Abstract
Inclusion of pyrazinamide in the tuberculosis drug regimen in the 1970s allowed reduction of treatment duration from 12 to 6 months. Pyrazinamide has this remarkable effect in patients despite displaying poor potency against Mycobacterium tuberculosis in vitro. The pharmacological basis for the drug’s in vivo sterilizing activity has remained obscure and its bacterial target controversial. Recently it was shown that pyrazinamide penetrates necrotic caseous TB lung lesions and kills non-growing, drug tolerant bacilli. Furthermore, it was uncovered that the drug inhibits bacterial Coenzyme A biosynthesis. Pyrazinamide may block this pathway by triggering degradation of its target aspartate decarboxylase. The elucidation of the drug’s pharmacological and molecular mechanisms provides the basis for the rational discovery of the next generation pyrazinamide with improved in vitro potency while maintaining attractive pharmacological properties.
Keywords: tuberculosis, pyrazinamide, lesion penetration, drug tolerance, target degradation
The Pyrazinamide enigma and the importance of decoding it
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is now the largest infectious disease killer worldwide and a major cause of death for people living with Acquired Immunodeficiency Syndrome. In immune-competent populations, drug therapy and immunity join forces to win the lengthy war against Mtb. In HIV-positive individuals on the other hand, chemotherapy must fully sterilize all infection sites. With the rampant spread of drug resistance, and in order to shorten the 6 to 24 months long treatment regimens for drug-susceptible and multi-drug-resistant TB, there is an urgent medical need to develop more potent sterilizing drugs.
In 1952, the anti-TB activity of pyrazinamide (PZA) was discovered in animal models [1],i and was shortly thereafter confirmed in humansii. The drug displayed remarkable sterilizing activity in mouse tissues [2] and its inclusion in the regimen of drug-susceptible patients in the 1970s resulted in significant shortening of therapy duration from 12 to 6 months. The present-day ‘short-course’ regimen was finalized in the 1980s, comprising of an initial 2-months intensive phase of isoniazid, rifampicin, PZA, and ethambutol, followed by a 4-month treatment continuation phase of only isoniazid and rifampicin [3],iii. Owing to its relapse-preventing properties, PZA is also included in most new drug combinations that are in clinical and preclinical development for both drug sensitive and multidrug resistant TB [4, 5].
The basis of PZA’s remarkable sterilizing activity in patients remains puzzling since this fragment-size drug (MW = 123 g/mol) exhibits poor in vitro potency (MIC = 30 to 100 μg/mL), concentrations that are barely exceeded in plasma and lesions [6, 7]. PZA being a structural analog of niacin, a vitamin with anti-inflammatory properties [8], it was suggested that it may exert host-directed effects [9]. This was explored by Almeida et al., who hypothesized that if PZA has host-directed activity, this should be detectable in mice infected with PZA resistant Mtb. However, no such activity could be detected, pointing to a lack of significant host-directed activity [10].
In addition to the mysterious in vivo – in vitro disconnect of PZA activity, the antibacterial mechanism of action of the drug remained elusive for decades, despite intense research. PZA is a prodrug which is converted to its bioactive form pyrazinoic acid (POA) by the bacterial pyrazinamidase PncA as well as by host enzymes [11–15]. Several mechanisms of action of POA were proposed, only to be later questioned or disproven (see below). Understanding the drug’s pharmacological and molecular mechanisms is critical to rationally discover the next generation PZA with improved sterilizing activity. This opinion discusses major recent advances in this field. Pharmacological explanations for the unique sterilizing property of PZA have been proposed based on lesion-centric analyses in animal models, and a target has been identified based on genetic and biochemical evidence. At the same time, the modest on-target potency of PZA constitutes a clear limitation and points to an obvious area for improvement. Together, these findings provide the basis for a rational mechanism-based chemical optimization to discover more efficacious next generation PZA analogs.
The pharmacological mechanisms underlying PZA’s sterilizing activity
A spectrum of lesion types exists in human pulmonary TB: cellular lesions comprising primarily immune cells, caseous (cheese-like) granulomas containing a necrotic core of bacterial and host cell debris, and cavities where lesions erode into major airways. In these lesions, Mtb can reside either intracellularly within various immune cells or extracellularly. Within these different compartments, the bacilli can either be growing or quiescent and non-growing. Caseous lesions are believed to harbor mostly extracellular non-growing bacteria. The response of these physiologically different bacterial populations to antibiotic treatment is proposed to be heterogeneous, with non-growing bacilli assumed to display phenotypic drug tolerance, i.e. they are not as readily killed by antibiotics [16]. We hypothesized that the two key elements in PZA’s clinical efficacy may be that the drug i) reaches the bacilli residing in all these different lesions and ii) is able to kill non-growing drug tolerant forms of the pathogen.
PZA penetrates well in all human lung lesion types, including difficult-to-penetrate caseous lesions.
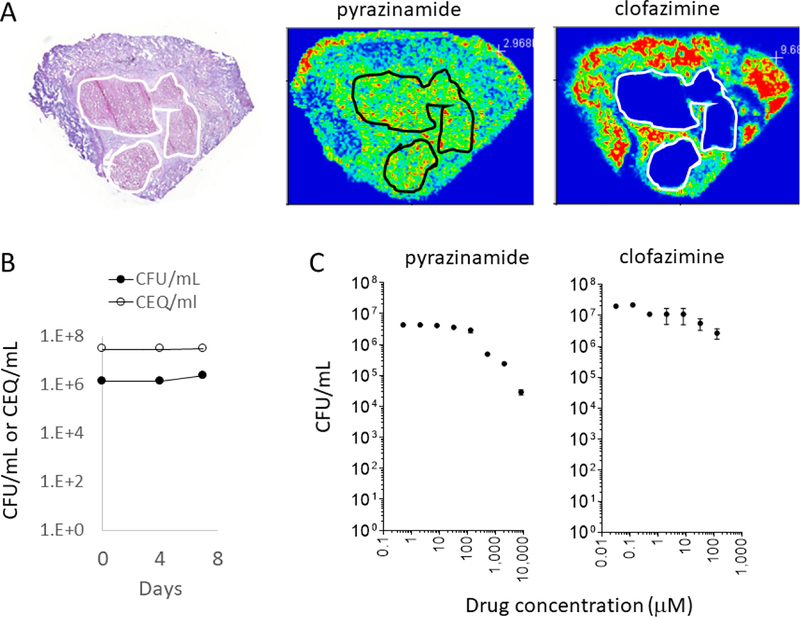
Using MALDI mass spectrometry-based imaging of drug distribution in TB infected human lung tissue, we found that distribution of drugs into the various lesion types is drug and lesion type dependent. Most drugs do not penetrate all compartments in all lesions. In contrast, PZA distributes rapidly and homogenously into all lesions types, both cellular and caseous lesions, where larger and more hydrophobic drugs diffuse less effectively [17] (Fig. 1A). Thus, PZA can reach pathogens residing within the various sites of infection within the lungs of TB patients.
Figure 1.
Pharmacological basis of PZA’s sterilizing activity. (A) MALDI mass spectrometry ion maps of PZA and clofazimine, showing that PZA effectively penetrates the cellular and necrotic compartments of TB lesions, at concentrations identical to those achieved in plasma, in contrast to clofazimine which diffuses poorly into non-vascularized caseum. The left panel shows hematoxylin & eosin staining of the adjacent tissue section revealing the underlying lesion architecture and histology. The necrotic or caseous core of the lesion is highlighted in white or black contour lines. (B) Mtb bacilli do not replicate in ex vivo caseum for the 7-day duration of the cidal assay. Bacterial burden is shown as black circles, and cumulative burden measured as chromosome equivalent is shown as empty circles, indicating that the observed no-net growth is true no-growth rather than the result of balanced death and growth. (C) PZA kills non-growing drug tolerant persister Mtb present in necrotic caseous lesion material ex vivo. Clofazimine is shown as an example of a drug that is not active against caseum Mtb. Experimental details are described in.
PZA kills non-growing drug tolerant persister Mtb in ex vivo caseum.
To determine whether Mtb in caseum is indeed non-growing and drug tolerant, we developed an ex vivo assay with caseum isolated from Mtb-infected rabbits. Indeed, under these conditions, Mtb does not grow (Fig. 1B) and displays extreme drug tolerance to most drugs [18]. PZA was found to display bactericidal activity in caseum, albeit at rather high concentrations, suggesting that it not only reaches drug tolerant bacilli but also kills them [18] (Fig. 1C).
PZA reduces bacterial burden in all lesion types, including caseous lesions, in an animal model presenting the spectrum of human lesion types.
Despite these attractive pharmacokinetic and pharmacodynamic properties, efficacy studies in different mouse and guinea pig models have yielded contradicting results [19–21]. To resolve these conflicting data, we used the rabbit model of active TB, a disease model which recapitulates the major lung lesions observed in human TB [22]. With this model we confirmed that PZA not only penetrates all lesion types, including caseous necrotic compartments, but also reduces bacterial burden and sterilizes both cellular and necrotic lesions [7]. In-depth analyses of bacterial burden and cumulative burden at the lesion level revealed that the onset of bacterial killing by PZA is slow, suggesting that the rather weak in vitro and ex vivo activities of PZA indeed limit its efficacy in vivo at clinically approved dosage [7].
Human pharmacokinetic – pharmacodynamic modeling confirms suboptimal target attainment in lesions.
Pharmacokinetic – pharmacodynamic modeling of PZA penetration in human lesions [17] was carried out to identify lesion types and lesion compartments where PZA achieves target concentrations and for what fraction of the dosing interval. The model revealed that PZA performed sub optimally in all lesion types due to its high MIC, with only a small fraction of the dosing interval above adequate concentrations, and a significant proportion of ‘at–risk’ patients (those achieving relevant concentrations for less than 4h) [23]. Given the common side effects of PZA due to hepatotoxicity, higher dosage is not a viable option.
Taken together, the recent pharmacological findings provide an explanation for the clinically observed sterilizing and treatment shortening effects of PZA: the drug penetrates all TB lung lesion compartments, including difficult-to-penetrate lesions, and kills non-growing persisters that reside in caseum. However, consistent with its poor in vitro potency, concentrations required to kill Mtb in caseum are high and achieved for a small fraction of the dosing interval, and onset of lesion sterilization in vivo is slow. The finding that efficacy is potency-limited, suggests a way forward for the discovery of next generation PZA: improving potency, while maintaining existing lesion penetration properties. Structure-driven optimization of PZA requires elucidation of its molecular mechanism.
Molecular mechanisms underlying PZA’s sterilizing activity
Following early studies where PZA demonstrated improved potency in media adjusted to acidic pHiv, it became common belief that it is active only at acidic pH and that TB lung lesions are therefore acidicv. In this context, and since POA is a weak acid (pKa = 2.9), an ionophore model was proposed and became the most widely accepted hypothesis of PZA’s mechanism of action. POA, a carboxylic acid, shuttles protons from the extracellular acidic environment into the bacilli, causing collapse of membrane energetics and cytoplasmic acidification [24]. However, this model has been opposed by three lines of evidence. PZA and POA can inhibit growth of Mtb at neutral pH [13, 14, 25, 26], implying that acidic pH is not essential for antibacterial activity of the drug. TB lesions are not necessarily acidic, rather they present with a spectrum of pH from acidic to alkaline [7, 18, 19, 21, 27],vi. Importantly, unlike bona fide ionophores such as carbonyl cyanide m-chlorophenyl hydrazine, POA does not cause rapid collapse of membrane potential or cytoplasmic acidification in Mtb [25]. Thus, POA appears not to function as an ionophore in Mtb.
Based primarily on biochemical evidence, several discrete proteins were proposed to be targeted by POA (Table 1). POA was suggested to inhibit fatty acid synthesis via FAS-I [28] and trans-translation, a process that frees ribosomes stalled during translation, by binding to ribosomal protein S1 / RpsA [29, 30]. However, these suggested mechanisms have been put into question or directly contradicted (Table 1, [31, 32]). Thus, FAS-I and RpsA appear not to present significant targets for POA. Recently, GpsI, a probable guanosine pentaphosphate synthase, was shown to bind POA at high (mM) concentrations. A polymorphism in the corresponding gene was identified in two clinical isolates [33]. Whether GpsI is indeed involved in POA’s mechanism of action remains to be determined. A summary of the currently proposed mechanisms of action of PZA/POA with key evidence arguing against these mechanisms is presented in Table 1. We also refer to recent review by Anthony and colleagues in which the authors discuss evidence for and against the various proposed mechanisms [34].
Table 1.
Proposed mechanisms of action of POA against M. tuberculosis
| Model | Proposed mechanism | References supporting model | Key evidence opposing model | References opposing model |
|---|---|---|---|---|
| 1. POA acts as an ionophore | POA acts as a protonophore and shuttles protons from an acidic environment to the cytoplasm resulting in cytoplasm acidification and collapse of membrane potential | [24],vii |
|
[13, 14, 25, 26, 36] |
| 2. POA targets fatty acid synthase FAS-I | POA inhibits synthesis of fatty acids via inhibition of fatty acid synthase I | [28] |
|
[31, 36, 38, 44] |
| 3. POA targets ribosomal protein S1 / RpsA | POA inhibits trans-translation via binding to the ribosomal protein S1 / RpsA | [29, 30] |
|
[32, 36, 38, 44] |
| 4. POA targets guanosine pentaphosphate synthase GpsI | POA inhibits GpsI involved in nucleic acid and ppGpp metabolism | [33] |
|
[36, 38] |
| 5. POA targets Quinolinic acid phosphoribosyl transferase (QAPRTase) | POA, a structural analog of Quinolinic acid, inhibits the catalytic activity of QAPRTase and thus, de novo NAD biosynthesis | [54] |
|
[36, 38, 44] |
| 6. POA targets aspartate decarboxylase PanD | Inhibition of bacterial Coenzyme A biosynthesis by POA- induced degradation of PanD via ClpC1-ClpP. | [36–39, 44, 45, 47, 48] |
|
[45] |
A validated approach to identify the mechanism of action of an antibacterial is to isolate spontaneous resistance mutants on solid medium. However, various attempts to isolate POA resistant Mtb mutants failed [28, 35]. Based on the prevailing ‘PZA requires acidic pH’ concept discussed above, acidic pH agar was used in these experiments. With the re-discoveries that PZA/POA is also active at neutral pH and that TB lung lesions are not necessarily acidic, we hypothesized that the failure to isolate POA-resistant mutants was due to the acidic pH used in the selection media and that it may be possible to isolate POA resistant mutants on neutral pH agar.
Missense mutations in the Coenzyme A biosynthetic pathway gene panD encoding aspartate decarboxylase cause PZA resistance in vitro and in vivo.
Indeed, PZA/POA-resistant mycobacteria could be recovered on neutral pH agar [14, 34, 36, 37]. Whole genome sequencing revealed missense mutations in the aspartate decarboxylase PanD involved in the biosynthesis of the essential cofactor Coenzyme A [36, 37]. The resistant mutant selection experiment was repeated in Mtb-infected mice treated with POA, which again delivered PZA/POA resistance mutations in panD, providing in vivo evidence for the relevance of the observed panD related resistance mechanism [38]. Interestingly, the frequency of panD polymorphisms in clinical isolates is very low ([39–41], see Table 2), the reason of which remains to be determined.
Table 2.
Reported PZA/POA resistance mutations and proposed resistance mechanisms in M. tuberculosis
| Gene (Rv number) | Gene function | Nature of resistance mutationsa | Proposed mechanism(s) of resistance | Polymorphisms associated with PZA resistance in clinical isolates |
|---|---|---|---|---|
| pncA (Rv2043c) | Pyrazinamidase PncA | Prevents activation of prodrug PZA to bioactive component POA [11] | Yes [55, 56] | |
| panD (Rv3601c) | Aspartate decarboxylase PanD, involved in Coenzyme A biosynthesis | Prevents POA binding to its target PanD [44] | Yes [39], No [40, 41] | |
| clpC1 (Rv3596c) | Substrate recognition and unfoldase component ClpC1 of caseinolytic protease complex ClpP |
|
Prevents POA-induced degradation of PanD [46] | ndb |
| rpsA (Rv1630)c | 30S Ribosomal Protein S1 / RpsA | Missense mutation ΔA438 in 1 PZA-resistant clinical isolate [29, 30]c,d | Prevents POA binding to RpsA [29, 30]c | No [41, 57–59] |
| gpsI (Rv2783c) | Bifunctional protein GpsI | Missense mutation Asp67Asn in 2 PZA-resistant clinical isolates [33]d | Prevents POA binding to GpsI [33] | nd |
| mas/ppsA-E (Rv2940c/Rv2931-Rv2935) | Mycocerosic acid synthase Mas/ phenolpthiocerol synthesis type-I polyketide synthases PpsA-E involved in phthiocerol dimycocerosate (PDIM) synthesis | Frameshift loss-of-function mutations [36] | Affects cell envelope permeability, induces drug- tolerant phenotype, reduces Coenzyme A utilization [34, 36, 60] | ndb |
| tap (Rv1258c) | Integral membrane transport protein Tap | Missense mutations identified in clinical isolates [61, 62]d | Prevents intracellular drug accumulation [63, 64] | nd |
| IprG (Rv1411c) | Lipoprotein LprG | Nonsense mutation Trp224Stop [64] | Increases TAG accumulation, leading to drug tolerance [65] | nd |
| fadD2 (Rv0270)e | Fatty acid CoA ligase FadD2 | Loss-of-function mutations Note: mutations cause hyper-susceptibility to PZA/POA [53] | Wild type FadD2 detoxifies cytotoxic fatty acids accumulated within Mtb cells due to disruption of Coenzyme A biosynthesis by POA [53] | na |
nd, not reported; na, not applicable.
Nature of PZA/POA resistance mutations isolated in vitro are shown, unless indicated otherwise. Note: For panD and clpC1 resistance mutations were also isolated in vivo from Mtb infected mice and POA-treated mice [38].
PZA/POA resistance mutations cause reduced fitness in vivo [38].
No in vitro or in vivo isolated spontaneous PZA-resistant mutants contain mutations in RpsA/GpsI/Tap [36, 38, 47].
FadD2 is responsible for intrinsic PZA resistance. Mutations in fadD2 cause hyper-susceptibility, not resistance, to POA/PZA [53].
POA blocks bacterial Coenzyme A biosynthesis by binding to aspartate decarboxylase PanD.
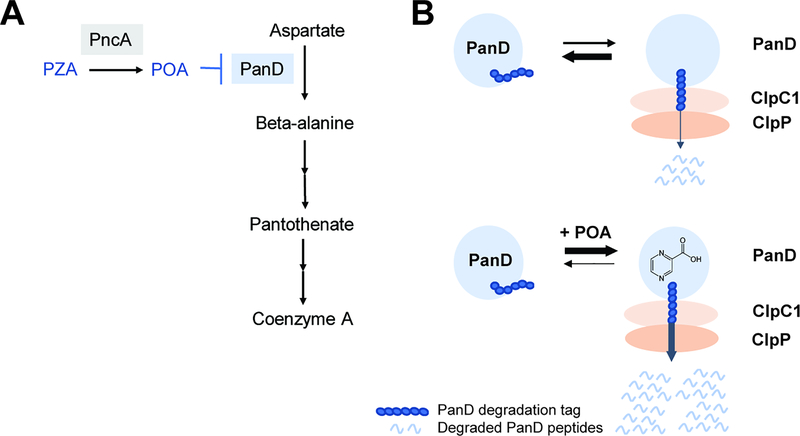
Coenzyme A biosynthesis is essential in Mtb in vitro and in vivo [42, 43]. Thus, based on our genetic data, we hypothesized that POA may bind to PanD and inhibit the corresponding catalytic step in the Coenzyme A pathway (Fig. 2A, Key Figure). Resistance caused by missense mutations in PanD would then be due to lack of drug binding. Indeed, biophysical analyses showed that POA binds recombinant wild type PanD but not proteins containing POA resistance mutations [44]. Metabolomic analyses showed that POA treatment resulted in a collapse in concentrations of Coenzyme A biosynthetic metabolites downstream of the PanD catalyzed step, starting with beta-alanine, the product of PanD, and all the way to Coenzyme A, the end product of the pathway [36, 44]. Finally, exogenous supplementation of media with pantothenate, a Coenzyme A precursor downstream of PanD, phenocopied panD resistance [36, 37, 45]. Taken together, these results show that POA inhibits bacterial Coenzyme A biosynthesis by binding to PanD (Fig. 2A). Consistent with poor whole cell activities of PZA/POA, affinity of POA for PanD was in the μM range, suggesting room for improvement [44].
Figure 2, Key Figure.
Proposed antibacterial mechanism of action of PZA. (A) PZA, once converted into POA by the bacterial amidase PncA, blocks synthesis of the essential cofactor Coenzyme A at the aspartate decarboxylase PanD-catalyzed step. (B) PanD level is post-translationally regulated by the caseinolytic protease complex ClpC1-ClpP and binding of POA to PanD triggers increased degradation of the protein. Upper part: PanD (blue circle) contains a C-terminal protease degradation tag. The tag is recognized by ClpC1 (light orange) which unfolds PanD for degradation by the ClpP protease (dark orange). Lower part: Binding of POA to PanD causes conformational changes resulting in increased exposure of the degradation tag and hence increased degradation of PanD by ClpC1-ClpP.
POA triggers degradation of its target aspartate decarboxylase PanD.
When we set out to characterize the biochemical inhibition of the PanD-catalyzed reaction (the conversion of aspartate to beta-alanine) by POA, we were surprised to find that POA is not a bona fide inhibitor of PanD’s enzymatic activity [46]. If POA does not inhibit the activity of the aspartate decarboxylase, how does binding of POA to the enzyme affect this enzymatic step in coenzyme A synthesis inside the bacterium? We had previously isolated, both in vitro and in vivo, POA resistant mutants in ClpC1, a component of Mtb’s caseinolytic protease complex [38, 47, 48]. ClpC1 is an unfoldase that recognizes proteins containing various C-terminal tags and delivers these proteins for degradation to the ClpP protease [49, 50]. The ClpC1-ClpP complex is involved in proteome house-keeping and regulation of the protein level of certain proteins [50]. Protein overexpression experiments suggested that ClpC1 mutations confer resistance to POA by an indirect mechanism [48]. Interestingly, the resistance level conferred by ClpC1 mutations was the same as the resistance level conferred by mutations in PanD. Could there be a link between aspartate decarboxylase and ClpC1? Analyses employing Mtb reporter strains revealed that PanD contains a C-terminal degradation tag and is a substrate of the ClpC1-ClpP protease, which regulates PanD levels post-translationally [46] (Fig. 2B). POA treatment studies with Mtb reporter strains further showed that POA binding to PanD accelerates degradation of PanD, possibly via the activity of ClpC1-ClpP [6]. Thus, rather than inhibiting the biochemical activity of its target (the usual on-target mechanism of drugs), POA triggers degradation of PanD (Fig. 2B).
Taken together, mechanism of action studies support that POA may act as a bacterial target ‘degrader’, a new event-driven pharmacology paradigm in the field of anti-infectives. POA may promote a derailing sequence of events that result in uncontrolled degradation of an essential enzyme, PanD, by Mtb’s ClpC1-ClpP protein degradation machinery. It is interesting to mention that targeted protein degradation has gained momentum in recent years as a conceptually novel drug discovery approach. PROTACs, heterobifunctional molecules, which contain binding moieties for the protein of interest and for E3 ligase, make use of the human proteasome system to specifically degrade tagged proteins [51]. Our mechanistic findings with a clinically used drug validate drug-induced target degradation as a new approach in drug discovery.
Concluding remarks
Thanks to recent discoveries, PZA-associated enigmas are becoming less enigmatic. POA, the bioactive component of PZA, binds to aspartate decarboxylase PanD and triggers its degradation by the bacterium, thus blocking biosynthesis of the essential Coenzyme A. These data demonstrate druggability of this cofactor pathway via PanD and its vulnerability in vitro and in vivo [42, 43]. It is interesting to note that the intrabacterial PanD protein level is extremely low, which may contribute to making this enzymatic step exquisitely vulnerable for chemotherapeutic intervention [46]. Furthermore, PanD presents an attractive target for antibacterial discovery as it catalyzes the first committed step in the pathway and has been shown to be tightly regulated in E. coli by a negative feedback mechanism [52].
POA displays weak (μM) binding affinity to PanD, providing an explanation for the poor MIC [44, 46]. This is compensated, but only to some extent, by excellent lesion penetration properties, and cidal activity against non-growing drug tolerant persister Mtb residing in caseous lesions, consistent with the slow onset of its sterilizing activity [7].
Despite the advances in understanding the pharmacological and molecular mechanisms behind the sterilizing activity of pyrazinamide a range of questions remain (see “Outstanding Questions”).
Outstanding questions.
Will it be possible to develop high affinity PanD degraders while maintaining the physico-chemical properties critical for effective lesion penetration?
How does POA exactly bind PanD and what are the conformational changes induced by this binding?
Are there targets of POA other than PanD?
Does Mtb produce additional bacterio-toxic PZA or POA metabolites?
Why is the frequency of PZA resistance due to panD mutations low in clinical isolates?
Can methods be developed to target specific bacterial proteins of interest for degradation, thus applying target degradation as a novel approach to anti-bacterial drug discovery?
Collectively, our findings, together with the corresponding pharmacological, microbiological and biophysical tools, provide the basis for the rational discovery of more potent PZA analogs. Will it be possible to develop high affinity PanD degraders while maintaining the physico-chemical properties critical for effective lesion penetration? Lead optimization programs will answer this question. If PZA’s excellent lesion penetration properties can be maintained during optimization, this could deliver a next-generation PZA with improved efficacy.
To enable structure-based lead optimization the exact interactions of POA with PanD protein need to be elucidated. POA binds to PanD and appears to trigger its degradation by the caseinolytic protease ClpC1-ClpP, presumably by exposing PanD’s C-terminal protease recognition sequence. How does POA bind its target and what are the conformational changes induced by this binding? Biophysical analyses will answer these questions.
Although strong evidence suggests that PanD is a major target via which POA exerts its anti-mycobacterial activity, it cannot be excluded that POA has additional molecular mechanisms of action [45]. Furthermore, it is possible that POA is metabolized by Mtb to generate additional bacterio-toxic derivatives [53]. Considering that POA is a small, fragment-sized drug, and thus likely a promiscuous binder and substrate [6], it is conceivable and even likely that POA applies polypharmacology and modulates several Mtb targets. There are several hints that polypharmacology properties of PZA/POA may indeed contribute to the drug’s whole cell activity. For instance, low level loss-of-function POA-resistant mutants were identified in the biosynthetic pathway that produces phthiocerol dimycocerosates located in the cell wall (see Table 2 for a summary of currently reported Mtb mutations proposed to be associated with PZA/POA resistance). Whether POA targets this and other processes directly or indirectly remains to be determined. It was also shown that PZA exerts antibacterial activity against a panD loss-of-function auxotrophic mutant Mtb that was supplemented with pantetheine [45]. Further metabolomic, genetic and biochemical work may reveal additional, PanD-independent mechanisms of action of POA. How much - if anything - possible additional mechanisms may contribute to the whole cell anti-Mtb activity of the drug is an open question that can be answered by the generation and profiling of a - yet to be discovered – high affinity PanD degrader.
Perhaps the biggest remaining puzzle around the mechanism of action / resistance of PZA is that resistant mutations in panD can be readily isolated in vitro and in vivo from Mtb-infected mice. However, panD polymorphisms associated with PZA resistance are rare in clinical isolates of Mtb. Why is the frequency of panD mutations low in isolates derived from human sputum? Infection studies with mice showed that PZA resistant PanD mutations do not affect fitness of Mtb in mouse lungs [38]. However, the pathology of mouse TB differs substantially from human TB. Do POA resistant PanD mutants display fitness costs in specific human lesions not present in mouse TB, such as cavities from which sputum Mtb is derived? Studies of POA resistant PanD mutant Mtb in animal models that recapitulate human lesion types will answer these questions.
PZA is the first anti-bacterial to act as a selective target degrader, a novel drug discovery paradigm currently exploited in lead finding programs against human targets. The first human target degraders were - similar to PZA - discovered unintentionally: it turned out that some drugs ‘accidentally’ promote degradation of their target by the human cellular proteolytic machinery upon binding to their target. Methods were developed to target specific human proteins of interest for degradation, thus enabling the rational discovery of target degraders. The application of targeted protein degradation has so far been limited to human cells. Increasing anti-bacterial resistance makes the discovery of novel antibiotics more urgent than ever. Can target degradation as a general approach also be developed for the discovery of new antibiotics?
Highlights.
PZA is able to penetrate all TB lung lesion types, including necrotic caseous granulomas, where it kills non-growing drug tolerant Mtb, explaining the drug’s sterilizing activity in vivo.
POA, the bioactive component of the prodrug PZA, inhibits bacterial synthesis of Coenzyme A by binding to aspartate decarboxylase PanD and blocking this enzymatic step.
Rather than inhibiting PanD’s catalytic activity, we propose that binding of POA triggers the degradation of its target by the bacterial caseinolytic protease.
The hypothesis that PZA’s bioactive component acts as a target degrader supports selective target degradation as a novel antibacterial drug discovery approach.
Identification of the pharmacological and molecular mechanisms of PZA’s activity provide the basis for the rational discovery of the next generation PZA with improved in vitro potency while maintaining the drug’s attractive lesion penetration properties.
Acknowledgements
We would like to thank Courtney Aldrich, University of Minnesota, for discussions. Research reported in this publication is supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number 2R01AI106398-05. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research was also supported by the Singapore Ministry of Health’s National Medical Research Council under NMRC/TCR/011-NUHS/2014 and NMRC/CG/013/2013 and is part of the SPRINT-TB program led by Nick Paton. TD holds a Toh Chin Chye Visiting Professorship at the Department of Microbiology and Immunology, National University of Singapore.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Resources
- i).Malone L et al. (1952) The effect of pyrazinamide (aldinamide) on experimental tuberculosis in mice. American review of tuberculosis 65 (5), 511–518. http://europepmc.org/abstract/MED/14924173 [PubMed] [Google Scholar]
- ii).Yeager RL et al. (1952) Pyrazinamide (aldinamide*) in the treatment of pulmonary tuberculosis. Transactions of the annual meeting. National Tuberculosis Association 48, 178–201. http://europepmc.org/abstract/MED/13038888 [PubMed] [Google Scholar]
- iii).Fox W et al. (1999) Studies on the treatment of tuberculosis undertaken by the British Medical Research Council Tuberculosis Units, 1946–1986, with relevant subsequent publications. The International Journal of Tuberculosis and Lung Disease 3 (2), S231–79. https://www.ingentaconnect.com/contentone/iuatld/ijtld/1999/00000003/A00210s2/art00001 [PubMed] [Google Scholar]
- iv).McDermott W and Tompsett R (1954) Activation of pyrazinamide and nicotinamide in acidic environments in vitro. American review of tuberculosis 70 (4), 748–54. https://www.atsjournals.org/doi/abs/10.1164/art.1954.70.4.748 [DOI] [PubMed] [Google Scholar]
- v).Zhang Y and Mitchison D (2003) The curious characteristics of pyrazinamide: a review. The International Journal of Tuberculosis and Lung Disease 7 (1), 6–21. https://www.ingentaconnect.com/content/iuatld/ijtld/2003/00000007/00000001/art00004 [PubMed] [Google Scholar]
- vi).Koller F and Leuthardt F (1934) Nekrose und autolyse beitrag zur kenntnis der dystrophischen verkalkung. Klinische Wochenschrift 13, 1527–1529. https://link.springer.com/article/10.1007%2FBF01779121 [Google Scholar]
- vii).Zhang Y et al. (1999) Role of Acid pH and Deficient Efflux of Pyrazinoic Acid in Unique Susceptibility of Mycobacterium tuberculosis to Pyrazinamide. Journal of Bacteriology 181 (7), 2044–2049. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC93615/ [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Solotorovsky M et al. (1952) Pyrazinoic acid amide; an agent active against experimental murine tuberculosis. Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine (New York, N.Y.) 79 (4), 563–565. doi: 10.3181/00379727-79-19447 [DOI] [PubMed] [Google Scholar]
- 2.McCune RM et al. (1956) THE FATE OF MYCOBACTERIUM TUBERCULOSIS IN MOUSE TISSUES AS DETERMINED BY THE MICROBIAL ENUMERATION TECHNIQUE : II. THE CONVERSION OF TUBERCULOUS INFECTION TO THE LATENT STATE BY THE ADMINISTRATION OF PYRAZINAMIDE AND A COMPANION DRUG. The Journal of Experimental Medicine 104 (5), 763–802. doi: 10.1084/jem.104.5.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchison DA (1985) The action of antituberculosis drugs in short-course chemotherapy. Tubercle 66 (3), 219–225. doi: 10.1016/0041-3879(85)90040-6 [DOI] [PubMed] [Google Scholar]
- 4.Diacon AH et al. (2012) 14-day bactericidal activity of PA-824, bedaquiline, pyrazinamide, and moxifloxacin combinations: a randomised trial. The Lancet 380 (9846), 986–993. doi: 10.1016/S0140-6736(12)61080-0 [DOI] [PubMed] [Google Scholar]
- 5.Tasneen R et al. (2011) Sterilizing Activity of Novel TMC207- and PA-824-Containing Regimens in a Murine Model of Tuberculosis. Antimicrobial Agents and Chemotherapy 55 (12), 5485–5492. doi: 10.1128/aac.05293-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopal P and Dick T (2014) Reactive dirty fragments: implications for tuberculosis drug discovery. Current Opinion in Microbiology 21, 7–12. doi: 10.1016/j.mib.2014.06.015 [DOI] [PubMed] [Google Scholar]
- 7.Blanc L et al. (2018) Impact of immunopathology on the antituberculous activity of pyrazinamide. The Journal of Experimental Medicine 215 (9). doi: 10.1084/jem.20180518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walocko FM et al. (2017) The role of nicotinamide in acne treatment. Dermatologic Therapy 30 (5), e12481. doi: 10.1111/dth.12481 [DOI] [PubMed] [Google Scholar]
- 9.Mendez S et al. (2009) The Antituberculosis Drug Pyrazinamide Affects the Course of Cutaneous Leishmaniasis In Vivo and Increases Activation of Macrophages and Dendritic Cells. Antimicrobial Agents and Chemotherapy 53 (12), 5114–5121. doi: 10.1128/AAC.01146-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almeida DV et al. (2014) Revisiting Anti-tuberculosis Activity of Pyrazinamide in Mice. Mycobacterial diseases : tuberculosis & leprosy 4, 145. doi: 10.4172/2161-1068.1000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scorpio A and Zhang Y (1996) Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nature Medicine 2, 662. doi: 10.1038/nm0696-662 [DOI] [PubMed] [Google Scholar]
- 12.Yadon AN et al. (2017) A comprehensive characterization of PncA polymorphisms that confer resistance to pyrazinamide. Nature Communications 8 (1), 588. doi: 10.1038/s41467-017-00721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Via LE et al. (2015) Host-Mediated Bioactivation of Pyrazinamide: Implications for Efficacy, Resistance, and Therapeutic Alternatives. ACS Infectious Diseases 1 (5), 203–214. doi: 10.1021/id500028m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lanoix J-P et al. (2016) High Systemic Exposure of Pyrazinoic Acid Has Limited Antituberculosis Activity in Murine and Rabbit Models of Tuberculosis. Antimicrobial Agents and Chemotherapy 60 (7), 4197–4205. doi: 10.1128/aac.03085-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naftalin CM et al. (2017) Co-administration of allopurinol to increase anti-mycobacterial efficacy of pyrazinamide: evaluation in a whole-blood bactericidal activity model. Antimicrobial Agents and Chemotherapy. doi: 10.1128/AAC.00482-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dartois V (2014) The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells. Nature Reviews Microbiology 12, 159. doi: 10.1038/nrmicro3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prideaux B et al. (2015) The association between sterilizing activity and drug distribution into tuberculosis lesions. Nature medicine 21 (10), 1223–1227. doi: 10.1038/nm.3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarathy JP et al. (2018) Extreme Drug Tolerance of Mycobacterium tuberculosis in Caseum. Antimicrobial Agents and Chemotherapy 62 (2). doi: 10.1128/aac.02266-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanoix J-P et al. (2016) Selective Inactivity of Pyrazinamide against Tuberculosis in C3HeB/FeJ Mice Is Best Explained by Neutral pH of Caseum. Antimicrobial Agents and Chemotherapy 60 (2), 735–743. doi: 10.1128/aac.01370-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad Z et al. (2011) Dose-Dependent Activity of Pyrazinamide in Animal Models of Intracellular and Extracellular Tuberculosis Infections. Antimicrobial Agents and Chemotherapy 55 (4), 1527–1532. doi: 10.1128/aac.01524-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irwin SM et al. (2016) Bedaquiline and Pyrazinamide Treatment Responses Are Affected by Pulmonary Lesion Heterogeneity in Mycobacterium tuberculosis Infected C3HeB/FeJ Mice. ACS Infectious Diseases 2 (4), 251–267. doi: 10.1021/acsinfecdis.5b00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subbian S et al. (2011) Chronic pulmonary cavitary tuberculosis in rabbits: a failed host immune response. Open Biology 1 (4). doi: 10.1098/rsob.110016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strydom N et al. (2019) Tuberculosis drugs’ distribution and emergence of resistance in patient’s lung lesions: A mechanistic model and tool for regimen and dose optimization. PLOS Medicine 16 (4), e1002773. doi: 10.1371/journal.pmed.1002773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y et al. (2003) Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. Journal of Antimicrobial Chemotherapy 52 (5), 790–795. doi: 10.1093/jac/dkg446 [DOI] [PubMed] [Google Scholar]
- 25.Peterson ND et al. (2015) Uncoupling Environmental pH and Intrabacterial Acidification from Pyrazinamide Susceptibility in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy 59 (12), 7320–7326. doi: 10.1128/aac.00967-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Hertog AL et al. (2016) PZA is active against Mycobacterium tuberculosis cultures at neutral pH with reduced temperature. Antimicrobial Agents and Chemotherapy. doi: 10.1128/aac.00654-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kempker RR et al. (2017) Lung Tissue Concentrations of Pyrazinamide among Patients with Drug-Resistant Pulmonary Tuberculosis. Antimicrobial Agents and Chemotherapy. doi: 10.1128/aac.00226-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimhony O et al. (2000) Pyrazinamide inhibits the eukaryotic-like fatty acid synthetase I (FASI) of Mycobacterium tuberculosis. Nature Medicine 6, 1043. doi: 10.1038/79558 [DOI] [PubMed] [Google Scholar]
- 29.Shi W et al. (2011) Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis: a potential mechanism for shortening the duration of tuberculosis chemotherapy. Science (New York, N.Y.) 333 (6049), 1630–1632. doi: 10.1126/science.1208813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi W et al. (2019) Introducing RpsA Point Mutations Δ438A and D123A into the Chromosome of Mycobacterium tuberculosis Confirms Their Role in Causing Resistance to Pyrazinamide. Antimicrobial Agents and Chemotherapy 63 (6), e02681–18. doi: 10.1128/aac.02681-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boshoff HI et al. (2002) Effects of Pyrazinamide on Fatty Acid Synthesis by Whole Mycobacterial Cells and Purified Fatty Acid Synthase I. Journal of Bacteriology 184 (8), 2167–2172. doi: 10.1128/JB.184.8.2167-2172.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dillon NA et al. (2017) Anti-tubercular Activity of Pyrazinamide is Independent of trans-Translation and RpsA. Scientific Reports 7 (1), 6135. doi: 10.1038/s41598-017-06415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Njire M et al. (2017) Pyrazinoic Acid Inhibits a Bifunctional Enzyme in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy. doi: 10.1128/AAC.00070-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anthony RM et al. (2018) ‘Happy the man, who, studying nature’s laws, Thro’ known effects can trace the secret cause.’† Do we have enough pieces to solve the pyrazinamide puzzle? Journal of Antimicrobial Chemotherapy 73 (7), 1750–1754. doi: 10.1093/jac/dky060 [DOI] [PubMed] [Google Scholar]
- 35.Scorpio A et al. (1997) Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy 41 (3), 540–543. doi: 10.1128/AAC.41.3.540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gopal P et al. (2016) Pyrazinamide Resistance Is Caused by Two Distinct Mechanisms: Prevention of Coenzyme A Depletion and Loss of Virulence Factor Synthesis. ACS Infectious Diseases 2 (9), 616–626. doi: 10.1021/acsinfecdis.6b00070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi W et al. (2014) Aspartate decarboxylase (PanD) as a new target of pyrazinamide in Mycobacterium tuberculosis. Emerging Microbes & Infections 3 (8), e58. doi: 10.1038/emi.2014.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gopal P et al. (2017) In Vivo-Selected Pyrazinoic Acid-Resistant Mycobacterium tuberculosis Strains Harbor Missense Mutations in the Aspartate Decarboxylase PanD and the Unfoldase ClpC1. ACS Infectious Diseases 3 (7), 492–501. doi: 10.1021/acsinfecdis.7b00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werngren J et al. (2017) Non-pncA Gene-Mutated but Pyrazinamide-Resistant Mycobacterium tuberculosis: Why Is That? Journal of Clinical Microbiology 55 (6), 1920–1927. doi: 10.1128/JCM.02532-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maslov DA et al. (2015) Resistance to pyrazinamide in Russian Mycobacterium tuberculosis isolates: pncA sequencing versus Bactec MGIT 960. Tuberculosis 95 (5), 608–612. doi: 10.1016/j.tube.2015.05.013 [DOI] [PubMed] [Google Scholar]
- 41.Coll F et al. (2018) Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nature Genetics 50 (2), 307–316. doi: 10.1038/s41588-017-0029-0 [DOI] [PubMed] [Google Scholar]
- 42.Sambandamurthy V et al. (2002) A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nature Medicine 8, 1171–4. doi: 10.1038/nm765 [DOI] [PubMed] [Google Scholar]
- 43.Evans JC et al. (2016) Validation of CoaBC as a Bactericidal Target in the Coenzyme A Pathway of Mycobacterium tuberculosis. ACS Infectious Diseases 2 (12), 958–968. doi: 10.1021/acsinfecdis.6b00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gopal P et al. (2017) Pyrazinoic Acid Inhibits Mycobacterial Coenzyme A Biosynthesis by Binding to Aspartate Decarboxylase PanD. ACS Infectious Diseases 3 (11), 807–819. doi: 10.1021/acsinfecdis.7b00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dillon NA et al. (2014) Pantothenate and Pantetheine Antagonize the Antitubercular Activity of Pyrazinamide. Antimicrobial Agents and Chemotherapy 58 (12), 7258–7263. doi: 10.1128/AAC.04028-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gopal P et al. (2019) Pyrazinamide triggers degradation of its target aspartate decarboxylase. bioRxiv, 674416. doi: 10.1101/674416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yee M et al. (2017) Missense Mutations in the Unfoldase ClpC1 of the Caseinolytic Protease Complex Are Associated with Pyrazinamide Resistance in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy 61 (2). doi: 10.1128/AAC.02342-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S et al. (2017) Mutation in clpC1 encoding an ATP-dependent ATPase involved in protein degradation is associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerging Microbes &Amp; Infections 6, e8. doi: 10.1038/emi.2017.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada Y and Dick T (2017) Mycobacterial Caseinolytic Protease Gene Regulator ClgR Is a Substrate of Caseinolytic Protease. mSphere 2 (2). doi: 10.1128/mSphere.00338-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raju RM et al. (2012) Bacterial proteolytic complexes as therapeutic targets. Nature Reviews Drug Discovery 11, 777. doi: 10.1038/nrd3846 [DOI] [PubMed] [Google Scholar]
- 51.Salami J and Crews CM (2017) Waste disposal—An attractive strategy for cancer therapy. Science 355 (6330), 1163–1167. doi: 10.1126/science.aam7340 [DOI] [PubMed] [Google Scholar]
- 52.Monteiro Diana C. et al. (2015) The Structure of the PanD/PanZ Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A. Chemistry & Biology 22 (4), 492–503. doi: 10.1016/j.chembiol.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosen BC et al. (2017) Long-Chain Fatty Acyl Coenzyme A Ligase FadD2 Mediates Intrinsic Pyrazinamide Resistance in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy 61 (2), e02130–16. doi: 10.1128/AAC.02130-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim H et al. (2014) Biochemical Characterization of Quinolinic Acid Phosphoribosyltransferase from Mycobacterium tuberculosis H37Rv and Inhibition of Its Activity by Pyrazinamide. PLoS ONE 9 (6), e100062. doi: 10.1371/journal.pone.0100062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitfield MG et al. (2015) A Global Perspective on Pyrazinamide Resistance: Systematic Review and Meta-Analysis. PLOS ONE 10 (7), e0133869. doi: 10.1371/journal.pone.0133869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miotto P et al. (2014) Mycobacterium tuberculosis Pyrazinamide Resistance Determinants: a Multicenter Study. mBio 5 (5). doi: 10.1128/mBio.01819-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexander DC et al. (2012) Gene Sequencing for Routine Verification of Pyrazinamide Resistance in Mycobacterium tuberculosis: a Role for pncA but Not rpsA. Journal of Clinical Microbiology 50 (11), 3726–3728. doi: 10.1128/jcm.00620-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simons SO et al. (2013) Role of rpsA Gene Sequencing in Diagnosis of Pyrazinamide Resistance. Journal of Clinical Microbiology 51 (1), 382–382. doi: 10.1128/JCM.02739-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan Y et al. (2014) Role of pncA and rpsA Gene Sequencing in Detection of Pyrazinamide Resistance in Mycobacterium tuberculosis Isolates from Southern China. Journal of Clinical Microbiology 52 (1), 291–297. doi: 10.1128/JCM.01903-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Torrey HL et al. (2016) High Persister Mutants in Mycobacterium tuberculosis. PLoS ONE 11 (5), e0155127. doi: 10.1371/journal.pone.0155127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J et al. (2018) Mutations in efflux pump Rv1258c (Tap) cause resistance to pyrazinamide and other drugs in M. tuberculosis. bioRxiv. doi: 10.1101/249102 [DOI] [Google Scholar]
- 62.Kanji A et al. (2017) Single nucleotide polymorphisms in efflux pumps genes in extensively drug resistant Mycobacterium tuberculosis isolates from Pakistan. Tuberculosis 107, 20–30. doi: 10.1016/j.tube.2017.07.012 [DOI] [PubMed] [Google Scholar]
- 63.Liu J et al. (2019) Mutations in Efflux Pump Rv1258c (Tap) Cause Resistance to Pyrazinamide, Isoniazid, and Streptomycin in M. tuberculosis. Frontiers in Microbiology 10 (216). doi: 10.3389/fmicb.2019.00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi W et al. (2018) Identification of novel mutations in LprG (rv1411c), rv0521, rv3630, rv0010c, ppsC, cyp128 associated with pyrazinoic acid/pyrazinamide resistance in Mycobacterium tuberculosis. Antimicrobial Agents and Chemotherapy, 00430–18. doi: 10.1128/aac.00430-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinot AJ et al. (2016) Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis. PLOS Pathogens 12 (1), e1005351. doi: 10.1371/journal.ppat.1005351 [DOI] [PMC free article] [PubMed] [Google Scholar]