to be involved in the nuclear import of histones and chromatin assembly24, however, many questions remain about the potential role of this diacetyl mark and Hat1 in these processes. Histone H3 and H4 are transported to the nucleus with the aid of karyopherin proteins. More established models claim that H3:H4 form complexes with HAT1, the histone chaperone Asf1 and importin-4 (IPO4) in the cytoplasm25, while recently published data suggest that H3 and H4 might be imported to the nucleus by IPO4 as monomers where they associate with nuclear HAT126. The nuclear localization signals (NLS) of histones H3 and H4 can be found at their positively charged N-terminal tails27. Analysis of the crystal structure of the Kluyveromyces lactis IPO4 homologue (Kap123) revealed that the H3-NLS and H4-NLS are competing for Kap123 binding: the H3K14, H3K23 and H4K16 residues are recognized by lysine binding pockets of Kap12328. Lysine to glutamine mutations of H4K16 or H4K5/K12 reduced the affinity of H4 towards Kap123, suggesting that positive charge at these positions facilitates karyopherin binding28. In Saccharomyces, mutation analysis of the H4 N-terminus in the absence of the H3-NLS showed that simultaneous mutations of H4 K5/K8/K12, and to a lesser extent, K5/K12 to glutamine resulted in cytoplasmic mislocalization of H4, while mutations to arginine did not have a similar effect13. However, deletion analysis of H3-NLS and H4-NLS indicated that in yeast the N-terminal tails of H3 and H4 are redundant for nuclear transport13. Furthermore, in Hat1 null mouse embryonic fibroblast (MEF) cells there was no change in the level of either cytosolic or nuclear histone H4 suggesting that Hat1 dependent acetylation is not essential for the nuclear import of H429. The data above suggest that while positive charge at the N-terminal tail of H4 promotes NLS function and its acetylation state influences nuclear transport of H4 specific post-translational modifications might play a redundant role in the import process. Our data corroborate these observations: we found that His4r proteins (which have the same amino acid sequence as H4) were localized to the nucleus in Hat1Δ57 mutants as well as when the His4r K5 and K12 residues were simultaneously mutated to arginine. In a recent article by Apta-Smith et al. the authors argue that Hat1 is mostly nuclear26 that would revert the timing of Hat1 dependent acetylation and nuclear import. Furthermore, as previous studies found that H4 K5/K12 acetylation rather impairs nuclear import than promotes it13,28 acetylation by Hat1 might be important for the release of H4 from the karyopherin inside the nucleus or binding to chromatin assembly factors that must precede the incorporation of H4 to chromatin.
Studies aimed at deciphering the role of H4 K5/K12 diacetylation in chromatin assembly also provided unambiguous results. In yeast, the entire N-terminal domains of both histone H3 and H4 in (H3-H4)2 tetramers were found to be dispensable for chromatin assembly factor-1 (CAF-1) dependent nucleosome formation on newly replicated DNA in vitro12 and mutation analysis also indicated that K5/K12 acetylation is not essential for nucleosome assembly11. Correspondingly, histone levels did not decrease on newly replicated DNA in Hat1 null mouse cells29. However, H4K5 and K12 were found to be involved in Asf1 dependent chromatin re-assembly following DNA double-strand break repair in yeast30, while in chicken cells these residues were found to be required for correct centromeric localization of CENP-A as it was more frequently mis-incorporated to non-centromeric regions in H4-K5R-K12R point mutants6. Similarly, in Drosophila Hat1 is a subunit of one of the CENP-A loading complexes and its knock-down resulted in significantly reduced CENP-A incorporation into chromatin17. The above delineated data showing that H4 K5/K12 diacetylation contributes to but is not absolutely necessary for either nuclear transport or general chromatin assembly are in line with our results. We found that even though Hat1 is responsible for H4K5 and H4K12 acetylation in embryos it is a non-essential protein in Drosophila. Loss of Hat1 did not affect fertility and only moderately reduced viability making it unlikely that Hat1 plays an essential, non-redundant role either in the transport of histones or in their assembly to chromatin. Redundancy in these processes can arise from several sources, including overlapping enzymatic activities, redundant acetylation positions on histones or redundant NLS functions of histone tails.
By analyzing the effects of loss of Hat1 on the transcriptome of 6–12 hours old embryos we found that more than two thousand genes were mis-regulated, nearly two-third of these genes were up-regulated while one-third were down-regulated. By cross-referencing our differential expression dataset with developmental RNA-Seq data of modENCODE we found that most of the genes up-regulated in Hat1 mutant embryos have the highest expression levels in early embryos, while most of the down-regulated genes have the highest expression levels in older embryos. From this we assume that the majority of the transcriptional effects we detected in the absence of Hat1 are rather due to developmental delay caused by negative effects on replication and/or chromatin maturation than direct effects on the transcriptional machinery or its regulation. Hat1 dependent acetylation was implied to have a modulatory role in replication in human MCF7 cells as ATAD2 (ATPase family AAA domain-containing protein 2), a chromatin regulator shown to be required for proper chromatin localization of PCNA and efficient replication, binds H4 diacetylated at K5/K12 and is recruited to nascent chromatin31. Furthermore, nascent chromatin of Hat1 null MEF cells are depleted of several bromodomain proteins while are enriched in Topoisomerase 2α and 2β suggesting that Hat1 has a role in chromatin maturation29. Defects in either of these processes might lead to the observed delay in the developmentally determined transcriptional program.
An interesting finding of this study is that reduced Hat1 ameliorates retinal neurodegeneration in a Drosophila model of Huntington’s Disease. In HD and in at least eight other neurodegenerative Polyglutamine (polyQ) disorders the primarily cause of pathogenesis is an elongated polyQ repeat in the disease proteins32. PolyQ containing aberrant proteins disturb important cellular processes, including epigenetic control of the genome32. Several factors affecting histone protein acetylation were tested previously with the conclusion that the activity of specific histone acetyltransferases (HAT) promoting open chromatin and gene expression and the level of histone acetylation are reduced in HD33–36, and that inhibition of histone deacetylases (HDAC) that remove acetyl marks and promote closed chromatin suppresses polyQ pathology37–39. However, not all acetylation related changes are linked to transcription or genome function, for example, inhibition of HDAC6 increases acetylation of α-tubulin and compensates for intracellular transport deficit observed in HD40. The effect of reduced Hat1 levels in Hat1Δ57 heterozygotes in the HD model is the opposite that was observed in the case of nuclear HATs35,36 suggesting that Hat1 might influence HD pathology by alternative means. First, instead of regulating transcription by acetylating nucleosomal histones on gene regulatory regions Hat1 might affect chromatin structure by modulating histone transport and deposition, as detailed above. Second, similarly to some other HAT enzymes Hat1 also has factor acetyltransferase activity i.e. its targets are not exclusively histones. For example, Hat1 regulates NF-κB signaling by acetylating the transcriptional regulator protein PLZF that leads to the recruitment of a histone deacetylase containing repressor complex41. At present there is a very limited body of knowledge about non-histone targets of Hat1 and detailed studies would be required to further the understanding of the role of Hat1 in the cell and the reasons of its evolutionary conservation.
Methods
Drosophila stocks and crosses
Stocks were maintained and crosses were done on standard cornmeal-yeast-agar Drosophila medium at 25 °C. The y1 w67c23; P{EPgy2}Hat1EY21697 strain was obtained from the Bloomington Drosophila Stock Center. To generate Hat1 revertants y1 w67c23; P{EPgy2}Hat1EY21697 males were crossed to w; Dr Δ2-3/TM3, Sb Δ2-3 females, then w; P{EPgy2}Hat1EY21697/TM3, Sb Δ2-3 jump-starter male offspring were mated with w; TM3/TM6 females. From the progeny of these crosses white eyed w; Hat1rev/TM6 male revertants were selected and backcrossed individually to w; TM3/TM6 females to generate revertant strains. Hat1 revertant strains were genotyped by PCR analysis of the heterozygous w; Hat1rev/TM6 founding males after they procreated.
For viability analysis, w; Hat1Δ57 females were crossed to w; Hat1Δ57/ZH-attP-86FB males in 20 vials, and the number of homozygous and heterozygous progeny that eclosed in each vial were recorded. Eclosion rates were calculated as the number of progeny of a given genotype divided by the number of all progeny in the vial.
For fertility assays, 3 days old w; Hat1Δ57, w; Hat1rev or da-GAL4 / Hat1EY21697 females were mated with w1118 males in 20 vials, 5 females and 3 males per vial. Vials were passed daily and the number of eggs laid in a 24 hour period was recorded.
For heat-stress tolerance assays freshly eclosed Hat1rev16 and Hat1Δ57 females and males were kept at 25 °C for 7 days then sorted to empty vials (20–25 flies / vial) and heat-shocked at 40 °C for 25, 30 or 35 minutes. After het-shock, flies were transferred to 25 °C and the number of survivors was scored after a 12 hours recovery period. At least 120 flies per treatment were tested.
To measure neurodegeneration w; Httex1p-Q120 females were crossed with elav-GAL4; Hat1rev16/Sb or elav-GAL4; Hat1Δ57/Sb males and the elav-GAL4/w; Httex1p-Q120/+; Hat1rev16/+ and elav-GAL4/w; Httex1p-Q120/+; Hat1Δ57/+ progeny selected for pseudopupil assay.
Activity monitoring
7-day-old Hat1rev16 and Hat1Δ57 male flies that were raised under 12:12 LD cycles were sorted to 5 mm diameter pyrex glass tubes (one fly per tube) and the activity of individual flies measured for five consecutive days under 12:12 LD condition at 25 °C in a DAM2 Drosophila Activity Monitor (TriKinetincs Inc). The DAM2 monitor records movement when a fly blocks a beam of infrared light hitting a sensor. A total of 38 flies per genotype were measured in three separate experiments. Monitor data were collected with DAMSystem and filtered with DAMFileScan programs (TriKinetics Inc) then analyzed with pySolo.
Genotyping by PCR
Individual revertant males were homogenized in 50 μl homogenization buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 25 mM NaCl, 0.2 μg/μl Proteinase K), then incubated for 60 minutes at 37 °C followed by 15 minutes at 85 °C. 5 μl homogenates were used as templates in PCR reactions using Hat1.F and Hat1.R primers (Supplementary Table S1) and DreamTaq DNA polymerase (Thermo Fisher Scientific). Those revertants that gave a shorter amplicon than the 2 kbp one expected from wild-type were selected as deletion lines.
Cloning and mutagenesis of His4r and generation of transgenic flies
His4r was cloned using nested primers. First, genomic region containing the His4r gene was PCR amplified from w1118 genomic DNA template with His4r.gF and His4r.gR primers (Supplementary Table S1) using Q5 High-Fidelity DNA polymerase (New England Biolabs) and cloned to pJET1.2 vector using CloneJET PCR cloning kit (Thermo Fisher Scientific) following the manufacturer’s recommendations. The gene body of His4r from the start codon to the codon before the stop codon was PCR amplified from the pJET1.2-His4r template using His4r.E3C.F and His4r.E3C.R primers (Supplementary Table S1) with Q5 DNA polymerase and reinserted to pJET1.2 with CloneJET PCR cloning kit. After digestion with FastDigest KpnI and EcoRI (Thermo Fisher Scientific) the His4r fragment was isolated and ligated to pENTR3C Gateway entry vector cut with the same enzymes.
K12R and K5R mutations were introduced to the His4r sequentially utilizing mutagenic PCR with modified primers. First, the His4r-K12R mutation was generated by PCR amplification of pENTR3C-His4r using Q5 DNA polymerase with His4r.K12R.F and His4r.K12.R primers (Supplementary Table S1). PCR fragments were isolated using Agencourt AMPure XP beads (Beckman Coulter), phosphorylated with T4 polynucleotide kinase (Thermo Fisher Scientific), circularized with T4 DNA ligase (Thermo Fisher Scientific) and template contamination removed by DpnI (Thermo Fisher Scientific) digestion. Next, K5R mutation was introduced similarly to pENTR3C-His4r-K12R construct using His4r.K5R.F and His4r.K5.R PCR primers (Supplementary Table S1) resulting in pENTR3C-His4r-K5R-K12R.
The inserts from pENTR3C-His4r and pENTR3C-His4r-K5R-K12R entry clones were subcloned to pTWFattB, a modified pTWF Gateway destination vector that contains a φC31 attB region cloned in the NsiI restriction site, with Gateway LR Clonase Enzyme Mix (Thermo Fisher Scientific) following the manufacturer’s protocol. Transgenic flies were generated by injecting the pTWFattB-His4r and pTWFattB-His4r-K5R-K12R constructs to embryos carrying the attP-zh86Fb φC31 docking site.
Pseudopupil assay
7-day-old female flies were decapitated and their heads immobilized with nail polish on a microscope slide. The heads were covered with immersion oil and the number of visible rhabdomeres per ommatidium in the compound eye were counted using a Nikon Eclipse 80i microscope with a 50× Nikon oil immersion lens. At least 20 ommatidia per eye and at least 10 eyes per genotype were scored.
Immunoblotting
For sample preparation Drosophila embryos were homogenized with a plastic pestle in sonication buffer (50 mM Tris-HCl pH7.9, 2 mM EDTA, 50 mM NaCl, 0.5 mM DTT, 10 mM Na-butyrate and 1x Protease inhibitor cocktail set I (Calbiochem)), then sonicated on “high” energy setting for four cycles of 30 sec ON/30 sec OFF in a Bioruptor sonicator (Diagenode). Homogenized samples were boiled for 10 minutes in 2x Laemmli sample buffer containing 5% β-mercaptoethanol, then centrifuged for 10 minutes at 13000 RPM. Sample supernatants containing 30 μg protein were separated by 10% Tris-Tricine-SDS PAGE and electrotransferred to Amersham Protran Premium 0.45 μm nitrocellulose membrane (GE Healthcare Life Sciences). Membranes were blocked in 5% nonfat milk and incubated with the following primary and secondary antibodies in the indicated dilutions: anti-acetyl-H4K5 (ab61236, Abcam, 1:500), anti-acetyl-H4K8 (ab15823, Abcam, 1:1000), anti-acetyl-H4K12 (ab61238, Abcam, 1:1000), anti-acetyl-H3K18 (ab1191, Abcam, 1:500), anti-acetyl-H3K23 (ab47813, Abcam, 1:1000) anti-H3 (ab1791, Abcam, 1:4000), goat-anti-rabbit IgG-HRP (P0448, Dako, 1:4000). Immunoblots were developed with Immobilon Western Chemiluminescent HRP substrate (Millipore) and recorded with a C-DiGit chemiluminescent blot scanner (Li-Cor Biosciences). Band intensities were quantitated with Image Studio software (Li-Cor Biosciences).
Immunohistochemistry
Salivary glands of wandering L3 larvae were dissected, fixed for 20 minutes in fixative solution (4% formaldehyde in PBS), washed for 3 × 10 minutes in PBS then permeabilized for 1 hour in PBS containing 1% Triton X-100. After blocking for 50 minutes with 5% BSA in PBST (0.1% Tween 20 in PBS) samples were incubated at 4 °C overnight with monoclonal anti-FLAG M2 antibody (F3165, Sigma, 1:750) in PBST containing 1% BSA. After 6 × 10 minutes washes in PBST samples were incubated with goat anti-mouse IgG Alexa Fluor 488 secondary antibody (R37120, Thermo Fisher Scientific, 1:1500) for 1.5 hours at RT. After 4 × 10 minutes washes in PBST samples were stained with 0.5 μg/ml DAPI (PanReac AppliChem), mounted in Fluoromount (Sigma) and visualized with an Olympus FV10i Confocal Laser Scanning Microscope.
RNA-sequencing
Total RNA samples were isolated from ~15 mg 6–12 hours old synchronized w1118 and w1118; Df(2L)Hat1Δ57 embryos with Qiagen RNeasy Plus Mini kit, three biological replicates each. Sample purity was assessed by measurement of OD260/280 ratio with a NanoDrop ND-1000 spectrophotometer; RNA concentrations were determined using fluorometric Qubit RNA HS Assay Kit (Thermo Fisher Scientific); and RNA integrity was verified by capillary gel electrophoresis with a Bioanalyzer 2100 (Agilent) instrument using Agilent RNA 6000 nano kit. RNA-seq libraries were prepared from 1 µg total RNA samples using TruSeq RNA sample prep kit v2 (Illumina) following the low sample (LS) protocol provided by the manufacturer. This, in short, includes purification and fragmentation of poly-A mRNA, synthesis of double-stranded cDNA with SuperScript II reverse transcriptase (Invitrogen), ligation of indexed sequencing adapters and limited amplification by PCR. Sequencing libraries were validated and average fragment length determined by capillary gel electrophoresis with a Bioanalyzer 2100 instrument using Agilent High Sensitivity DNA kit. Library concentrations were calculated based on average fragment length and quantitative-PCR measurement using NEBNext Library Quant Kit for Illumina (New England Biolabs). After denaturing libraries were diluted to 15 pM and were sequenced with an Illumina MiSeq sequencer using MiSeq Reagent Kit V3–150 providing 2 × 75 bp paired-end sequence reads.
Sequencing data analysis
Primary data analysis (base-calling, fastq generation, demultiplexing) was done by MiSeq Control Software v2.6/Real-Time Analysis software v1.18.54. Fastq files were quality trimmed with Trimmomatic v0.33 in paired-end mode, then paired fastq files were aligned to the dmr6.13 Drosophila melanogaster reference genome with TopHat242. Alignment files were sorted and deduplicated with SAMtools, then differential expression analysis was performed with Cufflinks42 using corresponding transcript annotation (dmr6.13.gtf) from FlyBase. Gene Ontology enrichment analysis was performed using PANTHER Overrepresentation Test (Released 20181003, Test type: Fisher’s Exact with FDR correction) with GO Ontology database (Released 2018-09-06) using all Drosophila melanogaster genes as reference list.
Statistical methods
For statistical calculations R 3.4.3 was used. Data are given as mean ± standard deviation. We used Shapiro-Wilk test for normality testing and F-test to compare variances. In case of not normally distributed data Wilcoxon Rank Sum Test was used for pairwise comparison of unpaired samples, Wilcoxon Signed Ranks Test was used for pairwise comparison of paired samples, and Kruskal-Wallis Test was used for multiple comparisons followed by Wilcoxon Rank Sum Test with Bonferroni correction. For pairwise comparison of normally distributed data Student’s t-test was used if sample groups had equal variances while Welch’s t-test was used if sample groups had unequal variances. For multiple comparisons we applied ANOVA with post-hoc Tukey HSD test.
Supplementary information
Acknowledgements
We thank the Drosophila microinjection service of the Biological Research Centre of the Hungarian Academy of Sciences for transgenesis, Dr Péter Deák and Ágota Nagy (Department of Genetics, University of Szeged) for their help with confocal microscopy, and J. Lawrence Marsh (University of California, Irvine) for Drosophila lines. This study was supported by Hungarian National Research, Development and Innovation Office (NKFIH) grants K-112294, GINOP-2.3.2-15-2016-00032 and GINOP-2.3.2-15-2016-00034. László Bodai was supported by the ÚNKP-19-4-SZTE-8 New National Excellence Program of the Ministry for Innovation and Technology and by the János Bolyai Research Scholarship (BO/00522/19/8) of the Hungarian Academy of Sciences. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author contributions
J.V. participated in study design, acquisition, analysis and interpretation of data and revising the manuscript. S.K. acquired and analyzed data and revised the manuscript. A.N. acquired and analyzed data. N.Z. participated in designing experiments, acquiring, analysis and interpretation of data and revising the manuscript. L.B. participated in study conception and design of experiments, acquiring, statistical analysis and interpretation of data and drafting the manuscript. All authors read and approved the final manuscript.
Data availability
RNA-Seq data are deposited to the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under accession PRJNA551569. Other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54497-0.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.McGinty RK, Tan S. Nucleosome Structure and Function. Chem. Rev. 2015;115:2255–2273. doi: 10.1021/cr500373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gates LA, Foulds CE, O’Malley BW. Histone Marks in the ‘Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem. Sci. 2017;42:977–989. doi: 10.1016/j.tibs.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 5.Ruiz-Carrillo A, Wangh LJ, Allfrey VG. Processing of newly synthesized histone molecules. Science. 1975;190:117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- 6.Shang, W.-H. et al. Acetylation of histone H4 lysine 5 and 12 is required for CENP-A deposition into centromeres. Nat. Commun. 7 (2016). [DOI] [PMC free article] [PubMed]
- 7.Annunziato AT, Seale RL. Histone deacetylation is required for the maturation of newly replicated chromatin. J. Biol. Chem. 1983;258:12675–12684. [PubMed] [Google Scholar]
- 8.Scharf AND, Barth TK, Imhof A. Establishment of Histone Modifications after Chromatin Assembly. Nucleic Acids Res. 2009;37:5032–5040. doi: 10.1093/nar/gkp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagarajan P, et al. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013;9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma XJ, Wu JS, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibahara K, Verreault A, Stillman B. The N-terminal domains of histones H3 and H4 are not necessary for chromatin assembly factor-1-mediated nucleosome assembly onto replicated DNA in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:7766–7771. doi: 10.1073/pnas.97.14.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackwell JS, Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational Analysis of H3 and H4 N Termini Reveals Distinct Roles in Nuclear Import. J. Biol. Chem. 2007;282:20142–20150. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- 14.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 15.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/S0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 16.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998;8:96–108. doi: 10.1016/S0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 17.Boltengagen M, et al. A novel role for the histone acetyltransferase Hat1 in the CENP-A/CID assembly pathway in Drosophila melanogaster. Nucleic Acids Res. 2016;44:2145–2159. doi: 10.1093/nar/gkv1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barth TK, et al. Identification of novel Drosophila centromere-associated proteins. PROTEOMICS. 2014;14:2167–2178. doi: 10.1002/pmic.201400052. [DOI] [PubMed] [Google Scholar]
- 19.Adewoye AB, Kyriacou CP, Tauber E. Identification and functional analysis of early gene expression induced by circadian light-resetting in Drosophila. BMC Genomics. 2015;16:570. doi: 10.1186/s12864-015-1787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graveley BR, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bates GP, et al. Huntington disease. Nat. Rev. Dis. Primer. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 22.Sandler JE, Stathopoulos A. Quantitative Single-Embryo Profile of Drosophila Genome Activation and the Dorsal–Ventral Patterning Network. Genetics. 2016;202:1575–1584. doi: 10.1534/genetics.116.186783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ciurciu A, Komonyi O, Pankotai T, Boros IM. The Drosophila histone acetyltransferase Gcn5 and transcriptional adaptor Ada2a are involved in nucleosomal histone H4 acetylation. Mol. Cell. Biol. 2006;26:9413–9423. doi: 10.1128/MCB.01401-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parthun MR. Histone Acetyltransferase 1: More than Just an Enzyme? Biochim. Biophys. Acta. 2012;1819:256–263. doi: 10.1016/j.bbagrm.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarez F, et al. Sequential establishment of marks on soluble histones H3 and H4. J. Biol. Chem. 2011;286:17714–17721. doi: 10.1074/jbc.M111.223453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apta‐Smith MJ, Hernandez‐Fernaud JR, Bowman AJ. Evidence for the nuclear import of histones H3.1 and H4 as monomers. EMBO J. 2018;37:e98714. doi: 10.15252/embj.201798714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soniat M, Cağatay T, Chook YM. Recognition Elements in the Histone H3 and H4 Tails for Seven Different Importins. J. Biol. Chem. 2016;291:21171–21183. doi: 10.1074/jbc.M116.730218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An S, Yoon J, Kim H, Song J-J, Choi U. Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123. Elife. 2017;6:e30244. doi: 10.7554/eLife.30244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia PAA, et al. Identification of multiple roles for histone acetyltransferase 1 in replication-coupled chromatin assembly. Nucleic Acids Res. 2017;45:9319–9335. doi: 10.1093/nar/gkx545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge Z, et al. Sites of Acetylation on Newly Synthesized Histone H4 Are Required for Chromatin Assembly and DNA Damage Response Signaling. Mol. Cell. Biol. 2013;33:3286–3298. doi: 10.1128/MCB.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo SJ, et al. ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget. 2016;7:70323–70335. doi: 10.18632/oncotarget.11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen-Carmon D, Meshorer E. Polyglutamine (polyQ) disorders. Nucleus. 2012;3:433–441. doi: 10.4161/nucl.21481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 34.Nucifora FC, Jr., et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 35.Taylor JP, et al. Aberrant histone acetylation, altered transcription, and retinal degeneration in a Drosophila model of polyglutamine disease are rescued by CREB-binding protein. Genes Dev. 2003;17:1463–1468. doi: 10.1101/gad.1087503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bodai L, Pallos J, Thompson LM, Marsh JL. Pcaf Modulates Polyglutamine Pathology in a Drosophila Model of Huntington’s Disease. Neurodegener. Dis. 2012;9:104–106. doi: 10.1159/000330505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington’s disease. Hum. Mol. Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia H, et al. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington’s disease. Neurobiol. Dis. 2012;46:351–361. doi: 10.1016/j.nbd.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mielcarek M, et al. SAHA Decreases HDAC 2 and 4 Levels In Vivo and Improves Molecular Phenotypes in the R6/2 Mouse Model of Huntington’s Disease. Plos One. 2011;6:e27746. doi: 10.1371/journal.pone.0027746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dompierre JP, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J. Neurosci. Off. J. Soc. Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadler AJ, et al. The acetyltransferase HAT1 moderates the NF-κB response by regulating the transcription factor PLZF. Nat. Commun. 2015;6:6795. doi: 10.1038/ncomms7795. [DOI] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data are deposited to the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under accession PRJNA551569. Other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.