Abstract
Early diagnosis of leprosy is challenging, particularly its inflammatory reactions, the major cause of irreversible neuropathy in leprosy. Current diagnostics cannot identify which patients are at risk of developing reactions. This study assessed blood RNA expression levels as potential biomarkers for leprosy. Prospective cohorts of newly diagnosed leprosy patients, including reactions, and healthy controls were recruited in Bangladesh, Brazil, Ethiopia and Nepal. RNA expression in 1,090 whole blood samples was determined for 103 target genes for innate and adaptive immune profiling by dual color Reverse-Transcription Multiplex Ligation-dependent Probe Amplification (dcRT-MLPA) followed by cluster analysis. We identified transcriptomic biomarkers associated with leprosy disease, different leprosy phenotypes as well as high exposure to Mycobacterium leprae which respectively allow improved diagnosis and classification of leprosy patients and detection of infection. Importantly, a transcriptomic signature of risk for reversal reactions consisting of five genes (CCL2, CD8A, IL2, IL15 and MARCO) was identified based on cross-sectional comparison of RNA expression. In addition, intra-individual longitudinal analyses of leprosy patients before, during and after treatment of reversal reactions, indicated that several IFN-induced genes increased significantly at onset of reaction whereas IL15 decreased. This multi-site study, situated in four leprosy endemic areas, demonstrates the potential of host transcriptomic biomarkers as correlates of risk for leprosy. Importantly, a prospective five-gene signature for reversal reactions could predict reversal reactions at least 2 weeks before onset. Thus, transcriptomic biomarkers provide promise for early detection of these acute inflammatory episodes and thereby help prevent permanent neuropathy and disability in leprosy patients.
Subject terms: Predictive markers, Biomarkers
Introduction
Leprosy is a chronic infectious disease caused by Mycobacterium leprae, a bacillus with tropism for skin and peripheral nerves. Despite decades of programs using multidrug therapy (MDT), leprosy remains persistently endemic or re-emerging in some regions where it predominantly affects poor and marginalized people. A featuring aspect regarding leprosy diagnosis is the plateauing annual new case detection rates of roughly 200,000 worldwide1. The pathology of leprosy is complex as it presents as a spectral disease in which immunity against M. leprae matches the clinical manifestations after infection with the bacterium. At one pole of the spectrum, the disease manifests as tuberculoid leprosy (TT), characterized by strong pro-inflammatory cellular immunity including Th1 and Th17 cells2,3, granuloma formation and elimination of bacteria. At the other pole, lepromatous leprosy (LL) is characterized by humoral immunity against M. leprae along with Th2 cells but almost no protective cell mediated immunity, allowing accumulation of high numbers of bacilli around foamy macrophages4–8. Nonetheless, the majority of individuals present unstable borderline phenotypes (BT, BB and BL) between the two poles5.
A major challenge in leprosy control is the prevention of permanent disability due to nerve damage. Although leprosy is curable by MDT, nerve damage cannot always be avoided. Dynamic and unpredictable episodes of increased inflammation, leprosy reactions, can occur before, during and even after treatment, with a higher likelihood to occur in adults than in children9,10. These immunological complications are the principal cause of leprosy-associated irreversible neuropathy and are experienced by 30–50% of leprosy patients one or more times, mostly in the unstable borderline lepromatous patients with substantial bacterial loads11. Two types of reactions are recognized: reversal reactions or type 1 (RR) and erythema nodosum leprosum (ENL). RRs are caused by changes in the host immune response against M. leprae which is upgrading from borderline to the TT pole characterized by an enhanced cell-mediated immunity, inflammation12,13. These reactions can occur spontaneously but are also linked to shifts from Th2 to Th1, e.g. occurring during anti-helminth treatment of co-infected leprosy patients14–17, HIV highly active antiretroviral therapy (HAART) and at the end of extensive anti-TNF-α therapy10,13 and even BCG vaccination18.
Prompt diagnosis and treatment of reactions significantly favors successful recovery9,19. Unfortunately, reactions are often late- or misdiagnosed, in part due to decreased expertise within integrated health services19 which urges the need for new diagnostic tools. Delays in diagnosis of reactions directly translate into negative clinical outcomes, as associated neuropathy not properly diagnosed or treated within the first 6 months of symptoms will likely become permanent20 alongside the disabilities it may later initiate via recurrent ulcers and other related pathologies21. Despite recent scientific progress with respect to complement22,23 and serum-proteins, particularly CXCL10 (IP-10), as biomarkers associated with onset of reactions15–17,24–26, discovery of accurate, clinically useful prognostic biomarkers remains elusive, leaving early diagnosis of reactions a currently unmet need.
Since host transcriptomic biomarkers reflect early stages of or ongoing biological processes, they have been widely used to profile the host transcriptome for diagnostics of tuberculosis (TB)27–30. Moreover, multicomponent host biomarker signatures have been described that predict development of disease in retro- and prospective cohorts31,32. In this respect dual color Reverse-Transcription Multiplex Ligation-dependent Probe Amplification (dcRT-MLPA) has proven to be a valuable tool for monitoring gene expression profiles in large cohorts29,33. Techniques such as RNA-Seq and microarray are costly, technically challenging and require high RNA concentrations which limits their application for large cohorts. Therefore, a selection of genes related to immune-mediated inflammatory pathways, which play a role in the immunopathology of leprosy can be assessed by dcRT-MLPA29,34.
Many reactions occur during MDT, with the highest rates reported within the first 6 months of treatment11,19,35. To identify transcriptomic signatures for applications to surveillance of leprosy reactions, whole blood RNA of leprosy patients was monitored during MDT. To accommodate worldwide applicability, this study was executed in four prospective cohorts in Asia, Africa and South America. Improved knowledge on longitudinal fluctuations of RNA expression associated with reactions will promote identification of patients with imminent reactions leading to timely interventions that can impact nerve damage in affected individuals.
Materials and Methods
Participants
Patients and controls were recruited on a voluntary basis between February 2008 and March 2015 (Table 1) in four leprosy endemic populations: in Bangladesh (International Centre for Diarrhoeal Disease Research Bangladesh, Dhaka), Brazil (National Reference Centre for Sanitary Dermatology and Leprosy, Uberlandia and Leprosy Laboratory, Oswaldo Cruz Institute, Oswaldo Cruz Foundation (FIOCRUZ), Rio de Janeiro), Ethiopia (ALERT hospital and Health Centre, Addis Ababa) and Nepal (Mycobacterial Research Laboratories, Kathmandu). Leprosy was diagnosed based on clinical, bacteriological and histological observations and classified by skin biopsies according to Ridley-Jopling36. Clinical monitoring for reactions was performed during monthly clinic visits. Participant information was collected with emphasis on standardizing data collection and definition of reactions between all cohorts37,38. Endemic controls (EC) were living in the same area without known contact with leprosy or TB patients and were assessed for the absence of clinical signs and symptoms of leprosy and TB. Staff of leprosy or TB clinics and laboratory staff were excluded. Healthy household contacts (HHC) were defined as adults living in the same household as leprosy patients for at least the preceding six months.
Table 1.
Samples of participants included in cross-sectional analysis.
| Site | Categorya | LRb | Timepointc | Number |
|---|---|---|---|---|
| Bangladesh | EC | na | na | 61 |
| BB/BL/LL (MB) no LR t = 0 | no LR | t = 0 | 62 | |
| BB/BL/LL (MB) no LR t = end | no LR | t = end | 26 | |
| TT/BT (PB) no LR t = 0 | no LR | t = 0 | 36 | |
| TT/BT (PB) no LR t = end | no LR | t = end | 9 | |
| BB/BL/LL (MB) RR t = 0 | RR | t = 0 | 5 | |
| BB/BL/LL (MB) RR t = x | RR | t = x | 30 | |
| BB/BL/LL (MB) RR t = end | RR | t = end | 42 | |
| TT/BT (PB) RR | RR | t = 0 or = x, or = end | 9 | |
| ENL/Neuritis | ENL/Neuritis | t = 0 or = x, or = end | 11/0 | |
| HHC | na | na | 38 | |
| Brazil | EC | na | na | 46 |
| BB/BL/LL (MB) no LR t = 0 | no LR | t = 0 | 26 | |
| BB/BL/LL (MB) no LR t = end | no LR | t = end | 20 | |
| TT/BT (PB) no LR t = 0 | no LR | t = 0 | 52 | |
| TT/BT (PB) no LR t = end | no LR | t = end | 38 | |
| BB/BL/LL (MB) RR t = 0 | RR | t = 0 | 17 | |
| BB/BL/LL (MB) RR t = x | RR | t = x | 20 | |
| BB/BL/LL (MB) RR t = end | RR | t = end | 20 | |
| TT/BT (PB) RR | RR | t = 0 or = x, or = end | 28 | |
| ENL or Neuritis | ENL/Neuritis | t = 0 or = x, or = end | 22/18 | |
| HHC | na | na | 14 | |
| Ethiopia | EC | na | na | 51 |
| BB/BL/LL (MB) no LR t = 0 | no LR | t = 0 | 83 | |
| BB/BL/LL (MB) no LR t = end | no LR | t = end | 9 | |
| TT/BT (PB) no LR t = 0 | no LR | t = 0 | 16 | |
| TT/BT (PB) no LR t = end | no LR | t = end | 1 | |
| BB/BL/LL (MB) RR t = 0 | RR | t = 0 | 2 | |
| BB/BL/LL (MB) RR t = x | RR | t = x | 36 | |
| BB/BL/LL (MB) RR t = end | RR | t = end | 12 | |
| TT/BT (PB) RR | RR | t = 0 or = x, or = end | 6 | |
| ENL or Neuritis | ENL/Neuritis | t = 0 or = x, or = end | 11/1 | |
| HHC | na | na | 33 | |
| Nepal | EC | na | na | 42 |
| BB/BL/LL (MB) no LR t = 0 | no LR | t = 0 | 14 | |
| BB/BL/LL (MB) no LR t = end | no LR | t = end | 5 | |
| TT/BT (PB) no LR t = 0 | no LR | t = 0 | 19 | |
| TT/BT (PB) no LR t = end | no LR | t = end | 8 | |
| BB/BL/LL (MB) RR t = 0 | RR | t = 0 | 6 | |
| BB/BL/LL (MB) RR t = x | RR | t = x | 12 | |
| BB/BL/LL (MB) RR t = end | RR | t = end | 7 | |
| TT/BT (PB) RR | RR | t = 0 or = x, or = end | 29 | |
| ENL or Neuritis | ENL/Neuritis | t = 0 or = x, or = end | 2/0 | |
| HHC | na | na | 6 | |
| Netherlands | NEC first time point | na | na | 19 |
| NEC second time points | na | na | 10 | |
| total | 1090 |
aEC: endemic control; BB/BL/LL: borderline borderline/borderline lepromatous/lepromatous leprosy; TT/BT: tuberculoid leprosy/borderline tuberculoid leprosy; bLR: leprosy reaction; RR: reversal reaction; NEC: non-endemic control; na: not applicable; ct = 0: time point of enrolment before initiation of multidrug (MDT) therapy; t = x: time point of LR; t = end: time point of completion of MDT and/or steroid therapy.
Recruitment
Newly diagnosed, untreated leprosy patients without clinical reactions were enrolled and blood was drawn before initiation of MDT(t = 0) as previously described15. Patients with reactions within initiation of three months of therapy were excluded. Patients were often diagnosed with RR at first clinic visits, leading to a low frequency of untreated cases without RR at their first visits that subsequently developed RR during this study. If patients presented with reactions after more than three months of MDT, blood was drawn again before initiation of anti-reactional therapy (t = x). Patients diagnosed with RR at their first clinic visits were also recruited (t = x) but blood was collected after completion of MDT and/or after steroid therapy (t = end). For patients with RR this was done at least one month after completion of steroid therapy. Patients were assessed for absence of reactions one year after t = end. For patients showing clinical signs of reactions within three months after t = end, this time point was excluded from analyses. Thus, analyses included two samples of each patient without reactions [before (t = 0) and after treatment (t = end)] and three of each patient who developed RR [in the absence of clinical signs of reactions, ≥2 months before RR diagnosis (t = 0); at RR diagnosis, before steroid-treatment (t = x); after RR, at least one month after ending steroid-treatment (t = end)]. Patients with leprosy relapse and pure neural leprosy were excluded from the analysis.
Ethics
This study was performed according to the Helsinki Declaration. Written informed consent was obtained before enrolment. Patients received treatment according to national guidelines. Ethical approval of the study-protocol was obtained through Ethical Review Committee of ICDDR,B (#PR-10032; #PR-2007-069); Brazilian National Council of Ethics in Research (CONEP) and the Fiocruz Ethical research Council CEP (# 555/10) or UFU Research Ethics Committee (#499/08); National Health Research Ethical Review committee Ethiopia (NERC # RDHE/127-83/08); Nepal Health Research Council (NHRC #751).
RNA isolation
RNA from PAXgene tubes was extracted using PAXgene Blood RNA kits (BD Biosciences, Franklin Lakes, NJ) according to the manufacturers’ protocol. RNA yield was determined by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Dual color reverse-transcription multiplex ligation-dependent probe amplification (dcRT-MLPA) assays
dcRT-MLPA assay was performed as previously described33,39. In short, for each target-specific sequence, a specific RT primer was designed located downstream of the half-probe target sequences (Sigma-Aldrich, Saint Louis, MO). RNA (2.5 μl of a 50 ng/μl solution) was reverse transcribed with 1x MMLV reverse transcriptase buffer, dNTPs (0.4 mM of each nucleotide), and 80 nM of the target-specific RT primers in a final volume of 4.5 μl. After heating for 1 min to 80 °C and incubation for 5 min at 45 °C, 30U MMLV reverse transcriptase (Promega, Madison, WI) was added and incubated for 15 min at 37 °C before heat inactivation of the enzyme for 2 min at 98 °C. Subsequently, half-probes (6 nM) were added to the reaction, heat denatured for 1 min at 95 °C followed by hybridization for 16 h at 60 °C. Ligation of the annealed half-probes was performed for 15 min at 54 °C by ligase-65 followed by heat inactivation for 5 min at 98 °C. Ligation products were amplified by PCR. Thermal cycling conditions encompassed: 33 cycles of 30 s/95 °C, 30 s/58 °C, and 60 s/72 °C, followed by 1 cycle of 20 min/72 °C. PCR products were diluted 1:10 in HiDi formamide containing 400 HD ROX size standards and analyzed on an Applied Biosystems 3730 capillary sequencer in GeneScan mode (Applied Biosystems, Foster City, CA). MLPA reagents were from MRC-Holland (Amsterdam, The Netherlands).
Data were analyzed using GeneMapper software 5 (Applied Biosystems). The areas of each assigned peak (in arbitrary units) were exported for further analysis in R-Project, normalized to GAPDH and log2 transformed. Signals below the threshold value for noise cut-off (peak area ≤ 7.644) were assigned the threshold value for noise cut-off. Pathway analysis was performed using Ingenuity Pathway Analysis (Qiagen, Hildern, Germany).
Statistical analyses dcRT-MLPA
Wilcoxon signed-rank test was applied for paired samples. For cross-sectional comparisons of different test groups at comparable time points, a Mann–Whitney test was used. P-values were corrected for multiple comparisons using the Benjamini-Hochberg method40. To classify BL/LL patients according to their likelihood to develop RR a logistic regression model was fitted with RR (yes/no) as outcome (dependent variable), and gene expression values at time t = 0 as covariates (risk factors). Genes were grouped based on correlation of their expression using a hierarchical clustering analysis (average linkage) based on absolute correlation difference. The global test (version 5.32.0)41 in R (version 3.4.1) was performed on these groups. Genes that were significantly differentially expressed between the test groups (inheritance <0.05) after multiple testing correction, constituted a biomarker signature for prediction of RR. To avoid overfitting, the biomarker signature was evaluated using a leave-one-out cross-validation (LOOCV): during the training of the model, one subsample (n-1) was reserved in each iteration for evaluation of the accuracy of the model on a sample excluded from the training set. The gene selection for the predictive signature was redone at every fold. The predictive biomarker signature was thus assessed in observations, which were not used to build the model. To evaluate the risk of developing RR, the cut-off for gene expression was determined based on the Youden Index. A score of 0 or 1*(weight of gene) was given per gene based on the association to RR (as indicated by the cut-off). The weight of gene was based on the results from the global test. All the scores from the significant genes were added and divided by the sum of weights to calculate the risk to develop RR. Area Under the Receiver Operating Characteristic curve (ROC-AUC) was calculated using GraphPad Prism (version 7.02).
Results
Prospective cohorts
To identify correlates of risk (CoR) for leprosy and RR, blood of 1,090 samples was obtained longitudinally in Bangladesh, Brazil, Ethiopia and Nepal (Tables 1 and 2).
Table 2.
Demographics and clinical characteristics of reactional patients recruited longitudinally§.
| # | area | RJ | LR | BI* | PGL-I* | sex | age* |
|---|---|---|---|---|---|---|---|
| 1 | Bangladesh | BL | RR | 2+ | 0.95 | male | 36 |
| 2 | Bangladesh | BL | RR | 2+ | 0.27 | female | 39 |
| 3 | Bangladesh | BL | RR | 0 | 1.82 | male | 36 |
| 4 | Bangladesh | BL | RR | 2+ | 1.17 | male | 32 |
| 5 | Brazil | BL | RR | 3.2+ | 0,43 | male | 35 |
| 6 | Brazil | BL | RR | 2.42+ | 0.07 | male | 29 |
| 7 | Brazil | BL | RR | 4.28+ | 1.02 | female | 42 |
| 8 | Nepal | BT | RR | 0.25+ | 0.07 | female | 40 |
| 9 | Ethiopia | BB | RR | 0 | 0.15 | male | 33 |
| 10 | Netherlands | BL | RR | 5+ | 1.83 | male | 17 |
§Blood samples were collected from 10 patients at 3 time points: at diagnosis of leprosy in the absence of any clinical signs of reactions, at diagnosis of reactions and after treatment; * at recruitment before treatment.
RJ: Ridley Joplin classification; LR: Leprosy Reaction; RR: reversal reaction.
BI: bacterial index; PGL-I: OD at 450 nm in anti-PGL-I antibody ELISA (threshold for positivity: OD450 = 0.2).
Leprosy-specific RNA-profiles
We first analyzed gene expression in ex vivo blood samples of newly diagnosed leprosy patients (irrespective of classification) without reactions (n = 359) from Bangladesh, Brazil, Ethiopia and Nepal compared to EC from these regions (n = 200). To this end, 103 target genes associated with innate and adaptive immunity29,33,39 were analyzed by dcRT-MLPA (Table S1). A substantial variety (36 genes) was observed to significantly differ between patients and EC at all sites. Expression of 13 genes was upregulated and 23 genes were downregulated in patients compared to EC (Table S2). When comparing leprosy patients to HHC (Table S3), 16 genes showed significantly different expression for leprosy patients (increased: FCGR1A, IL6, IL15, LRKK2, MBP, MSR1, PACRGv1, TLR1, TLR4; decreased: CAMTA, CD3E, CTLA4, CXCL13, GATA3, LAG3, TFGB). Importantly, whilst most of these genes were also differently expressed in leprosy patients compared to EC, MBP, MSR1, TLR1, CAMTA, CXCL13 and TFGB were differentially expressed in leprosy patients exclusively when compared to HHC. Such genes are potential CoR for leprosy in contacts of leprosy patients who are highly exposed to M. leprae. Being part of innate/adaptive and macrophage signaling pathways42, these genes, are not restricted to leprosy but are also relevant in rheumatoid arthritis and Crohn’s disease (Table S4).
RNA-profiles for leprosy classification
To identify genes applicable for classification of different types of leprosy, the 103 target genes (Table S1) were assessed similarly by dcRT-MLPA in blood from newly diagnosed, untreated BL/LL (n = 228) as well as TT/BT (n = 131) leprosy patients from the four-different leprosy endemic populations. After correction for multiple comparisons40 seven genes remained significantly different. Expression levels of IL2 and TLR6 were increased in TT/BT compared to BL/LL whereas CD46, CXCL10, FCGR1A, HDAC2 and TLR4 expression levels were increased in BL/LL (Fig. 1; Table S5).
Figure 1.
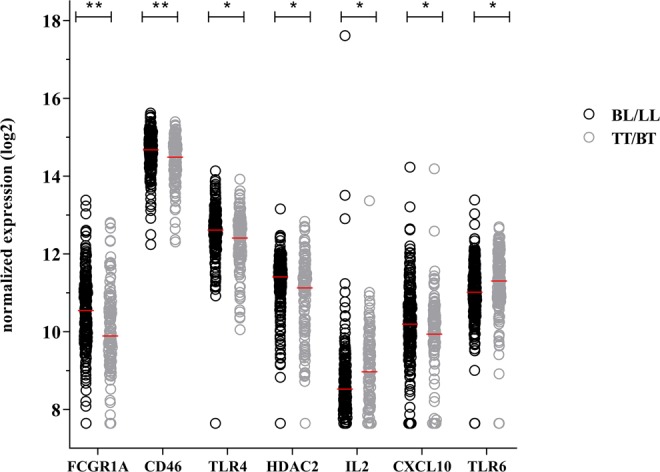
Transcriptional profiles of BL/LL and TT/BT patients without reactions. Gene expression levels of 103 target genes were assessed by dual-color RT-MLPA performed on ex vivo RNA isolated from whole blood of newly diagnosed, untreated BL/LL (n = 228; black circles) and TT/BT (n = 131; grey circles) leprosy patients without reactions from Bangladesh, Brazil, Ethiopia and Nepal. Log2-transformations of peak areas (normalized to the housekeeping gene GAPDH) of genes that were significantly differentially expressed between BL/LL and TT/BT are shown on the y-axis. Raw p-values were calculated using the Mann–Whitney test and adjusted for multiple comparisons using the Benjamini-Hochberg correction40. *adjusted p-values < 0.05; **adjusted p-values < 0.01; (see Table S5).
RNA-profiles associated with exposure to M. leprae
Since HHC of leprosy patients have a higher risk to develop leprosy than the general population in an endemic area43, biomarker profiles indicating this risk could help decision making on who needs preventive antibiotic treatment44. These genes represent potential transcriptomic tools to identify individuals substantially exposed to M. leprae. Thus, RNA-expression profiles of HHC (n = 83) were compared to EC (n = 200) (Table S6). We identified 10 differentially expressed genes with either significantly higher expression in HHC (Fig. 2; FOXP3, TGFB and CCL3) or in EC (CCR6, GZMA, HDAC2, IL22RA1, PTPRCv2, TLR1 and TLR7).
Figure 2.
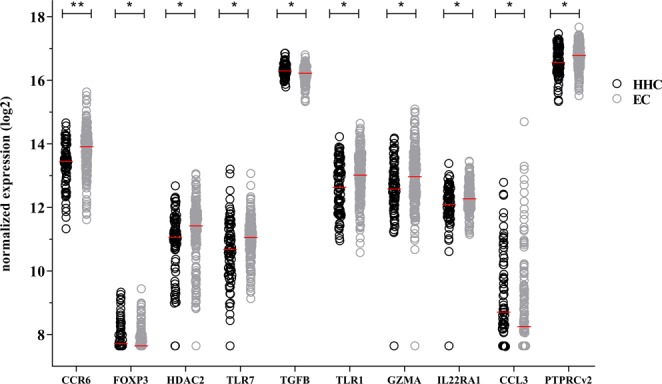
Transcriptional profiles of healthy household contacts and endemic controls. Gene expression levels of 103 target genes were assessed by dual-color RT-MLPA performed on ex vivo RNA isolated from whole blood of healthy household contacts (HHC; n = 83; black circles) and endemic controls (EC; n = 200; grey circles) from Bangladesh, Brazil, Ethiopia and Nepal. Log2-transformations of peak areas (normalized to GAPDH) of genes significantly differentially expressed between HHC and EC are shown on the y-axis. Raw p-values were calculated using the Mann–Whitney test and adjusted for multiple comparisons using the Benjamini-Hochberg correction40. *adjusted p-values < 0.05; **adjusted p-values < 0.01; (see Table S6).
Transcriptomic risk factors for development of RR
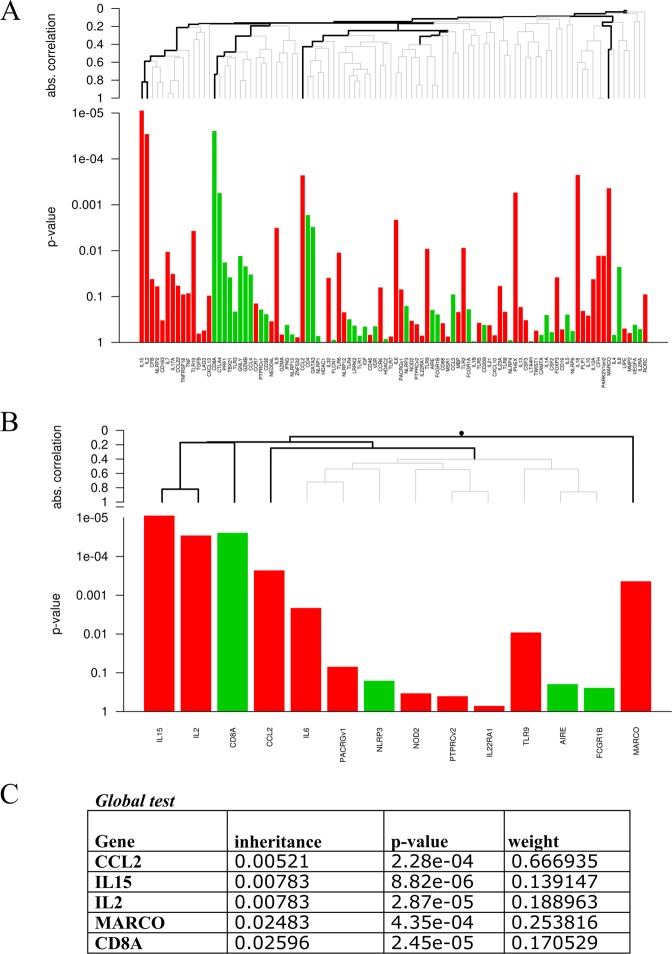
To assess whether RNA-expression levels can be used as a predictive tool for reactions during MDT, cross-sectional comparison of gene expression levels was performed for samples at the time of leprosy diagnosis in the absence of clinical signs of reactions (t = 0). Gene expression from BL/LL patients who developed RR ≥ 2 months later in the study (n = 30) was compared to BL/LL patients who did not develop reactions at all (n = 184) using the Mann–Whitney test. Transcriptomic profiles of the two groups of BL/LL patients resulted in a decreased expression of CTLA4 and GATA3 at diagnosis in patients who would later develop RR (Fig. 3), whereas nine genes (CCL2, IL2, IL15, IL18, MARCO, PHEX, TLR2, TLR6 and TLR10) were significantly increased (Fig. 3; Table S7).
Figure 3.
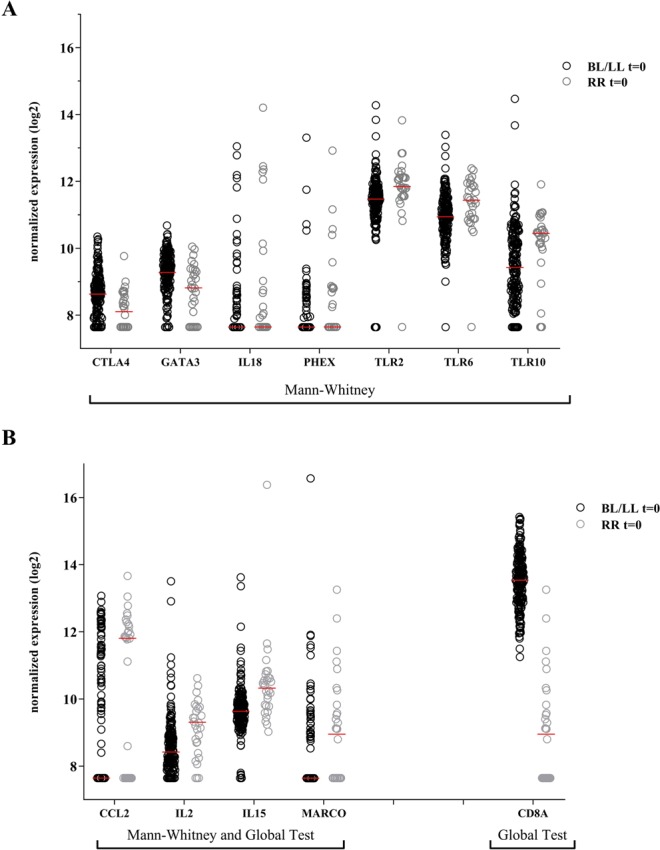
Identification of biomarker risk signature for developing reversal reactions. Gene expression levels of 103 target genes were assessed by dual-color RT-MLPA performed on ex vivo RNA isolated from whole blood from BL/LL patients who developed reversal reactions (RR) at least two months later during the study (n = 30; grey circles) and BL/LL who did not develop reactions (n = 184; black circles) from Bangladesh, Brazil, Ethiopia and Nepal. Samples were analyzed at t = 0: in the absence of clinical signs of reactions. Log2-transformations of peak areas (normalized for GAPDH expression) of genes with significantly different expression between both groups (at t = 0) are shown on the y-axis. (A) Genes with a significant (p-value < 0.05) different expression only using the Mann-Whitney test are show. P-values were adjusted for multiple comparisons using the Benjamini-Hochberg correction40 (see Table S7). (B) Genes with a significant different expression in the global test and Mann-Whitney or the global test only are shown.
Next, we also determined the biomarker signature with the least number of genes and the highest discriminatory power to categorize BL/LL patients according to their likelihood to develop RR using the global test41 (Fig. 4; p = 1.28*10−5). Four genes (CCL2, IL2, IL15 and MARCO) remained significantly (inheritance <0.05) associated with occurrence of RR at the time of recruitment, whereas CD8A was negatively associated with RR (Fig. 3B). Although expression of CD8A was not statistically significant using Mann–Whitney, it contributed significantly to the transcriptomic global test-signature. From the five genes CCL2 contributed most to the model (Fig. 4B).
Figure 4.
Identification of a minimal biomarker risk signature for developing reversal reactions. Biomarker signature to assess the risk of BL/LL patients to develop reversal reactions (RR). Gene expression data obtained by dual-color RT-MLPA of RNA isolated from whole blood of BL/LL patients from Bangladesh, Brazil, Ethiopia and Nepal at t = 0 were analyzed using the global test cluster analysis41. The global test is a cluster analysis based on absolute correlation difference and average linkage developed for data sets in which many covariates (or features) have been measured for the same subjects, together with a response variable. Graphs (A,B) indicate genes that are higher expressed in future RR patients (red) or in non-reactional BL/LL patients (green). In (A) all genes analyzed are shown and in (B) only significant branches are shown. Table (C) shows values for the 5 genes that were statistically significant after correction for multiple testing (inheritance <0.05), representing the output signature of the global test shrinkage model.
To evaluate the prediction a LOOCV was performed in which in every iteration a subsample (n = 1) was excluded in the global test to evaluate the model performance on unseen samples. The overall classifying ability of the biomarker-signature is indicated as a ROC-AUC = 0.80 (Fig. S1). Thus, transcriptomic profiles can prospectively differentiate, at the time of leprosy diagnosis in the absence of any clinical symptoms of reactions, patients who will develop RR.
Longitudinal transcriptomic changes: monitoring RR onset and treatment
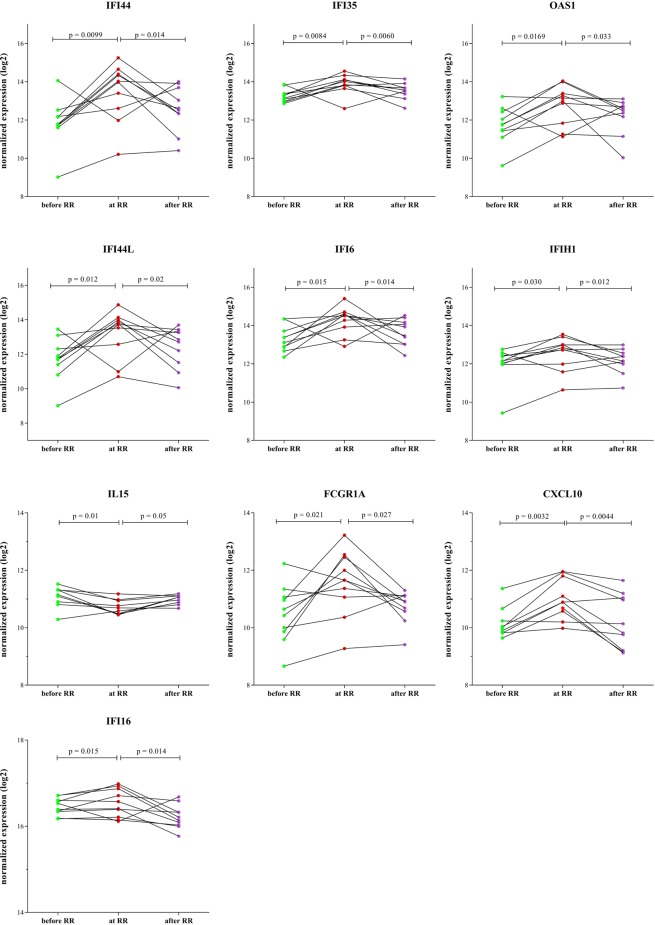
Cross-sectional analysis of gene expression at different time points amongst patients with RR showed only a significant increase in IL10 at RR compared to before RR (Supplementary results; Tables S8 and S9; Figs. S2 and S3).
Since RNA expression levels of genes may vary over time, RNA expression was also assessed longitudinally at three time points in whole blood of 10 leprosy patients (Table 2) developing RR during the study (Figs. 5, S4 and S5). This included besides 103 immune-associated genes, 38 IFN-induced genes (Table S1) previously identified as markers for mycobacterial disease45–49. Expression of ten genes (CXCL10, FCGR1A, IFI16, IFI44, IFI35, IFI44L, IFI6, IFIH1, IL15 and OAS1) significantly differed when comparing time points before RR vs at RR as well as at RR vs after RR. All genes except IL15 consistently increased with development of RR, and normalized after treatment, whereas IL15 decreased at onset of RR (Fig. 5). Most of the genes identified longitudinally are directly connected to the genes identified as predictive markers before clinical symptoms of RR (Fig. S6).
Figure 5.
Intra-individual longitudinal expression of patients who developed RR. Direct ex vivo RNA expression values were assessed by dual-color RT-MLPA on whole blood of 10 leprosy patients who developed RR during this study. Blood was analyzed at three time points: in the absence of any clinical signs of reactions and at least two months before RR (t = 0), at RR diagnosis before steroids (t = x) or after MDT and at least one month after end of steroids, in the absence of reactions (t = end). Log2-transformations of peak areas (normalized for GAPDH expression) are shown on the y-axis. Wilcoxon signed-rank test was performed. Genes with a significant difference (p-value < 0.05) in expression between before RR-at RR and at RR-after RR are shown.
Five genes (IL10, PRF1, CCL2, CCL3 and BMP6) were significantly different only at onset of RR compared to before RR (Fig. S4), while nine (IFITM3, IFIT3, GBP5, GBP1, GBP2, OAS3, STAT1, STAT2 and TAP1) only displayed a significant effect upon treatment (Fig. S5).
These data indicate that onset of RR can be monitored, based on differential expression of inflammatory genes in whole blood, allowing early detection and subsequent treatment of RR. Thus, helping to reduce irreversible nerve damage and associated disabilities.
Discussion
In the current state of leprosy elimination, reactions persist as a major problem since patients remain at risk due to M. leprae antigens that can persist for years post-MDT. RRs can occur at any time during, before or after MDT12 and although several factors have been associated with reactions, the underlying mechanism is not completely known10. Genetic susceptibility to reactions but also treatment for other diseases or co-morbidities have been suggested to play a role by causing an immunological shift from Th2 to Th110,13–17. In addition, Th17 cells have been proposed to play a role in RR pathogenesis as well50. Still, no validated biomarker signature is available at the moment. The implementation of diagnostics tests for reactions in leprosy health care could make significant differences in clinical outcomes and help reduce nerve damage. Transcriptomic biomarker signatures provide indications as to how immune responses are oriented and instructed to develop reactions31,32. Due to the available samples set we have focused on the analysis of biomarkers for RR; biomarkers for CoR of leprosy, leprosy classification and M. leprae exposure have also been investigated.
This is the first multi-site, prospective study analyzing unstimulated whole blood-derived RNA expression profiles of four endemic populations using dcRT-MLPA, a focused gene expression profiling platform for monitoring gene expression in large cohorts29,34. We showed that host RNA expression levels discriminate various stages of disease thereby offering potential for leprosy diagnostics. Since differentiating high exposure to M. leprae and early leprosy is difficult using serum proteins51, the here identified transcriptomic biomarkers provide potential to identify cases among contacts in high endemic regions. A limitation of the current study is the lack of follow-up of HHC in order to validate the identified biomarkers of M. leprae infection as CoR for leprosy. However, this is currently addressed in ongoing studies. Moreover, validation in an independent cohort will provide more evidence on whether transcriptomic signatures may be applied to guide prophylactic strategies by discriminating contacts highly exposed to M. leprae and at risk of developing disease.
We have identified genes differentially expressed between leprosy patients and EC or HHC. Whilst most of the genes overlap between the two comparisons, some genes discriminate leprosy patients from HHC exclusively (MBP, MSR1, TLR1, CAMTA, CXCL13 and TFGB). Thus, these genes could be correlated to development of disease. Additionally, we observed that a classification of leprosy patients is possible using transcriptomics since a set of genes were increased in BL/LL patients (CD46, CXCL10, FCGR1A, HDAC2 and TLR4) and TT/BT showed a higher expression of IL2 and TLR6.
Importantly, we identified a five-gene CoR signature (CCL2, CD8A, IL2, IL15, MARCO) for RR differentiating those developing RR ≥ 2 months prior to clinical symptoms. CCL2, was downregulated in leprosy patients without reactions compared to EC, in line with lower CCL2 expression observed in nerves of leprosy patients compared to patients with non-leprous peripheral neuropathy52. We observed a higher expression of CCL2 in future RR patients compared to patients who did not develop RR, as well as a longitudinal increase during RR onset. Similarly, a Brazilian study analysing 90 immune related genes, showed that CCL2 had the highest fold change in expression levels between leprosy patients with and without RR53 and increased CCL2 expression in RR patients has also been described in other studies35,54. CCL2 is associated with excessive deposit of extracellular matrix and macrophage recruitment55 which can be due to an increase of M. leprae antigens presented to the immune system after MDT. This leads to activation of pro-inflammatory cytokines and attraction of CD4+ T cells as confirmed by the similarly increased IL2 expression in future RR patients. Upregulation of IL2 and IL15, as well as downregulation of CTLA4 and GATA3 decreases regulation of Th2 and the lack of regulation leads to exacerbation of Th1, which is common in RR. Higher expression of MARCO in future RR patients is also in line with increased antigen presence. Differences in the expression of this scavenger receptor were also identified in several other diseases such as giant cell arteritis56, enthesitis-related arthritis57 and lupus58 suggesting a general role for antigenic triggers in respective disease etiology. IL15 which encodes a cytokine important for cytotoxic T-cell proliferation and increases GZMB expression59, was higher expressed in future RR patients, but decreased during RR in the longitudinal analysis. Upregulation of IL15 in patients who will develop RR may lead to an increase of cytotoxic T-cells and tissue destruction. CD8A is found on cytotoxic T lymphocytes, macrophages and dendritic cells leading to tissue damage in leprosy60. Even though we did not find significant difference in the expression before and at RR, previous longitudinal analysis showed increased expression at RR onset39. Thus, the decreased expression of CD8A as observed here in future RR patients when they still lack reactional symptoms (cross-sectional comparison) may indicate the number of CD8+ cells like cytotoxic T cells are increasing during RR development.
To evaluate the role of reaction-associated biomarkers as etiological and early disease prediction targets, temporal associations are implicit to indicate the utility of novel biomarkers for application in diagnostic tests. In this multi-site study, RNA expression was therefore also analyzed longitudinally: before, during reaction and after treatment. IFN-induced genes (IFI44, IFI35, IFI44L, IFI6, IFIH1, CXCL10 and FCGR1A) showed high expression at RR which decreased during reactional treatment; whereas IL15 expression decreased at RR. These biomarkers corresponding with increased inflammation can be useful when monitoring patients during their monthly dose of rifampicin helping to timely detect incipient RR. However, larger longitudinal cohorts should be studied prospectively to confirm our findings.
Previously, using longitudinal analysis of blood and skin samples of one leprosy patient who developed RR, a candidate blood-derived CoR for RR was identified composed of genetic host factors associated with T-cell cytotoxicity, regulation, vasculogenogenesis and IFN-signaling15. An interferon-dominant signature has also been identified for TB, first in 201045 and in multiple subsequent studies47,49,61. In line with our findings, higher levels of CXCL10 have also been reported in association with episodes of RR16,54. FCGR1A can differentiate between active and latent tuberculosis, suggesting a major role in the immune response against mycobacterial diseases34. Both CXCL10 and FCGR1A showed higher RNA expression during reaction as well as in BL/LL patients compared to TT/BT patients. These markers of innate immunity (CXCL10) and infection (FCGR1A) thus are increased in two different phases in leprosy associated either with M. leprae-specific T cell anergy and high bacterial load (BL/LL) or with a highly inflammatory state (RR). CXCL10 is associated with Th1 responses occurring during RR7,13,16. However, CXCL10-producing monocytes are induced during mycobacterial infections62, in line with the observed increased expression in BL/LL patients with high bacillary load. Therefore, monitoring transcriptomic changes in an individual is relevant as one marker in various conditions may reflect a different disease process.
In this study 103 target genes for innate and adaptive immune profiling were investigated in view of the immune mediated nature of the pathology of leprosy which strongly correlates with individuals’ immune responses against the bacterium. This selection could limit the discovery of novel genes that could potentially be used as biomarkers for RR. However, in contrast to studies focused at identification of disease mechanisms, for diagnostic purposes only a limited amount of discriminating genes is sufficient, leaving out genes which strongly correlate and hence do not have added value to the signature. Further in-depth transcriptomic analysis of the samples described in this study have been performed by RNASeq in a separate study with the aim to identify additional genes providing increased insight into mechanisms and pathways involved in RR63.
Although geographic differences were observed, our study showed that in prospective analyses of 4 different leprosy endemic areas, several genes are associated with onset of reactions at each site. Further analysis of the diagnostic signatures in extended cohorts worldwide need to be performed to validate the performance of the genes signatures.
Transcriptomic analyses have shown that TB31,32 disease can be characterized by 1664 to as few as 2–4 genes28. A gene set signature of reversal reaction consisting of 44 genes was previously described for the Vietnamese population using M. leprae-sonicate stimulated whole blood35. Their set signature included pro-inflammatory regulator genes such as CCL2 or IL1A in RR. We observed a decreased expression of IL1A after treatment for RR, however we did not find significant differences in patients who later developed RR compared to patients who did not have a reaction. We did not find significant differences either in other genes of that set such as IL1B, IL6, IL23A, CCL3 or CCL4. Another study instead using PBMC of leprosy patients with and without reactions identified a role for complement-associated genes54. They also found that CCL2 and MARCO, which are part of our signature biomarker, were differentially expressed in RR patients, as well as an interferon-γ significant upregulation during RR, which is in line with the increased expression of IFN-signaling genes we found longitudinally and our RNAseq data63, which shows an enrichment at timepoint of reaction for both IFN- γ and IFN-β pathways.
The here identified biomarker signature is based on unstimulated whole blood and identifies development of RR ≥ 2 months prior to clinical symptoms using a five-gene signature across four leprosy endemic populations on 3 different continents. Unstimulated whole blood and a signature with a small number of markers is more suitable for a point-of-care (POC) use as a field-friendly test. Early diagnosis and treatment of reactions are currently the primary research targets for reducing permanent neuropathy and disability development in future patients. The ability to predict reactions ≥2 months before development of clinical symptoms, using POC diagnostic tests detecting transcriptomic biomarkers for RR, would represent momentous advancements in global leprosy health care. Next steps include prospective evaluation and translation to clinically useful tools that can be implemented by clinicians. The challenge for the academic community and industry is to develop innovative methods to translate multi-transcript signatures into low-cost tests for leprosy diagnostics, suitable for use in health facilities in leprosy endemic areas.
Supplementary information
Acknowledgements
The authors gratefully acknowledge all patients for their participation. AHRI, Fiocruz, ICDDR,B, LUMC and MRL Anandaban are part of the IDEAL (Initiative for Diagnostic and Epidemiological Assays for Leprosy) Consortium. We are indebted to Mr. Kapil Dev Neupane (Anandaban), Dr. J.A. Nery, A.M. Sales, Dr. Sarno (Fiocruz), Dr. Sheikh Abdul Hadi (Dhaka), S/r Genet Amare (Addis Ababa), S/r Haregewoin Yetesha, Mr. Alemayehu Kifle and Dr. Saba M. Lambert for recruitment of study participants and sample collection. This study was supported by the Order of Malta-Grants-for-Leprosy-Research (MALTALEP), the Heiser Program for Research in Leprosy in The New York Community Trust (P13-000392), the Q.M. Gastmann-Wichers Foundation, the UNICEF/UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (TDR) and the Netherlands Leprosy Relief Foundation (NLR; ILEP#: 702.02.68, 7.01.02.48 and 701.02.49) together with the Turing Foundation.
Author contributions
Designed research: A.G., M.T. and T.O. Performed or contributed to research: A.H., J.P., K.B., M.T. and R.T. Contributed reagents/tools/samples/recruited patients: D.H., I.G., M.H., M.M., S.B., S.K., P.T., C.K., Y.B., R.O. and A.A. Analyzed the data: A.G., A.H., E.Z., J.P., K.B., M.T., R.M., R.T. and J.G. Wrote the manuscript: A.G. and M.T. Agree with manuscript: All authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-54213-y.
References
- 1.WHO. Global leprosy update, 2017: reducing the disease burden due to leprosy. Weekly epidemiological record. 2018;93:11. [Google Scholar]
- 2.Barnes PF, et al. Cytokine production at the site of disease in human tuberculosis. Infect. Immun. 1993;61:3482–3489. doi: 10.1128/iai.61.8.3482-3489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saini C, Ramesh V, Nath I. CD4+ Th17 cells discriminate clinical types and constitute a third subset of non Th1, Non Th2 T cells in human leprosy. PLoS Negl Trop Dis. 2013;7:e2338. doi: 10.1371/journal.pntd.0002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modlin RL, et al. Learning from lesions: patterns of tissue inflammation in leprosy. Proc. Natl. Acad. Sci. USA. 1988;85:1213–1217. doi: 10.1073/pnas.85.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scollard, D. M. In International textbook of leprosy (eds D. M. Scollard & T. P. Gillis) (2016).
- 6.Misch E, Berrington W, Vary J, Hawn T. Leprosy and the Human Genome. Microbiology and molecular biology reviews: MMBR. 2010;74:589–620. doi: 10.1128/MMBR.00025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geluk A. Biomarkers for leprosy: would you prefer T (cells)? Leprosy review. 2013;84:3–12. [PubMed] [Google Scholar]
- 8.Geluk A. Challenges in immunodiagnostic tests for leprosy. Expert opinion on medical diagnostics. 2013;7:265–274. doi: 10.1517/17530059.2013.786039. [DOI] [PubMed] [Google Scholar]
- 9.Lockwood DN, Saunderson P. Nerve damage in Leprosy: a continuing challenge for scientists, clinicians and service providers. Int Health. 2012;4:77–85. doi: 10.1016/j.inhe.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Scollard DM, et al. Risk factors for leprosy reactions in three endemic countries. Am J Trop Med Hyg. 2015;92:108–114. doi: 10.4269/ajtmh.13-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antunes DE, et al. Number of leprosy reactions during treatment: clinical correlations and laboratory diagnosis. Revista da Sociedade Brasileira de Medicina Tropical. 2016;49:741–745. doi: 10.1590/0037-8682-0440-2015. [DOI] [PubMed] [Google Scholar]
- 12.Ridley DS, Radia KB. The histological course of reactions in borderline leprosy and their outcome. International journal of leprosy and other mycobacterial diseases: official organ of the International Leprosy Association. 1981;49:383–392. [PubMed] [Google Scholar]
- 13.Geluk A. Correlates of immune exacerbations in leprosy. Seminars in immunology. 2018;39:111–118. doi: 10.1016/j.smim.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Hagge DA, et al. Opening a Can of Worms: Leprosy Reactions and Complicit Soil-Transmitted Helminths. EBioMedicine. 2017;23:119–124. doi: 10.1016/j.ebiom.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khadge S, et al. Longitudinal immune profiles in type 1 leprosy reactions in Bangladesh, Brazil, Ethiopia and Nepal. BMC infectious diseases. 2015;15:477. doi: 10.1186/s12879-015-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scollard DM, et al. Increased CXC ligand 10 levels and gene expression in type 1 leprosy reactions. Clin Vaccine Immunol. 2011;18:947–953. doi: 10.1128/CVI.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corstjens PL, et al. Field-Friendly Test for Monitoring Multiple Immune Response Markers during Onset and Treatment of Exacerbated Immunity in Leprosy. Clin Vaccine Immunol. 2016;23:515–519. doi: 10.1128/CVI.00033-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardus RA, et al. Clinical manifestations of leprosy after BCG vaccination: an observational study in Bangladesh. Vaccine. 2015;33:1562–1567. doi: 10.1016/j.vaccine.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Raffe SF, et al. Diagnosis and treatment of leprosy reactions in integrated services - the patients’ perspective in Nepal. PLoS Negl Trop Dis. 2013;7:e2089. doi: 10.1371/journal.pntd.0002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Croft RP, et al. A clinical prediction rule for nerve function impairment in leprosy patients-revisited after 5 years of follow-up. Leprosy review. 2003;74:35–41. [PubMed] [Google Scholar]
- 21.Assis BPN, Lyon S, Grossi MAF, Rocha M. Risk factors for physical disability upon release from multidrug therapy in new cases of leprosy at a referral center in Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo. 2019;61:e13. doi: 10.1590/s1678-9946201961013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahia El Idrissi N, et al. Complement activation in leprosy: a retrospective study shows elevated circulating terminal complement complex in reactional leprosy. Clinical and experimental immunology. 2016;184:338–346. doi: 10.1111/cei.12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Negera E, et al. Complement C1q expression in Erythema nodosum leprosum. PLoS Negl Trop Dis. 2018;12:e0006321. doi: 10.1371/journal.pntd.0006321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fava VM, Sales-Marques C, Alcais A, Moraes MO, Schurr E. Age-Dependent Association of TNFSF15/TNFSF8 Variants and Leprosy Type 1 Reaction. Frontiers in immunology. 2017;8:155. doi: 10.3389/fimmu.2017.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stefani MM, et al. Potential plasma markers of Type 1 and Type 2 leprosy reactions: a preliminary report. BMC infectious diseases. 2009;9:75. doi: 10.1186/1471-2334-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer A, et al. Serum levels of interferon-gamma, tumour necrosis factor-alpha, soluble interleukin-6R and soluble cell activation markers for monitoring response to treatment of leprosy reactions. Clinical and experimental immunology. 2007;150:210–216. doi: 10.1111/j.1365-2249.2007.03485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maertzdorf J, et al. Concise gene signature for point-of-care classification of tuberculosis. EMBO Mol Med. 2015;8:86–95. doi: 10.15252/emmm.201505790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sutherland JS, et al. Differential gene expression of activating Fcgamma receptor classifies active tuberculosis regardless of human immunodeficiency virus status or ethnicity. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2014;20:O230–O238. doi: 10.1111/1469-0691.12383. [DOI] [PubMed] [Google Scholar]
- 29.Haks MC, Goeman JJ, Magis-Escurra C, Ottenhoff TH. Focused human gene expression profiling using dual-color reverse transcriptase multiplex ligation-dependent probe amplification. Vaccine. 2015;33:5282–5288. doi: 10.1016/j.vaccine.2015.04.054. [DOI] [PubMed] [Google Scholar]
- 30.Anderson ST, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. The New England journal of medicine. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sloot R, et al. Biomarkers Can Identify Pulmonary Tuberculosis in HIV-infected Drug Users Months Prior to Clinical Diagnosis. EBioMedicine. 2015;2:172–179. doi: 10.1016/j.ebiom.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zak DE, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet (London, England) 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joosten SA, et al. Identification of biomarkers for tuberculosis disease using a novel dual-color RT-MLPA assay. Genes Immun. 2012;13:71–82. doi: 10.1038/gene.2011.64. [DOI] [PubMed] [Google Scholar]
- 34.Gebremicael G, et al. Host Gene Expression Kinetics During Treatment of Tuberculosis in HIV-Coinfected Individuals Is Independent of Highly Active Antiretroviral Therapy. J Infect Dis. 2018;218:1833–1846. doi: 10.1093/infdis/jiy404. [DOI] [PubMed] [Google Scholar]
- 35.Orlova M, et al. Gene set signature of reversal reaction type I in leprosy patients. PLoS Genet. 2013;9:e1003624. doi: 10.1371/journal.pgen.1003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis. 1966;34:255–273. [PubMed] [Google Scholar]
- 37.Roche PW, Le MJ, Butlin CR. Risk factors for type 1 reactions in leprosy. International journal of leprosy and other mycobacterial diseases: official organ of the International Leprosy Association. 1997;65:450–455. [PubMed] [Google Scholar]
- 38.Walker SL, et al. Development and validation of a severity scale for leprosy type 1 reactions. PLoS Negl Trop Dis. 2008;2:e351. doi: 10.1371/journal.pntd.0000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geluk A, et al. Longitudinal immune responses and gene expression profiles in type 1 leprosy reactions. Journal of clinical immunology. 2014;34:245–255. doi: 10.1007/s10875-013-9979-x. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 41.Goeman JJ, van de Geer SA, de KF, van Houwelingen HC. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 42.Zhang FR, et al. Genomewide association study of leprosy. The New England journal of medicine. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 43.Moet FJ, Meima A, Oskam L, Richardus JH. Risk factors for the development of clinical leprosy among contacts, and their relevance for targeted interventions. Lepr. Rev. 2004;75:310–326. [PubMed] [Google Scholar]
- 44.Gillini L, Cooreman E, Wood T, Pemmaraju VR, Saunderson P. Global practices in regard to implementation of preventive measures for leprosy. PLoS Negl Trop Dis. 2017;11:e0005399. doi: 10.1371/journal.pntd.0005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maertzdorf J, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12:15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 47.Ottenhoff TH, et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PloS one. 2012;7:e45839. doi: 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cliff JM, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis. 2013;207:18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 49.Teles RM, et al. Type I interferon suppresses type II interferon-triggered human anti-mycobacterial responses. Science (New York, N.Y.) 2013;339:1448–1453. doi: 10.1126/science.1233665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaitanya VS, et al. Cortisol and proinflammatory cytokine profiles in type 1 (reversal) reactions of leprosy. Immunology letters. 2013;156:159–167. doi: 10.1016/j.imlet.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 51.van Hooij A, et al. Quantitative lateral flow strip assays as User-Friendly Tools To Detect Biomarker Profiles For Leprosy. Scientific reports. 2016;6:34260. doi: 10.1038/srep34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guerreiro LT, et al. Gene expression profiling specifies chemokine, mitochondrial and lipid metabolism signatures in leprosy. PloS one. 2013;8:e64748. doi: 10.1371/journal.pone.0064748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rego JL, et al. Whole blood profiling of leprosy type 1(reversal) reactions highlights prominence of innate immune response genes. BMC infectious diseases. 2018;18:422. doi: 10.1186/s12879-018-3348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupnik KM, et al. Transcriptional changes that characterize the immune reactions of leprosy. J Infect Dis. 2015;211:1658–1676. doi: 10.1093/infdis/jiu612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medeiros MF, et al. CXCL10, MCP-1, and other immunologic markers involved in neural leprosy. Applied immunohistochemistry & molecular morphology: AIMM. 2015;23:220–229. doi: 10.1097/PAI.0000000000000074. [DOI] [PubMed] [Google Scholar]
- 56.Burja B, et al. Utility of serological biomarkers for giant cell arteritis in a large cohort of treatment-naive patients. Clinical rheumatology. 2019;38:317–329. doi: 10.1007/s10067-018-4240-x. [DOI] [PubMed] [Google Scholar]
- 57.Gaur P, Myles A, Misra R, Aggarwal A. Intermediate monocytes are increased in enthesitis-related arthritis, a category of juvenile idiopathic arthritis. Clinical and experimental immunology. 2017;187:234–241. doi: 10.1111/cei.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han S, et al. Journal of immunology (Baltimore, Md.: 1950) 2017. A Novel Subset of Anti-Inflammatory CD138(+) Macrophages Is Deficient in Mice with Experimental Lupus; pp. 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balin Samuel J., Pellegrini Matteo, Klechevsky Eynav, Won Sohui T., Weiss David I., Choi Aaron W., Hakimian Joshua, Lu Jing, Ochoa Maria Teresa, Bloom Barry R., Lanier Lewis L., Stenger Steffen, Modlin Robert L. Human antimicrobial cytotoxic T lymphocytes, defined by NK receptors and antimicrobial proteins, kill intracellular bacteria. Science Immunology. 2018;3(26):eaat7668. doi: 10.1126/sciimmunol.aat7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ottenhoff TH, Mutis T. Role of cytotoxic cells in the protective immunity against and immunopathology of intracellular infections. Eur. J. Clin. Invest. 1995;25:371–377. doi: 10.1111/j.1365-2362.1995.tb01716.x. [DOI] [PubMed] [Google Scholar]
- 61.Maertzdorf J, et al. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joosten SA, et al. Mycobacterial growth inhibition is associated with trained innate immunity. The Journal of clinical investigation. 2018;128:1837–1851. doi: 10.1172/JCI97508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teles RMB, et al. Identification of a systemic interferon-γ inducible antimicrobial gene signature in leprosy patients undergoing reversal reaction. PLoS Negl Trop Dis. 2019;13:e0007764. doi: 10.1371/journal.pntd.0007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Levin M, Kaforou M. Predicting active tuberculosis progression by RNA analysis. Lancet (London, England) 2016;387:2268–2270. doi: 10.1016/S0140-6736(16)00165-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.