Abstract
Cyanobacteria have immense prospective as a platform for renewable energy; however, a major barrier in achieving optimal productivity is the low lipid yield. Fremyella diplosiphon, a model cyanobacterium, is an ideal biofuel agent due to its desirable fatty acid methyl esters (FAMEs). To enhance lipid content, we overexpressed the sterol desaturase (SD) gene in F. diplosiphon B481 wild type by genetic transformation. This effort resulted in a transformant (B481-SD) with a 64-fold increase in SD gene at the mRNA transcript level, with no loss in growth and pigmentation. The transformant was persistently grown for over 28 generations indicating long-term stability and vitality. We observed 27.3% and 23% increases in total lipid content and unsaturated FAMEs respectively in B481-SD transesterified lipids with methyl octadecadienoate as the most abundant unsaturated component. In addition, we detected an 81% increase in FAME composition in the transformant compared to the wild type. Theoretical physical and chemical properties confirmed a FAME profile with very high cetane number (65.972–67.494) and oxidative stability (50.493–18.66 h) in the engineered strain. Results of the study offer a promising approach to augment F. diplosiphon total lipid content and unsaturated FAMEs, thus paving the way to enhance biofuel capacity of the organism.
Introduction
Fossil fuels are a primary source of energy worldwide; however, increasing environmental consequences of its overuse, diminishing oil reserves, and rising costs have driven scientists to develop renewable sources of energy [1]. In recent years, cyanobacteria have gained great importance as third generation biofuel feedstock due to their rapid biomass production and lipids such as monogalactosyldiacylglycerol (MGDG), glycolipids, and digalactosyl diacylglycerol (DGDG) in their thylakoid membrane [2, 3, 4, 5]. In particular, fatty acids are a major class of lipids and essential biomolecules in the cyanobacterial cell membrane which determine important chemical and physical properties such as viscosity and permeability [1, 5].
Fatty acid desaturase enzymes such as acyl-lipid desaturase and sterol desaturase (SD) affect cyanobacterial lipid metabolism by synthesizing MGDG and DGDG, which are two major classes of galactolipids desirable for biofuel production [6, 7, 8]. However, insufficient lipid and fatty acid yields are key obstacles that hinder industrial-scale production of cyanobacteria-based biofuel. In addition, the high capital extraction/conversion cost poses a significant challenge and limits energy productivity of these biofuels [9]. Critical engineering breakthroughs to enhance lipid yield per harvest volume, which will reduce the cost of extraction per unit product are thus crucial to unlock the possibility of selected strains to serve as cost-effective biofuels.
Of the various cyanobacterial species, Fremyella diplosiphon has great potential as a commercial biofuel agent due to the generation of desirable fatty acid methyl esters (FAMEs) when its lipids are subjected to transesterification [10]. While few studies have been reported to enhance F. diplosiphon stress tolerance and photosynthetic efficacy [11, 12, 13], there has been no effort to increase its lipid and essential fatty acids. The objective of the present study was to overexpress the SD gene in F. diplosiphon via electroporation-mediated transformation. Level of gene overexpression was quantified and its effect on total lipid content and FAME composition/abundance characterized to confirm the role of SD gene in fatty acid synthesis.
Materials and methods
Strain and culture conditions
F. diplosiphon strain B481 obtained from the UTEX algal repository was used as the wild type (WT). Cultures were grown in BG11 liquid medium containing 20 mM HEPES at 170 rpm and 28°C for nine days under continuous white light adjusted to 30 μmol m−2s−1. For transformation studies, Escherichia coli FB5α competent cells (Thermo Fisher Scientific) were grown at 37°C in Luria-Bertani (LB) broth or agar plates supplemented with 80 mg L−1 ampicillin as the selective antibiotic.
RNA isolation and cDNA synthesis
F. diplosiphon WT was grown to an optical density of 0.6 at 750 nm (OD750) under culture conditions as mentioned above. Total RNA was extracted using Tri Reagent (Molecular Research Center, Inc.) according to manufacturer’s protocol. The purity of extracted RNA was tested on an agarose gel and A260/280 absorbance ratio measured using a Nanodrop 2000 (Thermo Fisher Scientific). Complementary DNA (cDNA) was reverse transcribed using the High Capacity RNA to cDNA kit (Life Technologies) and used for transformation.
Identification, cloning, and expression of F. diplosiphon sterol desaturase gene
Homologs of the SD gene were amplified from 50 ng F. diplosiphon WT cDNA using specific primers designed according to the gene sequence (JGI/IMG # 2501541024) and contained a HindIII restriction site added to the 5′end and BamHI to the 3′end (Table 1). Amplifications were performed using a C1000 Touch Thermocycler (Bio-Rad) under the following conditions: 95°C for 2 min; 40 cycles at 95°C for 30 s and an annealing temperature of 59°C for 30 s, followed by a final elongation step at 72°C for 45 s. Amplified products were electrophoresed on a 1.2% agarose gel, bands excised at the expected size ranges, and cDNA extracted using the Gel Recovery kit (Zymo Research). Amplified gene products were double digested with HindIII and BamHI, and pGEM-7Zf (+) vector triple digested with HindIII, BamHI, and ScaI restriction enzymes (Promega). Purified inserts were ligated into the vector at the HindIII and BamHI restriction sites with T4 DNA ligase (New England BioLabs), and the pGEM-7Zf (+)-SD expression plasmid constructed to overexpress the sterol desaturase gene. The plasmids were then cloned into E. coli FB5α competent cells via heat shock at 42°C for 20 s. Transformed competent cells were plated on LB agar containing 80 mg L−1 ampicillin and incubated for 16 h at 37°C. Resistant single colonies were selected, confirmed with X-gal staining for detection of β-galactosidase, and grown overnight in liquid LB medium containing 80 mg L−1 ampicillin. Ten random colonies were selected and plasmids extracted using the Zyppy Plasmid Miniprep kit (Zymo Research). Of these extracted plasmids, two were randomly selected and used for electroporation-mediated transformation. The presence of the insert was checked by PCR and further confirmed by sequencing.
Table 1.
Primers for the sterol desaturase gene used in plasmid construction and reverse transcription-quantitative PCR assay.
| Primer | Sequence (5’−3’) |
|---|---|
| Sterol desaturase (forward) | ATCAAGCTTGATGTTGACTTTTGACTTCTTCATAGCGGG |
| Sterol desaturase (reverse) | GCGGGATCCGTTAGTTCAAAGATTGAACCTTTTTA |
| Sterol desaturase (qPCR forward) | CGGGACTAGCGCTGATAAAA |
| Sterol desaturase (qPCR reverse) | TTTACCAAGCGAGGGGACAT |
Electroporation-mediated transformation
Expression plasmids containing SD gene were transformed into F. diplosiphon WT according to electroporation parameters described by Kehoe and Grossman [14]. The transformed strain was grown in 10 ml BG11/HEPES liquid medium for 16 h and transferred to BG11/HEPES agar containing 80 mg L−1 ampicillin. To verify the insertion of plasmid contained SD gene, PCR was performed using gene-specific primers as mentioned above (Table 1) and products visualized on a 1.2% agarose gel with a GeneRuler 100 bp Plus DNA marker (Thermo Scientific).
Quantification of F. diplosiphon gene expression levels using RT-qPCR
Total RNA in WT and transformant were extracted, cDNA synthesized as mentioned above, and RT-qPCR performed to quantify gene overexpression. Gene-specific primers were designed (Table 1) according to the gene sequence and real-time amplifications performed using SYBR green master mix (Applied Biosystems) in a Thermal Cycler CFX96 Real-Time machine (Bio-Rad). The reactions were performed in 20 μl volume containing 10 μl SYBR green Master Mix and 10 ng cDNA template. Amplifications were performed under the following conditions: 95°C for 20 s; 95°C for 20 s; and 40 cycles at 50.9°C for 30 s. Four replicates were maintained for each treatment type. Relative quantification (RQ) data of the transformant was analyzed using the ΔCt method with CFX Manager 3.1 (Bio-Rad) with the WT serving as the calibrator (RQ set to 1).
Evaluation of transformant growth
Stability of the transformant was tested on liquid BG11/HEPES media containing 80 mg L−1 ampicillin over a ten day-period under culture conditions described above. Wild type strain grown in the absence of ampicillin served as control. Growth (OD750) was measured every 24 h and stability of the strain confirmed after 32 generations by RNA extraction, cDNA synthesis, and PCR as mentioned above.
Comparison of wild type and transformant growth and photosynthetic pigment levels
We compared WT and transformant growth over ten days to determine the effect of SD gene overexpression. Culture conditions were maintained as described above with three biological replicates and growth rate calculated as doublings per day using the Least Fitting Squares method. The mean growth (OD750) and growth rate were calculated and statistical significance determined using one-way analysis of variance (ANOVA) and Tukey’s honest significant differences post-hoc test at 95% confidence intervals (P< 0.05). The single factor, fixed-effect ANOVA model, Yij = μ + αGi + εij, was used where Y is the growth (or growth rate) in strain i and biological replicate j. The μ represents mean growth with adjustments from the effects of strain (αG), and εij is the experimental error from strain i and biological replicate j.
We assessed photosynthetic efficacy of the transformant by extracting and quantifying chlorophyll a (chla), carotenoids and phycobiliprotein (phycoerythrin (PE), phycocyanin (PC), and allophycocyanin (AP)) pigments. Carotenoids and chla absorption spectra were measured at A470 and A665 respectively, while phycobiliproteins were quantified at A565, A620, and A650 to estimate photosynthetic capacity of the transformed strain [15, 16, 17].
Extraction and quantification of total lipids in F. diplosiphon WT and transformant
We compared total lipid content in WT and transformant using chloroform: methanol extraction method and gravimetric analysis based on Folch et al. [18] reported in Wahlen et al. [19]. Three biological replicates were maintained and the experiment repeated once. Significance among cumulative treatment means was determined using ANOVA and Tukey’s honest significant differences post hoc test at 95% confidence intervals (P< 0.05). The single factor, fixed-effect ANOVA model, Yij = μ + αSi + εij, was used where Y is the total lipid content in strain i and biological replicate j. The μ represents overall total lipid content mean with adjustments from the effects of strain (αS), and εij is the experimental error from strain i and biological replicate j.
Gas chromatography-mass spectrometry of transesterified product for FAME analysis
Total lipids in F. diplosiphon WT and transformant were extracted and converted to FAMEs via direct transesterification, a reaction that combines extraction and in situ biofuel production as described by Wahlen et al. [19] and modified by Tabatabai et al. [10]. We determined FAME composition of transesterified material using a Shimadzu GC17A/QP5050A GC-MS combination (Shimadzu Instruments) at the Mass Spectrometry Facility (Johns Hopkins University, Baltimore, MD) as previously described [5, 20]. GC/MS peaks were identified by comparing mass spectra to the Lipid Web Archive of FAME Mass Spectra. Three biological replicates of each treatment were maintained and the experiment repeated once. Theoretical chemical and physical properties of the FAME profiles (w%) were calculated using the BiodieselAnalyzer© software Version 2.2 [21].
GC × GC-TOFMS analysis of total transesterified lipids
High-resolution two dimensional gas chromatography-time of flight mass spectrometry (GC × GC-TOFMS) was used to identify FAME components. Total lipids from the WT and transformant strains were extracted, subjected to direct transesterification as mentioned above [19], and GC × GC-TOFMS (LECO) carried out as described by Tabatabai et al. [10].
Results
Identification of sterol desaturase homologs in Fremyella diplosiphon
Sterol desaturase gene-specific primer amplification revealed a single discrete band at the expected size of 1314 base pairs (bp). PCR product subjected to Sanger sequencing and NCBI BLAST confirmed identity of the SD gene with an open reading frame of 1314 bp and its encoded proteins with 437 amino acids. Sequence alignment identified a 94% match to SD gene in F. diplosiphon, thus confirming identity of the gene. The SD amino acid sequence revealed 93%, 86%, 77%, and 77% identities to Nostoc carneum NIES-2107, Calothrix sp. NIES-2100, Nodularia sp. NIES-3585, and Fortiea contorta, respectively. The SD gene sequence was deposited at NCBI Genbank with an accession number of MH329183.
Cloning and transformation of sterol desaturase gene in F. diplosiphon
Amplified SD gene and pGEM-7Zf (+) vector were successfully double and triple digested, followed by ligation of purified inserts at the corresponding restriction sites. The plasmid containing the SD gene (pGEM-7Zf-SD) was successfully transformed into WT via electroporation and gene insertion confirmed by PCR and Sanger sequencing.
RT-qPCR analysis confirms overexpression of sterol desaturase gene in B481-SD
We observed a significant increase in transcript abundance in the transformant. Relative quantification values revealed a 64-fold increase in SD gene expression of the transformant compared to WT (Fig. 1). The sterol desaturase-overexpressing F. diplosiphon strain was designated as B481-SD.
Fig. 1.
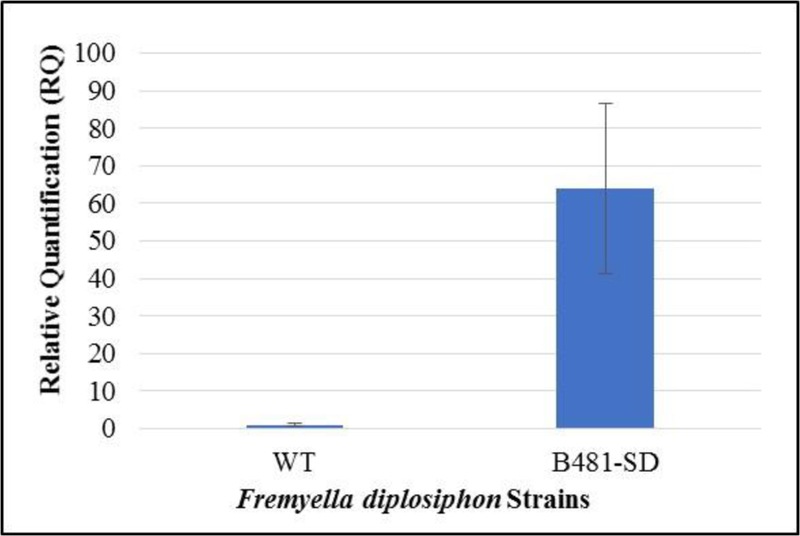
Reverse transcription-quantitative polymerase chain reaction relative quantification (RQ) of sterol desaturase transcript levels in Fremyella diplosiphon wild type (WT) and transformant (B481-SD). Error bars represent the standard error of ΔCt values at a 95% confidence interval across four replicates.
Evaluation of growth and photosynthetic pigment accumulation in the transformant
We did not observe significant differences (P> 0.05) in growth and growth rate of B481-SD grown in liquid BG11/HEPES medium compared to WT (Fig. 2 and 3). No growth was observed in WT exposed to 80 mg L−1 ampicillin. Concurrent analysis of photosynthetic pigments (Fig. S1 and S2) and phycobiliproteins (Fig. S3) revealed no significant differences (P> 0.05) between WT and transformant. We were able to grow the transformant persistently in BG11/HEPES solid media containing 80 mg L−1 ampicillin for 32 generations. Results of PCR revealed a single discrete band at the expected size of 1314 bp for B481-WT as well as B481-SD (Fig. 4).
Fig. 2.
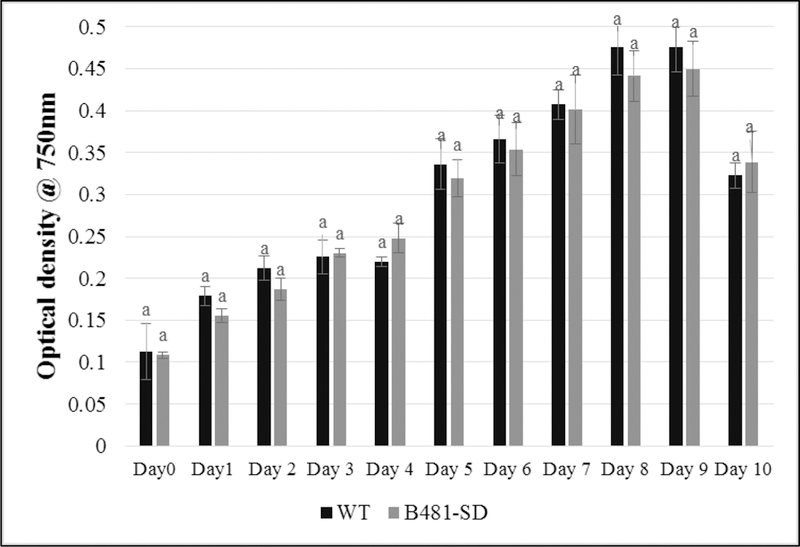
Growth of Fremyella diplosiphon wild type (WT) and transformant (B481-SD) strains in BG11/HEPES medium. Average optical density at 750 nm (±standard error) for three biological replicates is shown for each time point. Different letters above final time point indicate significance among treatment means (P<0.05).
Fig. 3.
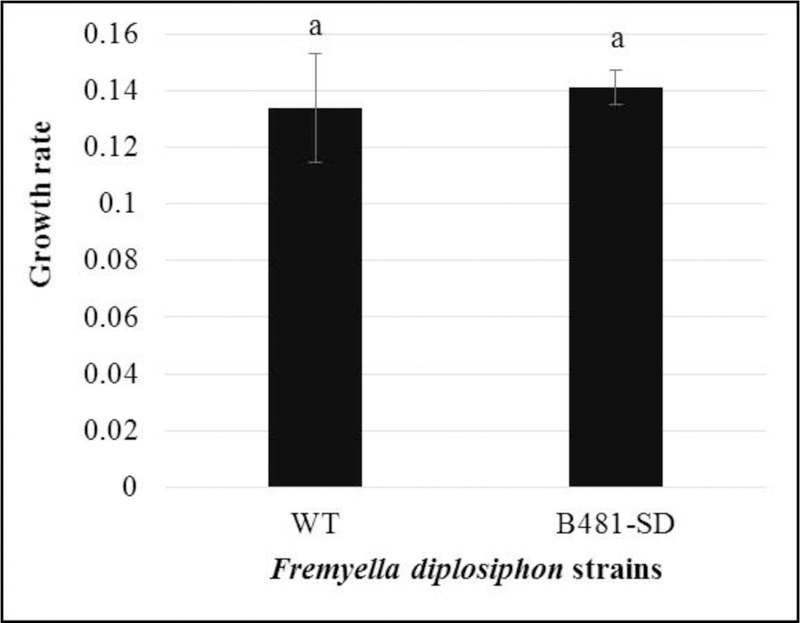
Effect of sterol desaturase gene overexpression on Fremyella diplosiphon wild type (WT) and transformant (B481-SD) growth rate in liquid BG11/HEPES medium over a nine day-period. Growth rate was denoted as doublings per day using the Least Fitting Squares method calculator.
Fig 4.

PCR amplification of sterol desaturase gene in Fremyella diplosiphon B481-wild type (WT) and B481-SD after 32 generations with a 100 bp ladder (L).
Comparison of total lipid content in B481-SD and WT by gravimetric analysis
We identified a significant increase (P< 0.05) in B481-SD total lipid content when compared to the WT (Fig. 5). While 23.6% total lipid content of cellular dry weight was detected in B481-SD, 18.5% lipid content was recovered in WT. Thus, a 27.2% increase in B481-SD total lipid content relative to WT was observed.
Fig. 5.
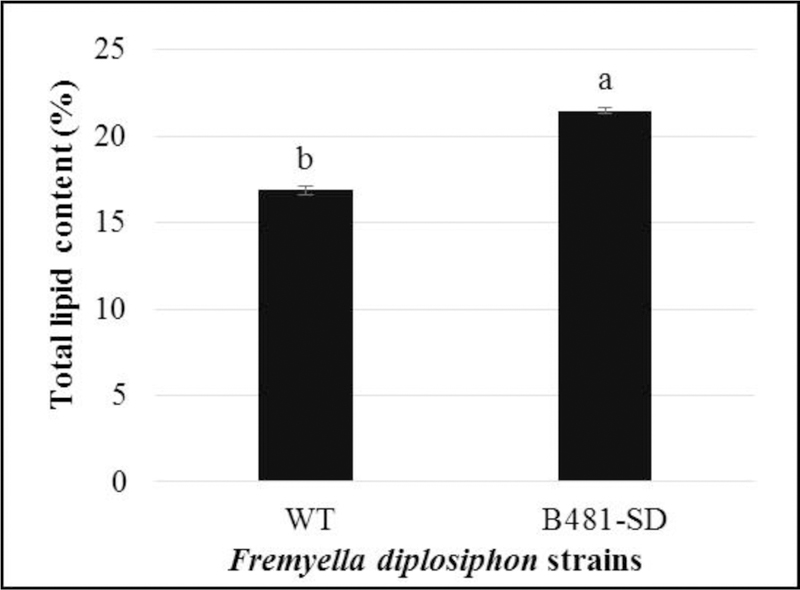
Comparison of total lipid content in wild type (WT) and transformant (B481-SD) Fremyella diplosiphon. Average percentage lipid content of cell dry weight (% w/cdw) (± standard error) of three biological replicates for each strain is shown. Different letters above bars indicate significance among treatment means (P<0.05).
Characterization of FAMEs in WT and B481-SD by GC-MS
We identified methyl palmitate as the dominant FAME component, which accounted for 76.35% and 65.93% of total FAMEs produced from WT and B481-SD total lipids respectively (Table 2). In addition, we identified the following FAME species: methyl tetradecanoate (C14:1), methyl hexadecenoate (C16:1), methyl octadecanoate (C18:0), methyl octadecenoate (C18:1), and methyl octadecadienoate (C18:2) (Table 3). Results of the study also revealed significant increases (P< 0.05) in methyl octadecenoate (C18:1), and methyl octadecadienoate (C18:2) levels in B481-SD transesterified lipids, while no significant differences (P> 0.05) were observed in other FAME components (Fig. 6 and 7). Calculation of theoretical chemical and physical properties revealed high cetane number (65.97–67.49) and oxidative stability (50.49–18.67 h) in the FAME produced from B481-SD. In addition, calculated values for density (0.867–0.868 g/cm3), viscosity (3.781–3.817 mm2/s), and iodine (17.723–25.590 g I2/100 g) (Table 4) were higher than minimum American and European fuel standards (22).
Table 2.
Saturated and unsaturated fatty acid methyl ester (FAME) proportions in wild type (WT) and transformant (B481-SD) Fremyella diplosiphon.
| FAME Type (%) | Ratio of FAME | ||
|---|---|---|---|
| Strain | Saturated | Unsaturated | Saturated/Unsaturated |
| WT | 80.99 | 19.01 | 4.26 |
| B481-SD | 76.62 | 23.38 | 3.27 |
Table 3.
Quantitative composition of fatty acid methyl ester in transesterified lipids of Fremyella diplosiphon wild type (WT) and transformant (B481-SD) strains.
| WT | B481-SD | |||||||
|---|---|---|---|---|---|---|---|---|
| :0† | :1 | :2 | SUM | :0 | :1 | :2 | SUM | |
| C14* | - | 3.07 | - | 3.07 | - | 3.21 | - | 3.21 |
| C16 | 72.39 | 11.66 | - | 84.05 | 65.67 | 7.67 | - | 73.34 |
| C18 | 8.60 | 1.79 | 2.49 | 12.88 | 10.95 | 5.16 | 7.34 | 23.45 |
| SUM | 80.99 | 16.52 | 2.49 | 100 | 76.62 | 16.04 | 7.34 | 100 |
Column represents length of the carbon chain.
Row represents degree of saturation (number of double bonds in chain).
Fig. 6.
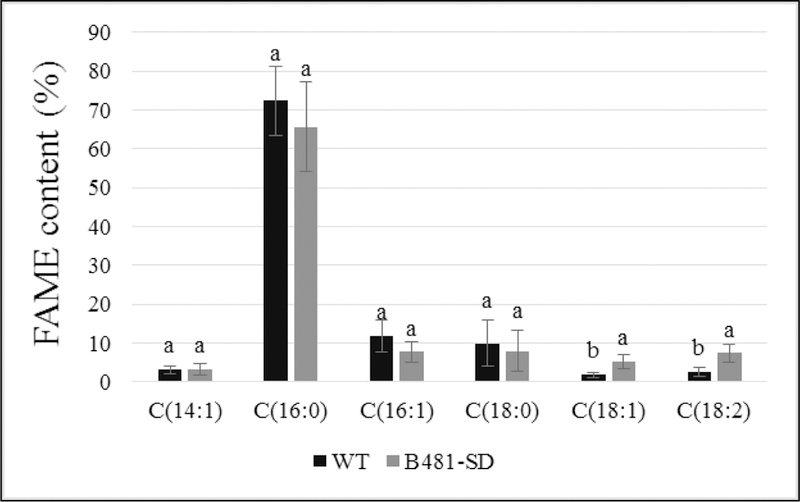
Comparison of fatty acid methyl ester (FAME) composition of wild type (WT) and transformant (B481-SD) Fremyella diplosiphon total lipids subjected to direct transesterification. Average percent FAME content (± standard error) for three biological replicates of each strain is shown. Different letters above bars indicate significance among treatment means (P<0.05).
Fig. 7.
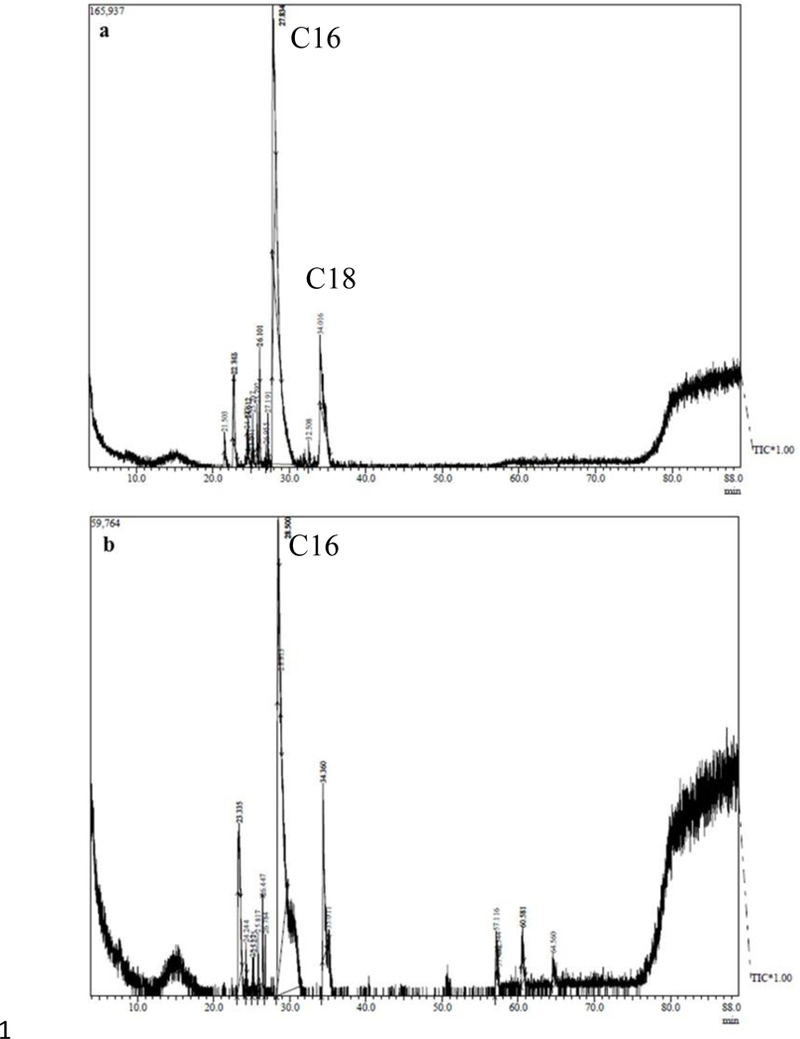
Representative one dimensional gas chromatogram of Fremyella diplosiphon strains (a) WT and (b) B481-SD total lipids subjected to direct transesterification.
Table 4.
Theoretical biodiesel properties of Fremyella diplosiphon wild type (WT) and transformant (B481-SD) transesterified lipids.
| Biodiesel Properties | WT | B481-SD |
|---|---|---|
| Saponification Value (mg KOH/g fat) | 216.743 | 214.627 |
| Iodine Value (g I2/100 g) | 17.723 | 25.590 |
| Cetane number | 67.494 | 65.972 |
| Long Chain Saturated Factor | 11.540 | 12.044 |
| Cold Filter Plugging Point (°C) | 19.771 | 21.361 |
| Cloud Point (°C) | 33.087 | 29.550 |
| Pour Point (°C) | 29.097 | 25.258 |
| Allylic Position Equivalent | 6.722 | 19.836 |
| Bis-Allylic Position Equivalent | 2.462 | 7.336 |
| Oxidation Stability (h) | 50.493 | 18.666 |
| Higher Heating Value (mJ/kg) | 39.208 | 39.259 |
| Kinematic Viscosity (mm2/s) | 3.781 | 3.817 |
| Density (g/cm3) | 0.867 | 0.868 |
Lipid characterization in B481-SD by GC × GC-TOFMS
In addition to conventional GC-MS, we used high-resolution GC × GC-TOFMS to further separate polar, aromatic compounds. GC × GC-TOFMS analysis revealed the presence of FAMEs with carbon number from 12–18, as well as alkanes from C11 to C34. We observed FAME composition in B481-SD (80.92% transesterified lipids) to be significantly higher (P< 0.05) than WT (77.92% transesterified lipids) (Fig. 8 and 9). FAME components such as C12:0, C15:0, C18:3, and C18:4 not detected in conventional GC-MS were identified by GC×GC-TOFMS.
Fig. 8.
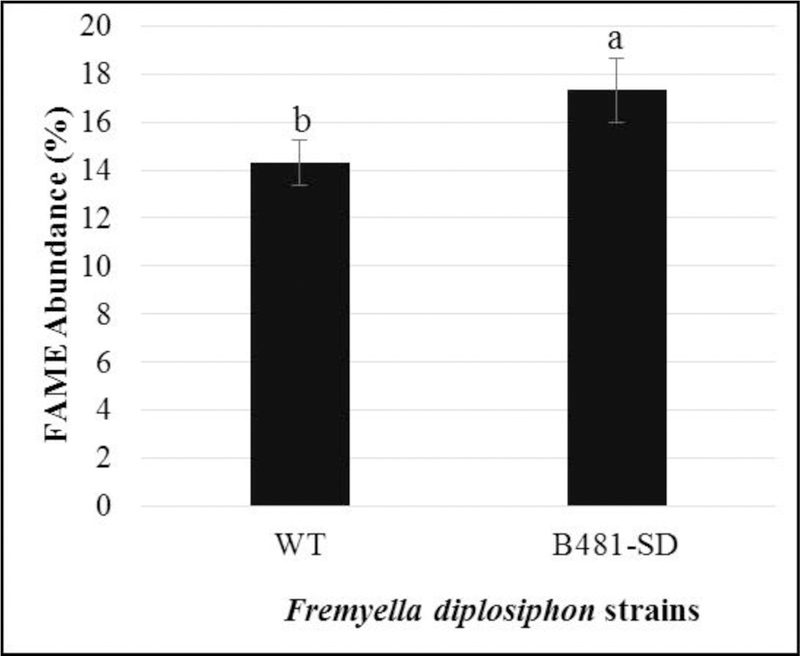
Fatty acid methyl ester (FAME) composition in transesterified extractable lipids of wild type (WT) and transformant (B481-SD) Fremyella diplosiphon strains determined using GC×GC-TOFMS. Average percent FAME content (± coefficient of variation) of each strain for three biological replicates is shown. Different letters above bars indicate significance among treatment means (P< 0.05).
Fig. 9.
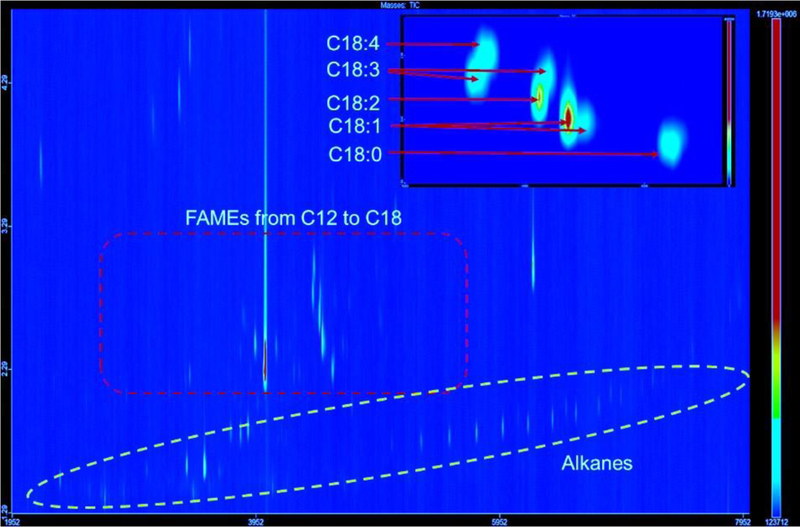
Representative GC×GC-TOFMS chromatogram of Fremyella diplosiphon B481-SD fatty acid methyl esters (FAMEs), alkanes, and other components. Compounds were separated by volatility which correlates to molecular weight (carbon number) on the first dimension and polarity which correlates to the degree of unsaturation (number of double bonds) on the second dimension.
Discussion
Although there is a great potential of cyanobacterial-based biofuels, major hurdles in terms of strain improvement and oil productivity exist before these fuels can be deployed into use. Efforts to overcome these challenges will significantly reduce the cost and progress the use of biofuel from promise to reality [9, 11, 12]. In the present study, overexpression of the SD gene in F. diplosiphon via homologous transformation resulted in a significant increase in total lipid content and essential unsaturated fatty acid (FA) composition. Homology of the cDNA-encoding gene exhibited high similarities (93% and 86%) to published sequences of these proteins in other cyanobacterial species, confirming their functional importance in lipid biosynthesis pathways. Quantification of gene expression in the transformant revealed a 64-fold increase in mRNA transcript levels (Fig. 1), suggesting successful integration of the pGEM-7Zf-SD plasmid in F. diplosiphon, thereby enhancing the robustness of the organism to produce lipids. A patent application describing this approach to enhance total lipid content and essential unsaturated fatty acid (FAs) composition has been filed (US Patent Application No.: 16/123,484; September 6, 2018).
Comparison of growth and growth rate of B481-SD and WT did not reveal significant differences indicating that overexpression of SD gene was not detrimental to the organism (Fig. 2 and 3). In addition, unaltered growth of the transformant and a clear single band at the expected size of 1314 pb after 32 generations confirmed stable integration of the gene in the pGEM-7Zf-SD plasmid. Further, we did not observe differences in pigmentation levels, confirming that photosynthetic activity of the transformant was unaltered by the transformation process (Fig. S1, S2, and S3). It is known that lipid production and fatty acid composition are impacted by cellular photosynthetic pathways in cyanobacteria [23], which indicate a direct correlation between cyanobacterial lipid and photosynthetic pigments. This antioxidative property of photosynthetic pigments is known to assist in a cellular protective response to oxidative stress induced by SD gene overexpression [7, 8, 24]. This protective mechanism has been known to significantly reduce reactive oxidative stress up to 82% in the cyanobacterium Spirulina platensis [24].
In cyanobacteria, sterol desaturase enzymes assist in biochemical processes such as reduction of NADP+ to NADPH [8] and lipid biosynthetic pathways inducing oxidative stress leading to increased lipids, specifically unsaturated fatty acids [25]. There were no significant differences in saturated FAs such as palmitic acid (C16:0) and octadecanoic acid (C18:0) between the WT and B481-SD; however, 8% and 65% reduction of these components was observed in the transformant suggesting that SD gene overexpression could be attributed to the desaturation of these components [26]. In addition, we detected methyl octadecadienoate (C18:1) and methyl octadecadienoate (C18:2), which are two most abundant desaturated FAME components known to yield high-quality biofuel. It has been reported that overexpression of the SD gene could increase unsaturated fatty acid levels such as oleic acid (C18:1) in Saccharomyces cerevisiae and Nicotiana tabacum BY-2 cells [7, 27]. We observed various high-value components such as FAMEs and alkanes using high-resolution GC × GC-TOFMS that were not identified by GC-MS, confirming additional benefits of F. diplosiphon-derived biodiesel.
Conclusion
This is the first report of SD overexpression resulting in enhanced lipid content and essential unsaturated fatty acids such as oleic and linoleic acids in F. diplosiphon, indicating that this approach is ideal to maximize biofuel production capacity of the organism. These findings offer a novel pathway for a highly efficient and cost-effective renewable fuel production system and pave the way for F. diplosiphon as a more efficient production-level agent. Future efforts will be aimed towards optimizing large-scale cultivation and lipid extraction/conversion to determine the viability of F. diplosiphon B481-SD for efficient and economical biofuel production.
Supplementary Material
Acknowledgments
The work was partially supported by the National Institutes of Health [UL1GM118973] awarded to Morgan State University and National Science Foundation [DMR 11–57490] awarded to the National High Magnetic Field Laboratory and the State of Florida.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Compliance with Ethical Standards
We confirm that this manuscript has not been published elsewhere and is not under consideration by other journals. All authors have approved the manuscript and agree with submission to Applied Biochemistry and Biotechnology.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Nozzi NE, Oliver JW, & Atsumi S (2013). Cyanobacteria as a platform for biofuel production. Frontiers in Bioengineering and Biotechnology, 1, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Los DA, & Mironov KS (2015). Modes of fatty acid desaturation in cyanobacteria: an update. Life, 5(1), 554–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murata N, Wada H, & Gombos Z (1992). Modes of fatty-acid desaturation in cyanobacteria. Plant and Cell Physiology, 33(7), 933–941. [Google Scholar]
- 4.Sharathchandra K, & Rajashekhar M (2011). Total lipid and fatty acid composition in some freshwater cyanobacteria. Journal of Algal Biomass Utilization, 2(2), 83–97. [Google Scholar]
- 5.Sato N, & Awai K (2016). Diversity in biosynthetic pathways of galactolipids in the light of endosymbiotic origin of chloroplasts. Frontiers in Plant Science, 7, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murata N, & Wada H (1995). Acyl-lipid desaturases and their importance in the tolerance and acclimatization to cold of cyanobacteria. Biochemical Journal, 308(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamthan A, Kamthan M, & Datta A (2017). Expression of C-5 sterol desaturase from an edible mushroom in fission yeast enhances its ethanol and thermotolerance. PloS One, 12(3), e0173381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramm A, Kisiela M, Schulz R, & Maser E (2012). Short-chain dehydrogenases/reductases in cyanobacteria. The Federation of European Biochemical Societies Journal, 279(6), 1030–1043. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen MA, & Hoang AL (2016). A review on microalgae and cyanobacteria in biofuel production. Economies and Finances, 1–37. [Google Scholar]
- 10.Tabatabai B, Chen H, Lu J, Giwa-Otusajo J, McKenna AM, Shrivastava AK, & Sitther V (2018). Fremyella diplosiphon as a Biodiesel Agent: Identification of Fatty Acid Methyl Esters via Microwave-Assisted Direct In Situ Transesterification. BioEnergy Research, 11(3), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabatabai B, Arumanayagam AS, Enitan O, Mani A, Natarajan SS, & Sitther V (2017). Overexpression of hlyB and mdh genes confers halotolerance in Fremyella diplosiphon, a freshwater cyanobacterium. Enzyme and Microbial Technology, 103, 12–17. [DOI] [PubMed] [Google Scholar]
- 12.Tabatabai B, Gharaie Fathabad S, Bonyi E, Rajini S, Aslan K, & Sitther V (2019). Nanoparticle-mediated impact on growth and fatty acid methyl ester composition in the cyanobacterium, Fremyella diplosiphon. BioEnergy Research, In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh SP, & Montgomery BL (2013). Salinity impacts photosynthetic pigmentation and cellular morphology changes by distinct mechanisms in Fremyella diplosiphon. Biochemical and Biophysical Research Communications, 433(1), 84–89. [DOI] [PubMed] [Google Scholar]
- 14.Kehoe DM, & Grossman AR (1998). Use of molecular genetics to investigate complementary chromatic adaptation: Advances in transformation and complementation. Methods in Enzymology, 297, 279–290. [Google Scholar]
- 15.Kahn K, Mazel D, Houmard J, De Marsac NT, & Schaefer MR (1997). A role for cpeYZ in cyanobacterial phycoerythrin biosynthesis. Journal of Bacteriology, 179(4), 998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Marsac NT, & Houmard J (1988). Complementary chromatic adaptation: physiological conditions and action spectra. Methods in Enzymology, 167, 318–328. [Google Scholar]
- 17.Whitaker MJ, Bordowitz JR, & Montgomery BL (2009). CpcF-dependent regulation of pigmentation and development in Fremyella diplosiphon. Biochemical and Biophysical Research Communications, 389(4), 602–606. [DOI] [PubMed] [Google Scholar]
- 18.Folch J, Lees M, & Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipids from animal tissues. Journal of Biological Chemistry, 226(1), 497–509. [PubMed] [Google Scholar]
- 19.Wahlen BD, Willis RM, & Seefeldt LC (2011). Biodiesel production by simultaneous extraction and conversion of total lipids from microalgae, cyanobacteria, and wild mixed-cultures. Bioresource Technology, 102(3), 2724–2730. [DOI] [PubMed] [Google Scholar]
- 20.Rosenberg JN, Kobayashi N, Barnes A, Noel EA, Betenbaugh MJ, & Oyler GA (2014). Comparative analyses of three Chlorella species in response to light and sugar reveal distinctive lipid accumulation patterns in the microalga C. sorokiniana. PloS One, 9(4), e92460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talebi AF, Tabatabaei M, & Chisti Y (2014). BiodieselAnalyzer: a user-friendly software for predicting the properties of prospective biodiesel. Biofuel Research Journal, 1(2), 55–57. [Google Scholar]
- 22.Ramos MJ, Fernandez CM, Casas A, Rodriguez L, & Perez A (2009). Influence of fatty acid composition of raw materials on biodiesel properties. Bioresource Technology, 100(1), 261–268. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Sheng J, & Curtiss R III (2011). Fatty acid production in genetically modified cyanobacteria. Proceedings of the National Academy of Sciences 108(17):6899–6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riss J, Décordé K, Sutra T, Delage M, Baccou JC, Jouy N, Brune JP, Oréal H, Cristol JP & Rouanet JM (2007). Phycobiliprotein C-phycocyanin from Spirulina platensis is powerfully responsible for reducing oxidative stress and NADPH oxidase expression induced by an atherogenic diet in hamsters. Journal of Agricultural and Food Chemistry, 55(19), 7962–7967. [DOI] [PubMed] [Google Scholar]
- 25.Sevcu A, El-Temsah YS, Joner EJ, & Cernik M (2009). Oxidative stress induced in microorganisms by zero-valent iron nanoparticles. Microbes and Environments, 26(4):271–81. [DOI] [PubMed] [Google Scholar]
- 26.Muller C, Binder U, Maurer E, Grimm C, Giera M, & Bracher F (2015). Fungal sterol C22-desaturase is not an antimycotic target as shown by selective inhibitors and testing on clinical isolates. Journal of Steroids, 101, 1–6. [DOI] [PubMed] [Google Scholar]
- 27.Zhang M, Barg R, Yin M, Gueta-Dahan Y, Leikin-Frenkel A, Salts Y, Shabtai S, & Ben-Hayyim G (2005). Modulated fatty acid desaturation via overexpression of two distinct ω−3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. The Plant Journal, 44(3), 361–371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


