Abstract
Non-SMC condensin I complex subunit H (NCAPH) is a vital gene associated with chromosome stability and is required for proper chromosome condensation and segregation. However, the mechanisms through which NCAPH affects pancreatic cancer (PC) and its molecular function remain unclear. In this study, we examined the role of NCAPH in PC cells. Our results showed that NCAPH was overexpressed in clinical PC specimens (GEPIA) and cell lines. In addition, in NCAPH-knockdown cells, colony formation and proliferation were inhibited, and the cell cycle was arrested at the S and G2/M phases owing to failure of mature chromosome condensation (MCC) in poorly condensed chromosomes. Increased cell death in NCAPH-knockdown cells was found to help initiate apoptosis through the activation of caspase-3 and PARP cleavage. Furthermore, NCAPH-knockdown cells showed an increase in chromosomal aberrations and DNA damage via activation of the DNA damage response (Chk1/Chk2) signaling pathways. These data demonstrated that NCAPH played an important role in cell cycle progression and DNA damage by maintaining chromosomal stability through progression of MCC from poorly condensed chromosomes. Ultimately, NCAPH knockdown induced apoptotic cell death, which was partially mediated by caspase-dependent pathways. These findings highlight the potential role of NCAPH as a therapeutic target for PC.
Subject terms: Targeted therapies, Apoptosis, Chromosome segregation
Introduction
More than 90% of pancreatic cancer (PC) cases are classified as pancreatic ductal adenocarcinoma (PDA), one of the most lethal human malignant tumors with a poor prognosis1. The 5-year relative survival rate for patients with PC was less than 8% during the most recently reported study period (2007–2013)2. Unfortunately, compared with that for other malignancies, the survival rate of patients with PC has not improved in recent decades3. Typically, patients are diagnosed with PC at a late stage of the disease, and the spread of highly metastatic PC cells into the lymphatic system and adjacent organs limits the choice of effective treatments4. The poor prognosis of patients with PC is related to its late detection, high metastatic potential of the cancer, and resistance of the tumor cells to chemotherapy and radiation therapy5–8. Therefore, to improve clinical treatment for patients with PC, identifying new biomarkers and potential therapeutic targets is essential.
Reliable chromosome condensation is an essential cellular function that ensures compaction and separation of sister chromosomes, which is important for maintaining genomic integrity during mitosis. Multisubunit protein complexes, known as condensins, play a fundamental role in this process in cooperation with other chromosomal components9,10. The two types of condensin complexes present in multicellular organisms11,12, condensins I and II, share the same structural maintenance of chromosome (SMC) subunits but have different sets of non-SMC subunits; condensin complex I is comprised of NCAPD2, NCAPG, and NCAPH, whereas condensin complex II is comprised of NCAPG2, NCAPD3, and NCAPH29. Each subunit of condensins I and II is highly conserved in a variety of organisms from yeasts to humans11. SMC2 and SMC4 (SMC core subunits) belong to a large family of chromosomal ATPases9, and SMC proteins form a heterodimeric structure that adopts a V-shaped structure with two long coiled-coil arms12. One of the non-SMC subunits of each of the human condensins, NCAPH in condensin I and NCAPH2 in condensin II, belongs to the kleisin family of proteins11,13 and binds the ATP-binding cassette (ABC)-transporter-like ATPase domains at the other end of the coils14. A kleisin subunit is composed of conserved N- and C-terminal globular domains separated by a variable linker region in different organisms13. The other two non-SMC subunits of each of the human condensins, NCAPD2 and NCAPG in condensin I and NCAP-D3 and NCAPG2 in condensin II, share a structural motif called the HEAT repeat, belonging to HEAT repeat proteins15,16. The HEAT repeats are repetitive arrays of short amphiphilic α-helices. According to a recent study, the Ycg1-Brn1 (NCAPG/G2-NCAPH/H2) subcomplex of condensin binds directly to DNA and is essential for the stable association of condensin complexes with chromosomes and consequently condensin function14.
The two condensins differ in expression depending on dynamic cell changes during the cell division cycle17–20. Condensin II is predominately found in the nucleus during interphase and binds to chromosomes during mitosis19–21. Condensin I is located in the cytoplasm and binds to the chromosome after nuclear envelope breakdown22. However, recent studies have shown that a small fraction of condensin I remains in the nucleus during interphase and plays a role in gene regulation and chromosome condensation23. Condensins I and II differ in their localization within the chromatid axis; condensin II is centrally confined, whereas condensin I localizes to the chromatid diameter from the center, enabling achievement of maximum chromosome compaction upon sister chromatid segregation23,24.
Recent studies have also suggested that subunits of condensin complexes I and II can serve as new potential biomarkers and therapeutic targets for some human cancers25–28. In particular, the depletion of NCAPH inhibits the proliferation and migration of colon cancer (CC) cells in vitro and blocks CC xenograft tumor formation29. However, to the best of our knowledge, the effects of NCAPH on PC pathogenesis and its mechanisms have not been previously reported.
Accordingly, in this study, we aimed to evaluate differences in the expression of NCAPH in PC tissues and cell lines compared with that in human pancreatic duct epithelial (HPDE) cells, examined the roles of NCAPH in PC, and assessed the mechanisms through which NCAPH functions in this context.
Results
NCAPH expression was upregulated in PC tissues and cell lines
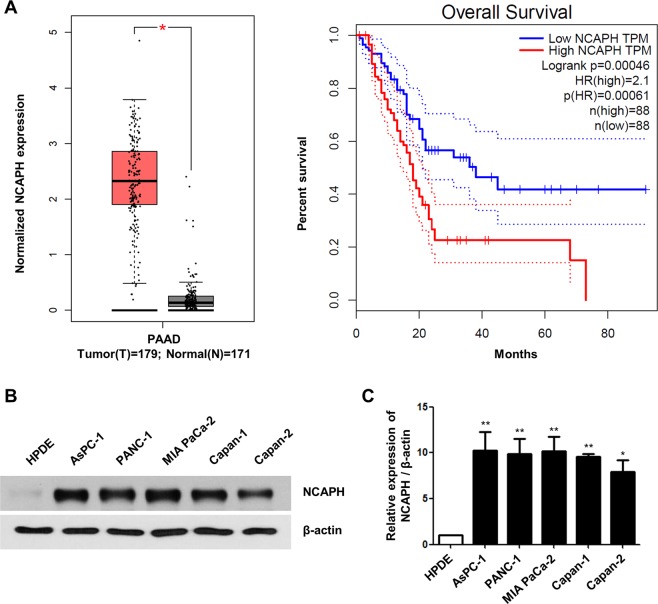
First, we analyzed the mRNA expression of all subunits of condensin I and II complexes in PC and normal tissues using a publicly available database (The Cancer Genome Atlas [TCGA]). According to the data obtained from TCGA, the expression of NCAPH was significantly higher than that of the condensin subunits in all pancreatic adenocarcinoma (PAAD) tumor types (n = 179) compared with that in their normal tissue counterparts obtained from TCGA and that obtained from GTEx data (n = 171; Fig. 1A and Supplementary Fig. 1A). In addition, the overall survival (OS) of patients with PC with high NCAPH expression was significantly lower than that of patients with low NCAPH expression (Fig. 1A). Among all subunits of condensins I and II, NCAPH elicited the greatest difference in OS, based on its mRNA expression (Fig. 1A and Supplementary Fig. 1A). We also examined condensin I (NCAPH, NCAPD2, NCAPG) and condensin II (NCAPH2, NCAPG2) protein levels in PC cell lines, including AsPC-1, PANC-1, MIA PaCa-2, Capan-1, and Capan-2 cells. The results showed that protein expression of condensin I rather than II was higher in various PC cell lines compared with its expression in HPDE cells, and NCAPH was markedly upregulated by approximately 10-fold (Fig. 1B,C, and Supplementary Fig. 1B).
Figure 1.
NCAPH expression is elevated in PC tissues and cell lines. (A) Expression levels of NCAPH in PC tissues (n = 179) and normal tissues (n = 171) and OS based on the TCGA database. Each dot represents sample expression. The OS of patients with low or high expression levels of NCAPH was analyzed by the Kaplan–Meier method and a log rank test in PAAD. The dotted high line is a sample with a higher expression level than the median value of TPM and is considered a high expression cohort, whereas the dotted low line is a sample with a lower expression level than the median value of TPM and is considered a low expression cohort. Median values are indicated by full lines. *P < 0.005. TCGA, The Cancer Genome Atlas; PAAD, Pancreatic adenocarcinoma; TPM, transcripts per million; OS, overall survival; HR, hazard ratio. (B) NCAPH protein expression was analyzed by western blotting in HPDE cells and different PC cell lines (AsPC-1, PANC-1, MIA PaCa-2, Capan-1, and Capan-2 cells). Cell lysates were immunoblotted with the indicated antibodies. (C) Frequency of NCAPH protein expression in HPDE and PC cell lines. The NCAPH/β-actin ratio was determined by densitometric analysis using ImageJ. Error bars represent standard deviations of the means of three biological replicates. Values represent means ± SEMs. *P < 0.005, **P < 0.001. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni’s multiple comparison tests.
NCAPH knockdown inhibited colony formation and proliferation in PC cells
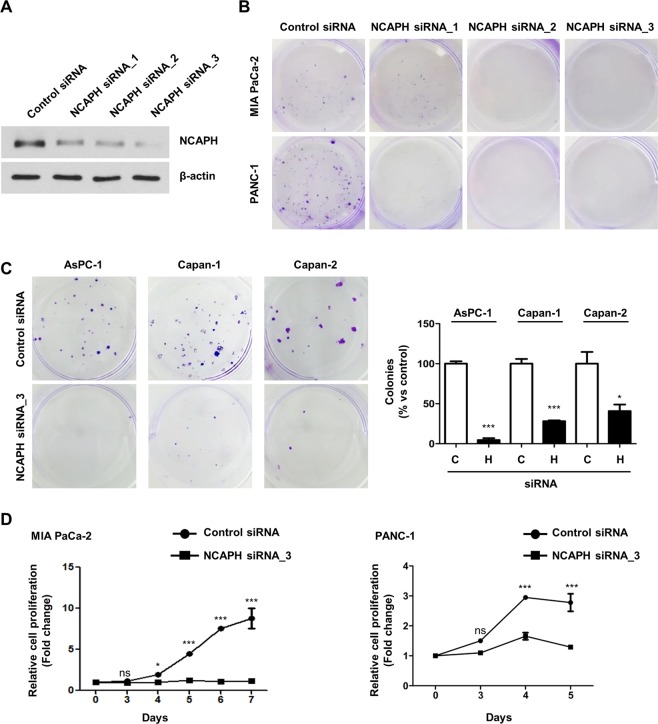
To evaluate the effects of NCAPH knockdown on the survival and growth of PC cells, we employed colony formation assays and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays using three NCAPH siRNAs specifically targeting NCAPH. Among these siRNAs, NCAPH siRNA_3 resulted in the most significant inhibition of NCAPH expression and colony formation in MIA PaCa-2 and PANC-1 cells (Fig. 2A,B, and Supplemental Fig. 2A) and was therefore selected for subsequent studies. Indeed, NCAPH knockdown significantly suppressed the colony formation ability of several PC cell lines (e.g., AsPC-1, Capan-1, and Capan-2 cells; Fig. 2C). NCAPH knockdown significantly inhibited the proliferation of PC cell lines, such as MIA PaCa-2 and PANC-1, as demonstrated by MTT assay 5–7 days after siRNA transfection (Fig. 2D). Additionally, we performed a reconstitution experiment after NCAPH siRNA transfection. For this experiment, we constructed a lentiviral vector and NCAPH. When NCAPH was depleted, the proliferation of PC cells was significantly reduced. In contrast, Lenti-NCAPH was able to partially recover the reduced PC cell proliferation (Fig. 2E). Expression of NCAPH protein was also restored by Lenti-NCAPH (Fig. 2F). These results indicated that NCAPH played essential roles in the colony formation and proliferation of PC cells.
Figure 2.
Effects of NCAPH on colony formation ability and proliferation of PC cells. (A) MIA PaCa-2 cells were transfected with three different siRNA constructs to knockdown NCAPH expression, and the expression levels of NCAPH protein were measured 48 h later by western blotting. Cell lysates were immunoblotted with the indicated antibodies. (B) Colony formation assays were performed on MIA PaCa-2 and PANC-1 cells transfected with the control and three different NCAPH siRNAs (NCAPH siRNA_1, siRNA_2, and siRNA _3). (C) Colony formation assays were performed in AsPC-1, Capan-1, and Capan-2 cells with control siRNA and NCAPH siRNA_3. The colonies were imaged and counted. Values represent means ± SEMs. Statistical analysis was performed using unpaired t-tests. ***P < 0.0001. (D) MTT assays were performed to determine proliferation of control and NCAPH-knockdown PC cells for 5 or 7 days. The curve was based on biological triplicates with values expressed as means ± SEMs. *P < 0.005, ***P < 0.0001. Statistical analysis was performed using two-way analysis of variance with Bonferroni’s multiple comparison tests.
NCAPH knockdown arrested cells in S and G2/M phases and promoted cell apoptosis
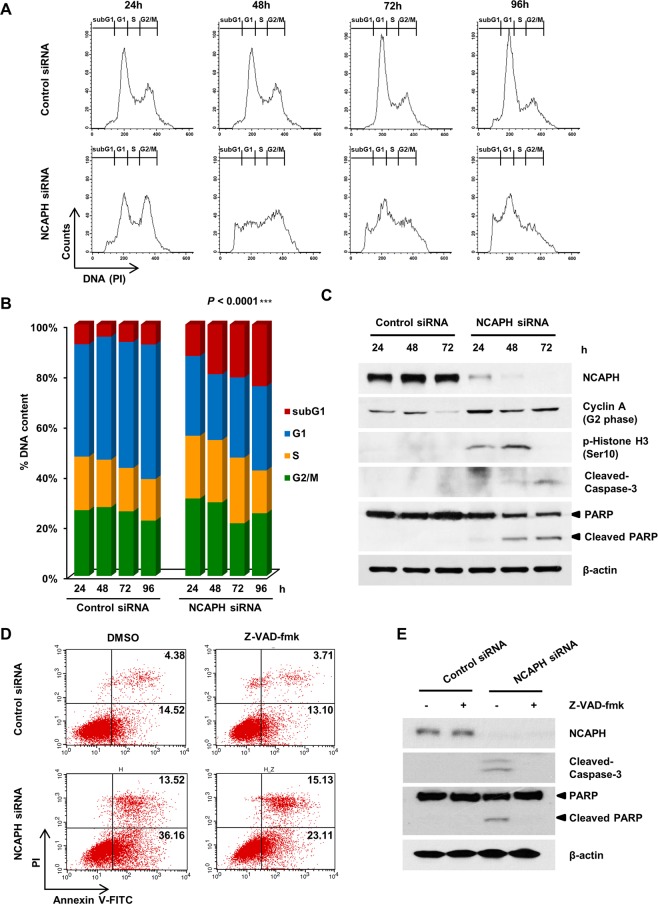
We performed flow cytometry analysis to investigate the effects of NCAPH on cell cycle regulation and apoptosis in PC cells. Quantification of cells at the subG1, G1, S, and G2/M phases of the cell cycle revealed that the number of NCAPH-knockdown cells accumulated in the S and G2/M populations and that the subG1 population (apoptotic cells) significantly increased compared with that in the control, whereas the G1 cell population was decreased in a concentration-dependent manner (Fig. 3A,B). In addition, we investigated the molecular mechanisms altered by NCAPH knockdown in cell cycle regulation by evaluating the expression of cyclin A and phospho-histone H3 (Ser10) proteins, which regulate the G2/M phase transition, by western blotting analysis. Cyclin A and phospho-histone H3 (Ser10) levels significantly increased in NCAPH-knockdown cells 24–48 h after transfection (Fig. 3C). Collectively, these observations suggested that NCAPH knockdown caused S and G2/M phase arrest, which could partly explain the reduced viability and enhanced cell death in PC cells.
Figure 3.
Knockdown of NCAPH induces S and G2/M phase arrest and apoptosis. (A) MIA PaCa-2 cells were transfected with control siRNA and NCAPH siRNA and then subjected to flow cytometric analysis at 24, 48, 72 and 96 h. The flow cytometry plots and data are representative of at least three separate experiments. (B) Bar graph showing the percentages of cells in sub-G1 (apoptosis, red), G1 (blue), S (orange), and G2/M (green) phases. The experiments were repeated three times, and mean values of the results are presented. ***P < 0.0001, two-way analysis of variance. (C) Western blot analysis was used to determine the levels of NCAPH, cyclin A, phospho-histone H3, caspase-3, PARP, and β-actin in MIA PaCa-2 cells after transfection with control siRNA or NCAPH siRNA for 24, 48, and 72 h. Cell lysates were immunoblotted with the indicated antibodies. (D) Annexin V staining was performed to measure apoptosis using flow cytometry. Z-VAD-Fmk (20 μM) was treated with cells for 24 h. LL (lower left) represents live, LR (lower right) represents early-apoptotic cells, UR (upper right) represents late stage apoptotic cells. (E) Western blot analysis was used to determine the levels of NCAPH, caspase-3, PARP, and β-actin in MIA PaCa-2 cells after transfection with control siRNA or NCAPH siRNA. Cells were treated with Z-VAD-Fmk (20 μM) for 24 h.
To further determine whether the cell death observed in NCAPH-knockdown cells was induced by apoptosis, apoptosis levels were investigated using the Annexin V-FITC/PI double staining technique. NCAPH knockdown by siRNA was observed in the early/late apoptosis induction effect (UR and LR quadrants; Fig. 3D). We also identified caspase-3 activity and poly-ADP ribose polymerase (PARP) cleavage events to ensure apoptotic cell death30 and found that caspase-3 activation and PARP cleavage were significantly increased in NCAPH-knockdown cells (Fig. 3C). To confirm these results, cells were incubated with 20 μM Z-VAD-Fmk, a pan-caspase inhibitor, for 24 h, which was able to suppress the early apoptosis induced by NCAPH knockdown (LR quadrant; Fig. 3D). Additionally, protein expression of caspase-3 and cleaved PARP was also significantly decreased by Z-VAD-Fmk (Fig. 3E). These data clearly indicated the partial involvement of caspase-dependent apoptosis in NCAPH knockdown-induced cell death.
NCAPH was involved in progression of mature chromosome condensation (MCC) from poorly condensed chromosomes
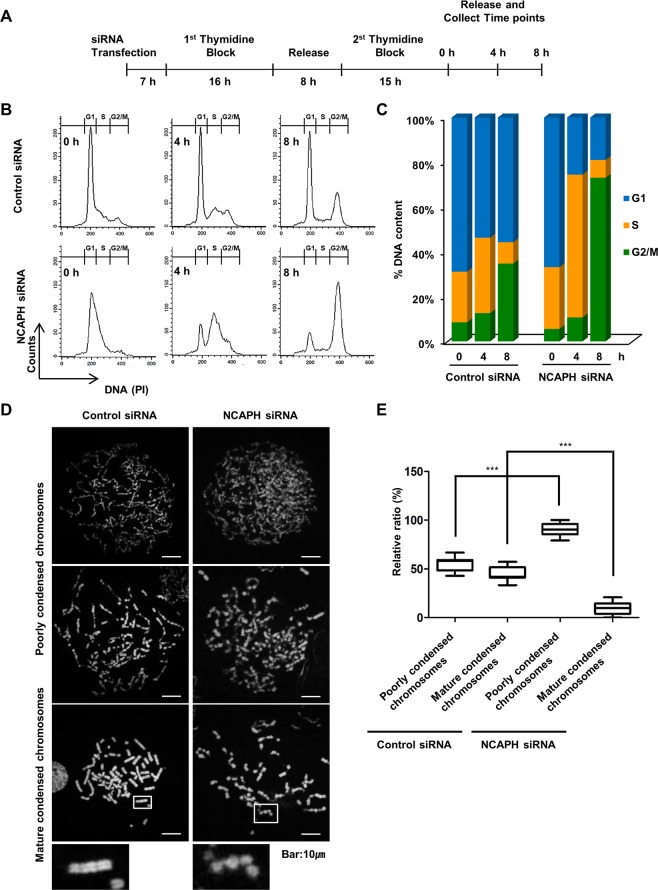
Accumulation of cell ratios at the S and G2/M phases indicated that these cells were not able to successfully undergo mitosis when the chromosomes could not be separated before the formation of two daughter cells. To better determine whether the S and G2/M phase accumulation previously observed in NCAPH-knockdown cells was a result of a delay or block in cell cycle progression, we investigated cell cycle progression over time after cell release from a double thymidine block (Fig. 4A). Thymidine is a pyrimidine deoxynucleoside that can be used to synchronize cells at the G1/S transition. Cell synchronization using double thymidine blocks in control cells allows entry into S phase (0–4 h) and promotes synchronous mitosis (G2/M phase: 7–8 h). NCAPH was depleted by siRNA, and then double thymidine block was used to prevent entry of cells into G1/S phase. Synchronized cells at different phases were harvested at 0, 4, and 8 h after release from the thymidine block (Fig. 4A). As expected, most cells in the control group had a DNA content consistent with being blocked at either G1 or S phase followed by double thymidine treatment (Fig. 4B,C), and the number of cells with a DNA content consistent with S phase increased (22.1% to 32.9%) at 4 h and they progressed to the G2/M phase (8.3% to 32.9%) at 8 h after release from the second thymidine block (Fig. 4B,C). In comparison, the cell cycle profile for NCAPH-knockdown cells showed an increase in cells with a DNA content consistent with S phase (26.8% to 62.7%) at 4 h, with a significant proportion of cells maintained at G2/M phase (5.4% to 65.8%) at 8 h after release from the second thymidine block (Fig. 4B,C). However, when we performed these experiments on HPDE cells, no differences in cell cycle progression between control siRNA and NCAPH siRNA treatment were observed, as observed in PC cells (Supplemental Fig. 3A). These results indicated that NCAPH may influence the S and G2/M phases in PC cells.
Figure 4.
NCAPH-knockdown cells fail to progress of mature chromosome condensation (MCC) from poorly condensed chromosomes. (A) General outline of the procedure followed for thymidine double block synchronization of MIA PaCa-2 cells and their cell cycle analysis. Cells were transfected with the indicated siRNAs, and after release from the second thymidine block, cells were assayed at different time points ranging from 0 to 8 h. (B) Cells were harvested for PI staining and analyzed by flow cytometry to determine the cell cycle fraction at 0, 4, and 8 h after release. Flow cytometry plots and data are representative of at least three separate experiments. (C) The percentages of cells in G1, S, and G2/M phases are indicated. (D,E) Frequencies of poorly condensed chromosomes and mature condensed chromosomes cells in control and NCAPH-knockdown cells. Cells were transfected with the indicated siRNA, treated with one thymidine block (36 h), released for 3 h and then harvested. For accurate quantification, more than 200 cells captured in at least three different fields were analyzed. Values represent means ± SEMs. *P < 0.005, ***P < 0.0001. Statistical analysis was performed using one-way analysis of variance followed by Bonferroni’s multiple comparison tests.
Next, to determine when structural chromosome aberrations occurred during cell division, we examined chromosomal breaks, segmentation, and aberrations in control and NCAPH-knockdown cells. In this experiment, thymidine block was performed in control or NCAPH-knockdown MIA PaCa-2 cells, and the chromosomal structure of cells released for 3 h was confirmed by metaphase chromosome spreading assays. Surprisingly, knockdown of NCAPH did not lead to the progression of MCC from poorly condensed chromosomes (Fig. 4D). Furthermore, in NCAPH-knockdown cells, only 9.7% of cells progressed to MCC from poorly condensed chromosomes, as compared with 45.8–54.1% of control cells (Fig. 4E). These results indicated that NCAPH was vital for the progression of MCC from poorly condensed chromosomes.
NCAPH knockdown promoted chromosomal aberrations and DNA damage
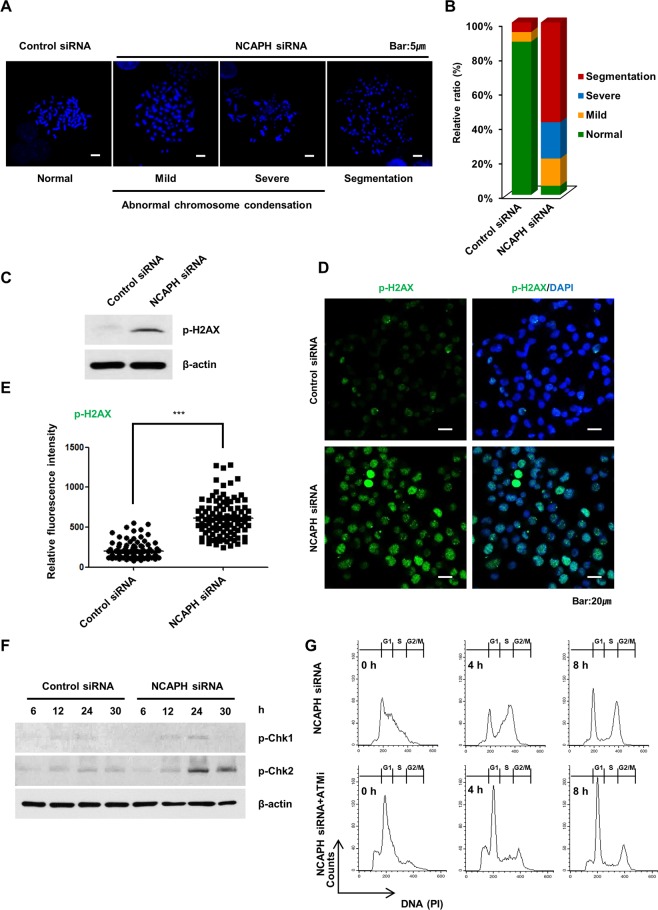
In Xenopus egg extracts, as condensins I and II are forced to be smaller, chromosomes become shorter and thicker. Condensin I is involved in lateral compaction, and condensin II is involved in axial shortening31. Additionally, in chicken DT40 cells, mitotic chromosomes are wide and short owing to depletion of condensin I, and chromosomes of condensin II-depleted cells appear to be more extended and lack axial stiffness32. To elucidate how mitotic chromosome structures are affected by NCAPH knockdown, we performed chromosome spreading assays in MIA PaCa-2 and HeLa cells. Similar to the previous report, shortening and thickening of chromosomes was observed in both types of cells (Supplementary Fig. 4A). However, upon specifically staining with anti-NCAPH antibodies and 4′,6-diamidino-2-phenylindole (DAPI) in MIA-PaCa-2 cells, NCAPH was detectable along the chromatid axis in cells of the control group but not in cells of the NCAPH-knockdown group, and the twisted and segmented chromosome morphology was observed in the NCAPH-knockdown group (Supplementary Fig. 4B). When measuring the number of structural chromosome aberrations in NCAPH-knockdown cells compared with those in control cells, we observed a significant increase (23.7% versus 75.2%, respectively; Supplementary Fig. 4C). To define chromosomal structures more clearly, we divided the state of the chromosomal structures into normal or abnormal chromosome condensations and classified them as mild, severe, or segmentation. The abnormal chromosome condensation (mild and severe) and segmentation type chromosome morphology were increased in NCAPH-knockdown cells (Fig. 5A,B). Additionally, we sought to determine whether the structural chromosome aberrations in NCAPH-knockdown cells were associated with DNA damage responses. To establish the presence of DNA damage, we monitored the appearance of DNA damage foci using antibodies detecting phosphorylated H2AX at S139 (phospho-H2AX), a marker of DNA double-strand breaks (DSBs). Western blot and immunofluorescence analyses showed that the levels of phospho-H2AX were higher in NCAPH-knockdown cells than in control cells (Fig. 5C–E). Moreover, phospho-H2AX was more abundant in NCAPH-knockdown cells than in control cells.
Figure 5.
Knockdown of NCAPH induces chromosomal aberrations and DNA damage. (A,B) To confirm the chromosome morphology, MIA PaCa-2 cells were transfected with control siRNA or NCAPH siRNA and arrested at metaphase by colcemid treatment for 4 h. The cells were spread onto slides, extracted, fixed, and stained with DAPI (blue). For accurate quantification, more than 50 cells captured in at three different fields were analyzed. Scale bar, 5 µm. (C) Western blot analysis of phospho-H2AX expression in control and NCAPH-knockdown cells. Cell lysates were immunoblotted with the indicated antibodies. (D) Phospho-H2AX fluorescence pattern (green) in control and NCAPH-knockdown cells was observed by confocal microscopy. DNA was stained using DAPI (blue). Scale bar, 20 µm. (E) Frequency of phospho-H2AX fluorescence intensity. For accurate quantification, more than 100 cells captured in at least two different fields were analyzed. Values represent means ± SEMs. ***P < 0.0001, two-way analysis of variance. (F) Western blot analysis showing levels of phospho-Chk1 and phospho-Chk2 activation in response to DNA damage in MIA PaCa-2 cells after transfection with control siRNA and NCAPH siRNA for 6, 12, 24, and 30 h. (G) Cells were harvested for PI staining and analyzed by flow cytometry to determine the cell cycle fraction after release from the second thymidine block at 0, 4, and 8 h. ATMi (13 μM) was treated with a second thymidine block.
Next, we determined whether G2/M phase cell cycle arrest was caused by DNA damage induced by NCAPH knockdown. The results showed that the levels of phospho-Chk1 and phospho-Chk2, which are DNA damage markers, were increased 24 and 30 h after NCAPH knockdown (Fig. 5F). In NCAPH-knockdown cells, the signals of phospho-Chk2 increased significantly compared with the signals of phospho-Chk1 (Fig. 5F). Moreover, to further define the role of ATM kinase in Chk2 phosphorylation in cell cycle defects induced by NCAPH knockdown, cell cycle changes were compared with those in cells when treated with thymidine double blocks alone or with a second thymidine block using an ATM inhibitor (KU-55933, ATMi). The result showed that the G2/M phase arrest in NCAPH-knockdown cells was significantly reduced by the ATM inhibitor (Fig. 5G). These results demonstrated that knockdown of NCAPH induced structural chromosome aberrations, which were associated with DNA damage, through the Chk1/Chk2 signaling pathway in PC cells.
Discussion
In this study, we reached the following conclusions: (1) NCAPH was upregulated in PC tissues and cell lines; (2) knockdown of NCAPH induced cell cycle arrest and cell apoptosis; (3) NCAPH was important for proper progression of MCC from poorly condensed chromosomes; and (4) NCAPH knockdown induced DNA damage though the Chk1/Chk2 signaling pathway. To the best of our knowledge, this is the first report demonstrating that chromosomal aberrations and DNA damage were induced in NCAPH-knockdown PC cells through the Chk1/Chk2 signaling pathway.
Recent studies have evaluated the relationships between condensin complexes and cancers25–29. For example, NCAPH knockdown inhibits proliferation and invasion of CC cells29. In hepatocellular carcinoma, lack of NCAPG reduces cell viability, increases apoptosis, and arrests cells in the S phase of the cell cycle26. In this study, we focused on NCAPH because its expression was increased in PC tissues and cell lines. Although NCAPH has been shown to be associated with cell invasion and proliferation in CC, its functional role in PC had not yet been evaluated. We found that cell cycle arrest and cell apoptosis were increased by NCAPH knockdown, and apoptosis occurred via the intrinsic apoptotic pathway with loss of mitochondrial potential, activation of caspase-3, and generation of the cleaved form of PARP-1, a typical caspase-3 substrate33. In addition, lack of NCAPD2, another subunit of condensin I, also inhibited colony formation by PC cells (Supplemental Fig. 2B,C), confirming that the condensin complex played an important role in PC growth.
To ensure proper chromosome condensation and segregation in mitosis, condensin II promotes the separation of replicated chromosomal loci and initiates structural reorganization of replicated chromosomes during S phase34. However, our study showed that when NCAPH (condensin I) was absent, cells were arrested in the S and G2/M phase of the cell cycle. Importantly, normal maintenance of the S phase in cell cycle progression could affect the structure of mitotic chromosomes. For example, when mammalian cells in the G2 phase are fused with G1-phase cells, all cells enter mitosis synchronously. However, the DNA from G1-phase cells passes through S phase faster than expected, resulting in less condensed mitotic chromosomes35. Importantly, when the G1-phase nucleus is fused with the mitotic nucleus, the detected chromosomes showed insufficiently condensed and much longer than normal chromatids35,36. These experiments have demonstrated that activities in mitotic cells result in condensation of interphase chromatin but that the structure of the condensed chromosomes is deficient. Moreover, lack of B-Myb, an Myb family transcription factor that plays an essential role during S phase, decreases S-phase progression and increases chromosomal fragmentation and other aberrations37. In any case, the completion of DNA replication is associated with proper chromosome condensation. Based on our observations after the knockdown of NCAPH, we hypothesize that NCAPH may have a major role in facilitating correct cell cycle progression through the S and G2/M phases and accompanying chromosome condensation. NCAPH knockdown resulted in chromosomal fragmentation and DNA damage responses, which affected the progression of MCC from poorly condensed chromosomes. We demonstrated that NCAPH-knockdown cells were arrested at S and G2/M phases of the cell cycle and blocked the progression of MCC from poorly condensed chromosomes, preventing the completion of proper chromosome condensation of PC cells. These findings suggested that NCAPH played an important role in the progression of MCC from poorly condensed chromosomes during the cell cycle process, revealing a novel mechanism through which progression of MCC from poorly condensed chromosomes is regulated by NCAPH.
When DNA damage occurs, cells spend more time undergoing DNA repair processes or apoptosis, particularly if the damage is severe. Therefore, cells have maintained genome integrity by developing complex machinery that includes DNA-damage responses, which detect and repair DNA damage. Generally, there are two types of DNA damage, DSBs and single-strand breaks (SSBs). DSBs are rapidly detected by the Mre11/Rad50/NBS1 complex, which subsequently promotes the activation of ataxia telangiectasia mutated kinase by autophosphorylation38. SSBs are sensed by the Rad9/Hus1/Rad1 complex, which activates ataxia telangiectasia and Rad3-related kinase39. Following this sensing step, Chk1 and Chk2 amplify the signals from the sensors, phosphorylating a variety of effectors40. Additionally, Chk1 monitors S phase replication and mitosis entry41,42, and Chk2 triggers the G1/S checkpoint and apoptosis43. In particular, SMC2 recruitment to mitotic chromosomes is controlled by DNA damage response checkpoints, such as Chk2, which facilitates effective repair of genomic damage44. Based on these findings, our data clearly indicated that NCAPH plays a critical role in the response to DNA damage through the increased phosphorylation of Chk1/Chk2 kinases. This finding was confirmed by the results of ATM treatment, which reduced the arrest of cells in G2/M phase caused by NCAPH knockdown because ATM is responsible for the phosphorylation of Chk2.
In conclusion, the results of this study confirmed that NCAPH knockdown induced chromosomal aberrations and DNA damage through the Chk1/Chk2 signaling pathway, ultimately resulting in selective cell death through partially caspase-dependent apoptotic signaling in PC cells. Based on our findings, we hypothesize that NCAPH may be associated with human carcinogenesis and could be a potential therapeutic target for human cancer. Further studies are needed to test this hypothesis.
Methods
Database analysis
Expression levels of the subunits of condensin I complex in PC and normal pancreas were achieved from Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/)45, which analyzed 179 PAAD tumors and 171 normal samples from TCGA tumor samples with paired adjacent TCGA normal samples and GTEx normal samples. Transcript per million (TPM) values were calculated, and the expression levels of genes were presented using the Log2 (TPM + 1) scale. The cutoff values were 1 for |Log2FC| and 0.01 for the P value. The OS of patients with PC was also analyzed.
Cell culture and siRNA knockdown
MIA PaCa-2 (American Type Culture Collection [ATCC] CRL-1420; ATCC, Manassas, VA, USA) and PANC-1 (ATCC CRL-1469) human PDAC cell lines were grown in high-glucose Dulbecco’s modified Eagle’s medium (DMEM). Human PDAC cell lines (AsPC-1, Capan-1, and Capan-2) were grown in RPMI medium. Noncancerous immortalized HPDE cells were obtained from Joo Kyung Park, MD (Samsung Medical Center, Seoul, South Korea). HPDE cells were grown in Defined K-SFM medium. All cell culture media contained 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, Life Technologies, Grand Island, NY, USA). To knockdown NCAPH expression, the cells were transfected with siRNA using the Lipofectamine RNAiMAX transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Three different NCAPH siRNAs were prepared46. The three NCAPH siRNAs and control siRNA were purchased from Cosmo Bio Co., Ltd. (Tokyo, Japan)
Cell proliferation and viability assay
Cell proliferation and viability assays were performed using MTT solution. MIA PaCa-2 and PANC-1 cells were seeded into 12-well plates at a density of 1 × 104 cells/well or 8 × 104 cells/well. After adhesion for 24 h, the cells were transfected with NCAPH siRNA or control siRNA and incubated for 5 or 7 days. For colony formation assays, cells were seeded into 6-well plates at a density of 500–1000 cells/well and incubated for 14 days. The colonies were then fixed in 100% methanol, stained with 10% crystal violet, and counted. Each assay was performed in triplicate.
Chromosome spreading assay
Cells were treated with colcemid (0.1 µg/mL) for 4 h and then harvested. After treatment with 0.075 M KCl, cells were fixed with a drop of freshly made fixative (methanol/acetic acid [3:1]) and placed on glass slides. The slides were dried at room temperature, stained with DAPI (100 ng/mL), and visualized under a confocal microscope (Zeiss LSM710, software ZEN; Zeiss, Germany).
Immunofluorescence
Cells were grown on Thermo Scientific Nunc Lab-Tek II Chamber Slides, permeabilized with 0.5% triton X-100 for 1 min, and fixed with 4% paraformaldehyde for 10 min. The fixed cells were incubated for 1 h at room temperature with blocking solution (1% bovine serum albumin) and then incubated overnight at 4 °C with primary antibodies. Cells were then incubated with secondary antibodies plus 100 ng/mL DAPI for 3 h. Samples were mounted in Prolong Gold Antifade reagent (Invitrogen), and the results were viewed under a confocal microscope (Zeiss LSM710, software ZEN). The primary antibodies were purchased from commercial sources, as follows: anti-NCAPH (Novus Biological, Littleton, CO, USA), and anti-phosphorylated H2AX at S139 (phospho-H2AX; Millipore, Temecula, CA, USA).
Western blotting and antibodies
Whole cells were harvested, and proteins were extracted using 1 × RIPA buffer. Protein concentrations were determined using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA). Protein extracts were resuspended in 5 × sample buffer (50 mM tris [pH 6.8], 100 mM dithiothreitol, 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, and 10% glycerol), boiled for 5 min, and subjected to SDS-polyacrylamide gel electrophoresis on 8–15% gels. Separated proteins were transferred to Trans-blot nitrocellulose membranes (Schleicher & Schuell, Keene, NH, USA), and membranes were blocked in 5% skim milk in TBST (10 mM Tris [pH 8.0], 150 mM NaCl, 0.05% tween 20) and incubated with several primary antibodies. The primary antibodies were purchased from commercial sources, as follows: anti-NCAPH (Novus Biological), anti-cyclin A (Cell Signaling Technology, Danvers, MA, USA), anti-phospho-histone H3 pS10 (Abcam, Cambridge, MA, USA), anti-phospho-Chk1 at Ser345 (Cell Signaling Technology), anti-phospho-Chk2 at Thr68 (Cell Signaling Technology), anti-caspase-3 (Cell Signaling Technology), anti-poly (ADP ribose) polymerase (PARP; Cell Signaling Technology), anti-phospho-H2AX (Millipore, Temecula, CA, USA) and anti-β-actin (Cell Signaling Technology). Protein expression was detected based on chemiluminescent signals activated by SuperSignal West Pico Chemiluminescent Substrate (Pierce, Thermo Scientific, USA) after reaction with horse radish peroxidase-tagged secondary antibodies (Jackson Immunoresearch Laboratories, West Grove, PA, USA).
Fluorescence-assisted cell sorting (FACS) analysis
For cell cycle and DNA content analysis, cultured cells were incubated in trypsin-ethylenediaminetetraacetic acid at 37 °C in an atmosphere containing 5% CO2, collected by centrifugation, and washed once with 1 × phosphate-buffered saline. Cells were centrifuged, supernatants were removed, and cells were stained with 50 μg/mL propidium iodide (PI; Sigma-Aldrich, St. Louis, MO, USA) plus 100 U ribonuclease A from bovine pancreas (Sigma-Aldrich). Analysis was performed with a FACSCalibur (BD Biosciences, San Diego, CA, USA) according to standard protocols.
Apoptosis was analyzed by FACS using FITC-conjugated annexin V (BD Biosciences, San Diego, CA, USA) and propidium iodide (PI; Sigma-Aldrich) staining. Analysis was performed with a FACSCalibur (BD Biosciences) according to standard protocols. Inhibition of apoptosis was performed with the pan-caspase inhibitor Z-VAD-FMK (20 µM; R&D Systems, Minneapolis, MN, USA).
Statistical analysis
All data represent average values obtained in form three independent experiments. Results were statistically analyzed using GraphPad Prism (version 5.0, GraphPad Software Inc., San Diego, CA, USA) and presented as means ± standard errors of the means (SEMs). Statistical analysis was performed using one-way or two-way analysis of variance followed by Bonferroni’s multiple comparison tests.
Supplementary information
Acknowledgements
This work was supported by grant nos. NRF-2017R1C1B2009106 and NRF-2017R1A2B2012191 from the National Research Foundation of Korea (NRF) and by Grant No. 00-2012-026 from the Seoul National University Hospital (SNUBH) Research Fund. We would like to thank Editage (www.editage.co.kr) for English language editing.
Author contributions
J.H.K. performed the experiments, carried out the data analysis, and wrote the manuscript. J.H.K. and J.H.H. conceived and designed the study and contributed to the manuscript revision. Y.Y. performed the reviewed experimental work. K.T.K. and J.G. provided the experimental material for the review. All of the authors discussed and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jae Hyeong Kim, Email: kingfish12@naver.com.
Jin-Hyeok Hwang, Email: woltoong@snu.ac.kr.
Supplementary information
is available for this paper at 10.1038/s41598-019-54478-3.
References
- 1.Zhao C, Gao F, Weng S, Liu Q. Pancreatic cancer and associated exosomes. Cancer biomarkers: section A of Disease markers. 2017;20:357–367. doi: 10.3233/cbm-170258. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, et al. Circulating biomarkers for early diagnosis of pancreatic cancer: facts and hopes. American journal of cancer research. 2018;8:332–353. [PMC free article] [PubMed] [Google Scholar]
- 4.Iovanna J, Mallmann MC, Goncalves A, Turrini O, Dagorn JC. Current knowledge on pancreatic cancer. Frontiers in oncology. 2012;2:6. doi: 10.3389/fonc.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falasca M, Kim M, Casari I. Pancreatic cancer: Current research and future directions. Biochimica et biophysica acta. 2016;1865:123–132. doi: 10.1016/j.bbcan.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Gharibi A, Adamian Y, Kelber JA. Cellular and molecular aspects of pancreatic cancer. Acta histochemica. 2016;118:305–316. doi: 10.1016/j.acthis.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hernandez YG, Lucas AL. MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions. World journal of gastrointestinal oncology. 2016;8:18–29. doi: 10.4251/wjgo.v8.i1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strunnikov AV. Condensin and biological role of chromosome condensation. Progress in cell cycle research. 2003;5:361–367. [PubMed] [Google Scholar]
- 10.Hirano T. Condensins: organizing and segregating the genome. Current biology: CB. 2005;15:R265–275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Hirano T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 12.Hirano T, Mitchison TJ. A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 13.Schleiffer A, et al. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Molecular cell. 2003;11:571–575. doi: 10.1016/S1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 14.Kschonsak M, et al. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell. 2017;171:588–600.e524. doi: 10.1016/j.cell.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome research. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeong FM, et al. Identification of a subunit of a novel Kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Current biology: CB. 2003;13:2058–2064. doi: 10.1016/j.cub.2003.10.032. [DOI] [PubMed] [Google Scholar]
- 17.Kagami Y, Yoshida K. The functional role for condensin in the regulation of chromosomal organization during the cell cycle. Cellular and molecular life sciences: CMLS. 2016;73:4591–4598. doi: 10.1007/s00018-016-2305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houlard M, et al. Condensin confers the longitudinal rigidity of chromosomes. Nature cell biology. 2015;17:771–781. doi: 10.1038/ncb3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. Journal of cell science. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- 20.Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Molecular biology of the cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeshima K, Laemmli UK. A two-step scaffolding model for mitotic chromosome assembly. Developmental cell. 2003;4:467–480. doi: 10.1016/S1534-5807(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 22.Gerlich D, Hirota T, Koch B, Peters JM, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Current biology: CB. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 23.Zhang T, et al. Condensin I and II behaviour in interphase nuclei and cells undergoing premature chromosome condensation. Chromosome research: an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2016;24:243–269. doi: 10.1007/s10577-016-9519-7. [DOI] [PubMed] [Google Scholar]
- 24.Ono T, et al. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/S0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 25.Davalos V, et al. Human SMC2 protein, a core subunit of human condensin complex, is a novel transcriptional target of the WNT signaling pathway and a new therapeutic target. The Journal of biological chemistry. 2012;287:43472–43481. doi: 10.1074/jbc.M112.428466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Su R, Shan C, Gao C, Wu P. Non-SMC Condensin I Complex, Subunit G (NCAPG) is a Novel Mitotic Gene Required for Hepatocellular Cancer Cell Proliferation and Migration. Oncology research. 2018;26:269–276. doi: 10.3727/096504017x15075967560980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W, et al. Overexpression of nonSMC condensin I complex subunit G serves as a promising prognostic marker and therapeutic target for hepatocellular carcinoma. International journal of molecular medicine. 2017;40:731–738. doi: 10.3892/ijmm.2017.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C, et al. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Scientific reports. 2016;6:34508. doi: 10.1038/srep34508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin L, et al. NCAPH plays important roles in human colon cancer. Cell death & disease. 2017;8:e2680. doi: 10.1038/cddis.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunez G, Benedict MA, Hu Y, Inohara N. Caspases: the proteases of the apoptotic pathway. Oncogene. 1998;17:3237–3245. doi: 10.1038/sj.onc.1202581. [DOI] [PubMed] [Google Scholar]
- 31.Shintomi K, Hirano T. The relative ratio of condensin I to II determines chromosome shapes. Genes & development. 2011;25:1464–1469. doi: 10.1101/gad.2060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green LC, et al. Contrasting roles of condensin I and condensin II in mitotic chromosome formation. Journal of cell science. 2012;125:1591–1604. doi: 10.1242/jcs.097790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science (New York, N.Y.) 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 34.Ono T, Yamashita D, Hirano T. Condensin II initiates sister chromatid resolution during S phase. The Journal of cell biology. 2013;200:429–441. doi: 10.1083/jcb.201208008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson RT, Rao PN. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 1970;226:717–722. doi: 10.1038/226717a0. [DOI] [PubMed] [Google Scholar]
- 36.Gollin SM, Wray W, Hanks SK, Hittelman WN, Rao PN. The ultrastructural organization of prematurely condensed chromosomes. Journal of cell science. Supplement. 1984;1:203–221. doi: 10.1242/jcs.1984.Supplement_1.13. [DOI] [PubMed] [Google Scholar]
- 37.Garcia P, Frampton J. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. Journal of cell science. 2006;119:1483–1493. doi: 10.1242/jcs.02870. [DOI] [PubMed] [Google Scholar]
- 38.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science (New York, N.Y.) 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 39.Nam EA, et al. Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. The Journal of biological chemistry. 2011;286:28707–28714. doi: 10.1074/jbc.M111.248914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer cell. 2003;3:421–429. doi: 10.1016/S1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 41.Sorensen CS, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer cell. 2003;3:247–258. doi: 10.1016/S1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 42.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature reviews. Molecular cell biology. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 43.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nature reviews. Molecular cell biology. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 44.Zhang T, Si-Hoe SL, Hudson DF, Surana U. Condensin recruitment to chromatin is inhibited by Chk2 kinase in response to DNA damage. Cell cycle (Georgetown, Tex.) 2016;15:3454–3470. doi: 10.1080/15384101.2016.1249075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research. 2017;45:W98–w102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Georges SA, et al. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer research. 2008;68:10105–10112. doi: 10.1158/0008-5472.can-08-1846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.