Abstract
Background
Currently, serodiagnosis of infection with the helminth parasite Onchocerca volvulus is limited to the Ov-16 IgG4 test, a test that has limited sensitivity and suboptimal specificity. In previous studies, we identified several linear epitopes that have the potential to supplement the diagnostic toolbox for onchocerciasis.
Methods
In this study three peptides, bearing in total six linear epitopes were transferred to a multiplex ELISA platform. This multiplex ELISA was used to assess the clinical utility of the peptide serology markers by analyzing sample sets from both O. volvulus endemic and non-endemic regions.
Results
The multiplex platform was shown to be reproducible and data obtained on the multiplex platform were comparable to the singleplex ELISA data. The clinical utility assessment showed that in a population of school-aged children from western Kenya, a virtually O. volvulus-free area, significant cross-reactivity with an as-yet to be determined immunogen was detected.
Conclusions
The observations made in this study invalidate the usefulness of the peptide serology markers for onchocerciasis detection. We discuss what could be the origin of this unexpected serological response, but also highlight the need for better characterized biobanks for biomarker discovery activities.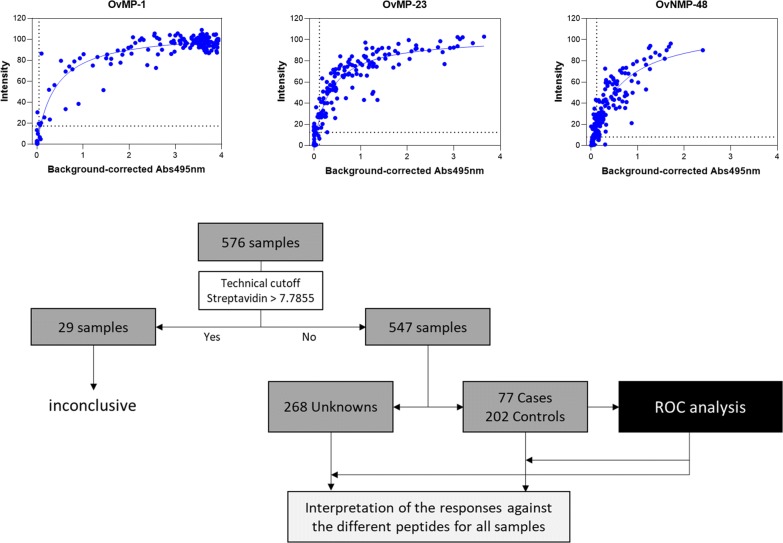
Keywords: Onchocerca volvulus, Peptide serology, Diagnostics, Multiplex ELISA
Background
Helminth infections make up a major part of the diseases listed on the World Health Organization (WHO) list of Neglected Tropical Diseases (NTD) [1–3]. One of these helminths, Onchocerca volvulus causes onchocerciasis (also known as river blindness). At least 120 million people are at risk of infection, the majority of them living in Africa [4, 5]. Efforts to control the spread of the disease are centered around mass drug administration (MDA) of the microfilaricidal agent ivermectin (Mectizan, Merck). Onchocerca infection is traditionally diagnosed by microscopic detection of microfilariae (mf) in skin biopsy samples (skin snips) and by serological assays detecting IgG4 antibodies against the Ov16 antigen [6, 7]. WHO guidelines for stopping MDA and verifying elimination of onchocerciasis require demonstration of < 0.1% seroprevalence of Ov-16 IgG4 in children under the age of 10 years [8]. The specificity of the Ov-16 rapid diagnostic test (RDT) ranges from 97% to 98%, which makes it virtually impossible to achieve the 0.1% threshold, and hence causes failure in recognizing interruption of transmission and stopping MDA [9–11].
One way to overcome the specificity challenge of the Ov16 IgG4 test, would be to introduce a serological confirmation test to confirm or refute a positive test result in a first line screening assay. The combination of peptides OvMP-1, OvMP-23 and OvNMP48 in a peptide-based multiplex ELISA test could possibly provide such second line confirmation capabilities [12–14].
In this study we demonstrate that a multiplex ELISA using these Onchocerca-derived peptides in the InfYnity Biomarkers platform confirms the previously demonstrated sensitivity [15, 16]. The clinical utility of these peptide antigens was not proven, since significant cross-reactivity with an as-yet to be determined immunogen was detected in a population of school-aged children in a virtually O. volvulus-free area of western Kenya. This observation invalidates the usefulness of these peptides as antigens to build a confirmation test for the Ov16 IgG4 RDT.
Methods
Study samples
Plasma samples from O. volvulus infected individuals, healthy controls, W. bancrofti-infected individuals, individuals with B. malayi infection and individuals infected with soil-transmitted helminths (STH) and/or Schistosoma mansoni have been described before [12, 13, 17]. An overview, including Ov16 IgG4 information is provided in Table 1.
Table 1.
Study populations included in this study
| Group | n | Origin | Helminth infection status | Ov16 IgG4-positive (%) |
|---|---|---|---|---|
| Nodule-positive | 84 | Ghana, Ashanti | O. volvulus-infected | 55 (65.5) |
| B. malayi | 20 | Indonesia, Central Sulawesi | B. malayi-infected | 0 (0) |
| W. bancrofti | 10 | Sri Lanka and Tahiti | W. bancrofti-infected | 0 (0) |
| Healthy controls | 84 | Belgium | No helminth infection | nd |
| South America | 96 | Brazil/South-America | No helminth infection (90 T. cruzi-seropositive) | nd |
| STH/S. mansoni | 24 | Ethiopia, Jimma | STH- and/or S. mansoni-infected | 0 (0) |
| Endemic controls | 50 | Ghana, Ashanti | No clinical signs of onchocerciasis | 25 (50.0) |
| Kenya, Kisumu | 100 | Kenya, Kisumu | 45 S. mansoni-infected, 3 STH-infected | 0 (0) |
| Kenya, Siaya | 108 | Kenya, Siaya | 43 STH-infected, 2 S. mansoni-infected | 1 (0.9) |
Plasma samples from a region in Kenya were collected as part of a field study in Kenya. This study was undertaken in the former Nyanza Province, in the southwest part of Kenya, with collections in the Kisumu county (high S. mansoni prevalence area) and Siaya county (high STH prevalence area). Stool samples were collected in order to determine the STH and S. mansoni infection status, based on microscopic egg counting by the Kato-Katz technique. A total of 208 subjects that had donated plasma samples were included in this study.
A set of 90 plasma samples from patients with chronic Chagas cardiomyopathy that were also Trypanosoma cruzi-seropositive were provided by InfYnity Biomarkers. A total of 48 samples were obtained from Barcelona’s blood bank from South-American immigrants and 42 samples were collected in Minas Gerais state in Brazil. Additionally, 6 samples from T. cruzi-uninfected South American immigrants were obtained from Barcelona’s blood bank.
Onchocerciasis IgG4 rapid test
The presence of IgG4 antibodies against the O. volvulus antigen Ov-16 was determined using the SD BIOLINE Onchocerciasis IgG4 test (Standard Diagnostics, Gyeonggi-do, Republic of Korea), according to the manufacturer’s instructions. Briefly, 10 µl of plasma was added to the round sample well on the lateral flow strip, immediately followed by the addition of 4 drops of assay diluent into the square assay diluent well. After 1 hour, tests were scored. Tests were considered positive only when both the test and control line were visible; faint lines were considered positive, as recommended by the manufacturer.
Multiplex peptide ELISA
Multiplex peptide ELISA plates were produced by InfYnity Biomarkers, Lyon, France (www.infynity-biomarkers.com). The sciFLEXARRAYER system (Scienion, Dortmund, Germany) was used to print C-terminally biotinylated peptides, linked to streptavidin beforehand, in duplicates in each well of a 96-well plate. In order to identify false positivity due to anti-streptavidin antibodies, a streptavidin control was printed in duplicate. In addition to these antigens, three spots of positive controls (PC) designed to check for the presence of human samples and enzyme conjugates were printed on the array. Additionally, low and medium control spots were embedded. A net intensity (mean value of duplicated spots intensity) score was obtained for each of the measured peptide-specific antibodies. To assess the reproducibility of microplates production, a non-destructive QC was performed consisting of visual inspection of printing consistency throughout the full production cycles.
The microplates were processed following the manufacturer’s instructions using the reagents provided in the kits. Briefly, the plasma samples diluted at 1:100 were incubated for 1 h at room temperature, then washed three times with PBST. The dilution parameter was optimized during the assay development. Next, horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibodies (CliniSciences, Nanterre, France) adequately diluted was added to the microplate for 1 h at room temperature. The microplates were then washed three times before adding a precipitating TMB formulation for 20 min at room temperature in the dark. Then, TMB solution was removed and plates were dried at 37 °C for 10 min. Each plate was then imaged using the sciREADER CL2 (Scienion). The software calculated the pixel intensity for each spot. In order to establish the net intensity for each antigen, we considered the mean value of the paired spots.
Malaria antibody ELISA
The presence of antibodies against recombinant antigens of P. falciparum and P. vivax was determined using the anti-malaria kit from Abcam PLC (Cambridge, UK). The tests were performed according to the manufacturer’s instructions using the reagents provided in the kit.
Statistical analysis
All analyses and plots were generated using the R environment (v3.5.2, https://www.R-project.org/) and R packages pROC (v1.13.0) and ggplot2 (v3.1.0) or GraphPad Prism v8.0.0).
Results
The multiplex ELISA
The three peptides with the best performance determined in our previous studies (OvMP-1, OvMP-23 and OvNMP-48) were spotted in the multiplex ELISA system. Reproducibility of the multiplex ELISA was measured using three independent production lots and tested with a total of 480 plasma samples. In total, 3 × 480 data points were used for each peptide to perform multiple linear regression and r2 values were calculated (Table 2). To avoid false positive results due to streptavidin-binding antibodies, we introduced a technical cut-off of 7.7855 spot intensity by which 95% of samples were found below this value for the streptavidin control spot. A total of 22 samples with signal intensity on the streptavidin control spot ≥ 7.7855 were excluded for further analysis. We concluded that the multiplex peptide ELISA can be produced and performed in a reproducible manner.
Table 2.
Parameters for Onchocerca volvulus peptides in the multiplex ELISA after testing on 480 plasma samples on 3 different production lots
| Peptide ID | Sequence | r2 | Intensity (Q1-Median-Q3) |
Spot intensity cut-off |
|---|---|---|---|---|
| OvMP-1 | VSVEPVTTQETVSV | 0.97 | 4.6–85.4–101.0 | 17.43 |
| OvMP-23 | VSVKDGEDKVSVQTSNLDVSV | 0.98 | 1.5–23.7–74.5 | 12.44 |
| OvMP-48 | VSVDNNGANFEVSVNLNANSNPNVSVEKDGKKVSV | 0.95 | 0.8–11.7–34.8 | 8.04 |
Note: Sequences in bold represent the different epitopes. Correlation coefficients r2 are based on multiple linear regression. The signal intensity levels of the individual peptides represent 1st quartile – median – 3rd quartile. “Spot intensity cut-off” threshold of all individual peptides was calculated based on receiver operating characteristic (ROC) analysis
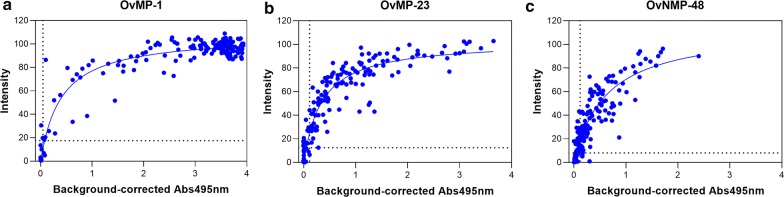
Comparing the results of the multiplex ELISA with the previously published singleplex ELISA [12, 13, 17] showed a correlation between the spot intensities (multiplex assay) and absorbance (singleplex assay). The intensity measured in the multiplex assay rapidly increased with increasing antibody levels and started to saturate at a level that corresponds with an absorbance of 1 (Fig. 1).
Fig. 1.
Correlation between spot intensity as measured in the multiplex ELISA and absorbance previously measured in singleplex ELISA for OvMP-1 (a), OvMP-23 (b) and OvNMP-48 (c). Regression lines were calculated using least squares fit. Spot intensity cut-offs are indicated by the horizontal dashed lines; absorbance cut-offs are indicated by the vertical dashed lines [12, 13]
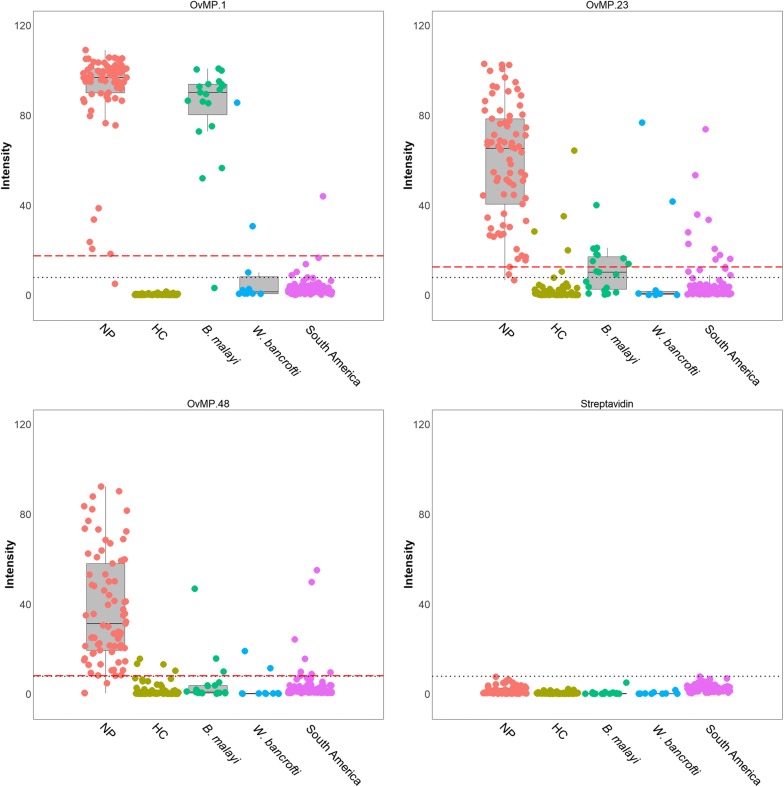
Besides the technical streptavidin-based cut-off, for each of the peptides a threshold for positivity was determined using receiver operating characteristic (ROC) analysis with data derived from testing confirmed onchocerciasis patients versus a control population from O. volvulus non-endemic regions (Fig. 2 and Table 3: known onchocerciasis status). The “spot intensity cut-offs” are given in Table 2. Occasionally, single peptide reactivity above the threshold was seen in control samples. A simple algorithm was designed: samples reactive on 0, 1 or 2 peptides were considered negative for O. volvulus infection, while 3-peptide positive reaction was necessary to confirm seropositivity. We concluded that the use of a 3-peptide multiplex ELISA resulted in a sensitivity of 93.5% and a specificity of 99.0%.
Fig. 2.
Individual data points and box plots of the samples from individuals with known onchocerciasis status for OvMP-1, OvMP-23, OvNMP-48 and streptavidin spot. Red dotted line indicates the threshold as calculated by ROC analysis, black dotted line is the technical cut-off. Boxplots represent the main statistical characteristics for each sample set. The lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles), the middle line corresponds to the median
Table 3.
Number of samples (and percentages) found to be positive for the three different peptide serology markers
| Sample set | Number (%) of plasma samples above “spot intensity cut-off” for peptide | |||
|---|---|---|---|---|
| OvMP-1 | OvMP-23 | OvNMP-48 | 3-peptide positives | |
| Known onchocerciasis status | ||||
| Nodule positive (Ghana, n = 77) | 76 (98.7) | 75 (97.4) | 74 (96.1) | 72 (93.5) |
| B. malayi (Indonesia, n = 19) | 18 (94.7) | 8 (42.1) | 3 (15.8) | 1 (5.3) |
| W. bancrofti (Sri Lanka, n = 8; Tahiti, n = 2) | 2 (20.0) | 2 (20.0) | 2 (20.0) | 1 (10.0) |
| Healthy controls (Belgium, n = 84) | 0 (0) | 4 (4.8) | 4 (4.8) | 0 (0) |
| South America (n=89) | 1 (1.1) | 10 (11.2) | 7 (7.9) | 0 (0) |
| Unknown onchocerciasis status | ||||
| Kenya, Kisumu (n = 98) | 86 (87.8) | 55 (56.1) | 45 (45.9) | 30 (30.6) |
| Kenya, Siaya (n = 98) | 83 (84.7) | 66 (67.3) | 69 (70.4) | 57 (58.2) |
| STH/S. mansoni (Ethiopia, n = 24) | 3 (12.5) | 5 (20.8) | 2 (8.3) | 0 (0) |
| Endemic controls (Ghana, n = 48) | 46 (95.8) | 46 (95.8) | 43 (89.6) | 40 (83.3) |
Performance of the multiplex peptide ELISA in populations with unknown infection
The seroreactivity against the three peptides was investigated in individuals with unknown O. volvulus infection status (Table 3: unknown onchocerciasis status). In agreement with what was published before for singleplex ELISA, most samples from Ethiopia positive for soil-transmitted helminths (STH) and/or S. mansoni infection were negative for the peptides, and none were found to be positive for all three peptide markers [18].
In two sample sets collected in an onchocerciasis-free area of Kenya (Kisumu county with high prevalence of S. mansoni; and Siaya county with high STH prevalence), more than 80% were positive for OvMP-1, both in Kisumu and Siaya county, and unexpected high seropositivity for OvMP-23 and OvNMP-48 was measured. The 3-peptide positives algorithm would label > 40% seropositive for O. volvulus in Kenya, a result that is in strong disagreement with the epidemiology of onchocerciasis. It was concluded that the three peptides bearing linear O. volvulus epitopes are recognized by antibodies induced by either/or O. volvulus and yet another immunogen, but not STH, S. mansoni, W. bancrofti, B. malayi, HIV, HCV, dengue, and possibly also not by the malaria parasite [12, 13, 18]. The majority of the endemic control samples from Ghana were found to be positive for peptides OvMP-1, OvMP-23 and/or OvNMP-48, with 83.3% of them positive for all three peptides.
The presence of antibodies against recombinant antigens of P. falciparum and P. vivax was determined in the 196 samples from Kenya (Table 4). Since in the 20 samples found to be negative for malaria, 16, 6 and 4 samples were found to be positive for OvMP-1, OvMP-23 and OvNMP-48, respectively, there is no evident causal relationship between infection with Plasmodium spp. and an immune response against these peptides.
Table 4.
Number of samples found to be positive for the three different peptide serology markers in the Kenyan population, stratified according to malaria serological status
| Malaria serostatus | n | Number (%) of plasma samples above “spot intensity cut-off” for peptide | |||
|---|---|---|---|---|---|
| OvMP-1 | OvMP-23 | OvNMP-48 | 3-peptide positives | ||
| Negative | 20 | 16 (80.0) | 6 (30.0) | 4 (20.0) | 2 (10.0) |
| Intermediate | 11 | 11 (100) | 7 (63.6) | 4 (36.4) | 3 (27.3) |
| Positive | 165 | 142 (86.1) | 108 (65.5) | 106 (64.2) | 82 (49.7) |
| Total | 196 | 169 (86.2) | 121 (61.7) | 114 (58.2) | 87 (44.4) |
Discussion
In this study, the performance of an InfYnity Biomarkers multiplex peptide ELISA was evaluated for the detection of antibodies against different linear epitopes of O. volvulus using a similar approach to what was already applied on Chagas disease serology [15, 16]. The panel of three peptides that was used is based on six different linear epitopes identified previously [12–14, 17, 18]. As previously observed, no IgG4 antibodies against these epitopes were detected, and therefore it was decided to determine epitope-specific total IgG in the multiplex ELISA [14].
First, the comparability of the multiplex and singleplex data was evaluated. Multiplex and singleplex data were comparable for all three peptides. Three independent batches of plates were produced on different days, using different solutions of peptides, and using a different piezo crystal (which pulses an electric current flow, resulting in a drop of the solution out of the nozzle [19]). A set of 480 plasma samples was simultaneously analyzed on the three different batches. For all three peptides a good correlation (r 2≥ 0.95) was found.
The multiplex ELISA was used to determine seroreactivity against the different O. volvulus epitopes in Kenyan individuals with unknown O. volvulus infection status. Kenya was the first country that used synthetic insecticides to eliminate the vector that transmits O. volvulus, in the case of Kenya being Simulium neavei [20, 21]. Already in 1955, vector control was completed and follow-up evaluations at 18 years after interruption of transmission demonstrated that microfilariae were no longer found in the skin, and onchocerciasis was considered to be eliminated in Kenya [22–24]. Two sets of plasma samples were collected: one set from Kisumu county (high prevalence of S. mansoni), the other set from Siaya county (high prevalence of STH). Only one out of 196 individuals had a positive Ov16 IgG4 response. We concluded that these populations were indeed onchocerciasis-free. In contrast however, 87 out of 196 (44.4%) samples were found to be triple-peptide positive.
Analysis of the evidence for candidates for this unexpected cross-reactivity falls into two groups: organisms to be excluded, and hypothetical candidates. First, the organisms that can be excluded are as follows: (i) STH and S. mansoni: individuals with infection from Ethiopia do not respond to the different peptides [18], and this is confirmed in this multiplex ELISA (Table 3); (ii) W. bancrofti (causing lymphatic filariasis): is confined to the Kenya coastal region and it is therefore unlikely to cause such high seroprevalence in Kisumu and Siaya counties [25]; (iii) Plasmodium: 84.2% of the Kenyan samples were seropositive for Plasmodium, while only half of them were triple-peptide positive. Furthermore, most of the 20 samples that were seronegative for Plasmodium were seropositive for at least one peptide (2 samples for all 3 peptides), making it unlikely that Plasmodium is causing these responses; and (iv) HIV, HCV and dengue virus: previous analyses using singleplex ELISA demonstrated that no response could be detected against any of the three peptides in individuals infected with one of these viruses.
Secondly, hypothetical causes for the cross-reactivity are as follows: (i) exposure to zoonotic Onchocerca species: O. gutturosa or O. fasciata can induce a cross-reactive immune response upon exposure [26–28], and cross-reactive antibodies were found in individuals living in areas where O. ochengi infects cattle [29]. This parasite requires Simulium species which are no longer present in Kenya [20, 21], hence this zoonotic hypothesis is unlikely; (ii) Mansonella perstans: probably the most prevalent known human filarial infection [30, 31] and possibly the most neglected filariasis [32]. It is estimated that more than 100 million people may be infected by M. perstans, predominantly in sub-Saharan Africa as well as in the northern part of the Amazon rainforest stretching from equatorial Brazil to the Caribbean coast of South America [32]. Historical reports however also indicated the presence of Mansonella in the Pacific region [33]. It was indicated before that mansonellosis parasites might interfere with some onchocerciasis immunodiagnostic assays [34]. In countries such as Ghana, Uganda and Cameroon, it was shown that in some communities, prevalence reaches levels above 70% [35–37]. The samples we tested from South America were from a non-endemic region in Brazil (Minas Gerais State) and the fact that most of them were seronegative for the three peptides at least does not contradict the hypothesis that M. perstans is causing the peptide specific responses. There is no information on mansonellosis in the Kisumu and Siaya counties, but it is thought to be endemic in Kenya [30]; (iii) Strongyloides stercoralis: current data indicate that S. stercoralis infections affect between 10% and 40% of the population in many tropical and subtropical countries. Community-based surveys in Cameroon, Ghana and Kenya showed a prevalence of 10.0%, 69.5% and 80.2% respectively [38]; and (iv) unknown: certain viral or bacterial infections or environmental stimuli might also induce the immune response directed against the peptides. In this respect, one might consider mining all the proteomes available in the databases. However, as we already demonstrated for OvMP1, OvMP-2 and OvMP-3, there are multiple proteins from several organisms that bear these epitope sequences. Although some of these proteins might even be antigenic in case the epitope is accessible in the native protein, no conclusion can be drawn on the immunogenicity of these protein sequences in vivo [14]. Furthermore, peptides may mimic essential features of epitopes (mimotopes) [39]. Therefore, the true immunogenic stimulus that resulted in the antibodies detected here, might even not contain the epitope sequence. Based on these considerations, it is unlikely that this approach will lead to the identification of the true immunogen.
Further evaluation of O. volvulus endemic control samples, i.e. from individuals living in a Ghana O. volvulus endemic area, but who had no visible signs of onchocerciasis, showed the majority (83%) to be triple-peptide positive, while 50% of these endemic controls were found to be positive for Ov-16 IgG4 antibodies (Table 1). Considering the above observations, we cannot conclude on the exposure status of the endemic control group beyond the Ov16 IgG4 antibody response. The data as currently available do not exclude the possibility that OvMP-1, OvMP–23, and OvNMP-48 react both with antibodies raised against O. volvulus, and/or solely to the as-yet to be identified immunogen.
It must be noted that two of the peptides evaluated in this study are designed to cover two or three different epitopes. Since a positive response against such peptide does not discriminate the response against the individual epitopes, there is still the possibility that some of the individual epitopes are specific for O. volvulus.
The observed cross-reactivity on the O. volvulus derived peptides opens an additional discussion on the quality check of the biobanks used for biomarker discovery activities. Since discovery of these peptides was performed on samples from Cameroon [14] and confirmation of their reactivity was done on samples from Ghana, Indonesia, Sri Lanka [12, 13] and Kenya, we came to the conclusion that, despite the careful selection of samples, we knew little or nothing of other filaria past or present co-infections. Therefore, validation of candidate biomarkers should be performed on extremely well-characterized samples for multiple infections. This would require a holistic view on biobanking, which would include biomaterial collection beyond the study objective. The presented work should also be seen as a call to the NTD community to collaborate precompetitively on biobanking initiatives prior to embarking on the biomarker discovery and validation.
Conclusions
This study demonstrates that a multiplex ELISA based on three O. volvulus peptides was successfully implemented. Assessment of the utility of this serology assay indicates that other, yet to be identified stimuli, significantly cross-react with these peptides. This observation invalidates the usefulness of the peptide serology markers for onchocerciasis detection.
Acknowledgements
We gratefully acknowledge Dr Alex Debrah and Dr Linda Batsa Debrah for providing plasma samples from O. volvulus infected individuals from Ghana, Professor Maria Yazdanbakhsh for providing plasma samples of B. malayi infected individuals from Indonesia and Professor B. Levecke (University of Ghent) for providing plasma samples of STH infected individuals from Ethiopia. We thank all study participants for providing samples to the biobanks. We thank Janssen Biobank for logistic support, Ann Verheyen and Kim Van Hoof for help with the multiplex peptide ELISA work, Will Colón (Janssen Global Public Health) for critically reviewing this manuscript, Martin Koldijk and Gerrit Jan Weverling (Janssen World Without Disease Accelerator) for helpful discussions on the statistical analysis and Benny Baeten and Marc Engelen from Janssen Global Public Health for programmatic support.
Abbreviations
- WHO
World Health Organization
- NTD
neglected tropical diseases
- MDA
mass drug administration
- RDT
rapid diagnostic test
- mf
microfilariae
- ELISA
enzyme-linked immunosorbent assay
- STH
soil-transmitted helminths
Authorsʼ contributions
OL, MO and LJS designed the study. OL, MZ and EG performed the research. OL and LJS performed the analysis of the data. OL and LJS wrote the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Data supporting the conclusions of this article are included within the article. The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008. All samples used in this study were de-identified before being provided and usage of these samples for research purposes was approved by an ethical committee or Institutional Review Board (IRB). For samples collected in Kenya, the study was approved by the KEMRI Scientific and Ethics Review Unit (SERU), Nairobi, Kenya (Protocol # KEMRI/SERU/CGHR/102/3554). Since all study participants were minors, informed consent forms were signed by parents/guardians of the study participants, and verbal assents were obtained from all study participants.
Consent for publication
Not applicable.
Competing interests
OL and LJS are current employees of Janssen Pharmaceutica NV, a Johnson and Johnson Company, and may own stock or stock options in that company. EG and MZ are current employees of InfYnity Biomarkers. MRO declares that he has no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ole Lagatie, Email: olagatie@its.jnj.com.
Elodie Granjon, Email: egranjon@infynity-biomarkers.com.
Maurice R. Odiere, Email: mauriceodiere@gmail.com
Maan Zrein, Email: mzrein@infynity-biomarkers.com.
Lieven J. Stuyver, Email: lstuyver@its.jnj.com
References
- 1.Holmes P, on behalf of the WHO Strategic and Advisory Group on Neglected Tropical Diseases Neglected tropical diseases in the post-2015 health agenda. Lancet. 2014;383:1803. doi: 10.1016/S0140-6736(14)60875-8. [DOI] [PubMed] [Google Scholar]
- 2.Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, Jacobson J. Helminth infections: the great neglected tropical diseases. J Clin Invest. 2008;118:1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO . Progress report on the elimination of human onchocerciasis, 2017–2018. In: Cantey P, editor. Releve epidemiologique hebdomadaire/Section dʼhygiene du Secretariat de la Societe des Nations = Weekly epidemiological record/Health Section of the Secretariat of the League of Nations. Geneva: World Health Organization; 2018. pp. 633–648. [Google Scholar]
- 4.Borup LH, Peters JS, Sartori CR. Onchocerciasis (river blindness) Cutis. 2003;72:297–302. [PubMed] [Google Scholar]
- 5.Denkert C, Budczies J, Kind T, Weichert W, Tablack P, Sehouli J, et al. Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 2006;66:10795–10804. doi: 10.1158/0008-5472.CAN-06-0755. [DOI] [PubMed] [Google Scholar]
- 6.Taylor HR, Munoz B, Keyvan-Larijani E, Greene BM. Reliability of detection of microfilariae in skin snips in the diagnosis of onchocerciasis. Am J Trop Med Hyg. 1989;41:467–471. doi: 10.4269/ajtmh.1989.41.467. [DOI] [PubMed] [Google Scholar]
- 7.Vlaminck J, Fischer PU, Weil GJ. Diagnostic tools for onchocerciasis elimination programs. Trends Parasitol. 2015;31:571–582. doi: 10.1016/j.pt.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 8.WHO. Guidelines for stopping mass drug administration and verifying elimination of human onchocerciasis: Criteria and procedures. In: WHO Guidelines Approved by the Guidelines Review Committee. Geneva: World Health Organization; 2016. [PubMed]
- 9.Steel C, Golden A, Stevens E, Yokobe L, Domingo GJ, de los Santos T, et al. Rapid point-of-contact tool for mapping and integrated surveillance of Wuchereria bancrofti and Onchocerca volvulus infection. Clin Vaccine Immunol. 2015;22:896–901. doi: 10.1128/CVI.00227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golden A, Steel C, Yokobe L, Jackson E, Barney R, Kubofcik J, et al. Extended result reading window in lateral flow tests detecting exposure to Onchocerca volvulus: a new technology to improve epidemiological surveillance tools. PLoS One. 2013;8:e69231. doi: 10.1371/journal.pone.0069231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lont YL, Coffeng LE, de Vlas SJ, Golden A, de Los Santos T, Domingo GJ, et al. Modelling Anti-Ov16 IgG4 antibody prevalence as an indicator for evaluation and decision making in onchocerciasis elimination programmes. PLoS Negl Trop Dis. 2017;11:e0005314. doi: 10.1371/journal.pntd.0005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagatie O, Verheyen A, Nijs E, Van Dorst B, Batsa Debrah L, Debrah A, et al. Evaluation of the diagnostic performance of Onchocerca volvulus linear epitopes in a peptide enzyme-linked immunosorbent assay. Am J Trop Med Hyg. 2018;98:779–785. doi: 10.4269/ajtmh.17-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagatie O, Verheyen A, Nijs E, Batsa Debrah L, Debrah YA, Stuyver LJ. Performance evaluation of 3 serodiagnostic peptide epitopes and the derived multi-epitope peptide OvNMP-48 for detection of Onchocerca volvulus infection. Parasitol Res. 2019;118:2263–2270. doi: 10.1007/s00436-019-06345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagatie O, Van Dorst B, Stuyver LJ. Identification of three immunodominant motifs with atypical isotype profile scattered over the Onchocerca volvulus proteome. PLoS Negl Trop Dis. 2017;11:e0005330. doi: 10.1371/journal.pntd.0005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granjon E, Dichtel-Danjoy ML, Saba E, Sabino E, de Oliveira L, Zrein M. Development of a novel multiplex immunoassay multi-cruzi for the serological confirmation of chagas disease. PLoS Negl Trop Dis. 2016;10:e0004596. doi: 10.1371/journal.pntd.0004596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zrein M, Granjon E, Gueyffier L, Caillaudeau J, Liehl P, Pottel H, et al. A novel antibody surrogate biomarker to monitor parasite persistence in Trypanosoma cruzi-infected patients. PLoS Negl Trop Dis. 2018;12:e0006226. doi: 10.1371/journal.pntd.0006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagatie O, Verheyen A, Van Dorst B, Batsa Debrah L, Debrah A, Stuyver LJ. Linear epitopes in Onchocerca volvulus vaccine candidate proteins and excretory-secretory proteins. Parasite Immunol. 2018;40:e12587. doi: 10.1111/pim.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlaminck J, Lagatie O, Verheyen A, Dana D, Van Dorst B, Mekonnen Z, et al. Patent infections with soil-transmitted helminths and Schistosoma mansoni are not associated with increased prevalence of antibodies to the Onchocerca volvulus peptide epitopes OvMP-1 and OvMP-23. Parasit Vectors. 2019;12:63. doi: 10.1186/s13071-019-3308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Rossignol F, Macdonald J. Inkjet printing for biosensor fabrication: combining chemistry and technology for advanced manufacturing. Lab Chip. 2015;15:2538–2558. doi: 10.1039/C5LC00235D. [DOI] [PubMed] [Google Scholar]
- 20.Mc MJ, Highton RB, Goiny H. The eradication of Simulium neavei from Kenya. Bull World Health Organ. 1958;19:75–107. [PMC free article] [PubMed] [Google Scholar]
- 21.Garnham PC, Mc MJ. The eradication of Simulium neavei Roubaud, from an onchocerciasis area in Kenya Colony. Bull Entomol Res. 1947;37:619–628. doi: 10.1017/S000748530003011X. [DOI] [PubMed] [Google Scholar]
- 22.Roberts JM, Neumann E, Gockel CW, Highton RB. Onchocerciasis in Kenya 9, 11 and 18 years after elimination of the vector. Bull World Health Organ. 1967;37:195–212. [PMC free article] [PubMed] [Google Scholar]
- 23.Crump A, Morel CM, Omura S. The onchocerciasis chronicle: from the beginning to the end? Trends Parasitol. 2012;28:280–288. doi: 10.1016/j.pt.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 24.WHO Onchocerciasis and its control. Report of a WHO Expert Committee on Onchocerciasis Control. World Health Organ Tech Rep Ser. 1995;852:1–104. [PubMed] [Google Scholar]
- 25.Moraga P, Cano J, Baggaley RF, Gyapong JO, Njenga SM, Nikolay B, et al. Modelling the distribution and transmission intensity of lymphatic filariasis in sub-Saharan Africa prior to scaling up interventions: integrated use of geostatistical and mathematical modelling. Parasit Vectors. 2015;8:560. doi: 10.1186/s13071-015-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarkson MJ. The species of Onchocerca in cattle in Kenya and Somalia. Ann Trop Med Parasitol. 1964;58:153–158. doi: 10.1080/00034983.1964.11686225. [DOI] [PubMed] [Google Scholar]
- 27.Round MC. The helminth parasites of domesticated animals in Kenya. J Helminthol. 1962;36:375–449. doi: 10.1017/S0022149X00019453. [DOI] [PubMed] [Google Scholar]
- 28.Orihel TC, Eberhard ML. Zoonotic filariasis. Clin Microbiol Rev. 1998;11:366–381. doi: 10.1128/CMR.11.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoch B, Wahl G, Enyong P, Lüder CG, Harnett W, Renz A. Onchocerciasis of cattle and man: serological recognition of parasite specific and cross-reactive antigens. Parasite. 1994;1:S14. doi: 10.1051/parasite/199401s1014. [DOI] [Google Scholar]
- 30.Simonsen PE, Onapa AW, Asio SM. Mansonella perstans filariasis in Africa. Acta Trop. 2011;120(Suppl. 1):S109–S120. doi: 10.1016/j.actatropica.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Downes BL, Jacobsen KH. A systematic review of the epidemiology of mansonelliasis. Afr J Infect Dis. 2010;4:7–14. doi: 10.4314/ajid.v4i1.55085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ta-Tang TH, Crainey JL, Post RJ, Luz SL, Rubio JM. Mansonellosis: current perspectives. Res Rep Trop Med. 2018;9:9–24. doi: 10.2147/RRTM.S125750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crainey JL, da Silva TR, Luz SL. Historic accounts of Mansonella parasitaemias in the South Pacific and their relevance to lymphatic filariasis elimination efforts today. Asian Pac J Trop Med. 2016;9:205–210. doi: 10.1016/j.apjtm.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 34.Luz SL, Crainey JL, Shelley AJ, Rubio M. Outstanding insecurities concerning the use of an Ov16-based ELISA in the Amazonia onchocerciasis focus. Mem Inst Oswaldo Cruz. 2014;109:506–508. doi: 10.1590/0074-0276140079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onapa AW, Simonsen PE, Baehr I, Pedersen EM. Rapid assessment of the geographical distribution of Mansonella perstans infections in Uganda, by screening schoolchildren for microfilariae. Ann Trop Med Parasitol. 2005;99:383–393. doi: 10.1179/136485905X361990. [DOI] [PubMed] [Google Scholar]
- 36.Debrah LB, Nausch N, Opoku VS, Owusu W, Mubarik Y, Berko DA, et al. Epidemiology of Mansonella perstans in the middle belt of Ghana. Parasit Vectors. 2017;10:15. doi: 10.1186/s13071-016-1960-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wanji S, Tayong DB, Layland LE, Datchoua Poutcheu FR, Ndongmo WP, Kengne-Ouafo JA, et al. Update on the distribution of Mansonella perstans in the southern part of Cameroon: influence of ecological factors and mass drug administration with ivermectin. Parasit Vectors. 2016;9:311. doi: 10.1186/s13071-016-1595-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schar F, Trostdorf U, Giardina F, Khieu V, Muth S, Marti H, et al. Strongyloides stercoralis: global distribution and risk factors. PLoS Negl Trop Dis. 2013;7:e2288. doi: 10.1371/journal.pntd.0002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deroo S, Muller CP. Antigenic and immunogenic phage displayed mimotopes as substitute antigens: applications and limitations. Comb Chem High Throughput Screen. 2001;4:75–110. doi: 10.2174/1386207013331309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the conclusions of this article are included within the article. The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.