Abstract
Double-positive (DP) thymocytes undergo positive selection to become mature single-positive CD4+ and CD8+ T cells in response to T cell receptor (TCR) signaling. Unlike mature T cells, DP cells must respond to low-affinity self-peptide-MHC ligands before full upregulation of their surface TCR expression can occur. Thus, DP thymocytes must be more sensitive to ligands than mature T cells. A number of molecules have been found that are able to enhance the strength of the TCR signal to facilitate positive selection. However, almost all of these molecules are also active in mature T cells. Themis (thymocyte expressed molecule involved in selection) and Tespa1 (thymocyte expressed positive selection associated 1) are two recently discovered molecules essential for optimal TCR signaling and thymocyte development. A deficiency in both molecules leads to defects in positive selection. Here, we compared the relative contributions of Themis and Tespa1 to positive selection in thymocytes. We show that Tespa1 deficiency led to more limited and specific gene expression profile changes in cells undergoing positive selection. In mixed bone marrow transfer experiments, Tespa1−/− cells showed more severe defects in thymocyte development than Themis−/− cells. However, Tespa1−/− cells showed a substantial degree of homeostatic expansion and became predominant in the peripheral lymphoid organs, suggesting that Tespa1 is a thymic-specific TCR signaling regulator. This hypothesis is further supported by our observations in Tespa1 conditional knockout mice, as Tespa1 deletion in peripheral T cells did not affect TCR signaling or cell proliferation. The different regulatory effects of Tespa1 and Themis are in accordance with their nonredundant roles in thymocyte selection, during which Tespa1 and Themis double knockouts showed additive defects.
Keywords: T cell development, Positive selection, TCR signaling
Subject terms: Thymus, T-cell receptor
Introduction
T cell precursors from bone marrow (BM) complete their developmental process in the thymus before becoming mature single-positive (SP) CD4+ and CD8+ cells. The developmental process of thymocytes can be divided into three main stages defined by the expression of the cell surface proteins CD4 and CD8: the double-negative (DN) stage without the expression of CD4 or CD8, the double-positive (DP) stage with the expression of both CD4 and CD8, and the SP stage with the expression of either CD4 or CD8. After expressing T cell receptors on their surface after successive VDJ recombination, DP thymocytes are subjected to positive selection, in which only cells that are able to react to self-peptide-MHC stimulation and generate a sufficient T cell receptor (TCR) signal intensity will survive.1 Upon TCR stimulation, tyrosine residues in the immunoreceptor tyrosine-based activation motifs of CD3 are phosphorylated by the protein tyrosine kinase Lck, which then recruits Zap-70 to activate phospholipase C γ1 (PLCγ1). PLCγ1 produces diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), which are essential for the subsequent signals that are needed to activate the transcription factors NFAT, NF-κB and AP-1.2
It has long been acknowledged that thymocytes undergo positive selection by low-affinity cross-reactive self-peptides. Positive selection is based on the fact that DP cells have higher TCR sensitivity than mature peripheral T cells, which allows them to respond to low-affinity self-peptide-MHC complexes.3 This phenomenon cannot be explained by the overall TCR level; however, DP thymocytes are immature T cells with lower numbers of TCRs expressed on their surfaces. The presence of thymic-specific TCR signaling-positive modulators has been proposed, which could account for the increased TCR sensitivity observed in this context. Over the past few decades, a variety of signal regulators with the capacity to augment TCR-mediated signaling have been discovered.4 To date, none have been defined as stage-specific positive selection regulators. However, the recent discoveries of Themis (thymocyte expressed molecule involved in selection) and Tespa1 (thymocyte expressed positive selection associated 1) have provided promising candidates for such regulators. The deficiency of Themis in mice results in impaired thymocyte selection.5–9 Themis has been shown to enhance TCR signaling by either promoting Vav1 activity and Grb2 stability or by inhibiting SHP1 activation.10,11 The latter study also suggested that Themis played a reactive oxygen species-dependent, thymic-specific regulatory role.12,13 In a previous study, we showed that Tespa1 plays an essential role in mediating TCR signaling in thymocytes. We found that Tespa1 bound the N-terminal regulatory domain of the ER calcium channel IP3R1 and recruited IP3R1 to the proximal region of the TCR, thus facilitating the optimal activation of IP3R1 and downstream calcium flux.14 Deficiency of Tespa1 in mice leads to defects in calcium flux and impaired positive selection.15 Although a substantial reduction in SP CD4+ and CD8+ thymocytes was observed in Tespa1 knockout mice, they were able to partially restore peripheral CD4+ and CD8+ cell levels as they aged, suggesting the restricted role of Tespa1 in positive selection.
In this study, we compared the relative contributions and stage specificity of Themis and Tespa1 during thymocyte development. We found that Tespa1 deficiency influences a group of genes in thymocytes that influence cells undergoing positive selection. In BM chimeras, Tespa1−/− T cells achieved improved recovery through homeostatic proliferation in the periphery instead of their more severe developmental defect in thymus when compared to Themis−/− T cells. Moreover, Tespa1 deficiency in peripheral T cells did not result in any defects in TCR signaling and TCR-induced proliferation, thereby indicating the stage-specific function of Tespa1 in the regulation of TCR signaling during positive selection.
Results
Differences in transcriptional patterns between Tespa1−/− and Themis−/− thymocytes
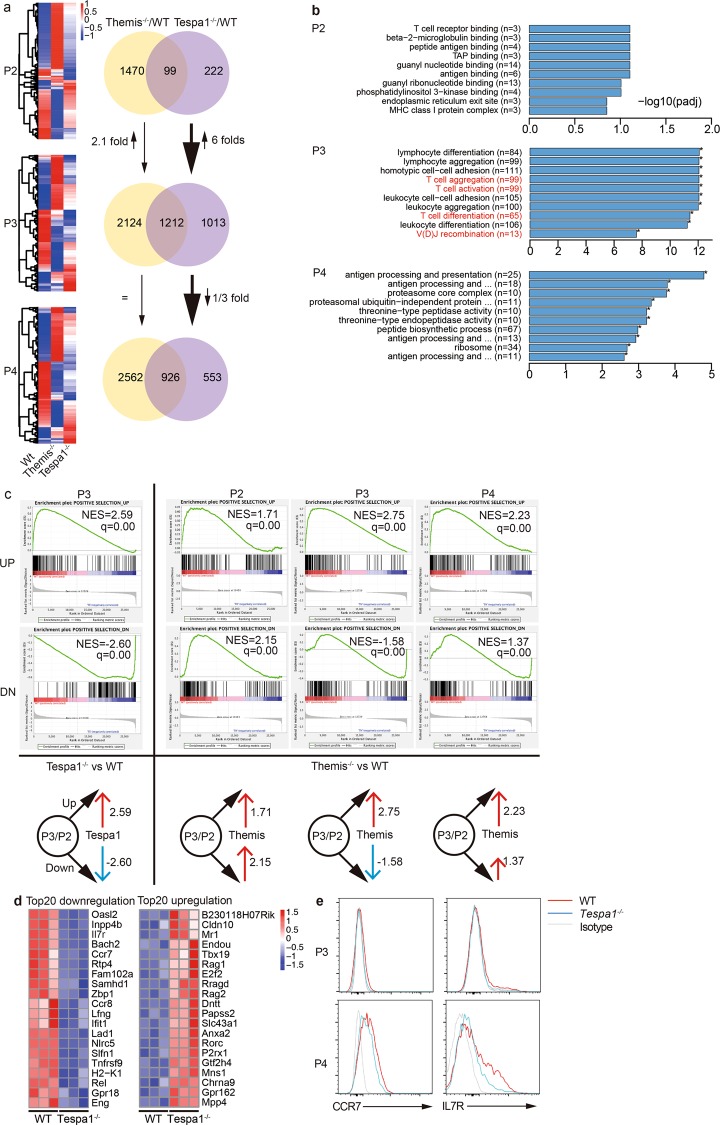
To pinpoint the role of Tespa1 during positive selection, we classified thymocytes according to the different stages of positive selection (P2, preselection; P3, selection; P4, postselection) for sorting (Supplementary Fig. 1a).5,7,16 We then compared the transcriptomes of thymocytes from wild-type (WT), Themis−/−, and Tespa1−/− mice. The validity of the sequencing data was indicated by the detection of two marker genes, CD5 and CD69, whose expression levels were related to different stages of positive selection (Supplementary Fig. 1b). Both CD5 and CD69 had low expression in the preselection stage, high expression in the selection stage, and decreased expression after the postselection stage, which was consistent with the results of previous studies.17,18 Significantly altered patterns of gene expression in WT, Themis−/−, and Tespa1−/− thymocytes from P2, P3, and P4 were observed (Fig. 1a). Compared to WT mice, we found that Themis-deficient mice showed more profound gene transcription changes (1569 vs. 321 in Tespa1−/− thymocytes) in the P2 stage, yet the changes in only 99 genes were found to be the same in both Themis−/− and Tespa1−/− thymocytes. In the P3 stage, the expression from more than 3000 genes was significantly different, while differences in the expression of an additional 1212 genes were shared by Themis−/− and Tespa1−/− thymocytes. Furthermore, the total number of genes with altered expression was approximately 6-fold higher in Tespa1−/− thymocytes in the P3 stage compared to those in the P2 stage, while in the equivalent comparison of Themis−/− thymocytes, only a 1.1-fold increase was observed in gene number. During comparison of the P3 and P4 stages, there was a decrease of one-third in the genes with changed expression in Tespa1−/− thymocytes, whereas the number of changed genes was nearly equal in the P3 and P4 stages in Themis−/− thymocytes (Fig. 1a). These results suggested a limited role of Tespa1 during thymocyte selection.
Fig. 1.
Differential transcriptional patterns in Themis (thymocyte expressed molecule involved in selection)- and Tespa1 (thymocyte expressed positive selection associated 1)-deficient thymocytes. a Heat maps (left) showing significantly changed gene expression (blue, low expression; red, high expression) in Themis−/− or Tespa1−/− thymocytes compared to that in wild-type (WT) thymocytes in the P2, P3, and P4 stages. The numbers in the Venn diagram (right) indicate genes with significantly changed expression only in Themis−/− (yellow) or Tespa1−/− (violet) thymocytes and genes with changed expression in both (overlap). b The top 10 GO enrichment terms most significantly associated with WT vs. Tespa1−/− cells in different stages. Gene ontogeny (GO) items in red text are related to T cell specific events. c Gene set enrichment analysis (GSEA) analysis: left, WT vs. Tespa1−/−; right, WT vs. Themis−/−. q specifies the adjusted p value. NES specifies normalized enrichment score. The diagrams below represent the relationship between the Tespa1/Themis and Positive gene sets. The black arrow denotes “Positive selection_UP” and “Positive selection_DN”. A red arrow denotes a positive correlation; a blue arrow denotes a negative correlation. The numbers denote the NES values. d Top 20 differentially expressed genes in Tespa1−/− thymocytes in the P3 stage. Left, the top 20 downregulated genes. Right, the top 20 upregulated genes. e Surface expression of CCR7 and IL7R in thymocytes in the P3 and P4 stages. The red line specifies WT; the blue line specifies Tespa1−/−; the gray line specifies the isotype. The samples used for sequencing were pooled from six WT, Themis−/−, and Tespa1−/− mice. Three biological replicates were sequenced
To investigate whether these changed genes are related to thymocyte development, we performed gene ontogeny (GO) enrichment analysis. We found that there were no GO terms that were significantly enriched in the P2 stage in Tespa1−/− samples, while four of the top ten GO enrichment terms were related to T cell function only in the P3 stage (Fig. 1b). In Themis−/− thymocytes, T cell function was not ranked within the top 10 enriched GO terms (Supplementary Fig. 1c), yet mitochondrion- and respiratory chain-related terms were significantly enriched, indicating the possible role of Themis in energy metabolism. To further confirm whether Tespa1 influences thymocyte development, we generated two gene sets consisting of 188 upregulated genes and 193 downregulated genes that were screened in both the Immgen database and our own WT thymocyte RNA-sequencing (RNA-seq) data (Supplementary Table 1). These two gene sets were referred to as “Positive selection_UP” and “Positive selection_DN”, respectively. We then performed GSEA (gene set enrichment analysis) with the “Positive selection” gene sets were generated. Compared with Themis−/− thymocytes, which showed the significant positive enrichment of both gene sets in the P2 and P4 developmental stages, we found that Tespa1−/− thymocytes did not show any significant enrichment of the “Positive selection_UP” gene set or any negative enrichment of the “Positive selection_DN” gene set in the P2 or P4 stages, which reflects the minimal influence of Tespa1 deficiency on thymocytes in the P2 and P4 stages (Fig. 1c). Both Tespa1 and Themis deficiency led to the significant positive enrichment of the “Positive selection_UP” gene set and negative enrichment of the “Positive selection_DN” gene set, which indicate the regulatory roles of both Tespa1 and Themis in the P3 stage. Nevertheless, Tespa1 deficiency led to the enrichment of more genes within these two gene sets than Themis deficiency, indicating the limited but influential role of Tespa1 in the P3 stage (Fig. 1c). Among the genes that made the greatest contribution to gene set enrichment in P3 Tespa1−/− thymocytes, the levels of Ccr7 and Il7r, which are essential for positive selection, were decreased, resulting in their lower expression in the P4 stage. Rag1/2 and Rorc, which are expressed in the preselection stage and should be suppressed during positive selection,19 were found to be increased. These results indicate the vital role of Tespa1 in positive selection in the P3 stage (Fig. 1d). To determine the biological state of Tespa1-deficient thymocytes, we performed an enrichment analysis by using hallmark gene sets to compare WT and Tespa1−/− thymocytes at different stages. The hallmark gene sets consist of 50 gene sets that characterize and represent specific well-defined biological states or processes and exhibit coherent expression.20 We found that the gene set named “TNFα_signal_via NFκB,” which was crucial to thymocyte survival in positive selection, was continuously shown to be enriched in the P2 stage and the P3 stage (Supplementary Fig. 1d) in Tespa1−/− thymocytes.21 Compared with Themis−/− thymocytes, the expression of the enriched gene sets were drastically increased in the P3 stage compared to those in the P2 stage in Tespa1−/− thymocytes (Supplementary Fig. 1d, e). These data imply that Tespa1 makes a distinct contribution in the P3 stage or during the transition from P2 to P3.
Tespa1 deficiency leads to a profound defect in positive selection
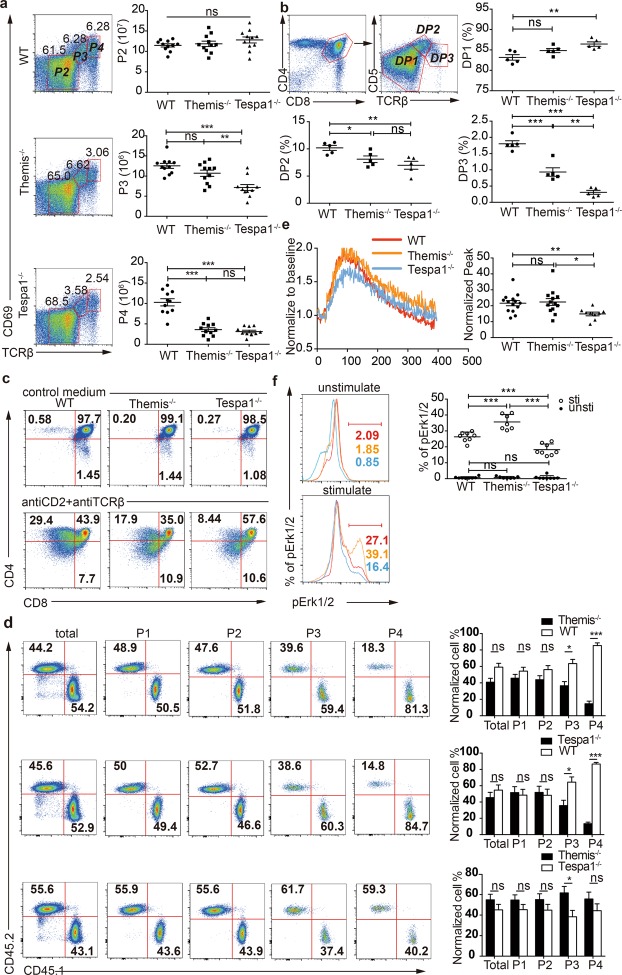
When we compared thymocytes from the P2, P3, and P4 stages from WT, Themis−/−, and Tespa1−/− mice, we found a more severe defect in terms of cell numbers in the P3 stage in Tespa1−/− thymocytes (Fig. 2a). We performed a more accurate classification by gating for the DP1 (CD5lo-intTCRβlo), DP2 (CD5hiTCRβint), and DP3 (CD5hi-intTCRβhi) populations within CD4+CD8+ thymocytes.22,23 It was found that the percentage of thymocytes was diminished in DP2 and DP3 Tespa1−/− and Themis−/− thymocytes (Fig. 2b). To ensure the same developmental conditions, we mixed Themis−/− and Tespa1−/− DP thymocytes for a two-stage differentiation assay in vitro to replicate the initial stage of positive selection in vivo.24 In contrast to WT DP thymocytes, both Themis−/− and Tespa1−/− DP thymocytes showed an impaired ability to develop into the CD4+CD8lo cells, and this defect was more severe in Tespa1−/− thymocytes7,11 (Fig. 2c, Supplementary Fig. 2a). BM competing chimeras were then constructed to compare the impairment of positive selection mediated by Themis and Tespa1 deficiency in vivo. After reconstitution, thymocytes from the chimeras were classified according to CD69 and TCRβ expression into P1 (CD69negTCRβneg-lo, mostly DN and DP cells), P2 (CD69negTCRβlo-int, preselected DP cells), P3 (CD69intTCRβint-hi, selected cells), P4 (CD69hiTCRβhi, postselected cells), and P5 (CD69loTCRβhi) subpopulations. As previously reported,5,7,15 both Themis and Tespa1 deficiency impaired positive selection, resulting decreased numbers of thymocytes in the P3 stage. Interestingly, Tespa1-deficient thymocytes exhibited more severe impairment compared to Themis-deficient thymocytes in the P3 stage (Fig. 2d). Since the TCR signal intensity is involved in positive selection and a decreased TCR signal intensity results in impaired positive selection,1 we studied two hallmarks of TCR signaling, calcium flux and MAPK phosphorylation25,26, to determine the TCR intensity via anti-CD3 stimulation in vitro. We found that calcium flux and the Erk pathway were impaired in Tespa1−/− thymocytes compared to those in Themis−/− thymocytes7,9,27 (Fig. 2e, f). These findings demonstrate that Tespa1 has a more profound effect than Themis during the positive selection of thymocytes.
Fig. 2.
Impaired positive selection in Themis−/− (thymocyte expressed molecule involved in selection) and Tespa1−/− (thymocyte expressed positive selection associated 1) thymocytes. a Surface staining of CD69 and TCRβ in wild-type (WT), Themis−/−, and Tespa1−/− thymocytes. The numbers adjacent to the outlined areas indicate the cell percentages. Right, quantification of the P2, P3, and P4 thymocyte populations from WT, Themis−/−, and Tespa1−/− mice. b Surface staining of CD5 and TCRβ in WT, Themis−/−, and Tespa1−/− thymocytes gated on CD4+ and CD8+ double-positive populations. The numbers adjacent to the outlined areas indicate the cell percentages. Lower right, quantification of the DP1, DP2, and DP3 thymocyte populations from WT, Themis−/− and Tespa1−/− mice. c Staining of CD4 and CD8 in sorted DP thymocytes from WT, Themis−/−, and Tespa1−/− mice. Top row, unstimulated DP cells cultured for 20 h in medium; bottom row, DP cells stimulated by anti-CD2 and anti-TCRβ for 20 h with additional culture for 12 h in medium without antibodies. Data are representative of four separate experiments in one mouse per group. Thymocytes were obtained from CD45.2 WT, CD45.2 Themis−/−, and CD45.1 Tespa1−/− mice. d Flow cytometry analysis of thymocytes from Rag1−/− bone marrow chimeras at different stages (P1, CD69negTCRβlow; P2, CD69lowTCRβlow; P3, CD69intTCRβint; P4, CD69hiTCRβhi) defined according to CD69 and TCRβ. Staining of CD45.1 and CD45.2 in thymocytes from chimeras transferred from WT and Themis−/− bone marrrow (BM) (upper row), WT and Tespa1−/− BM (middle row), and Tespa1−/− and Themis−/− BM (bottom row). The bar graph (right) shows the quantification of the populations indicated. Data from each stage were normalized to the percentages of cells measured preinjection; n = 3 for WT and Themis−/−, n = 4 for Tespa1−/− chimeras, and n = 5 for Themis−/− and Tespa1−/− chimeras. e Calcium flux in Fluo-4 AM-labeled preselection thymocytes from WT, Themis−/−, and Tespa1−/− mice. The fluorescence intensity of Fluo-4 was normalized to the unstimulated baseline. Right, statistics for the normalized peak values of the calcium flux assays. f Phospho-flow analysis of pErk levels in WT, Themis−/−, and Tespa1−/− preselection thymocytes. Right, the quantification of pErk-positive cells. Each symbol represents an individual mouse (a, b) or a biological and technical replicate (e, f). The small horizontal lines indicate the means (±SEM); ns = not significant; *p<0.05; **p<0.01; ***p<0.0001. Student’s t test was used for a, b, and d; the Mann–Whitney test was used for e, f
Tespa1 gene knockout has less effect than Themis gene knockout on peripheral T cells
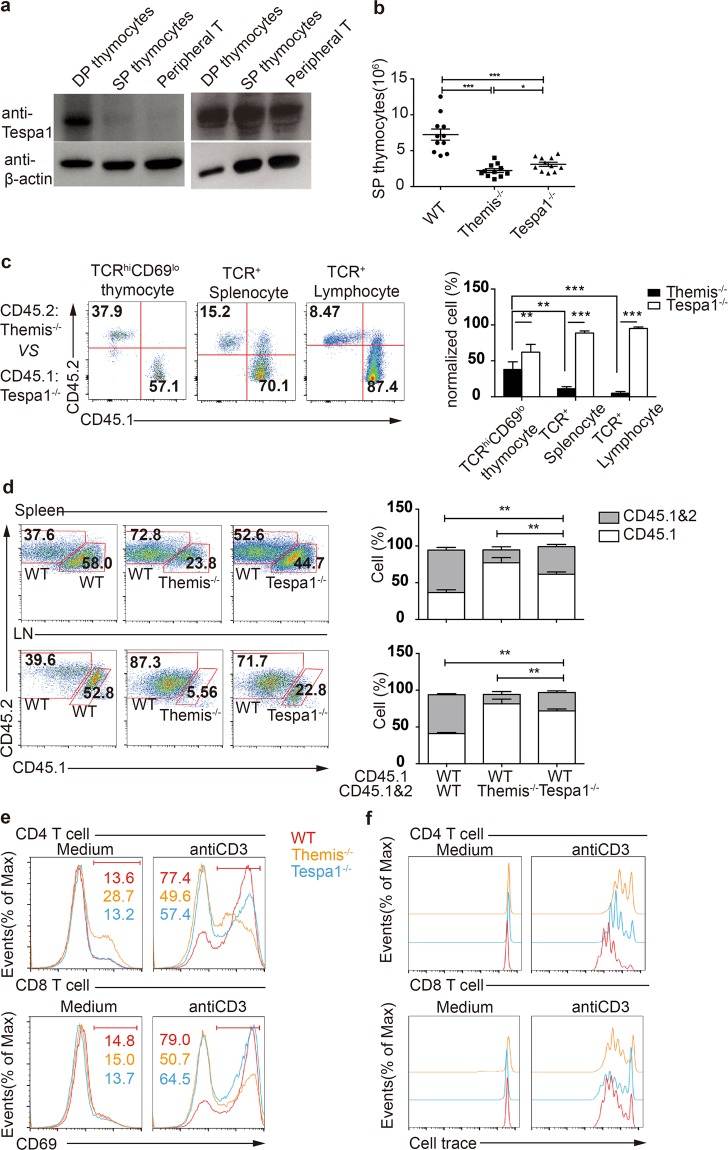
Tespa1 shows low expression in SP thymocytes and peripheral T cells (Fig. 3a), and thus it is necessary to clarify whether this expression pattern indicates the reduced function of Tespa1 in mature T cells. In contrast to DP thymocytes, we found that the reduction of SP cells in Tespa1−/− mice was less obvious than that in Themis−/− mice (Fig. 3b). Moreover, we found that the percentage of Tespa1−/− BM-derived SP thymocytes was greater than that of Themis−/− BM-derived SP thymocytes in peripheral T cells in mixed chimeras (Fig. 3c). The relative difference in the percentages of Tespa1−/− and Themis−/− peripheral T cells was further increased because of lymphopenia-induced homeostatic proliferation in Rag1−/− hosts (Fig. 3c, Supplementary Fig. 3a). To further confirm the decrease in the homeostatic proliferation of Tespa1−/− peripheral T cells, we performed proliferation assays in vivo by transferring equal numbers of naive CD4+ T cells into Rag1−/− mice. We found that both Themis−/− T cells and Tespa1−/− T cells showed impaired homeostatic proliferation relative to WT naive CD4+ T cells, while the impairment of Tespa1−/− naive CD4+ T cells was less significant than that of Themis−/− T cells (Fig. 3d). Since the TCR signaling intensity is crucial for homeostatic proliferation,28 we next investigated TCR signaling by detecting TCR-driven CD69 upregulation and proliferation. We found higher CD69 expression and CellTrace dilution in Tespa1−/− splenocytes than in Themis−/− splenocytes after anti-CD3 stimulation, although both were impaired relative to WT splenocytes (Fig. 3e, f). Collectively, these data indicate that Tespa1 deficiency is less important in regulating TCR signaling and homeostatic proliferation in peripheral T cells, which is likely due to the downregulated expression level of Tespa1 in these cells.
Fig. 3.
Less efficient function of Tespa1 (thymocyte expressed positive selection associated 1) in the periphery. a Western blot analysis of Tespa1 expression in double-positive (DP), single-positive (SP), and peripheral T cells. b Numbers of CD69loTCRβ+ SP thymocytes in wild-type (WT), Themis−/− (thymocyte expressed positive selection associated 1) and Tespa1−/− mice. c Flow cytometry analysis of the total number of Themis−/− BM-derived T cells and Tespa1−/− BM-derived T cells in chimeras. Staining of CD45.1 and CD45.2 in CD69loTCRβ+ thymocytes (left), TCRβ+ splenocytes (middle), or lymph node cells (right). The bar graph shows the quantification of the populations indicated. d Proliferation of naive polyclonal CD4+ T cells from the spleen (upper row) and lymph nodes (bottom row). Rag1−/− mice were injected with WT, Themis−/− and Tespa1−/− CD4+ T cells as indicated. The bar graphs (right) show the quantification of the populations indicated; n = 4 for each group. e CD69 staining in gated CD4+ and CD8+ cells in WT, Themis−/−, and Tespa1−/− total splenocytes. Data are representative of three independent experiments. f Flow cytometry analysis of the CellTrace dilution in WT, Themis−/−, and Tespa1−/− CD4+ and CD8+ splenocytes unstimulated (gray shaded curves) or stimulated for 72 h with anti-CD3. Data are representative of three independent experiments. The small horizontal lines in the bar graphs indicate the means (±SEM); **p < 0.01; ***p < 0.0001. Student’s t test was used for a–c
Tespa1 conditional gene knockout mice show normal TCR signaling
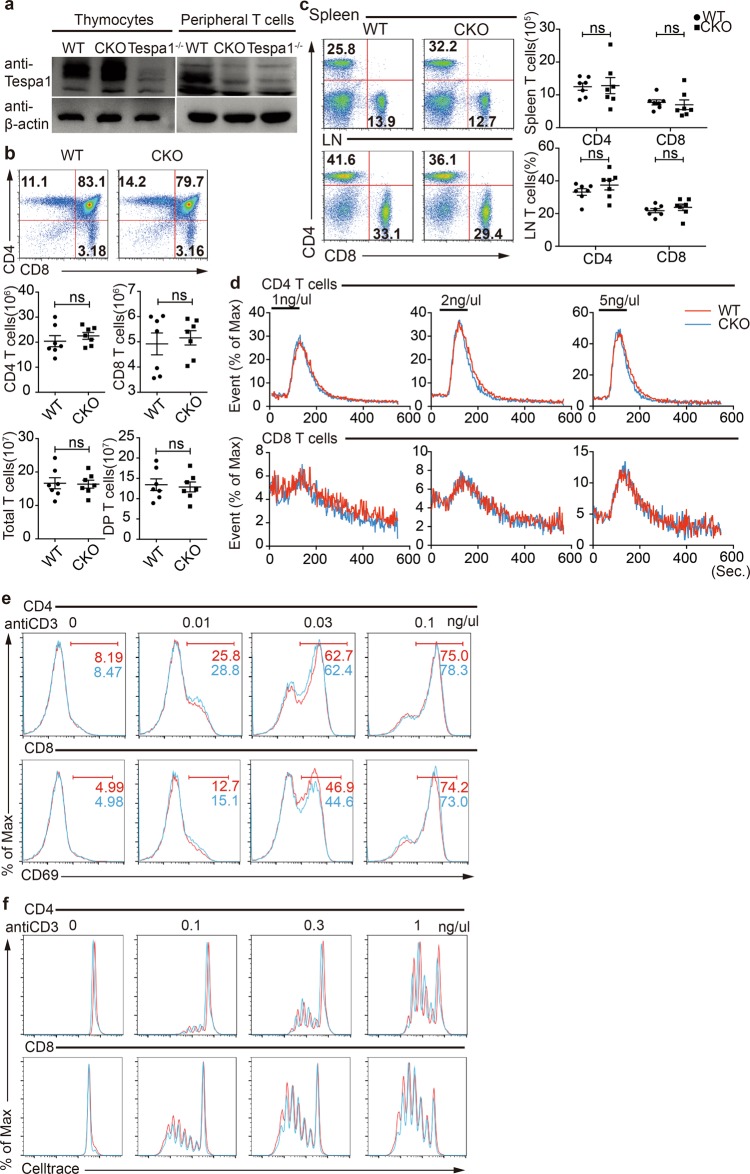
We wanted to further elucidate the function of Tespa1 in peripheral T cells without any possible influence from factors associated with the developmental stage. We thus generated Tespa1 conditional knockout mice. We acquired the floxed allele of the Tespa1 gene and used distal Lck-cre transgenic mice to eliminate Tespa1 in peripheral T cells (Fig. 4a, Supplementary Fig. 4a), and we referred to the generated mice as CKO mice. We found that the number of thymocytes in different subsets was normal in CKO mice (Fig. 4b), while the number of peripheral T cells in CKO mice was also normal in terms of quantities and percentages (Fig. 4c). As Tespa1−/− DP thymocytes exhibited impaired TCR signaling, we next determined TCR signaling in CKO peripheral T cells by measuring TCR-induced calcium flux and activation marker upregulation. We found no difference between WT and CKO T cells in either calcium flux or CD69 upregulation (Fig. 4d, e). Moreover, in an assay of in vitro proliferation, CKO CD4+ or CD8+ T cells showed an equivalent proliferative capacity relative to that of WT T cells (Fig. 4f). Overall, these data indicate that Tespa1 deficiency in peripheral T cells has no influence on TCR signaling, suggesting that Tespa1 is dispensable in peripheral T cells.
Fig. 4.
Phenotypic analysis of Tespa1 (thymocyte expressed positive selection associated 1) conditional knockout (CKO) mice. a Confirmation of Tespa1 deficiency in the periphery by Western blot analysis. b Flow cytometry analysis of CD4- and CD8-stained thymocytes from wild-type (WT) and CKO mice. The numbers in the squares indicate the percentages of the subpopulations. The cell numbers of each population are shown below. c Flow cytometry analysis of CD4+ T cells and CD8+ T cells in the spleen and lymph nodes from WT and CKO mice. The numbers in the squares indicate the percentages of the subpopulations. The cell numbers or cell percentages of each population are shown on the right. d T cell receptor (TCR) stimulation-induced calcium flux in CD4+ or CD8+ peripheral T cells was measured in different concentrations of anti-CD3. e TCR stimulation-induced CD69 upregulation of CD4+ or CD8+ peripheral T cells was measured in different concentrations of anti-CD3 . f TCR stimulation-induced proliferation of CD4+ or CD8+ peripheral T cells was detected in different concentrations of anti-CD3. Proliferation was indicated by the dilution of the CellTrace dye. Data are representative of three independent experiments. The small horizontal lines in the bar graphs indicate the means (±SEM); ns = not significant
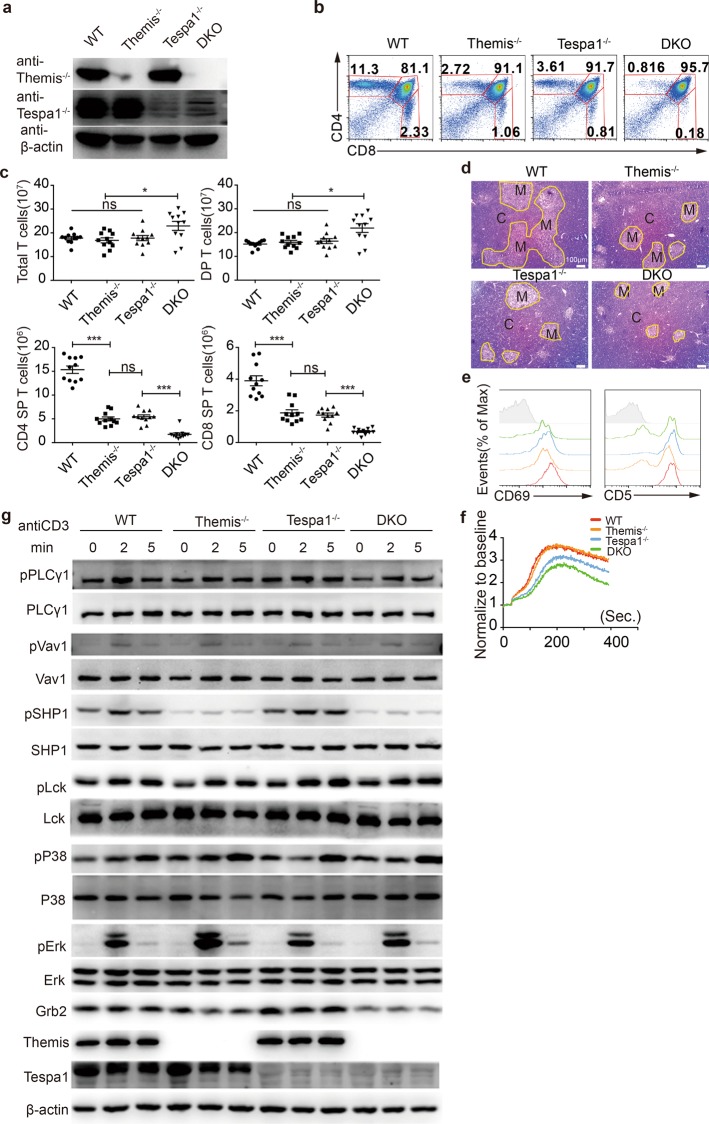
Additive defects in Tespa1 and Themis double knockout mice
Previous studies have suggested that Tespa1 and Themis have different functions in regulating TCR signaling.10,11,14 However, our data above indicated a limited role played by Tespa1 in the positive selection stage only. We thus reasoned that Tespa1 and Themis may work independently in regulating T cell development. To further determine whether there is any redundancy between Tespa1 and Themis, we generated Themis and Tespa1 double knockout mice (hereafter known as DKO mice). Immunoblotting analysis found no compensatory expression in Themis and Tespa1 in single knockout mice (Fig. 5a). DKO mice showed an additive reduction in both CD4+ and CD8+ SP thymocytes (Fig. 5b, c). Reduced thymocyte numbers are known to alter the structure of the thymus,29 so we investigated this phenomenon in thymus sections from DKO mice. We found a much smaller medulla area in the thymus in DKO mice compared to Themis−/− or Tespa1−/− mice (Fig. 5d). Impaired thymocyte development in the thymus will also induce a defect in the periphery, leading to lymphopenia. Therefore, we further investigated peripheral T cells in DKO mice. We found a reduced number of CD4+ and CD8+ T cells in both spleens and lymph nodes from DKO mice (Supplementary Fig. 5a). Lymphopenia can induce cell proliferation, leading to higher percentages of memory cells. The highest percentages of memory cells were observed in DKO spleens and lymph nodes, which was in line with the fact that the most severe lymphopenia was observed in DKO mice (Supplementary Fig. 5b, c). These data therefore indicate that DKO mice have an additive impairment in thymocyte development. Tespa1 and Themis have been shown to regulate thymocyte development through TCR signaling, and we assumed that the additive defect in thymocyte development might therefore result from the additive defect in TCR signaling. We thus investigated TCR signaling by measuring the expression of CD69 and CD5. We found that DKO CD4+CD8int thymocytes expressed the lowest levels of CD69 and CD5 during the initiation of positive selection (Fig. 5e). We also measured Ca2+ flux in DP thymocytes by anti-CD3 stimulation. Although Themis−/− thymocytes exhibited normal Ca2+ flux, DKO thymocytes showed lower TCR-induced Ca2+ flux than Tespa1−/− thymocytes (Fig. 5f). We then studied TCR signaling by immunoblotting. We found that Themis−/− thymocytes exhibited decreased phosphorylation of SHP1 and lower Grb2 expression, as previously reported,10,27 whereas Vav1 phosphorylation was not affected. The phosphorylation of Erk was higher in Themis−/− thymocytes and lower in Tespa1−/− thymocytes. Consistent with an additive defect in thymocyte development, TCR signaling in DKO thymocytes was influenced by defects resulting from the deficiency of both Themis and Tespa1 (Fig. 5g). In summary, all these data collectively indicate that Tespa1 and Themis DKO mice showed additive defects in thymocyte selection.
Fig. 5.
Additive impairment of thymocyte development in double knockout (DKO) mice. a Expression of Themis (thymocyte expressed positive selection associated 1) and Tespa1 (thymocyte expressed positive selection associated 1) in thymocytes from wild-type (WT), Themis−/−, Tespa1−/−, and DKO mice, as detected by immunoblot. b Flow cytometry analysis of thymocytes from WT, Themis−/−, Tespa1−/−, and DKO mice. The numbers in the outlined areas indicate the percentages of T cells in the CD4+ single positive (SP) (top left), CD8+ SP (bottom right), and double positive (DP) (top right) populations. c Quantification of the total thymocytes and of each population shown in b. d Thymus sections from WT, Themis−/−, Tespa1−/−, and DKO mice stained with hematoxylin and eosin. The medulla (M) is shown in the lighter outlined areas; the cortex (C) is represented by the darker areas; original magnification, ×100. The results are representative of two experiments. e Surface staining of CD69 and CD5 in gated DP and CD4+CD8int thymocytes from WT, Themis−/−, Tespa1−/−, and DKO mice. f Calcium flux analysis of DP thymocytes from WT, Themis−/−, Tespa1−/−, and DKO mice after stimulation with anti-CD3 and anti-CD4. g Immunoblot analysis of total and phosphorylated PLCg1, Vav1, SHP1, Lck, Erk1/2, P38, and Grb2 in sorted DP thymocytes from WT, Themis−/−, Tespa1−/−, and DKO mice stimulated by biotin anti-CD3 and streptavidin for the time indicated. Themis and Tespa1 serve as genotype markers; β-actin serves as a loading control. Data are representative of three independent experiments. Student’s t test (ns = not significant; *<0.05; ***<0.001) was used for c. The small horizontal lines indicate the means (±SEM)
Discussion
In this study, we showed that Tespa1−/− thymocytes and Themis−/− thymocytes exhibited different gene expression pattern changes compared to those in WT T cells during positive selection, including the preselection, selection, and postselection stages. In GO analysis, both Themis deficiency and Tespa1 deficiency affected T cell-related biological processes in the P3 stage. We observed an increase in Rag1/2 expression in both Tespa1−/− thymocytes and Themis−/− thymocytes, which reflects the enrichment of immature DP thymocytes. However, Tespa1, rather than Themis, played a predominant role in influencing T cell-related processes, as shown in Fig. 1. Although the defects in Themis and Tespa1 cause similar reductions in CD4+ and CD8+ SP cells, here, we observed a more stage-specific and severe impairment in the P3 stage (in cells under positive selection) in Tespa1-deficient mice through the direct comparison of the two types of KO cells in both in vitro and in vivo assays. This experimental evidence supports our hypothesis that Tespa1 is a more specific regulator of positive selection but that Themis might have a broader biological function.
The different functions of Tespa1 and Themis could be explained by their independent roles in regulating distinct intercellular signaling events. Tespa1-deficient DP thymocytes had substantial reductions in TCR-induced calcium signaling, which was not observed in Themis-deficient T cells. The most predominant signaling change resulting from Themis deficiency, we observed here is the reduction of SHP1 phosphorylation, which is consistent with previous reports.27 It has been reported that Themis functions through regulating SHP1 catalytic activity; thus, the broad functionality of SHP1 might account for the wide-ranging function of Themis, which was reflected by the wider impact in gene expression changes resulting from Themis deficiency in all stages of thymocyte selection.30
The precise function of Themis is still subject to debate. The additive defect during thymocyte development that we observed in our experiment could be explained in both cases. If Themis is a positive regulator of TCR signaling, the DKO cells will reflect greater signal loss, which would support positive selection. In contrast, the sequential actions of Tespa1 and Themis would be critical for such a phenotype if Themis plays a negative role in TCR signaling. During positive selection, Tespa1 deficiency might predominately account for the observed defect, as evidenced by a more profound reduction of P3 cell numbers in Tespa1−/− mice than in Themis−/− mice. Subsequently, the effects of the defect resulting from Themis deficiency gradually accumulated. Indeed, Themis KO alone results in a significant loss in cell numbers in the P4 stage, which occurs later than the P3 stage in Tespa1−/− mice. It has been recently shown that the knockdown of either Themis or Ptpn6 (encoding protein phosphatase SHP1) in Jurkat T cells could increase activation-induced cell death.31 It is thus also possible that the increased susceptibility to cell death caused by Themis deficiency could have an additive effect when combine with Tespa1 deficiency to further reduce the SP cell numbers in DKO mice.
In mixed BM chimeric mice, Tespa1 deficiency leads to a more profound loss in P3 cells compared to that in Themis−/− mice. However, the number of Tespa1−/− cells gradually surpasses the number of Themis−/− cells as they become SP cells. It is very likely that Tespa1−/−cells regain some of their normal functions (e.g., the capacity for homeostatic proliferation) once they manage to complete the developmental process, while the defective proliferative capability of Themis-deficient T cells still exists in the periphery. We compared TCR-mediated responses in peripheral T cells lacking Tespa1 or Themis. We found that Themis−/− T cells show a more profound defect in CD69 upregulation as well as proliferation than Tespa1−/− T cells after anti-CD3 antibody stimulation. Thus, we provided evidence to show that Themis has a more striking effect on TCR signaling in peripheral T cells than on that in DP thymocytes. Compared to WT T cells, the proliferative capability of Tespa1−/− cells is also lower but to a lesser degree than that of Themis−/− cells. These results suggested that the function of Tespa1 might be more limited in the developmental stage, but Themis plays a more enduring role in both developing and mature T cells. We next investigated whether the defective responses of Tespa1−/− cells is due to the residual effects of Tespa1 in peripheral T cells or to a carry-over defect from the developmental stage by analyzing Tespa1 conditional knockout mice. Normal TCR signaling and cell proliferation were observed in Tespa1 CKO mice, indicating the dispensability of Tespa1 in peripheral T cells.
The higher expression of Tespa1 in DP thymocytes compared with that in mature T cells could explain the stage-specific role of Tespa1 in thymocyte selection. However, our study only reached the limited conclusion that Tespa1 deficiency did not affect TCR signaling in peripheral T cells. This conclusion does not rule out the possibility that Tespa1 might play some other regulatory roles in other biological processes in mature T cells. It will be necessary to study the functions of Tespa1 in peripheral cell in the context of different processes, including infection and anti-tumor responses.
In conclusion, we propose that Tespa1 functions in the early positive selection stage and that its function is dispensable in peripheral T cells. However, Themis functions in the late positive selection stage, and its activity gradually increases as thymocytes mature into peripheral T cells. Therefore, we defined Tespa1 as a stage-specific regulator of positive selection, providing an explanation for the longstanding and essential question of why thymocytes are highly sensitive to low-affinity self-peptides.
Materials and methods
Mice
Tespa1−/− mice were generated by homologous recombination-mediated gene targeting at the Shanghai Research Center for Model Organisms, as previously described.15 Mice from a mixed 129×C57BL/6 background were back-crossed onto the C57BL/6 background for 6–8 generations. Themis−/− mice were a gift from Guo Fu (Xiamen University). Rag1−/− mice were obtained from the Model Animal Research Center of Nanjing University. Tespa1 CKO mice were purchased from the International Mouse Phenotyping Consortium (https://www.mousephenotype.org/data/genes/MGI%3A1914846). Age-matched littermate mice used for experimental purposes were between 4 and 6 weeks of age. All experimental animal protocols were approved by the Review Committee of Zhejiang University School of Medicine and in compliance with institutional guidelines.
Antibodies
The biotin anti-mouse CD3 (145-2C11), purified anti-mouse CD3 (145-2C11), purified anti-mouse TCRβ (H57-597), and purified anti-mouse CD2 (RM2-5) antibodies were purchased from BioLegend (San Diego, CA, USA). The phospho-PLCg1 (Tyr783) antibody (#2821, pAb, 1:1000), Vav1 antibody (#2502, pAb, 1:1000), SHP-1 antibody (#3759, monoclonal antibody (mAb), 1:1000), phospho-SHP-1 (Tyr564) antibody (#8849, mAb, 1:1000), phospho-pLck (Tyr505) antibody (#2751, pAb, 1:1000), p44/42 MAPK (Erk1/2) antibody (#9102, pAb, 1:1000), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (#9101, pAb, 1:1000), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) antibody (#4377, mAb, 1:50), p38 MAPK antibody (#9212, pAb, 1:1000), and phospho-p38 MAPK (Thr180/Tyr182) antibody (#9211, pAb, 1:1000) were obtained from Cell Signaling Technology (Danvers, MA, USA). The anti-PLCγ1 (EP1898-7Y, mAb, 1:1000), anti-Vav1 (phospho Y174) (EP510Y, mAb, 1:1000), anti-Themis (EPR7353, mAb, 1:1000), and anti-Grb2 (Y237, mAb, 1:1000) antibodies were purchased from Abcam (Cambridge, MA, USA). The anti-Lck (#06-583, pAb, 1: 1000) antibody was obtained from Merck Millipore (Billerica, MA, USA). The polyclonal antibody against mouse Tespa1 was used at a 1:500 dilution and was generated by HuaAn Biotechnology using a glutathione-S-transferase-fused mouse Tespa1 recombinant protein fragment (amino acids 2–182).
RNA sequence data analysis
Three subpopulations of thymocytes were sorted from thymi. Each subpopulation (4 ×106) was pooled from several mice. Samples were lysed in Trizol for RNA extraction. Then, the RNA samples were transcribed to complementary DNA and sequenced by using an Illumina MiSeq Next Generation Sequencer (Novogene Bioinformatics Technology Co., Ltd). The genes with significantly changed expression (P value <0.05, fold change >2) were identified by DGEseq2 analysis. GO analysis was conducted by Novomagic V2.0 (Novogene Bioinformatics Technology Co., Ltd). For GSEA, individual normalized datasets (T_DP_69-_e17_Th_v2, T_DP69+_Th_v2, T_DP_Th, T_DP69+_Th) were downloaded from Immgen (http://www.immgen.org/index_content.html). Expression data from WT DP 69−(P2) and WT DP 69+(P3) cells were obtained from our RNA-seq data analysis. Genes whose expression increased at least two-fold between DP 69− and DP 69+ cells were selected. The overlapping parts of these three different gene sets were identified as the “Positive selection” gene set. This gene set was further separated into “Positive selection_UP” and “Positive selection_DN”. Cells were sorted by a BD FACS Aria II flow cytometer.
In vitro assay of DP to CD4+CD8int development
Sorted DP thymocytes were resuspended in RPMI-1640 containing 10% charcoal-stripped fetal bovine serum (FBS) and 50 μM 2-mercaptoethanol. Then, 5 × 105 cells were seeded onto culture plates coated with or without 10 ng/μl anti-TCRβ and 5 ng/μl anti-CD2 antibodies and cultured for 20 h. Then, the cells were harvested and extensively washed with phosphate-buffered saline (PBS) and transferred into medium without stimulation. After another 12 h of culture, the cells were washed and stained with the indicated antibodies for flow cytometry analysis. Cells were sorted with a BD FACS Aria II flow cytometer and analyzed by an ACEA NovoCyte TM flow cytometer. Data were plotted by the FlowJo software (TreeStar).
Ca2+ flux
Thymocytes or enriched peripheral T cells (3x106) in suspension were first labeled with 4 µg/ml Fluo-4 AM and 0.02% F-127 (Invitrogen) for 0.5 h at 37 °C and then washed with ice-cold PBS and resuspended in PBS. The cells were surface labeled with PerCP/Cy5.5-conjugated anti-CD4 (or CD69), APC-conjugated anti-CD8 (or TCRβ), and biotinylated anti-CD3 (5 µg/ml) for 30 min on ice. The labeled cells were warmed for 10 min to room temperature and then crosslinked with streptavidin (25 µg/ml) before flow cytometry analysis. The mean fluorescence ratios were plotted after analysis with the FlowJo software (TreeStar).
pERK measurement via flow cytometry
Thymocytes (3 × 106) were preincubated with PerCP/Cy5.5-conjugated anti-CD69, APC-conjugated anti-TCRβ, and 30 ng/μl biotinylated anti-CD3ε for 15 min at 4 °C. The cells were then washed with RPMI-1640 and resuspended in 50 μl LCM (RPMI-1640 medium containing 10% FBS, penicillin (100 U/ml), streptomycin (10 mg/ml), 2 mM glutamine, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol, and 10 mM HEPES) before incubation with 50 μl streptavidin (20 μg/ml final concentration) for 2 min. Stimulation was terminated by incubation with 100 μl BD Fix/Perm solution (BD Biosciences) for 20 min at room temperature. The cells were then permeabilized by adding 200 μl of −20 °C methanol and incubated on ice for 10 min. The cells were washed with PBS and resuspended in 50 μl diluted anti-pERK antibody and incubated at room temperature for 45 min. The samples were washed once and stained with Alexa Fluor 488-conjugated anti-IgG at room temperature for 30 min. The cells were washed and resuspended in PBS for analysis. The cells were analyzed by an ACEA NovoCyteTM flow cytometer. The data were plotted by the FlowJo software (TreeStar).
BM chimeras
BM cells harvested from femurs and tibias were processed into single-cell suspensions. Recipient mice (Rag1−/−, CD45.2) were irradiated (400rad) and injected with 5 × 106 BM cells from CD45.1 WT and CD45.2 Themis−/− or Tespa1−/− mice mixed at a 1:1 ratio. Recipient mice were treated with acidified antibiotic water containing 100mg/L neomycin sulfate and 10mg/L polymyxin B for 2 weeks after irradiation. The mice were sacrificed and analyzed 6–8 weeks after BM transfer. The cells were analyzed by a BD Fortessa flow cytometer. The data were plotted by the FlowJo software (TreeStar).
Homeostatic proliferation competition assay
CD4+ naive T cells (CD44loCD62Lhi) were purified from CD45.1 WT, CD45.1/2 Themis−/−, CD45.1/2 Tespa1−/−, and CD45.2 WT mice. CD4+ naive T cells (1 ×106) from the indicated mice were mixed at a 1:1 ratio and injected into Rag1−/− mice. Mice were sacrificed after 7 days for analysis. The cells were analyzed by a BD FACSCalibur flow cytometer. Data were plotted by the FlowJo software (TreeStar).
Splenocyte CD69 upregulation assay
Splenocytes (1 ×106) from WT, Themis−/−, or Tespa1−/− mice were stimulated for 5 h at 37 °C with or without different doses of plate-bound anti-CD3 in LCM. The cells were stained and analyzed after gating on CD4+ or CD8+ cells. The cells were analyzed by an ACEA NovoCyte TM flow cytometer. Data were plotted by the FlowJo software (TreeStar).
In vitro proliferation assay
CD4+ and CD8+ peripheral T cells from WT, Themis−/−, or Tespa1−/− mice were isolated and incubated with 5 μM CellTrace solution at 37 °C for 10 min according to the manufacturer’s protocol. Then, the cells were extensively washed with medium containing serum and resuspended in LCM. Subsequently, the cells were cultured for 72 h with or without different doses of plate-bound anti-CD3. T-cell proliferation was measured by analyzing CellTrace Violet dilution by flow cytometry. The cells were analyzed by an ACEA NovoCyteTM flow cytometer. Data were plotted by the FlowJo software (TreeStar).
Western blotting analysis
DP thymocytes (3 ×106) were purified and resuspended in 50 μl LCM. The cells were prewarmed at 37 °C and stimulated by adding 50 μl LCM containing 60 ng/μl biotin anti-CD3 and 40 ng/μl streptavidin for the time indicated. Stimulation was stopped by adding cold PBS. Then, the cells were resuspended and lysed in sodium doceyl sulfate (SDS) sample buffer by the addition of 1/4 volume of 5× SDS sample buffer directly into the cell suspension. The samples were then boiled for 5 min and separated by 10% SDS-polyacrylamide gel electrophoresis.
Statistical analysis
The group mean values were compared by a two-tailed Student’s t test or a Mann–Whitney U test. P values of <0.05 were considered significant (*P < 0.05; **P < 0.01; ***P < 0.001). The statistics were calculated using the GraphPad Prism 6 software (GraphPad Software Inc.).
Accession number
All RNA-seq data were deposited in the Sequence Read Archive (SRA) database under the accession number PRJNA522079.
Supplementary information
Acknowledgements
We thank Dr. Richard Sloan for helpful discussions and assistance with the manuscript editing. We are grateful to Yewei Li, Jin Chen, and Yingying Huang from the core facilities at the Zhejiang University School of Medicine for technical assistance in the FACS analysis. This work was supported by grants from the National Natural Science Foundation of China (31530019, 31770954, and 31325009 to L.L.), the National Key R&D Program of China (2018YFC1105102) and the Fundamental Research Funds for the Central Universities (2018XZZX001-12).
Author contributions
J.L., G.F., and L.L. designed the research; J.L., P.W., T.X., Y.S., Z.C., and M.Z. performed the experiments; J.L., G.F., and L.L. analyzed the data; J.L. and L.L. wrote the paper.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version of this article (10.1038/s41423-019-0259-4) contains supplementary material.
References
- 1.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 2.Brownlie RJ, Zamoyska R. T cell receptor signalling networks: branched, diversified and bounded. Nat. Rev. Immunol. 2013;13:257–269. doi: 10.1038/nri3403. [DOI] [PubMed] [Google Scholar]
- 3.Davey GM, et al. Preselection thymocytes are more sensitive to T cell receptor stimulation than mature T cells. J. Exp. Med. 1998;188:1867–1874. doi: 10.1084/jem.188.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gascoigne NR, Rybakin V, Acuto O, Brzostek J. TCR signal strength and T cell development. Annu. Rev. Cell Dev. Biol. 2016;32:327–348. doi: 10.1146/annurev-cellbio-111315-125324. [DOI] [PubMed] [Google Scholar]
- 5.Fu G, et al. Themis controls thymocyte selection through regulation of T cell antigen receptor-mediated signaling. Nat. Immunol. 2009;10:848–856. doi: 10.1038/ni.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson AL, et al. Themis is a member of a new metazoan gene family and is required for the completion of thymocyte positive selection. Nat. Immunol. 2009;10:831–839. doi: 10.1038/ni.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lesourne R, et al. Themis, a T cell-specific protein important for late thymocyte development. Nat. Immunol. 2009;10:840–847. doi: 10.1038/ni.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patrick MS, et al. Gasp, a Grb2-associating protein, is critical for positive selection of thymocytes. Proc. Natl. Acad. Sci. USA. 2009;106:16345–16350. doi: 10.1073/pnas.0908593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kakugawa K, et al. A novel gene essential for the development of single positive thymocytes. Mol. Cell. Biol. 2009;29:5128–5135. doi: 10.1128/MCB.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zvezdova E, et al. Themis1 enhances T cell receptor signaling during thymocyte development by promoting Vav1 activity and Grb2 stability. Sci. Signal. 2016;9:ra51. doi: 10.1126/scisignal.aad1576. [DOI] [PubMed] [Google Scholar]
- 11.Choi S, et al. THEMIS enhances TCR signaling and enables positive selection by selective inhibition of the phosphatase SHP-1. Nat. Immunol. 2017;18:433–441. doi: 10.1038/ni.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patrussi L, Baldari CT. Themis releases the brakes on TCR signaling during thymocyte selection by disabling SHP-1. Cell. Mol. Immunol. 2017;14:724–726. doi: 10.1038/cmi.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinoza JA, Jara EL, Kalergis AM. THEMIS, the new kid on the block for T-cell development. Cell. Mol. Immunol. 2017;14:721–723. doi: 10.1038/cmi.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang J, et al. Tespa1 regulates T cell receptor-induced calcium signals by recruiting inositol 1,4,5-trisphosphate receptors. Nat. Commun. 2017;8:15732. doi: 10.1038/ncomms15732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, et al. Tespa1 is involved in late thymocyte development through the regulation of TCR-mediated signaling. Nat. Immunol. 2012;13:560–568. doi: 10.1038/ni.2301. [DOI] [PubMed] [Google Scholar]
- 16.Yamashita I, Nagata T, Tada T, Nakayama T. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int. Immunol. 1993;5:1139–1150. doi: 10.1093/intimm/5.9.1139. [DOI] [PubMed] [Google Scholar]
- 17.Hare KJ, Jenkinson EJ, Anderson G. CD69 expression discriminates MHC-dependent and -independent stages of thymocyte positive selection. J. Immunol. 1999;162:3978–3983. [PubMed] [Google Scholar]
- 18.Azzam HS, et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat. Immunol. 2010;11:666. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberzon A, et al. The molecular signatures database hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voll RE, et al. NF-κB activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity. 2000;13:677–689. doi: 10.1016/S1074-7613(00)00067-4. [DOI] [PubMed] [Google Scholar]
- 22.Saini M, et al. Regulation of Zap70 expression during thymocyte development enables temporal separation of CD4 and CD8 repertoire selection at different signaling thresholds. Sci. Signal. 2010;3:ra23. doi: 10.1126/scisignal.2000702. [DOI] [PubMed] [Google Scholar]
- 23.Sinclair C, Bains I, Yates AJ, Seddon B. Asymmetric thymocyte death underlies the CD4:CD8 T-cell ratio in the adaptive immune system. Proc. Natl. Acad. Sci. USA. 2013;110:E2905–E2914. doi: 10.1073/pnas.1304859110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibotti R, Punt JA, Dash KS, Sharrow SO, Singer A. Surface molecules that drive T cell development in vitro in the absence of thymic epithelium and in the absence of lineage-specific signals. Immunity. 1997;6:245–255. doi: 10.1016/S1074-7613(00)80327-1. [DOI] [PubMed] [Google Scholar]
- 25.Gardner P. Calcium and T lymphocyte activation. Cell. 1989;59:15–20. doi: 10.1016/0092-8674(89)90865-9. [DOI] [PubMed] [Google Scholar]
- 26.Kortum RL, Rouquette-Jazdanian AK, Samelson LE. Ras and extracellular signal-regulated kinase signaling in thymocytes and T cells. Trends Immunol. 2013;34:259–268. doi: 10.1016/j.it.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu G, et al. Themis sets the signal threshold for positive and negative selection in T-cell development. Nature. 2013;504:441–445. doi: 10.1038/nature12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surh CD, Sprent J. Homeostatic T cell proliferation: how far can T cells be activated to self-ligands? J. Exp. Med. 2000;192:F9–F14. doi: 10.1084/jem.192.4.F9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canelles M, Park ML, Schwartz OM, Fowlkes BJ. The influence of the thymic environment on the CD4-versus-CD8 T lineage decision. Nat. Immunol. 2003;4:756–764. doi: 10.1038/ni953. [DOI] [PubMed] [Google Scholar]
- 30.Mehta M, et al. Themis-associated phosphatase activity controls signaling in T cell development. Proc. Natl. Acad. Sci. USA. 2018;115:E11331–E11340. doi: 10.1073/pnas.1720209115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paster W, et al. A THEMIS:SHP1 complex promotes T-cell survival. EMBO J. 2015;34:393–409. doi: 10.15252/embj.201387725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.