Abstract
During skeletal development, limb progenitors become specified as chondrocytes and subsequently differentiate into specialized cartilage compartments. We previously showed that the arginine dimethyl transferase, PRMT5, is essential for regulating the specification of progenitor cells into chondrocytes within early limb buds. Here, we report that PRMT5 regulates the survival of a separate progenitor domain that gives rise to the patella. Independent of its role in knee development, PRMT5 regulates several distinct types of chondrocyte differentiation within the long bones. Chondrocytes lacking PRMT5 have a striking blockage in hypertrophic chondrocyte differentiation and are marked by abnormal gene expression. PRMT5 remains important for articular cartilage and hypertrophic cell identity during adult stages, indicating an ongoing role in homeostasis of these tissues. We conclude that PRMT5 is required for distinct steps of early and late chondrogenic specialization and is thus a critical component of multiple aspects of long bone development and maintenance.
Keywords: PRMT5, Knee, chondrogenesis, chondrocyte differentiation, joint, articular cartilage, patella, progenitors, growth plate, homeostasis
INTRODUCTION
Starting with the initial specification of chondrocytes from mesodermal cells, a combination of extrinsic signals and cell-intrinsic responses drive successive steps of cartilage development. Ultimately, these result in the formation of a cartilage template for the skeleton as well as the formation of numerous types of specialized cartilage cells (reviewed in (1)). During the initial stages of development, nascent chondrocytes temporarily stop proliferating and start expressing early differentiation markers (Sox9 and Collagen2a1) as they change shape and coalesce to form the bone rudiments.
Chondrocyte proliferation subsequently resumes on the ends of the bones as chondrocytes in the center progressively differentiate into hypertrophic chondrocytes in part through a signaling loop driven by Indian hedgehog and Parathyroid hormone-related protein (2). This second phase of hypertrophic chondrocyte maturation is essential for normal long bone growth and ossification and requires the establishment of new genetic programs driven by RUNX2, FOXA2, and MEF2C which activate the transcription of terminal differentiation markers Collagen10a1 and Mmp13 (3-7). Dysregulation of these terminal differentiation programs leads to a wide range of cartilage and bone defects including skeletal dysplasias, characterized by increased cell hypertrophy and reduced/delayed bone formation (8,9). Hypertrophic chondrocytes ultimately have two fates, to either undergo apoptosis after establishing permissive conditions for osteoblast colonization or transdifferentiate into osteoblasts (10,11).
After the formation of the initial cartilage anlagen, the knee joint is generated by the addition of several different cell types. Externally-derived GDF5-expressing cells engraft onto or within the existing bone to form the synovial joint, and additional progenitor cells marked by Scleraxis and Sox9 give rise to the patella (12,13). These joint structures, along with the proliferative distal ends of the long bones subsequently differentiate and become ossified, generating a mature, multi-tissue knee joint (13-15). In contrast to the long bone itself, the origin and regulation of these additional joint progenitors are still poorly understood.
We recently found that the methyltransferase PRMT5 is important for chondrocyte specification. Using PrxCre to conditionally remove Prmt5 from the limb buds, we found that forelimb morphogenesis was profoundly impaired while the hindlimbs were relatively normal (16). Because PrxCre is expressed at comparatively earlier stages of forelimb than hindlimb development (17), it suggested that PRMT5 has a critical role in the initial specification of chondrocytes from progenitor cells, but is dispensable for later chondrocyte development. In addition to chondrocyte specification, PRMT5 has distinct roles in the development of many other organ systems, including the lung, muscles, oligodendrocytes and osteoclasts (18-21). Several of these roles implicate PRMT5 in the maintenance of stem/progenitor cells (22,23). While we previously demonstrated that PRMT5 is not necessary for maintaining nascent chondrocytes in limb buds (16), we hypothesized that it might be required for the transition to more differentiated types of cartilage as well as for the differentiation of later cartilage populations derived from different progenitor populations.
Here, we show that loss of Prmt5 at different developmental stages results in specific cartilage maturation defects in the knee joint and long bones. Consistent with our initial hypothesis, we find that PRMT5 is required for the survival of patella progenitor cells. PRMT5 is separately required for the differentiation of multiple cartilage domains within the long bones, including the growth plate and articular cartilage. PRMT5 continues to be required for cartilage homeostasis during adulthood, where it remains necessary for hypertrophic cartilage differentiation in the growth plate and maintains signature gene expression in the articular cartilage. We conclude that PRMT5 is essential for the transition of chondrocytes through multiple states of differentiation during chondrogenesis.
MATERIALS AND METHODS
Mice
Experiments involving mice were approved by the Institutional Animal Care and Use Committee at The University of Texas at Austin (AUP-2016-00255 and AUP-2018-00276). Prx-Cre (17) and Col2-Cre (24) mice were crossed with Prmt5tm2c(EUCOMM)Wtsi (referred to as Prmt5c/c) (16) females to generate control, PrxCre;Prmt5c/c, and Col2Cre;Prmt5c/c mice respectively. ATC-Cre (25) mice were crossed with Prmt5c/c females to generate control and ATC;Prmt5c/c mice. Doxycycline (Dox) was administered by intraperitoneal (IP) injections starting at 4 weeks of age (P28), once a week at 10 mg/kg body weight for four weeks. OC-Cre mice (26) were purchased from Jackson Laboratory (Stock# 019509) and crossed with Prmt5c/c females to generate control and OC-Cre;Prmt5c/c mice.
Micro-CT Analysis
Whole-mount skeletal preparation was performed as previously described (27). For Micro-CT analysis, hind limbs were harvested from control and PrxCre;Prmt5c/c mice at P21, fixed for three days in 10% buffered formalin and stored in 70% ethanol until imaging. Samples were scanned by the high-resolution X-ray CT facility at UT Austin on a Fein Focus High Power source, 140 kV, 0.16 mA, aluminum filter, with a Perkin Elmer detector.
Histology and immuno-staining
Following antigen retrieval in Tris-EDTA, pH 9 at 70°C for ten minutes, paraffin sections of adult limbs (decalcified in formic acid) were incubated with anti-PRMT5 (Abcam, ab109451, 1:100) or anti-RUNX2 (MBL International, D130-3, 1:100), processed using a Vectastain ABC HRP kit (Vector Labs, PK-4000) and developed using DAB substrate (Roche 11718096001). Visualization of PRG4 was similarly carried out using anti-PRG4 (Abcam, ab28484, 1:400) following antigen retrieval in 0.1mg/ml hyaluronic acid at 37 °C for 10 minutes. Immunofluorescent staining was performed on frozen tissues harvested and fixed for 1hour in 4% paraformaldehyde and subsequently embedded in OCT. 10μm-sections were then post fixed in 4% paraformaldehyde for 10 minutes at room temperature, permeabilized in 0.06% PBST (Triton-X) for 30 minutes, and blocked in 3% bovine serum albumin (BSA) and 5% normal goat serum (NGS)/PBST (1% Tween-20) for 1 hour at room temperature. Samples were then incubated with: anti-SOX9 (Millipore, AB5535, 1:200), anti-phospho-Histone H3 (Millipore, 06-570, 1:200), anti-GFP (Aves Labs, GFP-1020, 1:500). After overnight incubation at 4°C, the samples were visualized using the following secondary antibodies: Alexa 568 goat anti-rabbit secondary (Life Technologies, A11036, 1:250) and Cy2 anti-chicken secondary (Jackson Immuno Research, 703-225-155, 1:200). Apoptosis was detected by TUNEL staining was performed using an In Situ Cell Death, TMR detection kit (Roche, 12156792910). Fluorescent samples were counterstained for nuclei with 4’,6-diamidino-2-phenylindole (DAPI; Invitrogen, D1306, 1:5000).
Section In-Situ Hybridization
Paraffin sections (5μm) were permeabilized in Proteinase K (7.5μg/mL in PBST) for 5 minutes at room temperature, and post-fixed in 4% paraformaldehyde and 2% glutaraldehyde. Samples were incubated with Digoxygenin-labeled antisense riboprobes overnight at 68°C. Followed by incubation with a sheep anti-Dig-POD antibody (Sigma, 11207733910, 1:500) at 4°C overnight. Hybridization was detected using a tyramide-amplified fluorescent antibody (Perkin Elmer, NEL753001KT) incubated at room temperature for 10-20 minutes (incubation times varied depending on the probe).
Quantitative-RT-PCR
RNA was extracted from embryonic knee samples using Trizol Reagent (Life Technologies, 15596-026), DNase-treated, and 500ng was then used for cDNA synthesis (SuperScript II, Invitrogen 18064-014). Quantitative RT-PCR was performed using SensiFast SYBR Lo-Rox (Bioline BIO-94005) on a Via7 (ABI) platform. Gene expression was normalized to GAPDH, and fold change was calculated using the delta-CT method (28). The following qRT-PCR primers were used: Prmt5 (F) CATGAGAGCAAGCCCACAAA, (R) CCCCACCAGCATTTTCCTAA; Bmp4 (F) ACGTACTCCCAAGCATCACC, (R) GCACAATGGCATGGTTGGTT; Gapdh (F) GGTGAAGGTCGGTGTGAACG, (R) CTCGCTCCTGGAAGATGGTG.
Results
PRMT5 is required for the differentiation of distinct cartilage sub-types
We previously showed that Prmt5 is required for the initial specification of appendicular chondrocytes but is not required to maintain them after this specification (16). To determine if Prmt5 is subsequently important for later events in appendicular skeletal development, we examined 3 week old (P21) hindlimbs of PrxCre;Prmt5 conditional knockouts (PrxCKOs) by micro computed tomography (Micro-CT) and found that PrxCKOs had pronounced knee defects. Specifically, we observed sharp reductions in bone volume within the patella and the epiphyseal regions of the femur and tibia (Fig. 1A-C). We additionally noted a characteristic bowing of the tibia and fibula (Fig. 1B). These defects suggest that PRMT5 is required for the normal development of the knee and long bone.
Figure 1. PRMT5 is required for normal knee and bone development.
A-C. Micro-CT analysis of the knee joint in wild type (WT) and PrxCre;Prmt5c/c mutants at P21, false colored to show the patella (red), the epiphysis of the tibia (green) and the epiphysis of the femur (yellow). Bone volume of each joint tissue was quantified in sibling control (n=4) and mutants (n=4). D-E. Histological analysis of knee joint cross-sections at P21 in WT and PrxCre;Prmt5 c/c mice. Sections were stained with SafraninO and Fast green. F, I. Detailed histological analysis of articular cartilage in the tibia at P21. Sections were stained for Alcian Blue and eosin (3 control and 3 PrxCre;Prmt5c/c). G,J. Section fluorescent in situ hybridization for Prg4 expression (green), and nuclei visualized with DAPI (blue), in the articular cartilage of 3 WT and 3 PrxCre;Prmt5c/c mice at P21. H,K. Col2a1 expression (green) in the articular cartilage of the tibia in 3 WT and 3 PrxCre;Prmt5c/c mice at P21 (DAPI in blue). L,O. Histological analysis of growth plate cartilage layers in the tibia at P21. Sections from 5 control and 5 PrxCre;Prmt5c/c mice were stained with SafraninO and Fast Green. M,P. Section in situ hybridization for Col10a1 expression (green) in the growth plate of 3 WT and 3 PrxCre;Prmt5c/c mice at P21 (DAPI in blue). N,Q. Col2a1 expression in the growth plate of the tibia in 3 WT and 3 PrxCre;Prmt5c/c mice at P21 (DAPI in blue). R-T. Hypertrophic cell density (R), zone width (S) and surface area (T) measured across 5 control and 5 mutant mice. *p<0.05; **p<0.01; ***p<0.001, using an unpaired Student’s t-test. AC-articular cartilage; HC-hypertrophic chondrocytes (Scale = 100μm).
We next examined sections through the knee joint and observed severe reduction in the secondary ossification centers of PrxCKO mice (Fig. 1D,E), which was more pronounced in the tibia than in the femur. Cells within the most superficial layers of the epiphysis (which normally make up the articular cartilage) in PrxCKOs are more disorganized, forming small clusters of cells (Fig. 1I), in contrast to the stereotypic columnar organization in wild-type tissues (Fig. 1F). The articular chondrocyte marker Proteoglycan 4 (Prg4) which is normally restricted to the most superficial layers of articular cartilage cells, is mis-expressed throughout multiple layers of cartilage (Fig. 1G,J). Loss of/severe reductions in the secondary ossification center in PrxCKOs is further marked by the presence of Collagen2a1 (Col2a1)-expressing cells within the deeper articular layers (Fig. 1H, K).
In the growth plate, we observed a large expansion in the size of the hypertrophic cell domain (Fig. 1S). There is a ~2-fold increase in the average size of hypertrophic chondrocytes and a 2-fold decrease in their cell density (Fig. 1R,T). There was no significant change in the number of cells within this domain between control and PrxCKO samples (an average of 209 cells versus 175 cells, respectively; n=5, p=0.3114 Student’s t-test). The expanded domain was also marked by a striking increase in the domain of Collagen10a1 (Col10a1) expression and a corresponding reduction in Collagen2a1 (Col2a1) expression (Fig 1M-N;P-Q). Col10a1-expressing cells in PrxCKOs also expressed robust levels of Indian hedgehog (Ihh) (Supplemental Fig. 1A,C). Together, these findings indicate that PRMT5 is required for the development of multiple chondrogenic sub-types within the epiphysis.
PRMT5 is required for the survival of early patella progenitors
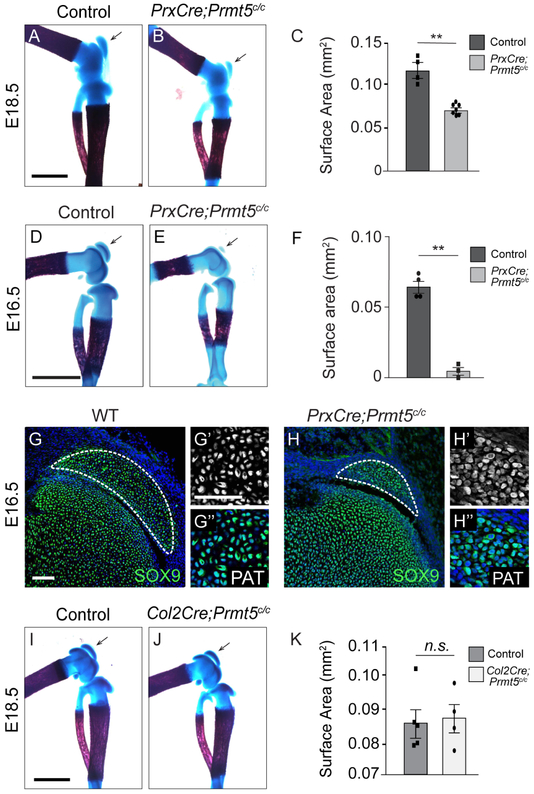
To determine at what stage PRMT5 first presents a phenotype, we examined wholemount skeletal preparations at embryonic timepoints. Compared to sibling controls, PrxCKO embryos have significantly reduced patellas at both E18.5 and E16.5 (Fig. 2A-F). Additionally, we examined SOX9 expression in the patella at E16.5 and observed that PrxCKOs contained a clearly reduced domain of SOX9-positive cells at E16.5 (Fig. 2 G,H). Although the mitotic index was unchanged in PrxCKOs (Supplemental Fig. 2A-C), there was a small but significant increase in cell death by TUNEL within the patella of PrxCKO mice, suggesting that cell death may contribute to this reduced domain (Supplemental Fig. 2D-F). It is presently unclear why the patella appears more reduced at E16.5 than at E18.5. As there is a clearly reduced domain of SOX9+ cells at this stage, as well as low levels of cell death, we speculate that the difference could be due to an initial delay in development due to lower cell numbers. To determine whether patella size reduction occurred before or after chondrocyte specification in PRMT5 knockouts, we also inactivated Prmt5 with Col2-Cre, which is first expressed in chondrocytes after specification (24). In contrast to PrxCKO, Col2-Cre;Prmt5c/c (Col2-CKO) embryos had normally sized patellas (Fig. 2I-K), suggesting that PRMT5 is not required for the differentiation of committed chondrocytes in the patella, but instead is required earlier during patella specification. If this is the case, reduced patella size in PrxCKOs could either be due to a reduction in the number of chondrocyte progenitors or a delay in the differentiation of chondrocytes from mesenchymal limb bud progenitors.
Figure 2. PrxCre-CKO embryos have a reduced patella.
A-C. Whole mount skeletal preparation of control and PrxCre;Prmt5c/c knees at E18.5, stained for alcian blue and alizarin red. Patella surface area was quantified across 4 control and 7 mutant embryos. (Scale = 1mm) D-F. Whole mount skeletal preparations of E16.5 sibling control (n=4) and PrxCre;Prmt5c/c (n=3) hindlimbs, stained for alcian blue and alizarin red. (Scale = 0.5mm) G-H. SOX9 expression (green; DAPI in blue) analysis in 3 control and 3 mutant knees at E16.5 by immunofluorescence. (Scale = 100μm) I-K. Whole mount skeletal preparation of control and Col2Cre;Prmt5c/c knees at E18.5, stained for alcian blue and alizarin red. Patella surface area was quantified across 5 control and 4 mutant limbs. PAT-patella (Scale = 1mm).
The patella is initially formed by a population of SOX9+SCX+ progenitor cells that coalesce on the articulating femur head at E13.5 and subsequently differentiate into patella-specific chondrocytes by E14.5 (29,30). Based on the reduction in patella size at E16.5, we hypothesized that PRMT5 might be similarly required for the normal commitment and survival of patella progenitor cells. To test this, we used the Scleraxis-GFP (Scx-GFP) allele in combination with antibodies against SOX9 to visualize SOX9+SCX+ progenitor cells in PrxCKOs (31). There were comparable numbers of SOX9+;SCX+ progenitor cells in control and mutant knees at E13.5 (72%; n=4 versus 69%; n=4 respectively; Fig. 3A-B). Strikingly, there was a 30% increase in cell death within the patella progenitors in mutants (Fig. 3C-D,I). We also noted increased TUNEL staining within the SCX-GFP+ connective tissues surrounding the early long bones. Elevated levels of cell death also occurred in PrxCKO patellas at E14.5 (Fig. 3G-H, I), highlighting a 24-hour window for PRMT5 regulation. We then examined the expression of Bmp4, which is upregulated in early limb buds in PrxCKOs (16). As expected, Prmt5 levels are substantially reduced. However, there were no significant differences in the expression Bmp4 within knee at E14.5 (Fig. 3J). We conclude that an early increase in patella progenitor death in PrxCKOs is the primary cause for the overall reduction in patellar size observed at later developmental stages.
Figure 3. PRMT5 is required for the survival of early patella progenitors.
A-B. Early committed patella cells were visualized by SOX9 (red), SCX-GFP (green) expression at E13.5 in the knee joint of 4 control and 4 PrxCre;Prmt5c/c embryos. C-D. TUNEL staining (white) of apoptosing cells in E13.5 control and mutant knees. E-F. Visualization of differentiating patella progenitor cells in the E14.5 knee joint of 3 control and 3 PrxCre;Prmt5c/c embryos. G-H. TUNEL staining (white) of apoptosing cells in E14.5 knees. Nuclei stained with DAPI (blue). I. Percentage of TUNEL-positive cells in E13.5/E14.5 was quantified across 4 control and 4 mutant knees at E13.5 and across 3 control and 3 mutant knees at E14.5. J. Gene expression of Prmt5 and Bmp4 were measured through quantitative-RT-PCR in 3 control and 3 mutant knee joints at E14.5. *p<0.05; **p<0.01; ***p<0.001 using an unpaired Student’s t-test (Scale = 50μm).
A postnatal requirement for PRMT5 in hypertrophic cartilage maturation in the long bones
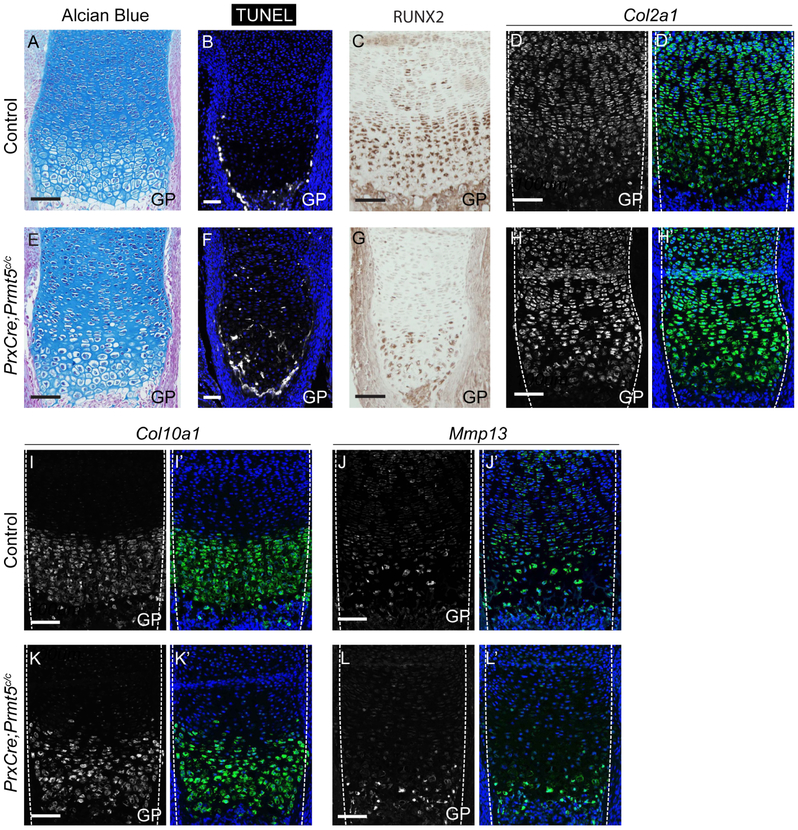
To determine when the cartilage differentiation defects observed within the long bones of P21 PrxCKOs first appeared, we assayed for morphological and gene expression changes in the growth plates of PrxCKO tibias at E18.5 (Fig. 4). At E18.5, there was a mild increase in cell size in the hypertrophic zone (Fig. 4A,E). This was accompanied by increased cell death both within the hypertrophic zone as well as along the ossification boundary in the growth plate (Fig. 4B,F). There was also a decrease in RUNX2 expression within the pre-hypertrophic/hypertrophic chondrocytes in PrxCKOs compared to controls (Fig. 4C,G). However, at this stage Col2a1 expression in the proliferating chondrocytes and Col10a1 expression in the hypertrophic chondrocytes was largely unaltered between controls and mutants (Fig. 4D,H,I,K). Similarly, expression of Mmp13 (a direct transcriptional target of RUNX2) is still expressed within the growth plate (Fig. 4J,L)(32), suggesting that the general organization of the growth plate into sub-type specific zones is unaffected by loss of Prmt5 from the limb at embryonic stages.
Figure 4. PRMT5 is partially required for the differentiation of embryonic cartilage.
A,E. Histological analysis of PrxCre;Prmt5c/c knees at E18.5 by alcian blue staining revealed subtle organizational defects in the growth plate cartilage layers of the long bones (n=5 controls; and 5 mutants). B,F Apoptosing cells in the tibia of 4 control and 4 PrxCre;Prmt5c/c embryos were visualized by TUNEL staining (white; DAPI in blue). C,G. Visualization of RUNX2 expression (brown) by immunohistochemical staining in the tibia of control and PrxCre;Prmt5c/c mice at E18.5. D,H. Visualization of Col2a1 expression (green) in the tibia of control and PrxCre;Prmt5c/c mice at E18.5 by fluorescent in situ hybridization (DAPI in blue). I,K. Visualization of Col10a1 expression (green) in the tibia of control and PrxCre;Prmt5c/c mice at E18.5 by fluorescent in situ hybridization (DAPI in blue). J,L. Visualization of Mmp13 expression (green) in the tibia of control and mutant embryos by fluorescent in situ hybridization (DAPI in blue). D-L. Gene expression studies were performed on 3 control and 3 mutant knees. GP-growth plate (Scale = 50μm).
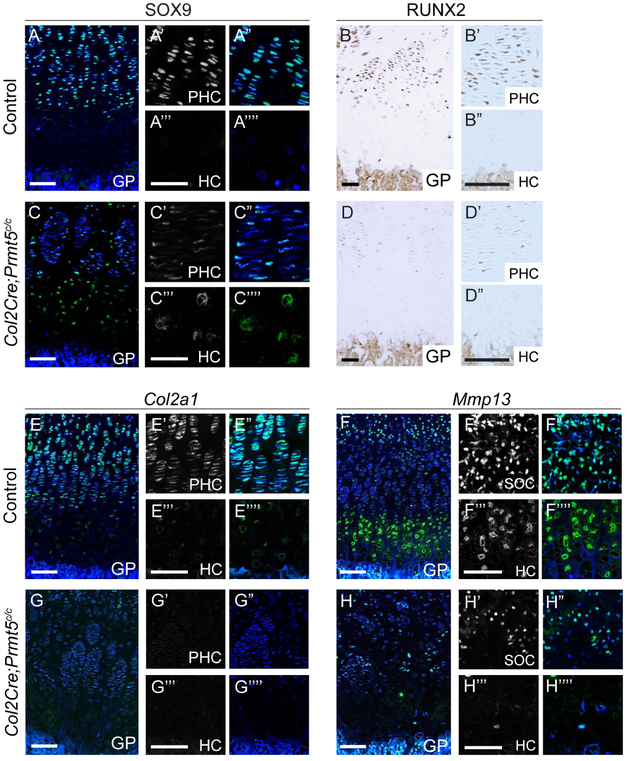
The relatively subtle changes in the growth plate at late gestation contrast with the pronounced abnormalities present at P21 (Fig. 1), indicating that defects within the epiphysis intensify during early postnatal development. To refine the timing, we assayed tibias from P10 pups using Col2-Cre to delete Prmt5 specifically within committed chondrocytes (Supplemental Fig. 3A-B). The proliferative domain of the growth plate has a reduced SOX9 expression (Fig. 5A,C). In contrast, there is persistent SOX9 expression in some hypertrophic chondrocytes in Col2-CKOs, a pattern which was not observed in control mice (Fig. 5A,C). We also found dramatic reductions in the SOX9 target gene Col2a1 within the proliferative zone (Fig. 5E,G). These changes in gene expression patterns are comparable to the initial growth plate defects seen in PrxCKO mice at P21 (Fig. 1N-Q). As chondrocytes transition into hypertrophy, RUNX2 activates terminal differentiation programs by downregulating SOX9 and activating Mmp13 (5,6,32,33). We observed reductions in RUNX2 expression within the growth plate (Fig. 5B, D) and a near absence of Mmp13 expression in Col2-CKO mutants (Fig. 5F, H), suggesting that PRMT5 is progressively important for maintaining RUNX2 expression within differentiating growth plate cells between E18.5 and P10. We also observed reduced expression of Mef2c expression, another regulator of hypertrophy, in the hypertrophic chondrocytes of the growth plate (Supplemental Fig. 1B, D) (4). Together these results indicate that PRMT5 becomes increasingly important for the terminal differentiation of hypertrophic chondrocytes during the early postnatal period.
Figure 5. PRMT5 is required for later differentiation of cartilage cells.
A,C. SOX9 protein expression (green) by immunofluorescence in the tibia of control and Col2Cre;Prmt5c/c mice at P10 (DAPI in blue). B,D. Visualization of RUNX2 expression (brown) by immunohistochemical staining in the tibia of control and Col2Cre;Prmt5c/c mice at P10. E,G. Visualization of Col2a1 expression (green) in the tibia of control and Col2Cre;Prmt5c/c mice at P10 by fluorescent in situ hybridization (DAPI in blue). F,H. Visualization of Mmp13 expression (green) in the tibia of control and Col2Cre;Prmt5c/c mice at P10 (DAPI in blue). Differentiation marker studies were performed on 3 control and 3 mutant knees at E18.5. GP-growth plate; PHC-pre-hypertrophic chondrocytes; HC-hypertrophic chondrocytes (Scale = 100μm).
Previous studies have implicated PRMT5 in regulating aspects of bone and osteoclast differentiation (19,34). Because the Col2Cre driver can directly or indirectly contribute to bone as well as chondrocytes (10,35), some aspects of the phenotypes within the epiphyseal tissues could be caused by a requirement for PRMT5 in the bone lineage. To address this, we crossed conditional Prmt5 mutants to the osteoblast-specific Osteocalcin-Cre (OC-Cre) (26) line (Supplemental Fig. 4A-B) and observed no detectable knee or bone phenotypes (Supplemental Fig. 4C-G), indicating that PRMT5 is not required in mature osteoblasts.
Prmt5 is required for maintenance of adult chondrocytes
To determine whether PRMT5 is further required for the maintenance of chondrocytes in the adult knee, we generated Prmt5 conditional knockout mice containing Acan enhancer-driver Tetracycline-inducible Cre (ATC-Cre). This Cre is active in all chondrocytes within the articular cartilage, and in pockets of growth plate chondrocytes (25). We induced recombination by administering Doxycycline (Dox) at 1 month of age and continuing treatment out to 2 months. The mice were then kept for 2 more months after the end of Dox administration prior to analysis (Fig. 6A). When examined by in situ hybridization, Prmt5 is nearly undetectable in the articular cartilage and is absent within patches of chondrocytes in the growth plate in ATC;Prmt5c/c (ATC-CKO) mice at 4 months, indicating the effectiveness of this conditional knockout approach (Supplemental Fig. 5A,B). ATC-CKO joints had no obvious morphological change in the organization of articular and growth plate chondrocytes compared to controls (Fig. 6B-D,F,I, K). However, in contrast to the embryonic deletion where we observed increases in Prg4, there was a decrease in PRG4 expression in the articular cartilage of Dox-induced ATC;Prmt5c/c mice (Fig. 6 E,G,H) (see Discussion). In addition, there is a significant expansion in the number of Col1Oa1-posiiive cells (Fig. 6J,L,M), similar to the phenotype observed in the embryonic deletion. We conclude that PRMT5 helps regulate the homeostasis of the articular cartilage and growth plate.
Figure 6. Conditional loss of Prmt5 from the adult knee results in altered gene expression.
A. ATC;Prmt5c/c mice were induced with Doxycycline starting at 1month for a total of 1 month. Knees from control and ATC;Prmt5c/c mice were collected at 4 months for analysis. B,C. Histological analysis of control and ATC;Prmt5c/c mice at 4 months by Safranin O and fast green staining. D,F,I,K. The articular (D,F) and growth plate cartilage (I,K) of control and ATC;Prmt5c/c mice are indicated by Safranin O and fast green staining. E,G. PRG4 protein expression (brown) in the articular cartilage of control and ATC;Prmt5c/c mice at 4 months was visualized by immunohistochemistry and quantified across 3 controls and 3 mutants (H). J,L. Col10a1 expression (green) in control and ATC;Prmt5c/c mice at 4 months was visualized by fluorescent in situ hybridization (DAPI in blue). M. The number of Col10a1-expressing cells was quantified across 3 controls and 3 mutants. All studies were performed on 3 control and 3 mutant mice. *p<0.05; **p<0.01 using an unpaired Student’s t-test. GP-growth plate; AC-articular cartilage; HC-hypertrophic chondrocytes (Scale = 100μm).
DISCUSSION
Differentiated chondrocytes encompass several specialized cell sub-sets which represent a divergence of developmental pathways from a more limited set of common progenitor cells. These pathways have multiple transient states, that are regulated by diverse mechanisms as they differentiate. We previously showed that PRMT5 is required during very early development for the survival of progenitor cells during the initial specification of SOX9-expressing chondrocytes (16). Here, we find that PRMT5 has a similar role in maintaining the survival of the different progenitor cells that give rise to the patella. Distinct from these roles, PRMT5 is also required for the differentiation of chondrocytes into some specific cell types, including articular cartilage and hypertrophic cartilage. Some of these roles are ongoing in adult bones, indicating that PRMT5 is involved in homeostasis.
Interestingly, conditional deletions of Prmt5 with PrxCre or Col2Cre resulted in different phenotypes. PrxCKOs have a reduced patella while the patella is not reduced in the chondrocyte-specific Col2-CKOs. The patella initially forms from a population of progenitors cells expressing SOX9 and SCX that reside adjacent to the femur head (29). PrxCre is active in the limb bud mesenchyme several days before the onset of Col2Cre. Thus, a likely explanation for the difference in phenotypes between the two Cre drivers lies in the transient requirement for PRMT5 in patella progenitor cells prior to their differentiation into patella-specific chondrocytes. In support of this possibility, SOX9+;SCX+ cells have high levels of apoptosis in PrxCKOs at E13.5-E14.5, during initial patella formation, almost certainly leading to the marked decrease in initial patella size. Although surviving patella progenitors are able to differentiate into SOX9 and Col2a1 -expressing chondrocytes, the domain size remains small, suggesting that patella size is limited by the initial number of progenitor cells. This function is analogous to the temporally-specific cell survival requirement for PRMT5 in progenitor cells that give rise to SOX9+ chondrocytes in the early limb bud (16).
Several previous studies have found that PRMT5 mutants have upregulated BMP signaling in different cell contexts, including the embryonic lung, early limb buds (16,36). In contrast to these findings, we do not observe elevated Bmp4 in mutants either during embryonic or postnatal stages (Fig. 3, Supplemental Fig. 6A,B). We cannot exclude the possibility that Bmp4 is upregulated in mutants at specific times that were missed in our analysis. Alternatively, PRMT5-dependent regulation of Bmp4 is context-dependent.
Deletion of Prmt5 causes a striking phenotype in the growth plate, where visibly hypertrophic cells appear to be blocked in the process of hypertrophic differentiation. Hypertrophic chondrocyte differentiation requires PRMT5 starting during late gestation and continuing into adulthood. Proliferative chondrocytes have reduced levels of SOX9. Col2a1 is a direct transcriptional target of SOX9 and thus the reduced levels of SOX9 likely account for the reduced levels of Col2a1 seen in these cells (37). Within the upper hypertrophic chondrocytes, there is a prominent expansion in the size of the Col10a1-expressing domain which is not due to an increase in cell number, but rather in size. Additionally, these cells contain persistent levels of SOX9 expression and a near absence of RUNX2. As SOX9 and MEF2C function as transcriptional activators of Col10a1(25), the continued presence of SOX9 and residual Mef2c suggests a possible mechanism for Col1Oa1 initiation. RUNX2 and MEF2C drive further hypertrophic chondrocyte differentiation, in part by activating Mmp13. Therefore, the near absence of RUNX2 and reduction in Mef2c suggest that hypertrophic chondrocytes arrest in a partially differentiated state in which they are unable to exit development (5,6,32,33). We note that the phenotype is similar to that observed in Mmp13 knockouts, which also have expanded hypertrophic domains expressing Col1Oa1 (38). As RUNX2, in combination with RUNX3, is required for embryonic Collagen lO activation (6), the expanded domain of Col1Oa1 was initially unexpected. However, RUNX2 becomes progressively depleted in hypertrophic chondrocytes (compare Figs 4G and 5D). Presumably, RUNX2 levels are initially sufficient to activate Col10a1 but are not required to sustain it during later developmental timepoints. This may also explain the lack of reduction in Ihh, which is initially dependent on RUNX2 (6).
Following initial joint development, Prmt5 continues to be required for ongoing chondrocyte homeostasis in the adult knee. The loss of Prmt5 results in reduced numbers of PRG4 expressing cells (Fig. 6E,G,H), a finding that is in contrast with the upregulation of Prg4 observed when Prmt5 is lost during developmental stages (Fig. 1J, 6E). The difference in these roles could highlight differences in the requirement for PRMT5 during developmental versus adult stages. PRG4 is a component of the cartilage extracellular matrix and synovial joint fluid, and its expression is both necessary and sufficient to prevent osteoarthritis (39-41). The loss of Prmt5 from adult articular cartilage tissues may result in loss of superficial cartilage identity and thus a depletion of PRG4 similar to phenotypes reported in response to cartilage degeneration (39,40,42). In future studies, it will be interesting to determine if PRMT5 activity has a pathological role in preventing degenerative bone diseases such as osteoarthritis. Prmt5 also has an ongoing role in regulating pre-hypertrophic cell identity in adults (Fig. 6L), suggesting that Prmt5 is broadly required for specialized chondrocyte identity throughout an organism’s lifetime.
The requirement for PRMT5 in mediating step-wise chondrogenic identity at distinct developmental timepoints raises interesting questions regarding its molecular mechanism. PRMT5 is dispensable for maintaining generic chondrocytes but is required for multiple distinct lineages at later timepoints. It is possible that PRMT5 represses distinct sets of target genes in these systems through epigenetic repression via histone modifications. In this case, perhaps a conserved suite of specification/differentiation genes that are broadly required for iterative steps in cartilage specialization, could require PRMT5-mediated activation and result in context-specific differentiation. PRMT5 could additionally influence cell fate decisions by regulating the activity of one or more target proteins through direct methylation. This possibility is supported by evidence that PRMT family members (PRMT4 and PRMT5) interact directly with- and confer di-methyl marks on SOX9 to regulate its activity (43,44). In this case, we predict that additional protein targets or regulatory mechanisms will also occur as the phenotypes we observe are inconsistent with post-natal reductions in SOX9 alone (43). A final possibility is that chondrocytes lacking Prmt5 may lack robustness to transition to different cell fates. This reduced robustness might be sufficient to allow chondrocytes to maintain the status quo but would make cell fate transitions particularly challenging.
Supplementary Material
A, C. Ihh expression (green) by fluorescent in situ hybridization in P21 control (n=3) and PrxCre;Prmt5c/c tibias (n=3). B, D. Visualization of Mef2c expression (green) by fluorescent in situ hybridization in the tibias of P21 3 control and 3 PrxCKO mice. Nuclei stained with DAPI (blue). GP-growth plate (Scale = 100μm).
A-C. Visualization of cell proliferation in the embryonic knee of wild type and PrxCre;Prmt5c/c mutants at E16.5, by phospho-Histone H3 (pHH3) immunostaining (green). The percentage of pHH3-positive cells was quantified across 3 control and 3 mutant embryos (C). D-F. Apoptosis in wild type and mutant knees was visualized through TUNEL staining (white) at E16.5, and the percentage of TUNEL-positive cells was quantified across 3 control and 5 mutant embryos (F). Nuclei stained with DAPI (blue). *p<0.05; **p<0.01 using an unpaired Student’s t-test (Scale = 100μm).
A-B. PRMT5 expression (brown) in the tibia of control and Col2Cre;Prmt5c/c mice at P10 (n=3 controls and 3 mutants). GP-growth plate; AC-articular cartilage; PHC-pre-hypertrophic chondrocytes; HC-hypertrophic chondrocytes (Scale = 100μm).
A-B. β-galactosidase activity (blue) for control and Osteocalcin-Cre (OC-Cre);Rosa-LacZ?c/+ tibias counterstained with eosin at P1 (n=2) (Scale = 100μm). β-galactosidase activity is present in the bone collar and in the trabeculated bone. C-D. X-Ray images of wild-type and OC-Cre;Prmt5c/c mice at P10 (Scale = 1mm). E-F. Alcian blue and eosin stains of the primary ossification center and bone collar in the tibia of 3 WT and 3 OC-Cre;Prmt5c/c mice at P10 (Scale = 100μm). G. Bone collar width was measured adjacent to the growth plate in 3 control and 3 mutant tibias.
A-B. Deletion of Prmt5 was induced with Doxycycline (Dox) at 1 month, and samples were collected for analysis at 4 months. Visualization of Prmt5 expression (green) in the articular cartilage and growth plate of control and ATC;Prmt5c/c mice at 4 months by fluorescent in situ hybridization. Nuclei stained with DAPI (blue). AC-articular chondrocytes; GP-growth plate (Scale = 100μm).
A-B. Visualization of Bmp4 expression in the tibia of control (n=3) and Col2Cre;Prmt5c/c (n=3) mice at P10 by fluorescent in situ hybridization (green). Nuclei stained with DAPI (blue). GP-growth plate; AC-articular cartilage; PHC-pre-hypertrophic chondrocytes; HC-hypertrophic chondrocytes (Scale = 100μm).
Highlights.
PRMT5 promotes the survival of patella progenitor cells before specification.
Loss of Prmt5 results in an expanded domain of hypertrophic chondrocytes.
Mutant hypertrophic chondrocytes undergo a blockage in terminal differentiation.
PRMT5 maintains articular cartilage gene expression in adults.
ACKNOWLEDGEMENTS
We would like to thank Dr. Mathew Hilton, Dr. Veronique Lefebvre, Dr. Fanxin Long, Dr. Susan Mackem, Dr. Francesca Mariani, Dr. John Schwarz, and Dr. Ronen Schweitzer for providing mouse strains and reagents. We thank Jann Uy for technical assistance and Rachel Lex for comments on the manuscript. This study was supported by NIH Grants R01-HD073151 (SV), R01-AR072009-01 (RG), F32-AR073648-01 (ZL), and a UT Austin Provost Graduate Excellence Fellowship (JR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142(5):817–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–6. [DOI] [PubMed] [Google Scholar]
- 3.Inada M, Yasui T, Nomura S, Miyake S, Deguchi K, Himeno M, et al. Maturational disturbance of chondrocytes in Cbfal-deficient mice. Dev Dyn. 1999;214(4):279–90. [DOI] [PubMed] [Google Scholar]
- 4.Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12(3):377–89. [DOI] [PubMed] [Google Scholar]
- 5.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89(5):755–64. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue KI, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18(8):952–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan Z, Niu B, Tsang KY, Melhado IG, Ohba S, He X, et al. Synergistic co-regulation and competition by a SOX9-GLI-FOXA phasic transcriptional network coordinate chondrocyte differentiation transitions. PLoS Genetics. 2018(14):1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89(5):773–9. [DOI] [PubMed] [Google Scholar]
- 9.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79(6): 1111–20. [DOI] [PubMed] [Google Scholar]
- 10.Yang L, Tsang KY, Tang HC, Chan D, Cheah KSE. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. PNAS. 2016. August 19;114(33): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes Transdifferentiate into Osteoblasts in Endochondral Bone during Development, Postnatal Growth and Fracture Healing in Mice. PLoS Genet. 2014;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shwartz Y, Viukov S, Krief S, Zelzer E. Joint Development Involves a Continuous Influx of Gdf5-Positive Cells. Cell Rep. 2016;15(12):2577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eyal S, Blitz E, Shwartz Y, Akiyama H, Schweitzer R, Zelzer E. On the development of the patella. Development. 2015;142(10):1831–9. [DOI] [PubMed] [Google Scholar]
- 14.Decker RS, Koyama E, Enomoto-Iwamoto M, Maye P, Rowe D, Zhu S, et al. Mouse limb skeletal growth and synovial joint development are coordinately enhanced by Kartogenin. Dev Biol. 2014;395(2):255–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, Hilton MJ, Keefe RJO. Cartilage-specific β-CATENIN signaling regulates chondrocyte maturation, generation of ossification centers, and perichondral bone formation during skeletal development. J Bone Miner Res. 2012; 27(8): 1680–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norrie JL, Li Q, Co S, Huang B-L, Ding D, Uy JC, et al. PRMT5 is essential for the maintenance of chondrogenic progenitor cells in the limb bud. Development. 2016;143(24):4608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the Developing Mouse Limb Bud Driven by a Prxl Enhancer. 2002;80:77–80. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Jiao J, Li H, Wan H, Zheng C, Cai J, et al. Histone arginine methylation by Prmt5 is required for lung branching morphogenesis through repression of BMP signaling. J Cell Sci. 2018;131(14):jcs217406. [DOI] [PubMed] [Google Scholar]
- 19.Dong Y, Song C, Wang Y, Lei Z, Xu F, Guan H, et al. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell Signal. 2017;34:55–65. [DOI] [PubMed] [Google Scholar]
- 20.Chen D, Zeng S, Huang M, Xu H, Liang L, Yang X. Role of protein arginine methyltransferase 5 in inflammation and migration of fibroblast-like synoviocytes in rheumatoid arthritis. J Cell Mol Med. 2017;21(4):781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanc RS, Richard S. Arginine Methylation: The Coming of Age. Mol Cell. 2017;65(1):8–24. [DOI] [PubMed] [Google Scholar]
- 22.Tee WW, Pardo M, Theunissen TW, Yu L, Choudhary JS, Hajkova P, et al. Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev. 2010;24(24):2772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, et al. Blimpl associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8(6):623–30. [DOI] [PubMed] [Google Scholar]
- 24.Long F, Zhang XM, Karp S, Yang Y, Mcmahon AP. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. 2001;128(9):5099–108. [DOI] [PubMed] [Google Scholar]
- 25.Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, et al. Sox9 Directs Hypertrophic Maturation and Blocks Osteoblast Differentiation of Growth Plate Chondrocytes. Dev Cell. 2012;22(3):597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Xuan S, Bouxsein ML, Von Stechow D, Akeno N, Faugere MC, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277(46):44005–12. [DOI] [PubMed] [Google Scholar]
- 27.Allen BL, Song JY, Izzi L, Althaus IW, Kang J-S, Charron F, et al. Overlapping roles and collective requirement for the co-receptors Gasl, Cdo and Boc in Shh pathway function. Dev Cell. 2011;20(6):775–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewandowski JP, Du F, Zhang S, Powell MB, Falkenstein KN, Ji H, et al. Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev Biol. 2015;406(1):92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyal S, Blitz E, Shwartz Y, Akiyama H, Schweitzer R, Zelzer E. On the development of the patella. Development. 2015;142(10):1831–9. [DOI] [PubMed] [Google Scholar]
- 30.Eyal S, Kult S, Rubin S, Krief S, Felsenthal N, Pineault KM, et al. Bone morphology is regulated modularly by global and regional genetic programs. 2019;1:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pryce BA, Watson SS, Murchison ND, Staverosky JA, Dunker N, Schweitzer R. Recruitment and maintenance of tendon progenitors by TGF signaling are essential for tendon formation. Development. 2009;136(8):1351–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiménez MJ, Balbín M, López JM, Alvarez J, Komori T, López-Otín C. Collagenase 3 is a target of Cbfal, a transcription factor of the runt gene family involved in bone formation. Mol Cell Biol. 1999;19(6):4431–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ionescu A, Kozhemyakina E, Nicolae C, Kaestner KH, Olsen BR, Lassar AB. FoxA Family Members Are Crucial Regulators of the Hypertrophic Chondrocyte Differentiation Program. Dev Cell. 2012;22(5):927–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kota SK, Roening C, Patel N, Kota SB, Baron R. PRMT5 inhibition promotes osteogenic differentiation of mesenchymal stromal cells and represses basal interferon stimulated gene expression. Bone. 2018;117(9):37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilton MJ, Tu X, Long F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev Biol. 2007;308(1):93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Q, Jiao J, Li H, Wan H, Zheng C, Cai J, et al. Histone arginine methylation by Prmt5 is required for lung branching morphogenesis through repression of BMP signaling. J Cell Sci. 2018;131(14):jcs217406. [DOI] [PubMed] [Google Scholar]
- 37.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alphal(II) collagen gene. Mol Cell Biol. 1997;17(4):2336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inada M, Wang Y, Byrne MH, Rahman MU, Miyaura C, Lopez-Otin C, et al. Critical roles for collagenase-3 (Mmpl3) in development of growth plate cartilage and in endochondral ossification. Proc Natl Acad Sci. 2004;101(49):17192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhee DK, Marcelino J, Baker M, Gong Y, Smits P, Lefebvre V, et al. The secreted glycoprotein lubricin protects cartilage surfaces and inhibits synovial cell overgrowth. J Clin Invest. 2005;115(3):622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coles JM, Zhang L, Blum JJ, Warman ML, Jay GD, Guilak F, et al. Loss of cartilage structure, stiffness, and frictional properties in mice lacking PRG4. Arthritis Rheum. 2010;62(6):1666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan M ZC, Erez A, Guse K, Dawson B, Bertin T, Chen Y, Jiang MM, Yustein J, Gannon F. Proteoglycan4 expression protects against the development of osteoarthritis. Sci Transl Med. 2013;5(176). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Decker RS, Um H Bin, Dyment NA, Cottingham N, Usami Y, Enomoto-Iwamoto M, et al. Cell origin, volume and arrangement are drivers of articular cartilage formation, morphogenesis and response to injury in mouse limbs. Dev Biol. 2017;426(1):56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun M, Hussain S, Hu Y, Yan J, Min Z, Lan X, et al. Maintenance of SOX9 stability and ECM homeostasis by selenium-sensitive PRMT5 in cartilage. Osteoarthr Cartil. 2019;2. [DOI] [PubMed] [Google Scholar]
- 44.Ito T, Yadav N, Lee J, Furumatsu T, Yamashita S, Yoshida K, et al. Arginine methyltransferase CARM1/PRMT4 regulates endochondral ossification. BMC Dev Biol. 2009;9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, C. Ihh expression (green) by fluorescent in situ hybridization in P21 control (n=3) and PrxCre;Prmt5c/c tibias (n=3). B, D. Visualization of Mef2c expression (green) by fluorescent in situ hybridization in the tibias of P21 3 control and 3 PrxCKO mice. Nuclei stained with DAPI (blue). GP-growth plate (Scale = 100μm).
A-C. Visualization of cell proliferation in the embryonic knee of wild type and PrxCre;Prmt5c/c mutants at E16.5, by phospho-Histone H3 (pHH3) immunostaining (green). The percentage of pHH3-positive cells was quantified across 3 control and 3 mutant embryos (C). D-F. Apoptosis in wild type and mutant knees was visualized through TUNEL staining (white) at E16.5, and the percentage of TUNEL-positive cells was quantified across 3 control and 5 mutant embryos (F). Nuclei stained with DAPI (blue). *p<0.05; **p<0.01 using an unpaired Student’s t-test (Scale = 100μm).
A-B. PRMT5 expression (brown) in the tibia of control and Col2Cre;Prmt5c/c mice at P10 (n=3 controls and 3 mutants). GP-growth plate; AC-articular cartilage; PHC-pre-hypertrophic chondrocytes; HC-hypertrophic chondrocytes (Scale = 100μm).
A-B. β-galactosidase activity (blue) for control and Osteocalcin-Cre (OC-Cre);Rosa-LacZ?c/+ tibias counterstained with eosin at P1 (n=2) (Scale = 100μm). β-galactosidase activity is present in the bone collar and in the trabeculated bone. C-D. X-Ray images of wild-type and OC-Cre;Prmt5c/c mice at P10 (Scale = 1mm). E-F. Alcian blue and eosin stains of the primary ossification center and bone collar in the tibia of 3 WT and 3 OC-Cre;Prmt5c/c mice at P10 (Scale = 100μm). G. Bone collar width was measured adjacent to the growth plate in 3 control and 3 mutant tibias.
A-B. Deletion of Prmt5 was induced with Doxycycline (Dox) at 1 month, and samples were collected for analysis at 4 months. Visualization of Prmt5 expression (green) in the articular cartilage and growth plate of control and ATC;Prmt5c/c mice at 4 months by fluorescent in situ hybridization. Nuclei stained with DAPI (blue). AC-articular chondrocytes; GP-growth plate (Scale = 100μm).
A-B. Visualization of Bmp4 expression in the tibia of control (n=3) and Col2Cre;Prmt5c/c (n=3) mice at P10 by fluorescent in situ hybridization (green). Nuclei stained with DAPI (blue). GP-growth plate; AC-articular cartilage; PHC-pre-hypertrophic chondrocytes; HC-hypertrophic chondrocytes (Scale = 100μm).