Abstract
We analyze the exome sequences of approximately 8,000 children with autism spectrum disorder (ASD) and/or attention-deficit/hyperactivity disorder (ADHD) and 5,000 controls, and we find that ASD and ADHD have a similar burden of rare protein-truncating variants in evolutionarily constrained genes, both significantly higher than controls. This motivates a combined analysis across ASD and ADHD, which identifies microtubule-associated protein 1A (MAP1A) as a novel exome-wide significant gene conferring risk for childhood psychiatric disorders.
Introduction
Autism spectrum disorder (ASD) and attention-deficit/hyperactivity disorder (ADHD) are substantially heritable1–3, but individuals with psychiatric diagnoses often do not have blood drawn as part of routine medical procedure, making it difficult to collect cohorts for genetic analysis—particularly for ADHD, which has not previously been the subject of a large-scale sequencing study. To overcome this challenge, we drew upon two Danish national resources: the Danish Neonatal Screening Biobank (DNSB) and the Danish Psychiatric Central Research Register (DPCRR).
As part of the iPSYCH research initiative4, we identified individuals with psychiatric diagnoses using the DPCRR, and we extracted DNA from their archived dried blood samples stored in the DNSB. Individuals were born in Denmark between 1981 and 2005 and were matched to diagnoses of ASD, ADHD, schizophrenia, bipolar disorder, affective disorder, and anorexia, as well as intellectual disability (ID), conferred by the end of 2016. We have previously validated the genotyping5 and sequencing6 of archived samples (Methods), and in this study we exome sequenced a subset of the DNA samples genotyped in recent common variant analyses of both ASD7 and ADHD8. After quality control, our dataset included 3,962 cases with ASD, 901 cases with both ASD and ADHD, 3,477 cases with ADHD, and 5,002 controls without any of the above diagnoses (Table 1).
Table 1: Phenotype breakdown of samples analyzed in this study.
Samples were matched to diagnoses of ASD, ADHD, schizophrenia, bipolar disorder, affective disorder, and anorexia, as well as intellectual disability (ID).
| Phenotype group | No diagnoses, no ID | 1 diagnosis, no ID | >1 diagnosis, no ID | ≥1 diagnosis, with ID | Total |
|---|---|---|---|---|---|
| ASD | - | 2,430 | 661 | 871 | 3,962 |
| ASD+ADHD | - | - | 684 | 217 | 901 |
| ADHD | - | 2,360 | 846 | 271 | 3,477 |
| Control | 5,002 | - | - | - | 5,002 |
Studies of de novo variants in ASD have found that the greatest excess of point mutations carried by affected children resides in protein-truncating variants (PTVs; e.g., nonsense, frameshift, and essential splice site mutations)9–13. Furthermore, this excess burden is almost exclusively carried by PTVs that are rare in the general population and that occur in likely haploinsufficient genes (i.e. probability of being loss-of-function intolerant, or pLI, of at least 0.9)14,15. Although we could not call de novo variants in our case-control data, we used these findings to guide our analysis. We defined as “rare” any variant with an allele count no greater than 5 across the combination of our dataset (n = 13,342) with non-Finnish Europeans from the non-psychiatric exome subset of the Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org/) (n = 44,779), a total population of 58,121 people, and we took special interest in genes with pLI ≥ 0.9, which we termed “constrained”.
Results
Rates of constrained rare variation
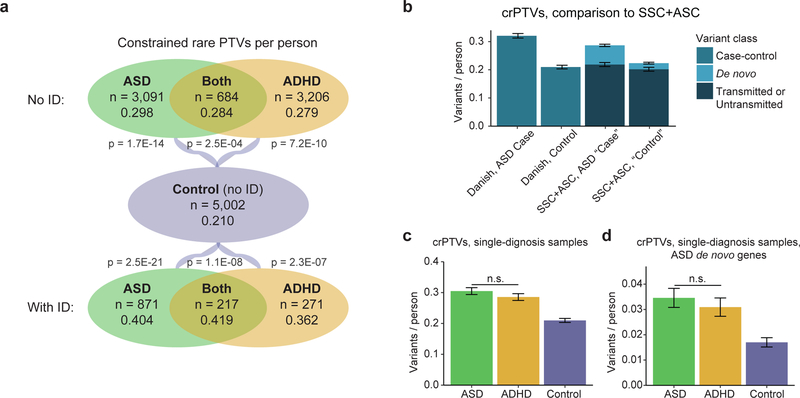
In samples without intellectual disability, we observed a significant excess of constrained rare PTVs (or “crPTVs”) in ASD cases (0.298/person, p = 1.7E-14 by logistic regression), cases with both ASD and ADHD (0.284/person, p = 2.5E-04), and ADHD cases (0.279/person, p = 7.2E-10) compared to controls (0.210/person) (Figure 1a; Figure S1a; Table S1). Consistent with previous observations, we also observed substantially higher rates of crPTVs in cases with comorbid ID compared to controls (0.404/person in ASD, p = 2.5E-21; 0.419 in ASD+ADHD, p = 1.1E-08; 0.362 in ADHD, p = 2.3E-07) (Figure 1a; Figure S1a). By contrast, none of our case categories had a significantly higher burden of rare PTVs in genes with pLI < 0.9 compared to controls (Figure S1b). Rates of constrained rare synonymous variation were similar across sample categories (with no case category significantly different from controls), showing that the excess crPTVs in cases did not result from technical differences in variant calling (Figure S1c). Rates of crPTVs were higher in females than in males across most phenotype groups (Table S1), consistent with a female protective effect16, although differences between the sexes were not significant. Most crPTVs were found in people with exactly one of them (Figure S2, Table S2).
Figure 1: Rates of constrained rare protein-truncating variants (crPTVs).
a) Mean rates of crPTVs across phenotypes, with and without intellectual disability (ID). “Constrained” denotes genes with pLI (probability of being loss-of-function intolerant) values at least 0.9. “Rare” denotes variants with an allele count of no greater than 5 across the 13,342 Danish samples analyzed in this study and the 44,779 non-Finnish Europeans in the non-psychiatric exome subset of gnomAD (58,121 total individuals). P values shown are for comparison to controls. Differences between case categories without ID are not significant (p = 0.49 for ASD vs ASD+ADHD; p = 0.91 for ADHD vs ASD+ADHD; p = 0.14 for ASD vs ADHD), nor are differences between case categories with ID significant (p = 0.59 for ASD vs ASD+ADHD; p = 0.60 for ADHD vs ASD+ADHD; p = 0.58 for ASD vs ADHD). b) Mean rates of crPTVs in Danish case-control data compared to crPTVs in Simons Simplex Collection (SSC) and Autism Sequencing Consortium (ASC) family-based data. From SSC+ASC data14, we constructed ASD “cases” using de novo variants from affected probands (n = 3,982) and transmitted variants from parents of probands (n = 4,319), and we constructed “controls” using de novo variants from unaffected children (n = 2,078) and untransmitted variants from parents of probands (n = 4,319). SSC+ASC variants were counted as “rare” if they had an allele count ≤ 5 across the SSC+ASC data and non-Finnish Europeans from the non-psychiatric exome subset of gnomAD. Danish data is from all individuals with an ASD diagnosis (including comorbid ADHD and/or intellectual disability, n = 4,863) and controls (n = 5,002), and “rare” is defined as in part a. c-d: Mean rates of crPTVs in ASD cases (n = 2,430) and ADHD cases (n = 2,360) with only a single diagnosis (i.e. no comorbid ASD+ADHD samples, no intellectual disability diagnosis, and no diagnoses of schizophrenia, bipolar disorder, affective disorder, or anorexia). “Rare” is defined as in part a, and the same controls (n = 5,002) are used. c) Rates in all constrained genes. ASD and ADHD rates are not significantly different from each other (p = 0.21), while both are significantly different from controls (OR = 1.46 for ASD based on 741 crPTVs, p = 1.12E-14; OR = 1.37 for ADHD based on 674 crPTVs, p = 2.26E-10; 1,049 crPTVs in controls). d) Rates in the 212 constrained genes with a published rare de novo PTV in ASD (“ASD de novo genes”)14. ASD and ADHD rates are again not significantly different from each other (p = 0.38), while both are significantly different from controls (OR = 2.19 for ASD based on 84 crPTVs, p = 5.39E-07; OR = 1.87 for ADHD based on 73 crPTVs, p = 1.40E-04; 85 crPTVs in controls). For a-d, all p values are by logistic regression (Methods), and all error bars are Poisson standard error. OR = odds ratio.
A similar trend to crPTVs was observed with rare missense variants, though the signal was less pronounced (e.g. 0.88/person in ASD cases without ID compared to 0.81 in controls, p = 4.1E-03 by logistic regression) (Figure S3; Figure S4a; Table S1). Here, we considered only missense variants with an MPC score (a measure of the deleteriousness of a missense variant based on a regional model of constraint17) of at least 2. A lower degree of enrichment was observed when considering rare missense variants with MPC < 2 (Figure S4b), with synonymous rates largely comparable across phenotype groups (Figure S4c).
To compare the results of our case-control study to those previously seen in de novo studies of the Simons Simplex Collection (SSC) and Autism Sequencing Consortium (ASC) datasets10,11,15, we examined genes with three or more published rare de novo PTVs in ASD. Combining all of our cases with an ASD diagnosis (including those with comorbid ADHD and/or ID), we observed a significantly enriched burden of rare PTVs in this set of 14 genes (Table 2; p = 1.6E-06 by logistic regression, OR = 6.4, n = 4,863 ASD cases vs 5,002 controls). The only rare PTVs observed in controls were in lysine demethylase 5B (KDM5B), which acts in a potentially recessive manner18; in the other 13 genes, we observed 37 rare PTVs in cases and none in controls. In addition, when applying our rarity threshold to the SSC+ASC data (Methods), the rate of crPTVs in the case-control Danish data was similar to the combined rates of published de novo and inherited crPTVs (Figure 1b).
Table 2: Rare PTV counts in genes with 3 or more published14 rare de novo protein-truncating variants in ASD.
Danish ASD data is from all individuals with an ASD diagnosis (including comorbid ADHD and/or ID, n = 4,863) and controls (n = 5,002). Danish variants were counted as “rare” if they had an allele count ≤ 5 across the Danish data and non-Finnish Europeans from the non-psychiatric exome subset of gnomAD. Published SSC+ASC variants were counted as “rare” if they had an allele count ≤ 5 across the SSC+ASC data and non-Finnish Europeans from the non-psychiatric exome subset of gnomAD. P values and odds ratios are for comparison to controls by logistic regression. OR = odds ratio. SE = standard error. PTV = protein-truncating variant. ID = intellectual disability.
| Gene | Published rare de novo PTVs in ASD | Published rare de novo PTVs in unaffected children | Danish rare PTVs: ASD, no ID (n = 3,775) | Danish rare PTVs: ASD, ID (n = 1,088) | Danish rare PTVs: ASD, total (n = 4,863) | Danish rare PTVs: Control (n = 5,002) |
|---|---|---|---|---|---|---|
| CHD8 | 6 | 0 | 1 | 1 | 2 | 0 |
| ARID1B | 5 | 0 | 3 | 0 | 3 | 0 |
| DYRK1A | 5 | 0 | 0 | 3 | 3 | 0 |
| SYNGAP1 | 5 | 0 | 0 | 4 | 4 | 0 |
| ADNP | 4 | 0 | 0 | 2 | 2 | 0 |
| ANK2 | 4 | 0 | 5 | 2 | 7 | 0 |
| DSCAM | 4 | 0 | 1 | 0 | 1 | 0 |
| SCN2A | 4 | 0 | 1 | 3 | 4 | 0 |
| ASH1L | 3 | 0 | 0 | 2 | 2 | 0 |
| CHD2 | 3 | 0 | 0 | 1 | 1 | 0 |
| GRIN2B | 3 | 0 | 0 | 4 | 4 | 0 |
| KDM5B | 3 | 2 | 7 | 1 | 8 | 8 |
| POGZ | 3 | 0 | 0 | 3 | 3 | 0 |
| SUV420H1 | 3 | 0 | 1 | 0 | 1 | 0 |
| Total, all genes | 55 | 2 | 19 | 26 | 45 | 8 |
| OR vs Control | - | - | 3.1 | 15.9 | 6.4 | - |
| OR +/− SE | - | - | 2.1–4.8 | 10.4–24.3 | 4.4–9.5 | - |
| p | - | - | 7.5E-03 | 9.1E-11 | 1.6E-06 | - |
Having observed similar rates of crPTVs between ASD and ADHD (e.g. Figure 1a), we decided to further explore the overlap of the two disorders. To rule out the possibility of a common comorbidity driving the signal, our next analyses included only those cases with a single diagnosis (e.g. no comorbid ASD+ADHD samples, no intellectual disability diagnosis, and no diagnoses of schizophrenia, bipolar disorder, affective disorder, or anorexia) (n = 2,430 for ASD and n = 2,360 for ADHD). As with the more inclusive sample groups, these single-diagnosis ASD cases and ADHD cases had similar burdens of crPTVs overall, and both were significantly greater than controls (Figure 1c; synonymous rates in Figure S5a; Table S1). We next considered the rates of crPTVs occurring in these samples in the set of 212 constrained genes with a published rare de novo PTV in ASD15. In this ASD-derived gene set, the ADHD cases again had a rate of crPTVs nearly as high as the ASD cases themselves (Figure 1d; synonymous rates in Figure S5b), with both case categories enriched above the control rate (OR = 2.19 for ASD, p = 5.39E-07 by logistic regression; OR = 1.87 for ADHD, p = 1.40E-04) but not significantly different from each other (p = 0.38).
Joint ASD and ADHD analysis
Given the similar crPTV burdens in ASD and ADHD cases, we used a c-alpha test19 to determine whether the sets of constrained genes with rare PTVs were similar or distinct in ASD and ADHD. The c-alpha test can be used to test whether two distributions of rare variants have been selected from the same underlying distribution20. Considering again only cases with a single diagnosis, the test did not find a significant difference between ASD and ADHD, but it did find a significant difference when comparing either case group and controls (Table 3; Table S3). This result suggests that the crPTVs in individuals with ASD or ADHD are not only occurring at similar rates, but also in similar sets of genes. The test did not find a significant difference in any pairwise comparison of ASD cases, ADHD cases, and controls when considering constrained rare synonymous variation (Table 3) or rare missense variation (MPC ≥ 2) (Table S4).
Table 3: c-alpha test results for constrained rare PTVs and constrained rare synonymous variants.
We tested ASD cases (n = 2,430) and ADHD cases (n = 2,360) with only a single diagnosis in pairwise comparisons against each other and against controls (n = 5,002) to determine whether the distributions of genes with crPTVs were significantly different between the phenotype groups. “Single” diagnosis refers to samples with only a diagnosis of ASD or ADHD (i.e. no comorbid ASD+ADHD samples, no intellectual disability diagnosis, and no diagnoses of schizophrenia, bipolar disorder, affective disorder, or anorexia). “Genes” column indicates number of genes in the comparison with at least one variant.
| Constrained rare PTVs | Constrained rare synonymous variants | |||
|---|---|---|---|---|
| Comparison | Genes | c-alpha p value | Genes | c-alpha p value |
| ASD vs ADHD | 932 | 0.93 | 2,947 | 0.83 |
| ASD vs Control | 1,102 | 5.7E-09 | 3,059 | 0.31 |
| ADHD vs Control | 1,064 | 1.3E-05 | 3,047 | 0.93 |
The finding that ASD and ADHD had similar burdens of crPTVs occurring in similar genes, and that both were distinct from controls, motivated pooling all of our ASD, ASD+ADHD, and ADHD cases (n = 8,340) for the purposes of gene discovery. To increase our control population, we included non-Finnish Europeans from the non-psychiatric exome subset of gnomAD, for a total of 49,781 controls. To ensure that these cohorts were comparable, we determined the portions of the exome that were well-covered in both the Danish exomes and the gnomAD exomes, and we only considered variants in this consensus high-confidence region (Methods). We then counted the number of rare protein-truncating, missense (MPC ≥ 2), and synonymous variants by gene and sample group, applying our definition of rare to variants in gnomAD as well, and used a two-tailed Fisher’s exact test to calculate case vs control p values for each class of variation in each gene. When combining datasets in this manner, the rate of rare variation within each dataset is an important consideration; in this analysis, we took the conservative approach of only considering genes with greater rates of synonymous variation in controls than cases as we searched for genes with greater rates of protein-truncating or missense (MPC ≥ 2) variation in cases than controls (Methods).
Among constrained genes, the top result in our PTV analysis was microtubule-associated protein 1A (MAP1A), in which we observed 11 rare PTVs in Danish cases (4 ASD without ID, 5 ADHD without ID, 1 ASD with ID, 1 ASD+ADHD with ID), none in Danish controls, and only 4 in gnomAD (Table 4; Table S5). With a case vs control p value of 4.11E-07, it survives Bonferroni correction for 17,903 genes and is exome-wide significant. MAP1A is highly expressed in the mammalian brain and is important for the organization of neuronal microtubules; a candidate gene study identified an excess of rare missense variants in MAP1A in ASD and schizophrenia21. Although our case-control study includes inherited variation and does not have the power of a de novo study to isolate high-penetrance PTVs, we do observe genes flagged by de novo studies—such as ANKRD11, which is associated with intellectual disability22, and SCN2A, which is associated with ASD13—among genes with a p value of less than 0.01. We also note RAI1, which is associated with Smith-Magenis syndrome23, among our top results. A quantile-quantile plot is shown in Figure S6a, and an analogous plot for synonymous variants (Figure S6b) shows little inflation. In the analysis based on missense variation (Figure S6c; Table S5; Table S6), no genes passed exome-wide significance.
Table 4: Top 15 constrained genes in rare PTV analysis, ranked by two-tailed Fisher’s exact p value comparing case (n = 8,340) total to combined control+gnomAD (n = 49,781) total variant counts.
Cases include all samples with an ASD and/or ADHD diagnosis, regardless of intellectual disability status. Controls include all control samples as well as non-Finnish Europeans from the non-psychiatric exome subset of gnomAD. Only genes with pLI ≥ 0.9 are shown. P values are also given for comparison of cases to Danish controls (n = 5,002) before combination with gnomAD. “ASD dn” denotes number of published rare de novo PTVs in ASD (SSC+ASC data, 3,982 probands)14. “DDD dn” denotes number of published rare de novo PTVs in the Deciphering Developmental Disorders study, which examines intellectual disability/developmental delay (4,293 probands)20. Note that SCN2A has 4 PTVs listed in Table 2 but only 3 listed here because one fell 2bp outside the consensus high-confidence region used when combining with gnomAD (Methods). OR = odds ratio.
| Gene | ASD (n = 3,962) | ASD+ADHD (n = 901) | ADHD (n = 3,477) | Control (n = 5,002) | p value (Danish) | gnomAD (n = 44,779) | p value (Combined) | OR | ASD dn | DDD dn |
|---|---|---|---|---|---|---|---|---|---|---|
| MAP1A | 5 | 1 | 5 | 0 | 9.21E-03 | 4 | 4.11E-07 | 16.4 | 0 | 1 |
| ZNF536 | 2 | 2 | 0 | 0 | 3.04E-01 | 0 | 4.24E-04 | Inf | 0 | 0 |
| SPTBN1 | 1 | 1 | 3 | 0 | 1.65E-01 | 2 | 9.90E-04 | 14.9 | 1 | 1 |
| ANKRD11 | 2 | 0 | 2 | 0 | 3.04E-01 | 1 | 1.88E-03 | 23.9 | 2 | 32 |
| MAGEL2 | 4 | 0 | 0 | 0 | 3.04E-01 | 1 | 1.88E-03 | 23.9 | 0 | 0 |
| RAP1GAP2 | 4 | 0 | 2 | 1 | 2.68E-01 | 4 | 2.10E-03 | 7.2 | 0 | 0 |
| SLC2A14 | 3 | 0 | 3 | 2 | 7.18E-01 | 3 | 2.10E-03 | 7.2 | 0 | 0 |
| RAI1 | 1 | 2 | 2 | 0 | 1.65E-01 | 3 | 2.33E-03 | 10.0 | 1 | 1 |
| TNRC6C | 1 | 2 | 4 | 0 | 5.04E-02 | 8 | 2.78E-03 | 5.2 | 0 | 0 |
| GLUL | 1 | 0 | 2 | 0 | 2.97E-01 | 0 | 2.95E-03 | Inf | 0 | 0 |
| SCN2A | 3 | 0 | 0 | 0 | 2.97E-01 | 0 | 2.95E-03 | Inf | 4 | 5 |
| STAT5B | 2 | 0 | 1 | 0 | 2.97E-01 | 0 | 2.95E-03 | Inf | 0 | 0 |
| ZEB2 | 2 | 1 | 0 | 0 | 2.97E-01 | 0 | 2.95E-03 | Inf | 0 | 1 |
| DYNC1H1 | 5 | 0 | 1 | 0 | 9.01E-02 | 6 | 3.69E-03 | 6.0 | 0 | 0 |
| HSPA12A | 1 | 1 | 4 | 1 | 2.68E-01 | 5 | 3.69E-03 | 6.0 | 0 | 0 |
Discussion
In summary, we used DNA from archived bloodspots to conduct an exome sequencing study of ASD and ADHD. To place our study in the context of previous de novo variant studies of ASD, we examined our rare PTVs in the top published ASD genes and found an overwhelming burden in ASD cases compared to controls, suggesting that we are at least partly tapping into the same signal. We also showed that rates of crPTVs in our ASD cases and controls were consistent with the sum of de novo and transmitted (or untransmitted) crPTV rates previously seen in SSC+ASC data. In our data, we observed a similar burden of crPTVs in ASD and ADHD, and this motivated a combined analysis for gene discovery. Using gnomAD as an additional control population, we identified MAP1A as significantly associated with ASD and ADHD. Because we observe rare MAP1A PTVs in cases both with and without intellectual disability—and because the genes near the top of our list are not exclusively those previously identified by de novo studies—our case-control findings may include genes where protein-truncating variants are relevant to psychiatric cases with milder or more behavioral profiles (and with contribution from inherited variation) in addition to those characterized by more profound neurodevelopmental symptomatology (and primarily driven by de novo variation).
Genetic connections between ASD and ADHD have been made previously24; for example, twin studies show that traits related to ASD significantly co-occur with traits related to ADHD25, and siblings of children with an ASD diagnosis are more likely to exhibit symptoms of ADHD and develop ADHD than the general population26. In the genotype data from our population sample, additional evidence comes from the finding that the two disorders are genetically correlated (rg = 0.36, p = 1.24E-12)7. This study, however, is the first to have such a large sample size of exome sequences to analyze in the two disorders, enabling comparisons such as the c-alpha test. The similar burden of crPTVs in ASD and ADHD is noteworthy, and it suggests that it is worth investigating whether study designs that have been successful in ASD could also be useful in ADHD. Our results also suggest that cross-disorder rare variant studies could allow investigators to increase power for gene discovery in a combined analysis, in addition to comparing the contribution of variants across disorders.
Methods
gnomAD
All references to gnomAD in this study refer to release 2.1 (beta) of the non-psychiatric/non-brain subset which has had samples from psychiatric studies removed (http://gnomad.broadinstitute.org/; the dataset in Hail 0.2 format is hosted on the Google cloud at gs://gnomad-public/release/2.1_beta/ht/).
Sample selection
Individuals in the iPSYCH cohort were born in Denmark between May 1, 1981 and December 31, 20054. Neonatal dried blood samples were stored in the Danish Neonatal Screening Biobank, which houses samples from nearly all individuals born in Denmark since 1982 (and some from 1981). The iPSYCH initiative considers six primary psychiatric diagnoses—ASD, ADHD, schizophrenia, bipolar disorder, affective disorder, and anorexia—and individuals were selected for inclusion in the cohort after matching them to psychiatric diagnoses in the Danish Psychiatric Central Research Register. At the time of sample selection, diagnoses were those conferred by the end of 2012; in this study, we use diagnoses conferred by the end of 2016. ASD cases include individuals with an ICD10 diagnosis code of F84.0, F84.1, F84.5, F84.8, or F84.9. ADHD cases include individuals with an F90.0 diagnosis. The intellectual disability designation was based on an individual having any diagnosis for intellectual disability, including mild, moderate, or severe (codes F70-F79).
Sample sequencing and validation
The extraction of DNA from archived DNSB blood samples for use in genetic analysis has been extensively described over the past decade. Publications which form the basis for this study include papers describing the extraction27, whole-genome amplification28, validation for use in genotyping arrays5, and validation for use in exome sequencing6 of DNA from archived DNSB blood samples. Hollegaard et al. (2013)6, for example, compared DNA from whole blood samples to DNA from the same individuals extracted from archived blood samples of two different ages (3 years and ~27 years) and found that the archived samples performed as well as the whole blood samples with regard to error rates in sequencing6.
The DNA used in this study had previously been extracted and whole-genome amplified for use in iPSYCH genotyping studies of common variants in ASD7 and ADHD8. The genotyped iPSYCH cohort consists of over 88,000 samples, and a subset of approximately 20,000 age- and ancestry-matched samples was selected for exome sequencing. A validation study was carried out to confirm that DNA from these samples would generate exome sequences of sufficient quality; Poulsen et al. (2016)29 examined variant calls based on DNA from archived DNSB blood samples vs whole blood samples from the same individuals, as well as whole blood samples vs whole blood samples, and found that concordance rates were similar and close to 100%. The Poulsen et al. analysis included samples sequenced at the Broad Institute in Cambridge, MA—which subsequently generated the sequences used in this study—and concluded that whole-genome amplified DNA from archived DNSB samples performed similarly in exome sequencing to DNA from high-quality whole blood samples29.
Following the Poulsen et al. study, sequencing for this study commenced at the Genomics Platform of the Broad Institute using an Illumina Nextera capture kit and an Illumina HiSeq sequencer. Sequencing was carried out in multiple waves, including a smaller pilot wave (“Pilot 1”) and two larger production waves (“Wave 1” and “Wave 2”). After the pilot wave (n = 586), heterozygote calls from the exome sequence data were compared to the genotype data for the same samples and found to be over 99.8% concordant. The next two waves were then sequenced.
Callset creation
Raw sequencing data was processed using the Genome Analysis Toolkit30 (GATK) version 3.4 to produce a VCF version 4.1 variant callset file. The VCF used as the starting point for this study included 586 samples from Pilot 1, 6,733 samples from Wave 1, and 12,532 samples from Wave 2.
Callset quality control
Most filtering steps downstream of GATK were performed in the scalable genomics program Hail (https://hail.is, https://github.com/hail-is/hail). After importing the VCF into Hail 0.1, ACMG genes31 (https://www.ncbi.nlm.nih.gov/clinvar/docs/acmg/) were removed from the dataset, per Danish regulations. Next, sex was imputed using the impute_sex() function, relatedness between samples was calculated over a set of 5,848 common variants using the ibd() function, and principal components were calculated on the same set of common variants using the pca() function. Samples were dropped from the dataset a) if they lacked complete phenotype information (30 samples), b) if their imputed sex did not clearly match their reported sex (28 samples), c) if they were a duplicate (or monozygotic twin) (13 samples), d) if they were not putatively European by PCA (1,981 samples), e) if they were a control (i.e. without a diagnosis of ASD, ADHD, schizophrenia, bipolar disorder, affective disorder, or anorexia) with a diagnosis of intellectual disability (44 samples), or f) if they had an estimated level of contamination (the “FREEMIX” column in the .selfSM file of the bam directory) above 5% (59 samples). A 5% chimeric reads threshold was also imposed, but this did not filter any samples. Variants were then removed if they did not pass GATK variant quality score recalibration (VQSR), if they fell outside the exome target, or if they fell in a low-complexity region.
Next, several genotype filters were used to remove calls of low quality:
Any call with a depth a) less than 10 or b) greater than 1000;
Homozygous reference calls with a) GQ less than 25 or b) less than 90% reads supporting the reference allele;
Homozygous variant calls with a) PL(HomRef) less than 25 or b) less than 90% reads supporting the alternate allele;
Heterozygote calls with a) PL(HomRef) less than 25, b) less than 25% reads supporting the alternate allele, c) less than 90% informative reads (e.g. number of reads supporting the reference allele plus number of reads supporting the alternate allele less than 90% of the read depth), d) a probability of drawing the allele balance from a binomial distribution centered on 0.5 of less than 1E-09, or e) a location where the sample should be hemizygous (e.g. calls on the X chromosome outside the pseudoautosomal region in a male).
Any call on the Y chromosome outside the pseudoautosomal region on a sample from a female.
Following the application of these genotype filters, three call rate filters were used: first the removal of variants with a call rate below 90%, then the removal of samples with a call rate below 95% (575 samples), then the removal of variants with a call rate below 95%. Between the sample call rate filter and the final variant call rate filter, one of each pair of related samples was removed using the ibd_prune() function in Hail, defining relatedness as a pi-hat value of 0.2 or greater (124 samples). Variants remaining in the dataset were annotated with the Variant Effect Predictor32, and one transcript for each variant was selected (prioritizing canonical coding transcripts) to assign a gene and a consequence to each variant. As a final quality control step, samples were removed (505 samples) if they were significantly different (after Bonferroni correction) from the observed mean of number of not-in-gnomAD singletons, based on the probability of drawing the observed number from a Poisson distribution. The purpose of this final step was to remove any of the remaining samples that may have gained noise during the time spent in archive.
Following the application of these filters, the dataset contained 16,492 individuals, and the remaining ASD (3,962), ASD+ADHD (901), ADHD (3,477), and control (5,002) samples were selected for use in this study, while samples with other diagnoses were set aside. ASD cases were 3,005 male and 957 female and had an average birth year of 1992; ASD+ADHD cases were 725 male and 176 female and had an average birth year of 1994; ADHD cases were 2,382 male and 1,095 female and had an average birth year of 1991; controls were 3,373 male and 1,629 female and had an average birth year of 1991 (see also Table 1). Allele counts used in comparisons to gnomAD—and the combination with it—were calculated within these 13,342 samples.
Statistics: P value and odds ratio calculations
For calculating p values and odds ratios for classes of variants (e.g. crPTV rates compared to controls, Figure 1a; Table S1), logistic regression was performed using the glm function in R (https://cran.r-project.org/). Covariates included in the logistic regression model were birth year, sex, the first ten principal components of the genetic data (of PCA carried out after dropping non-European samples), number of rare synonymous variants, percent of exome target covered at a read depth of at least 20, mean read depth at sites within the exome target passing VQSR, number of SNPs (of any population frequency) at sites within the exome target passing VQSR, and sequencing wave (one-hot encoded). For Figure S2, the R function chisq.test was used with observed frequencies and Poisson-expected probabilities based on the observed mean, and p values were simulated with 10,000 replicates. For the c-alpha tests, we utilized the R package AssotesteR (http://cran.r-project.org/web/packages/AssotesteR/index.html); we ran 10,000 permutations for each pairwise test and checked that the permutation-based p value was comparable to the reported asymptotic p value (Table S3). For calculating gene-level p values and odds ratios (e.g. Table 4, Table S5; Table S6), a two-tailed Fisher’s exact test was performed using the fisher.test function in R. In all analyses, PTV counts from iPSYCH samples were capped at one per person per gene to correct for the rare situation where one insertion or deletion event is labeled as two separate variants by the genotype caller. We note that although this filter removed only 0.2% of PTVs, both overall and within constrained genes, there remains the possibility that recessive variants were removed.
Comparison to SSC+ASC
For comparison to our data, we obtained de novo and inherited Simons Simplex Collection and Autism Sequencing Consortium data15. Inherited data was obtained directly from the first author of Kosmicki et al. (2017)15. To apply the definition of “rare” used in this study as closely as possible, variants in both the de novo and inherited sets of SSC+ASC data were annotated with allele counts from non-Finnish Europeans in the non-psychiatric exome subset of gnomAD, and variants with an allele count greater than 5 in the combined SSC+ASC+gnomAD group of samples were dropped. Counting the resulting number of rare de novo PTVs per gene gave the list of top genes used in Table 2, the list of 212 constrained genes with an ASD de novo PTV used in the analyses shown in Figure 1d and Figure S5b, and the ASD de novo PTV counts given in Table 4. Integrating de novo crPTV counts with inherited crPTV counts gave the “case” and “control” crPTV rates we constructed for SSC+ASC data in Figure 1b. Here, “case” SSC+ASC rates consist of de novo variants in ASD-affected probands (n = 3,982) and transmitted variants from parents of probands (n = 4,319), while “control” SSC+ASC rates consist of de novo variants from unaffected children (n = 2,078) and untransmitted variants from parents of probands (n = 4,319). Danish ASD data in Figure 1b is from all children with an ASD diagnosis (with or without ADHD and regardless of ID status, n = 4,863), and Danish control data is the same group of controls (n = 5,002) used throughout our analyses.
Combination with gnomAD
When combining our data with gnomAD for the purpose of gene discovery, variants were dropped if they fell outside of a consensus high-confidence region for the two datasets. This region was defined as the intervals where at least 80% of the samples in both datasets had at least 10x sequencing coverage (based on analysis of bam files for the Danish samples, and based on coverage summary tables for gnomAD). We considered 17,903 genes overall (after dropping the 59 ACMG genes as mentioned above), and this number was not changed by restricting to the consensus high-confidence region. We then counted the number of rare protein-truncating, missense, and synonymous variants by gene. To ensure that the comparison was not biased by differential variation rates between cases (entirely Danish) and controls (mostly gnomAD), we excluded all genes in which rare synonymous variation rates were higher in cases than controls (removed 1,615/17,903, or 9.0% of genes). In the PTV analysis, we then considered only genes with greater rates of rare truncating variation in cases than controls (retained 3,182/16,288, or 19.5% of genes). In the missense analysis, we likewise considered only genes with greater rates of rare missense variation (MPC ≥ 2) in cases than controls (retained 957/16,288, or 5.9% of genes). As can be seen from these filters, the vast majority of genes had higher rates of variation in controls than in cases, indicating that more rare variants were, on average, being called per sample in gnomAD (potentially due to more liberal QC thresholds for parameters like call rate)—a trend which any gene had to overcome in order to have a greater burden of PTVs or missense variants in cases than controls.
Intellectual disability de novo variants
Table 4 lists the number of published “rare” de novo PTVs from the Deciphering Developmental Disorders (DDD) study22 for each of the top 15 constrained genes in our gene discovery analysis. Since none of the published DDD de novo PTVs in these genes had an allele count greater than 5 between the DDD study and the non-Finnish Europeans in the non-psychiatric exome subset of gnomAD, we in fact deemed all of them “rare”.
q-q plots
The PTV q-q plot (Figure S6a) displays the 3,182 genes included in the PTV gene discovery analysis, as described above. The synonymous q-q plot (Figure S6b) displays all genes with greater rates of synonymous variation in cases than controls (retained 1,615/17,903, or 9.0% of genes). The missense q-q plot (Figure S6c) displays the 957 genes included in the missense gene discovery analysis.
Notes on study design
All laboratory processing was performed blind to phenotype. Sample selection was necessarily not performed blind to phenotype, but it was performed blind to an individual’s rare variant burden. Sample sizes were set at a number of cases similar to previous useful studies of ASD (e.g. Ref. 10). To control for downstream batch effects, samples were sequenced in blocks (waves) that included cases and controls matched by birth cohort. The only subjects excluded from this study were filtered due to data quality concerns (described above in the “Callset quality control” section) prior to analysis. No data points were excluded after beginning the analysis. Error bars in bar plots are Poisson standard error; as shown in Fig S2, crPTV distributions did not differ significantly from Poisson expectation. The samples used in this study are considered consented under Danish regulations because parents are informed in writing at the time of blood sampling that the samples will be stored in the DNSB and may be used for approved research; parents are also informed how to opt out of including the sample in research studies4. Further information on study design is available in the Nature Research Life Sciences Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
The iPSYCH project is funded by the Lundbeck Foundation (grant numbers R102-A9118 and R155–2014-1724) and the universities and university hospitals of Aarhus and Copenhagen. The Danish Neonatal Screening Biobank resource at the Statens Serum Institut was supported by the Novo Nordisk Foundation. Sequencing of iPSYCH samples was supported by grants from the Simons Foundation (SFARI 311789 to M.J.D.) and the Stanley Foundation. Other support for this study was received from the NIMH (5U01MH094432–02, 5U01MH111660–02, and U01MH100229 to M.J.D.). Computational resources for handling and statistical analysis of iPSYCH data on the GenomeDK and Computerome HPC facilities were provided by, respectively, Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to A.D.B.), and iPSYCH.
Competing interests
T.M.W. has acted as advisor and lecturer to the pharmaceutical company H. Lundbeck A/S. B.M.N. is a member of Deep Genomics Scientific Advisory Board and has received travel expenses from Illumina. He also serves as a consultant for Avanir Pharmaceuticals and Trigeminal Solutions, Inc.
Footnotes
Data availability statement
Supplementary data are available as supplementary files to this manuscript (Tables S1, S3, and S5) or at the iPSYCH download page: http://ipsych.au.dk/downloads/. For inquiries about more detailed data, contact iPSYCH lead investigator A.D.B. (anders@biomed.au.dk).
Code availability statement
Hail (0.1) and R scripts used to handle and analyze this data are available upon request. Contact F.K.S. (satterst@broadinstitute.org).
Human research statement
This study was approved by the Regional Scientific Ethics Committee in Denmark and the Danish Data Protection Agency.
References
- 1.Hallmayer J et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch. Gen. Psychiatry 68, 1095–1102 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaugler T et al. Most genetic risk for autism resides with common variation. Nat. Genet 46, 881–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larsson H, Chang Z, D’Onofrio BM & Lichtenstein P The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychol. Med 44, 2223–2229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen CB et al. The iPSYCH2012 case-cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol. Psychiatry 23, 6–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollegaard MV et al. Robustness of genome-wide scanning using archived dried blood spot samples as a DNA source. BMC Genet. 12, 58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollegaard MV et al. Archived neonatal dried blood spot samples can be used for accurate whole genome and exome-targeted next-generation sequencing. Mol Genet Metab. 110, 65–72 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Grove J et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demontis D et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet 51, 63–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neale BM et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rubeis S et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iossifov I et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samocha KE et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet 46, 944–950 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders SJ et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosmicki JA et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat. Genet 49, 504–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacquemont S et al. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am. J. Hum. Genet 94, 415–425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samocha KE et al. Regional missense constraint improves variant deleteriousness prediction. Preprint at http://www.biorxiv.org/content/early/2017/06/12/148353 (2017).
- 18.Martin HC et al. Quantifying the contribution of recessive coding variation to developmental disorders. Science 362, 1161–1164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neyman J & Scott E On the use of c(α) optimal tests of composite hypothesis. Bulletin of the International Statistical Institute 41, 477–497 (1966). [Google Scholar]
- 20.Neale BM et al. Testing for an unusual distribution of rare variants. PLoS Genet. 7, e1001322 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers RA et al. A population genetic approach to mapping neurological disorder genes using deep resequencing. PLoS Genet. 7, e1001318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deciphering Developmental Disorders Study. Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slager RE, Newton TL, Vlangos CN, Finucane B & Elsea SH Mutations in RAI1 associated with Smith-Magenis syndrome. Nat. Genet 33, 466–468 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Martin J et al. Biological overlap of attention-deficit/hyperactivity disorder and autism spectrum disorder: evidence from copy number variants. J. Am. Acad. Child Adolesc. Psychiatry 53, 761–770 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ronald A, Simonoff E, Kuntsi J, Asherson P & Plomin R Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J. Child Psychol. Psychiatry 49, 535–542 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Chien YL et al. ADHD-related symptoms and attention profiles in the unaffected siblings of probands with autism spectrum disorder: focus on the subtypes of autism and Asperger’s disorder. Mol. Autism 8, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannelius U et al. Phenylketonuria screening registry as a resource for population genetic studies. J. Med. Genet 42, e60 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollegaard MV et al. Whole genome amplification and genetic analysis after extraction of proteins from dried blood spots. Clin. Chem 53, 1161–1162 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Poulsen JB et al. High-quality exome sequencing of whole-genome amplified neonatal dried blood spot DNA. PLoS One 11, e0153253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalia SS et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med 19, 249–255 (2017). [DOI] [PubMed] [Google Scholar]
- 32.McLaren W et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.