Abstract
Acinetobacter baumannii is a frequent cause of the nosocomial infections. Herein, a novel isothermal amplification technique, multiple cross displacement amplification (MCDA) is employed for detecting all A. baumannii strains and identifying the strains harboring blaOXA-23-like gene. The duplex MCDA assay, which targets the pgaD and blaOXA-23-like genes, could identify the A. baumannii isolates and differentiate these isolates harboring blaOXA-23-like gene. The disposable lateral flow biosensors (LFB) were used for analyzing the MCDA products. A total of sixty-eight isolates, include fifty-three A. baumannii strains and fifteen non-A. baumannii strains, were employed to optimize MCDA methods and determine the sensitivity, specificity and feasibility. The optimal reaction condition is found to be 63 °C within 1 h, with limit of detection at 100 fg templates per tube for pgaD and blaOXA-23-like genes in pure cultures. The specificity of this assay is 100%. Moreover, the practical application of the duplex MCDA-LFB assay was evaluated using clinical samples, and the results obtained from duplex MCDA-LFB method were consistent with conventional culture-based technique. In sum, the duplex MCDA-LFB assay appears to be a reliable, rapid and specific technique to detect all A. baumannii strains and identify these strains harboring blaOXA-23-like gene for appropriate antibiotic therapy.
Subject terms: Applied microbiology, Clinical microbiology
Introduction
Acinetobacter. spp is a genus of gram-negative, strictly aerobic, non-fermenting, oxidase negative coccobacillus1. It is a diverse genus, several species of which belong to the normal flora on the skin and mucosa of human beings. Acinetobacter baumannii (A. baumannii), as one pathogenic species of Acinetobacter spp., is a frequent cause of the nosocomial infections, which manifest as pneumonia, bloodstream infection, skin or soft tissue infections, urinary tract infection and meningitis2. Numerous outbreaks caused by A. baumannii had been reported, which mainly occurred in the intensive care units (ICUs)2,3. With the widely occurrence of multi-drug resistance, A. baumannii is an increasing menace for the hospitalized patients, especially for the severely immuno-compromised ones in the ICUs. Besides, some A. baumannii strains are capable of forming biofilms and show tolerance to desiccation, which further contribute to their maintenance in the hospitals4,5.
Carbapenem antibiotics are the suggested candidates for the treatment of infections caused by the multidrug-resistant bacteria6. However, resistance to carbapenem is increasingly becoming a great concern, especially among the nosocomial strains belonging to the ESKAPE group of pathogens (i.e. Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, A. baumannii, Pseudomonas aeruginosa and Enterobacter species)7. Carbapenem resistance of A. baumannii is mainly mediated by the blaOXA genes (like the ISAbal-blaOXA-51-like, blaOXA-58-like, and blaOXA-40-like genes), which encode the carbapenem-hydrolyzing class D β-lactamases1. The blaOXA-23-like gene is one of the most prevalent β-lactamase genes on the genome (mostly on the plasmids) of carbapenem-resistant A. baumannii1,8. Specific and rapid identification of A. baumannii and the strains harboring blaOXA-23-like gene, will offer referential information on the therapeutic and control precautions for the nosocomial infections owing to the carbapenem-resistant A. baumannii.
Generally, reliable identification of target pathogen has been primarily relying on culture-based technique that often fails to provide valuable results in time9. Although molecular-based assays (such as PCR-based assays) are more sensitive and rapid than culture-based methods, these results are often not in conformity with culture-based methods and challenged by doubts on false-positive results10. Moreover, the nucleic acids-based methodologies often rely on expensive laboratory apparatus11. As progress is made towards better infection control, more rapid, simple and sensitive methods will be needed for reliably detect target A. baumannii and accurately identify carbapenem-resistant A. baumannii. Thus, further efforts are required for establishing next generation diagnostic technologies for the use in field, “on-site” and clinical settings, and replacing the existing diagnostic tools for target pathogen detection.
The recently established multiple cross displacement amplification (MCDA) assay was a powerful innovative nucleic acid amplification technique12. MCDA was based on strand displacement nucleic acid synthesis in the presence of Bst polymerase under isothermal conditions. A total of ten primers were employed to recognize ten distinct regions on the target gene. Given that this technique eliminated the use of a thermocycler, and did not require sophisticated training, thus MCDA showed the potential as a valuable diagnostic tool for field testing and point-of-care diagnosis13. Similar to other isothermal amplification methods like LAMP (loop-mediated isothermal amplification) and CPA (cross-priming amplification), the amplification products were a combination of different sequences with varying fragment sizes, which introduce an obstacle for the multiplex amplification14. The label-based lateral flow biosensor (LFB) makes it possible for the multiple identifications by detecting the amplification products labeled with different biomarkers3,14. The portable and dry-proof gold nanoparticle LFB is simple and reliable technique showing the amplification results within a few minutes of reaction. Thus, the LFB scheme was introduced in this study to identify the genus A. baumannii and its prevalent carbapenem resistant gene blaOXA-23-like at the same time. However, it was found that the classic double labeled primers for the LFB testing may introduce a false positive result owing to the hybridization between the labeled primers14. Recently, Wang et al. reported a new LFB technique of single labeling which can generate the double labeled amplicons as well and successfully eliminated the hybridization interaction of labeled primers14,15.
The aim of the study is to develop a rapid and simple duplex-MCDA-LFB assay for the detection of A. baumannii and differentiation of the strains harboring the prevalent carbapenem resistant gene blaOXA-23-like. The single labeling technique was introduced to avoid the hybridization between labeled primers. And sputum samples from clinical patients were used for the applicability evaluation.
Results
Validation and confirmation of pgaD- and blaOXA-23-like-MCDA products
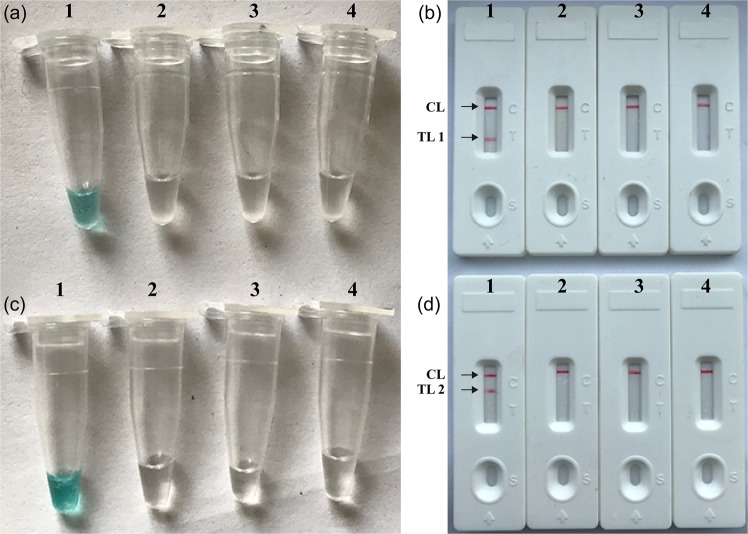
To demonstrate the feasibility of the pgaD- and blaOXA-23-like-MCDA primers, pgaD- and blaOXA-23-like-MCDA amplifications were conducted in the absence and presence of target DNAs for 60-min under the isothermal conditions (63 °C). As shown in Fig. 1, the positive pgaD- and blaOXA-23-like-MCDA tubes could be visualized with the unaided eye as being lake green, and the amplification vessels containing the blank control and negative control remained colorless (Fig. 1a,c). By lateral flow biosensor, two red bands (TL1 and CL) were observed in the positive pgaD-MCDA amplification (Fig. 1b), and two red lines (TL2 and CL) appeared on the lateral flow biosensor in the positive blaOXA-23-like-MCDA amplification (Fig. 1d). Particularly, only a red line (CL) was observed in the blank and negative controls (Fig. 1b,d). Together, our data verified that the pgaD- and blaOXA-23-like-MCDA primer sets were valuable candidates for development of the pgaD-MCDA-LFB, blaOXA-23-like-MCDA-LFB and duplex MCDA-LFB assays for target gene detection.
Figure 1.
Detection and confirmation of pgaD- and blaOXA-23-like-MCDA products. (a,c), Color change of pgaD- and blaOXA-23-like -MCDA tubes; (b,d), LFB applied for visual detection of pgaD- and blaOXA-23-like-MCDA products. Tube a1 (biosensor b1), positive amplification; tube a2 (biosensor b2), negative amplification (K. pneumoniae); tube a3 (biosensor b3), negative amplification (S. aureus); tube a4 (biosensor b4), negative control (DW); Tube c1 (biosensor d1), positive amplification; tube c2 (biosensor d2), negative amplification (K. pneumoniae); tube c3 (biosensor d3), negative amplification (S. aureus); tube c4 (biosensor d4), negative control (DW).
Screening of optimal amplification temperature
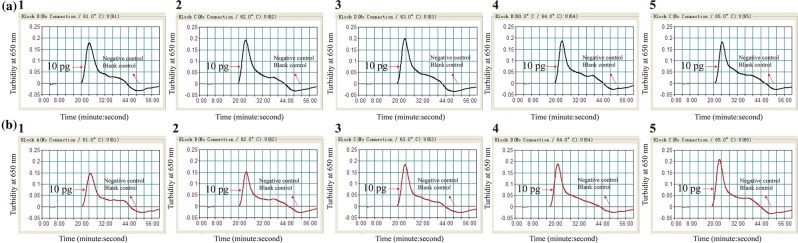
Then, we screened the optimal amplification temperatures of pgaD- and blaOXA-23-like- MCDA primer sets by observing the real time turbidity changes at different temperatures (61 °C to 65 °C with 1 °C interval) for 60 min. DNA templates of SG-AB001 at the level of 10 pg per tube were used for the optimal amplification temperature screening. As shown in Fig. 2, the optimal amplification temperatures for both pgaD- and blaOXA-23-like-MCDA primer sets were within the range of 62 °C to 64 °C. A amplification temperature of 63 °C was used in the following study.
Figure 2.
Optimal amplification temperature for pgaD- and blaOXA-23-like-MCDA primer sets. The MCDA amplifications for detection of pgaD (a) and blaOXA-23-like (b) were monitored by real-time measurement of turbidity and the corresponding curves of concentrations of templates were marked in the figures. The threshold value was 0.1 and the turbidity of >0.1 was considered as positive. Five kinetic graphs (1–5) were generated at various temperatures (61 °C-65 °C, 1 °C intervals) with target pathogens DNA. (a) graphs from 2 (62 °C) to 4 (64 °C) showed robust amplification; (b) graphs from 2 (62 °C) to 4 (64 °C) showed robust amplification.
Sensitivity of the single and duplex MCDA-LFB assay
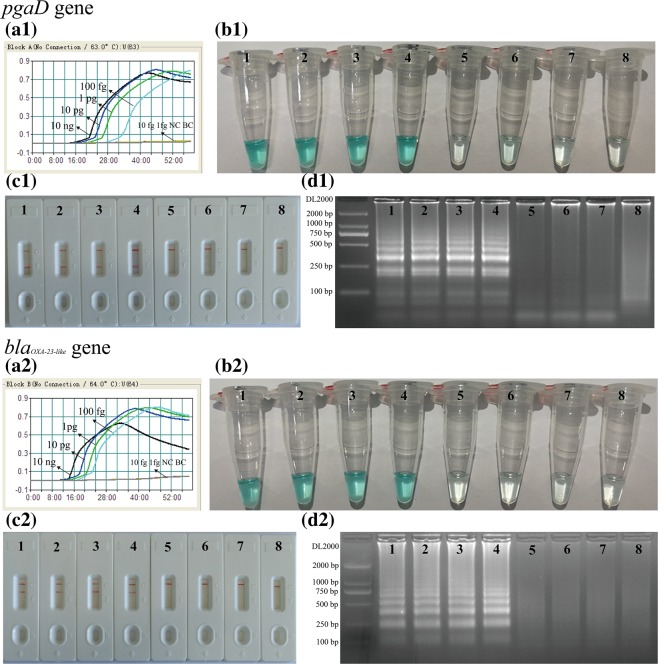
The limit of detection (LoD) of pgaD- and blaOXA-23-like-MCDA assays was firstly evaluated by examining the diluted DNA templates of SG-AB001 (10 ng/μL, 10 pg/μL, 1 pg/μL, 100 fg/μL, 10 fg/μL and 1 fg/μL). The reaction mixtures with DNA template of K. pneumoniae (ATCC2146) were used as negative control, and DW for blank control. The LoD for pgaD-MCDA assay was 100 fg (Fig. 3-a1, b1,c1, –d1). For positive amplifications of pgaD-MCDA assay, there were two red bands (TL1 and CL) appeared on LFB (Fig. 3-c1). Analytical sensitivity of blaOXA-23-like-MCDA assay was also 100 fg (Fig. 3-a2,b2,c2,d2), and two red bands (TL2 and CL) were seen in positive amplifications of blaOXA-23-like-MCDA reactions (Fig. 3-c2). By LFB, only a red line was visible in the negative and blank control for pgaD- and blaOXA-23-like-MCDA assays (Fig. 3-c1,c2). Analysis of pgaD- and blaOXA-23-like-MCDA results using LFB was in complete accordance with these monitoring techniques (real-time turbidity, color indicator and gel electrophoresis detection) employed in this report (Fig. 3).
Figure 3.
Detection of a single target in a MCDA reaction. Two sets of MCDA primers targeting the pgaD (a1,b1,c1,d1) and blaOXA-23-like (a2,b2,c2,d2) genes were used in different reactions and the serial dilutions (10 ng, 10 pg, 1 pg, 100 fg, 10 fg and 1 fg) of target templates were subjected to MCDA reactions. (a1) and (a2), real-time turbidity applied for analysis of pgaD- and blaOXA-23-like-MCDA products. (b1) and (b2), MG applied for analysis of pgaD- and blaOXA-23-like-MCDA products. (c1) and (c2), LFB applied for analysis of pgaD- and blaOXA-23-like-MCDA products. (d1) and (d2), gel electrophoresis applied for analysis of pgaD- and blaOXA-23-like-MCDA products. Signals/Tubes/Biosensors/Lanes 1–6: A. baumannii (SG-AB001) genomic templates (10 ng-1fg); Signal/Tube/Biosensor/Lane 7: negative control (K. pneumoniae); Signal/Tube/Biosensor/Lane 8: blank control (DW). The d1 and d2 were cropped from different gels, the full-length gels can be found as Supplementary Fig. S1.
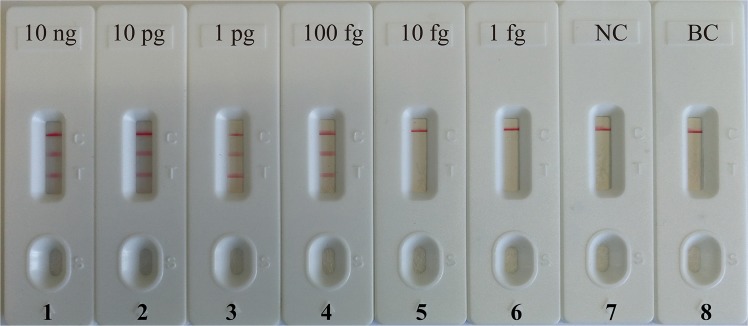
Following this, we examined the detection limit of the duplex MCDA reactions, which can simultaneously detect pgaD and blaOXA-23-like genes. The amplicons generated from duplex MCDA reactions were directly analyzed using the biosensor. As shown in Fig. 4, three red lines, including TL1, TL2 and CL, appeared on the LFB, indicating positive reactions for pgaD and blaOXA-23-like detection. However, only a red band (CL) appeared on the biosensor, reporting negative reactions at the concentration lower than 10 fg of templates per reaction, negative control and blank control. Analytical sensitivity of the duplexed MCDA-LFB method for simultaneous detecting pgaD gene and blaOXA-23-like gene was also 100 fg per reaction, which was consistent with the LoD of the single pgaD- and blaOXA-23-like-MCDA-LFB methods (Figs. 3 and 4).
Figure 4.
Detection of multiplex targets in a m-MCDA reaction. Two sets of MCDA primers targeting pgaD- and blaOXA-23-like-MCDA genes were simultaneously added to a reaction tube and the LoD of m-MCDA for simultaneously detecting pgaD and blaOXA-23-like genes was confirmed using LFB. Biosensors 1, 2, 3, 4, 5 and 6 represent DNA levels of 10 ng (A. baumannii SG-AB001), 10 pg (A. baumannii SG-AB001), 1 pg (A. baumannii SG-AB001), 100 fg (A. baumannii SG-AB001), 10 fg (A. baumannii SG-AB001) and 1 fg (A. baumannii SG-AB001); biosensor 7, negative control (K. pneumoniae); biosensor 8, blank control (DW). The LoD of m-MCDA assay for pgaD and blaOXA-23-like detection was 100 fg per vessel.
Specificity of the MCDA-LFB assay
Analytical specificity of duplex MCDA-LFB method is examined using extracted genomic DNA from 53 A. baumannii strains and 15 non-A. baumannii strains. All positive results were obtained only with the templates extracted from A. baumannii strains (Fig. 5 and Table 1). Three red bands (TL1, TL2 and CL) appeared on the biosensor, indicating the positive results for A. baumannii strains with the carbapenem resistance related gene blaOXA-23-like. TL1 and CL lines appeared on the biosensor, indicating the positive results for A. baumannii strains without the carbapenem resistance related gene blaOXA-23-like. Only CL line appeared on the biosensor, indicating the negative results for non-A. baumannii strains and negative control. The duplex MCDA-LFB assay could simultaneously detect and correctly identify pgaD and blaOXA-23-like genes in a single reaction system. No cross-reactions to non-A. baumannii isolates were obtained according the specificity analysis, thus the specificity of this assay was of 100% (Table 1). These data demonstrate that the duplex MCDA-LFB assay has high selectivity of A. baumannii strains.
Figure 5.
Specificity of duplex MCDA-LFB assay using different bacterial strains. The m-MCDA amplifications were carried out using different genomic DNA templates and the results were indicated using LFB. Biosensors 1–9, A. baumannii strains with blaOXA-23-like gene; Biosensors 10–12, A. baumannii strains without blaOXA-23-like gene; biosensors 13–27, Listeria monocytogenes, Bacillus cereus, Citrobacter braakii, Citrobacter freundii, Corynebacterium ammoniagenes, Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Providencia rettgeri, Pseudomonas aeruginosa, Serratia marcescens, Serratia marcescens, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus salivarius.
Table 1.
Bacterial strains used in the study.
| Strain ID | Strains | Sources of strains (No. of strains)a | Duplex MCDA-LFB b | |
|---|---|---|---|---|
| pgaD | blaOXA-23-like | |||
| 1 | Acinetobacter baumannii | SG-AB001 | P | P |
| 2–50 | Acinetobacter baumannii | Isolated strain(49) | P | P |
| 51–53 | Acinetobacter baumannii | Isolated strains (3) | P | N |
| 54 | Listeria monocytogenes | Isolated strains (1) | N | N |
| 55 | Bacillus cereus | Isolated strain (1) | N | N |
| 56 | Citrobacter braakii | Isolated strain (1) | N | N |
| 57 | Citrobacter freundii | Isolated strain (1) | N | N |
| 58 | Corynebacterium ammoniagenes | Isolated strain (1) | N | N |
| 59 | Escherichia coli | ATCC35218 | N | N |
| 60 | Klebsiella pneumoniae | ATCC2146 | N | N |
| 61 | Proteus mirabilis | Isolated strain (1) | N | N |
| 62 | Providencia rettgeri | Isolated strain (1) | N | N |
| 63 | Pseudomonas aeruginosa | ATCC27853 | N | N |
| 64 | Serratia marcescens | Isolated strain (1) | N | N |
| 65 | Serratia marcescens | Isolated strain (1) | N | N |
| 66 | Staphylococcus aureus | ATCC29213 | N | N |
| 67 | Staphylococcus epidermidis | Isolated strain (1) | N | N |
| 68 | Streptococcus salivarius | Isolated strain (1) | N | N |
aATCC, American Type Culture Collection; SG, Shougang Hospital; AB, Acinetobacter baumannii; bP, positive; N, negative. Only these strains belonging to the A. baumannii could be detected by the MCDA-LFB technique, indicating the extremely high selectivity of the method. Particularly, pgaD and blaOXA-23-like genes can be simultaneously detected and correctly identified in a reaction vessel.
The clinical application of the duplex-MCDA-LFB assay
In order to evaluate the clinical applicability of the duplex-MCDA-LFB assay, DNA templates extracted from 110 sputum samples were applied to the duplex-MCDA-LFB assay. As show in Table 2, a total of 28 samples were positive for A. baumannii strains, in which 26 samples were positive for blaOXA-23-likegenes. By the culture-method, 28 strains of A. baumannii were isolated from the 110 sputum samples, and 26 strains of A. baumannii possessed the blaOXA-23-likegene (Table 2). The 28 sputum samples were positive for A. baumannii by culture method, which was consistent with duplex MCDA-LFB detection.
Table 2.
Clinical evaluation of the MCDA-LFB assay.
| Methods | Sputum samples (n = 110) | |
|---|---|---|
| pgaD ( + ) | blaOXA-23-like ( + ) | |
| Culture | 28 | 26 |
| MCDA | 28 | 26 |
Discussion
The widely acquirement of lactamase production capability have jeopardized the treatment effect of most β-Lactams antibiotics, but few of the lactamase can hydrolyze the carbapenem molecules16. Thus, carbapenem antibiotics rank the last line antibiotics for the multi-drug resistant treatment, especially for the nosocomial infections in ICU6,16. However, resistance to the carbapenem antibioitcs among ESKAPE strains has become an increasingly concern in the past one decade16–18. Carbapenem resistant genes were acquired through horizontal transmission or mutation, which encodes carbapenemase and has the ability to hydrolyze the penicillin, cephalosporins and monobactams16–18. Among the ESKAPE strains, the carbapenem resistance is much more common among A. baumannii strains, the carbapenem resistant genes of which had been found to be able to transmit among different species19,20. Efficient identification of the carbapenem-resistant strains have been underlined to be one important strategy for the control of their dissemination and infection21,22.BlaOXA-23-like gene was one important carbapenem resistant gene, and many outbreaks were found to be caused by the blaOXA-23-like-harboring strains23,24. Thus, the prompt identification of A. baumannii from various samples and detection of carbapenem resistance is essential in cases of suspected infections.
Traditional detection methods, including culture-based techniques, colony morphology, micro-dilution resistance tests, and PCR-based assays (conventional PCR assays and real time PCR methods), are laborious and time-consuming. Herein, a rapid, simple, and specific technique is need for application in a hospital clinical laboratory and a basic laboratory. To obtain more such effective diagnostic tool, we developed a duplex MCDA-LFB method for the detection of A. baumannii specific gene pgaD and the carbapenem resistant gene blaOXA-23-like. In the MCDA-based system, the primer set of MCDA assay, which specially recognizes ten regions on pgaD and blaOXA-23-like genes, can offer a high degree of specificity (Figs. 5 and 6). The pgaD-MCDA primer set were designed based on the species-specific gene pgaD, which is unique to all A. baumannii isolates. Particularly, carbapenem-resistance in A. baumannii is primarily mediated by carbapenemase, which is encoded by blaOXA-23-like gene. Thus, blaOXA-23-like-MCDA primer set is designed using blaOXA-23-like gene, which is associated with carbapenem-resistance of A. baumannii strains. Analysis of MCDA assay was evaluated with the genomic templates extracted from 52 A. baumannii isolates and 15 non-A. baumannii strains, and the positive results were obtained from the assay of all A. baumannii strains but not for non-A. baumannii isolates. The duplex MCDA-LFB method targeting the pgaD gene identified A. baumannii with 100% specificity, and duplex-MCDA assay targeting the blaOXA-23-like gene associated with carbapenem resistance identified carbapenem-resistant A. baumannii with 100% specificity (Fig. 5). Importantly, the duplex MCDA-LFB assay developed here could detect all A. baumannii stains, and identify these strains with the carbapenem resistant gene blaOXA-23-like.
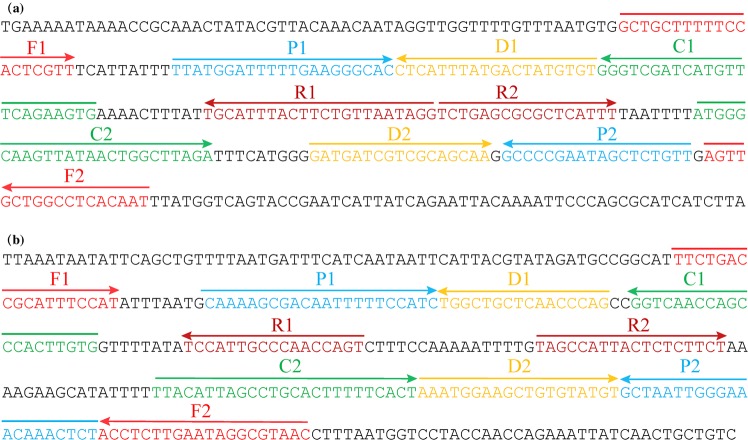
Figure 6.
Nucleotide sequence and location of pgaD and blaOXA-23-like genes used to design MCDA-LFB primers. The nucleotide sequences of the sense strands of pgaD (a) and blaOXA-23-like (b) are showed. The sites of primer sequences were underlined. Left arrows and right arrows showed complementary and sense sequences that are used.
In particular, the lateral flow biosensor (LFB) was employed to analyze the single and duplex MCDA products, because of its rapid results, simple operation, and ease of use in field setting and clinical laboratory. Detection of MCDA product using LFB is not only simpler and less error-prone, but also faster than detection using other analysis methods (such as real time turbidity and colorimetric indicator) employed in the current report. Moreover, lateral flow biosensor is more suitable than other monitoring techniques for simple, rapid and sensitive analysis of single and duplex MCDA products, because LFB eliminates the use of special reagent, instrument and process. LFB used in this study can simultaneously detect and correctly identify two targets in a single examination.
The results readout of the MCDA amplification using the LFBs are mainly based on two important steps: the capture of the amplification products on the test lines and its colorimetric indication25. The former step is based on the combination of the antibodies (anti-fluorescein or anti-digoxigenin antibodies embedded onto the test lines of LFBs) and antigens (FITC or digoxigenin labeled onto the amplification products, which will reach the test lines with the syphon reaction)25. Thus, the amplified MCDA products of pgaD (labeled with FITC and biotin) and blaOXA-23-like (labeled with digoxigenin and biotin) genes, can be specifically recognized and immobilized onto the TL1 and TL2 of LFBs, respectively. The gold nanoparticle being dyed with the crimson red will show red color when a high quantity of them were gathered together14. The high affinity of biotin (labeled onto the amplification products) and streptavidin (coated onto the gold nanoparticles) enable the gold-nanoparticles combined to the amplification products and show red bands on the test lines26. Biotins were usually labeled to the 5′-end of the primer, whereas there were occasions that the primer dimer or off-target effect may result in the false positivity of the LFBs14. Here we used the single-labeling method with biotins being labeled onto the biotin-14-dUTP in the reaction buffer, which removed the false positivity caused by the hybridization of labeled primers.
Analytical sensitivity of MCDA assays for independently identify pgaD and blaOXA-23-like genes were 100 fg of genomic templates per vessel, and LoD of LFB analysis for MCDA amplicons was in complete accordance with gel electrophoresis detection, colorimetric indicator (MG) analysis and real-time turbidity determination (Fig. 3). Analytical sensitivity of duplex MCDA method for simultaneously detecting pgaD and blaOXA-23-like genes were also 100 fg of DNA templates per reaction, which is conformity with the single pgaD-MCDA and blaOXA-23-like-MCDA detection (Figs. 3 and 4). The detection limit of the MCDA primers targeting the pgaD gene of A. baumannii is more sensitive than the LAMP-based assay for the rapid detection of A. baumannii targeting the blaOXA-51-likegene (50 pg per reaction)27. Besides, increasing number of studies reported that the blaOXA-51-like gene is harbored by other species of Acinetobacter spp., emphasizing that detection of the blaOXA-51-like gene on its own is not reliable for the identification of A. baumannii28. In this study, except for detecting the pgaD gene of A. baumannii, we developed the duplex MCDA detection for the prevalent carbapenem resistant gene blaOXA-23-like. These results indicate that the duplex-MCDA-LFB method can be good candidate for the rapid detection of A. baumannii and the strains harboring the blaOXA-23-like genes. The feasibility of duplex MCDA-LFB assay was successfully determined using clinical samples, and the results obtained from duplex MCDA-LFB assay are consistent with culture-based methods.
In sum, a duplex MCDA-LFB assay for simultaneous identification of A. baumannii strains and carbapenem-resistance based on pagD gene and blaOXA-23-like gene was successfully established. The duplex MCDA-LFB method established in this study displays high selectivity for target gene detection, and the LoD of the method is 100 fg per reaction with pure culture. Lateral flow biosensor is employed for analyzing the MCDA products, which was disposable, easy-to-use and objective. Hence, the duplex MCDA-LFB assay established here was a simple, rapid, sensitive and reliable method to detect all A. baumannii strains and identify carbapenem-resistance A. baumannii infection for appropriate antibiotic therapy.
Materials and Methods
Ethics
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Peking University Shougang Hospital.
Reagents and equipment preparation
The main reagents used in this study include the reaction buffer and Bst enzyme (Isothermal Amplification Kit, Haitaizhengyuan, Beijing), the Biotin-11-dUTP (Thermo Scientific, Shanghai), the colorimetric indicators (Haitaizhengyuan, Beijing), the nanoparticle LFB, the LFB running buffer, the Nano drop ND-1000 (Calibre, Beijing, China) and a Loopamp Realtime Turbidimeter LA-320C (Eiken Chemical Co., Ltd., Japan.
The LFB was constructed according to the instructions by Wang et al.’s report14. Briefly, the streptavidin-coated nanoparticles were adhered onto the conjugate pad. On the nitrocellulose membrane pad, there were three lines, including two test lines (conjugated with rabbit anti-fluorescein antibody and sheep anti-digoxigenin antibody, respectively) and one control line (conjugated with biotinylated bovine serum albumin). Finally, the assembled sample pad, conjugate pad, nitrocellulose membrane and the absorbent pad were cut (4 mm in width) and packaged in the plastic shells. The packaged biosensors were stored in dry environment at room temperature. The running buffer was the phosphate buffered saline (PBS) with the pH of 7.4.
Primers design and screening
Primer Primer 5.0 and PrimerExploer V4 were used for the design of MCDA primers targeting the A. baumannii specific gene pgaD (Accession No. FJ866500.1) and the carbapenem-resistant gene blaOXA-23-like (Accession No. NC_025109.1). The designed primers were then blasted on the NCBI to confirm their specificity. The oligomers were synthesized and purified by the company Ruiboxingke (Beijing, China).
Two sets of primers were designed for the two genes, respectively. A clinical-source strain of A. baumannii (SG-AB001), which harbors the blaOXA-23-like gene by the normal polymerase chain reaction (PCR) detection, was used for the primers screening both for the pgaD gene and the blaOXA-23-like gene. The location, the direction, the sequences and the modification of optimal primers were indicated in Fig. 6 and Table 3. Moreover, the genomic templates of Klebsiella pneumoniae (K. pneumoniae, ATCC2146) and Staphylococcus aureus (S. aureus) was used as the negative control, with distilled water as the blank control.
Table 3.
Primers used in the report.
| Gene | Primersa | Sequences and modifications (5′-3′)b | Lengthc |
|---|---|---|---|
| pgaD | p-F1 | GCTGCTTTTTCCACTCGTT | 19 nt |
| p-CP1 | CACTTCTGAAACATGATCGACCCTTATGGATTTTTGAAGGGCAC | 44 mer | |
| p-C1 | CACTTCTGAAACATGATCGACCC | 23 nt | |
| p-C1* | FITC-CACTTCTGAAACATGATCGACCC | 28 nt | |
| p-D1 | ACACATAGTCATAAATGAG | 19 nt | |
| p-R1 | CCTATTAACAGAAGTAAATGCA | 22 nt | |
| p-R2 | TCTGAGCGCGCTCATTT | 17 nt | |
| p-D2 | GATGATCGTCGCAGCAA | 17 nt | |
| p-C2 | ATGGGCAAGTTATAACTGGCTTAGA | 25 nt | |
| p-CP2 | ATGGGCAAGTTATAACTGGCTTAGAAACAGAGCTATTCGGGGC | 43 mer | |
| p-F2 | ATTGTGAGGCCAGCAACT | 18 nt | |
| blaOXA-23-like | b-F1 | TTCTGACCGCATTTCCAT | 18 nt |
| b-CP1 | CACAAGTGGGCTGGTTGACCCAAAAGCGACAATTTTTCCATC | 42 mer | |
| b-C1 | CACAAGTGGGCTGGTTGACC | 20 nt | |
| b-C1* | Dig-CACAAGTGGGCTGGTTGACC | 24 nt | |
| b-D1 | CTGGGTTGAGCAGCCA | 16 nt | |
| b-R1 | ACTGGTTGGGCAATGGA | 17 nt | |
| b-R2 | TAGCCATTACTCTCTTCT | 18 nt | |
| b-D2 | AAATGGAAGCTGTGTATGT | 19 nt | |
| b-C2 | TTACATTAGCCTGCACTTTTTCACT | 25 nt | |
| b-CP2 | TTACATTAGCCTGCACTTTTTCACTAGAGTTTGTTTCCCAATTAGC | 46 mer | |
| b-F2 | GTTACGCCTATTCAAGAGGT | 20 nt |
ap, pgaD gene; b, blaoxA-23-like gene; p-C1*, 5′-labeled with FITC when used in MCDA-LFB assay; b-C1*, 5′-labeled with Dig when used in MCDA-LFB assay;
bFITC, fluorescein isothiocyanate; Dig, digoxigenin;
cmer, monomeric unit; nt, nucleotide.
The reaction systems for the MCDA-LFB assays targeting the pgaD and blaOXA-23-like Genes
In order to examine the feasibility of two sets of MCDA primers, the single MCDA assay either for the pgaD gene and the blaOXA-23-like gene was conducted as the following description. Briefly, the 25 μL reaction system includes 1.6 μM each of CP1 and CP2, 0.4 μM each of F1 and F2, 0.8 μM each of C1*, C2, D1, D2, R1 and R2, 12.5 μL 2 × Reaction Buffer,0.4 mM of biotin-11-dUTP,1.25 μL Bst DNA polymerase (10 U) and 1 μL colorimetric indicator (Malachite Green, MG). The amplification mixture of standard MCDA was carried out at 63 °C and then at 85 °C for 5 min to stop the reaction.
For the duplex MCDA-LFB detection of the pgaD and the blaOXA-23-like genes, the 25 μL reaction system included 12.5 μL 2 × Reaction Buffer, 1.25 μL Bst DNA polymerase (10 U), 0.8 μM each of p-CP1 and p-CP2, 0.2 μM each of p-F1 and p-F2, 0.4 μM each of p-C1*, p-C2, p-D1, p-D2, p-R1 and p-R2, 1.6 μM each of b-CP1 and b-CP2, 0.2 μM each of b-F1 and b-F2, 0.8 μM each of b-C1*, b-C2, b-D1, b-D2, b-R1 and b-R2,0.4 mM of biotin-11-dUTP and 1 μl DNA template. The duplex MCDA mixtures were carried out at 63◦C, and amplification mixtures without the DNA template were used as the blank control. The lowest detectable template amount was tested in triplicate.
A total of four monitoring methods, including real-time turbidity (LA-C320), gel electrophoresis, colorimetric indicator (MG) and lateral flow biosensor (LFB) detection, were used for analyzing the single and duplex MCDA amplicons. During the isothermal amplification, turbidimeter was used to record the real time turbidity changes, which was higher than 0.1 for the positive tubes. By the colorimetric indicator, the positive reactions showed lake green, while the negative reactions faded into colorless. By gel electrophoresis, 3 μL of the MCDA amplification products were embedded into 2% agarose gel and run at 80 volt for 50 minutes. By the LFB, 0.5 μL of the MCDA products were deposited onto the sample pad of the LFB, followed by adding 80 μL of the running buffer (PBS), waiting for 2 to 5 minutes. During the siphon reaction on the LFB, the double strand DNA amplicons of pgaD gene, being labeled with FITC and biotin-SA- nanoparticles, will be caught by the first test line (TL 1) which then turns into red14. The double strand DNA amplicons of blaOXA-23-like, being labeled with digoxin and biotin-SA-DNPs, will be caught by the second test line (TL 2) and showing the red band. The third line referred to the control line, which will capture the streptavidin-coated nanoparticles, ensuring that the LFB is operating well.
Screening of optimal amplification temperature
In this report, we explored the optimal amplification temperature for pgaD-MCDA and blaOXA-23-like-MCDA primer sets. MCDA amplifications were performed at a constant temperature from 61 °C to 65 °C for 60 min, then incubated at 85 °C for 5 min to stop the MCDA reaction. MCDA mixtures with 1 μL double distilled water (DW) were used as blank control (BC), and 1 μL of DNA templates derived from K. pneumoniae and S. aureus were selected as negative controls (NC).
Sensitivity of the primers for MCDA test
DNA templates of strain SG-AB001, which harbor both the pgaD and blaOXA-23-like genes, was serially diluted with distilled water ranging from 10 ng to 1 fg. The single and duplex MCDA-LFB assays were conducted by adding 1 μL of the diluted DNA into the reaction systems. Analytical sensitivity evaluation was repeated for three times.
Specificity of the duplex MCDA assay
DNA templates extracted from 53 strains of A. baumannii, which were isolated from clinical patients, were firstly tested for the presence of blaOXA-23-like with PCR screening. The PCR tests were performed according to Li, et al.’s reports27. Then, these strains were applied for the duplex-MCDA-LFB test for pgaD and blaOXA-23-like genes. Moreover, another 15 strains of non-A. baumannii were tested for the duplex MCDA-LFB to verify the specificity of the primers for the pgaD and blaOXA-23-like genes. All the tests were repeated in triplicate.
The clinical application of the duplex-MCDA-LFB assay
In order to validate the clinical feasibility of the duplex MCDA technique developed in this report, DNA templates of 110 clinical sputum samples were used to evaluate the duplex MCDA-LFB assay. Particularly, these clinical sputum samples have been cultured for A. baumannii with the traditional clinical cultural-based methods. The results of the duplex MCDA-LFB were further confirmed with the single MCDA-LFB tests for the pgaD gene and blaOXA-23-like gene, independently. All the genomic-based tests were repeated twice independently.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Numbers 81760376), the Beijing Natural Science Foundation (grant number 7192240) and the Peking University Shougang Hospital (Grant Numbers SGYYQ201903, 2017-Yuan-LC-08).
Author contributions
S.H. designed the research and wrote the manuscript; L.N. performed the experiments; F.Z. and L.Y. analyzed the data; J.N., C.W., N.G., X.Z., L.W., T.B. and H.W. contributed the reagents and materials; J.G. supervised the study. All authors reviewed the manuscript.
Data availability
The datasets used during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shoukui Hu, Lina Niu, Fan Zhao and Linlin Yan.
Supplementary information
is available for this paper at 10.1038/s41598-019-54465-8.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a Successful Pathogen. Clinical Microbiology Reviews. 2008;21:538–582. doi: 10.1128/cmr.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianco A, et al. Control of carbapenem-resistant Acinetobacter baumannii outbreak in an intensive care unit of a teaching hospital in Southern Italy. BMC infectious diseases. 2016;16:747. doi: 10.1186/s12879-016-2036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molter G, et al. Outbreak of carbapenem-resistant Acinetobacter baumannii in the intensive care unit: a multi-level strategic management approach. Journal of Hospital Infection. 2016;92:194–198. doi: 10.1016/j.jhin.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on Dry Surfaces: Comparison of Outbreak and Sporadic Isolates. Journal of clinical microbiology. 1998;36:1938–1941. doi: 10.1128/jcm.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinal P, Martí S, Vila J. Effect of biofilm formation on the survival of Acinetobacter baumannii on dry surfaces. Journal of Hospital Infection. 2012;80:56–60. doi: 10.1016/j.jhin.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Kim HJ, Kim HS, Lee JM, Yoon SS, Yong D. Rapid Detection of Pseudomonas aeruginosa and Acinetobacter baumannii Harboring blaVIM-2, blaIMP-1 and blaOXA-23 Genes by Using Loop-Mediated Isothermal Amplification Methods. Ann Lab Med. 2016;36:15–22. doi: 10.3343/alm.2016.36.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush K. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Current Opinion in Microbiology. 2010;13:558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto N, et al. Clinical Specimen-Direct LAMP: A Useful Tool for the Surveillance of blaOXA-23-Positive Carbapenem-Resistant Acinetobacter baumannii. PloS one. 2015;10:e0133204. doi: 10.1371/journal.pone.0133204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, et al. Real-time fluorescence loop mediated isothermal amplification for the detection of Acinetobacter baumannii. PLoS One. 2013;8:e66406. doi: 10.1371/journal.pone.0066406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang HC, et al. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J Clin Microbiol. 2005;43:1632–1639. doi: 10.1128/JCM.43.4.1632-1639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins PG, Wisplinghoff H, Krut O, Seifert H. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin Microbiol Infect. 2007;13:1199–1201. doi: 10.1111/j.1469-0691.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, et al. Rapid and Sensitive Isothermal Detection of Nucleic-acid Sequence by Multiple Cross Displacement Amplification. Sci Rep. 2015;5:11902. doi: 10.1038/srep11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wang Y, Zhang L, Xu J, Ye C. Visual and multiplex detection of nucleic acid sequence by multiple cross displacement amplification coupled with gold nanoparticle-based lateral flow biosensor. Sensors and Actuators B: Chemical. 2017;241:1283–1293. doi: 10.1016/j.snb.2016.10.001. [DOI] [Google Scholar]
- 14.Wang Yi, Li Hui, Wang Yan, Xu Huaqing, Xu Jianguo, Ye Changyun. Antarctic thermolabile uracil-DNA-glycosylase-supplemented multiple cross displacement amplification using a label-based nanoparticle lateral flow biosensor for the simultaneous detection of nucleic acid sequences and elimination of carryover contamination. Nano Research. 2018;11(5):2632–2647. doi: 10.1007/s12274-017-1893-z. [DOI] [Google Scholar]
- 15.Wang Y, et al. Loop-mediated isothermal amplification using self-avoiding molecular recognition systems and antarctic thermal sensitive uracil-DNA-glycosylase for detection of nucleic acid with prevention of carryover contamination. Analytica Chimica Acta. 2017;996:74–87. doi: 10.1016/j.aca.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 16.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends in Molecular Medicine. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Rizek C, et al. Characterization of carbapenem-resistant Pseudomonas aeruginosa clinical isolates, carrying multiple genes coding for this antibiotic resistance. Annals of Clinical Microbiology and Antimicrobials. 2014;13:43. doi: 10.1186/s12941-014-0043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han JH, et al. Epidemiology of carbapenem-resistant Klebsiella pneumoniae in a network of long-term acute care hospitals. Clinical Infectious Diseases. 2017;64:839–844. doi: 10.1093/cid/ciw856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y-T, et al. Emergence of Carbapenem-Resistant Non-baumannii Species of Acinetobacter Harboring a blaOXA-51-Like Gene That Is Intrinsic to A. baumannii. Antimicrobial agents and chemotherapy. 2012;56:1124–1127. doi: 10.1128/aac.00622-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y-T, et al. First Identification of bla OXA-51-like in Non-baumannii Acinetobacter spp. Journal of Chemotherapy. 2009;21:514–520. doi: 10.1179/joc.2009.21.5.514. [DOI] [PubMed] [Google Scholar]
- 21.Ghaith DM, Hassan RM, Hasaninb AM. Rapid identification of nosocomial Acinetobacter baumannii isolated from a surgical intensive care unit in Egypt. Annals of Saudi medicine. 2015;35:440. doi: 10.5144/0256-4947.2015.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. International journal of antimicrobial agents. 2015;45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 23.Kohlenberg A, et al. Outbreak of carbapenem-resistant Acinetobacter baumannii carrying the carbapenemase OXA-23 in a German university medical centre. Journal of medical microbiology. 2009;58:1499–1507. doi: 10.1099/jmm.0.012302-0. [DOI] [PubMed] [Google Scholar]
- 24.Gokmen TG, Akcimen B, Kayar B, Marzi M, Koksal F. The outbreak of Acinetobacter baumannii producing OXA-23 and OXA-51 type carbapenemases in a state hospital. Journal of Experimental and Clinical Medicine (Turkey). 2016;33:157–161. doi: 10.5835/jecm.omu.33.03.006. [DOI] [Google Scholar]
- 25.Wang Y, et al. Loop-mediated isothermal amplification label-based gold nanoparticles lateral flow biosensor for detection of Enterococcus faecalis and Staphylococcus aureus. Frontiers in microbiology. 2017;8:192. doi: 10.3389/fcimb.2018.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chilkoti A, Tan PH, Stayton PS. Site-directed mutagenesis studies of the high-affinity streptavidin-biotin complex: contributions of tryptophan residues 79, 108, and 120. Proceedings of the National Academy of Sciences. 1995;92:1754–1758. doi: 10.1073/pnas.92.5.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P, et al. Rapid detection of Acinetobacter baumannii and molecular epidemiology of carbapenem-resistant A. baumannii in two comprehensive hospitals of Beijing, China. Frontiers in microbiology. 2015;6:997. doi: 10.3389/fmicb.2015.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zander E, Higgins PG, Fernández-González A, Seifert H. Detection of intrinsic blaOXA-51-like by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. International Journal of Medical Microbiology. 2013;303:88–89. doi: 10.1016/j.ijmm.2012.12.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.