Abstract
Background
Cervical cancer is one of the most frequent malignancies among females worldwide. Increasing evidence have indicated the participation of long noncoding RNAs (lncRNAs) in the progression and metastasis of cervical cancer. Our present study was conducted to explore the effects of lncRNA plasmacytoma variant translocation 1 (PVT1) on the progression of cervical cancer and the underlying mechanisms.
Materials and methods
Expressions of PVT1, miR-140-5p and Smad3 in cervical cancer cell lines were detected by qRT-PCR and western blotting. Bioinformatics analysis and luciferase assays were used to elucidate the potential correlations between PVT1, miR-140-5p and Smad3. The roles of PVT1 on the progression of cervical cancer cells were determined by transfecting sh-RNA through series function assays such as colony formation assay, wound healing assay, transwell assay.
Results
PVT1 and Smad3 were upregulated, and miR-140-5p was downregulated in cervical cancer cells. PVT1 could bind directly with miR-140-5p, and Smad3 was a downstream target of miR-140-5p. Inhibition of PVT1 could enhance expression of miR-140-5p, inhibit the expression of Smad3, significantly inhibited the proliferation, migration, invasion in cervical cancer cells. While transfection of miR-140-5p inhibitor could partially reverse the above changes in cervical cancer cells.
Conclusions
The results revealed that PVT1 could promote the proliferation and metastasis via increasing the Smad3 expression by sponging miR-140-5p, which might be a promising prognostic and therapeutic target for cervical cancer.
Key words: cervical cancer, lncRNA PVT1, miR-140-5p, Smad3
Introduction
Cervical cancer is the second most frequent malignancy-affecting women worldwide, with approximately over 570000 new cases diagnosed and 310000 deaths each year. Nearly 85% of the cases occur in underdeveloped and developing countries.1 Interactions between cervical cancer and the persistent infection of high-risk human papilloma virus (especially HPV16 and HPV18) have been indicated in epidemiological and molecular studies.2, 3, 4 Despite the improvement of diagnostic approaches and treatment strategies have been made, the morbidity and mortality rates remain high as well as the poor prognosis. Therefore, there is an urgent need to clarify the underlying molecular mechanisms of cervical cancer, which could improve the development of therapeutic strategies against cervical cancer.
Long noncoding RNAs (lncRNAs) are a group of noncoding RNAs longer than 200 nucleotides that participate in numerous biological and physiological processes including cell development, survival, differentiation and apoptosis.5, 6, 7 Accumulating evidence also proved that lncRNAs have pivotal roles in the progression of cervical cancer. Yang et al.8 showed that lncRNA CCHE1 could promote the proliferation of cervical cancer via upregulation the expression of PCNA. Han et al.9 discovered that lncRNA NEAT1 could strengthen the resistance of cervical cancer against radiotherapy through the regulation of CCND1. Up to now, plasmacytoma variant translocation 1 (PVT1) has been recognized as an oncogenic lncRNA, which participates in the migration and invasion of many kinds of cancer cells and the development of cancers.10, 11 Ding et al. found PVT1 was the most amplified gene in ovarian cancer patients, which was highly correlated with poor survival outcomes.12 It was also proven that high PVT1 expression correlates with poorer outcome in cervical cancer patients.13 However, the underlying mechanism on the effects of lncRNA PVT1 in cervical cancer remains for further investigation.
MicroRNAs (miRNAs) are another group of noncoding RNAs with length between 17-25 nucleotides that influence the pathogenesis of many human diseases including cancer.14, 15, 16 MiRNAs are normally dysregulated in human cancers and could serve as either oncogenes or tumor suppressors.17 Previous studies showed that expressions of miR-140-5p were downregulated in cancer tissues, which were negatively related to the prognosis of cancer18, 19, 20, indicating the tumor-suppressor role of miR-140-5p in the progression of cancer formation. Li et al.21 showed that miR-140-5p could inhibit the progression and invasion of colorectal cancer by targeting Smad3, indicating an interaction may exist between miR-140-5p and Smad3. However, whether miR-140-5p could regulate the metastasis of cervical cancer by targeting Smad3 remains to be determined. Bioinformatics analysis also revealed that targeted binding sites exist between lncRNA PVT1 and miR-140-5p. Therefore, we hypothesized that the elevated lncRNA PVT1 could upregulate the expression of Smad3 by targeting inhibition of miR-140-5p, thereby promoting the proliferation and migration of cervical cancer cells in cervical cancer.
In the present study, the role and the underlying molecular mechanism of lncRNA PVT1 in cervical cancer were illuminated. Our results showed that lncRNA PVT1 could promote the proliferation, migration and invasion through binding miR-140-5p, which may provide new insights into the therapeutic strategy against cervical cancer.
Materials and methods
Cell lines and cell culture
Human normal cervical epithelial cell line (End1/ E6E7) and human cervical cell lines (HeLa and SiHa) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The cells were maintained in Dulbecco’s minimal Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 6% fetal bovine serum (FBS; Gibico, Grand Island, NY, USA) and 1% penicillin/ streptomycin at 37°C in a humidified atmosphere containing 5% CO2.
Dual luciferase reporter assay
A bioinformatics website (http://www.rna-society.org/raid2/index.html) was used to identify the binding sites for miR-140-5p, lncRNA PVT1 and Smad3, and obtain the fragment sequences containing action sites. The bioinformatics software was then applied to predict the binding sites between miR-140-5p and PVT1 as well as between miR-140-5p and Smad3. Then the wild-type PVT1 (PVT1-WT) , mutant PVT1 (PVT1-MUT), wild-type Smad3 (Smad3-WT) and mutant Smad3 (Smad3-MUT) containing predicted miR-140-5 binding sites were synthesized and cloned into a pGL3 vector. Then, the above constructed vectors were transfected into cervical cancer cells in the presence of miR-NC or miR-140-5p inhibitor. Luciferase activity was detected using the Dual-Luciferase Reporter Assay System (Promega, Madison, USA).
Cell transfection
Sh-PVT1, negative control (sh-NC), miR-140-5p mimics and miR-140-5p inhibitor were synthesized by GenePharma (Shanghai, China). After reaching 60%-70% confluence, the cervical cancer cells were transfected by using Lipofectamine 2000 (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions.
Cell colony formation assay
After transfection, cells were seeded in 6-well plates at a density of 1×103 cells/well. The cells were then fixed and stained with 0.1% crystal violet after incubation for 14 days. The numbers of colonies were counted under microscope. This assay was performed at least three times.
Wound healing assays
Cells were incubated in 6-well plates at a density of 1 × 106 cells/well. After transfection, the cells were cultured with serum-free medium for 24 h. Wounds were created by using a sterile plastic pipette tip to scratch the cell layer. Then the cells were incubated with medium containing 10% FBS for 48 h. Images were acquired by microscope. This assay was performed at least three times.
Transwell assays
The transwell assays were determined using transwell chambers (Corning, NY, USA) as previously described.22 Briefly, transfected cells were suspended in serum-free medium and seeded into the upper chamber, while medium containing 10% FBS was added in the bottom chamber. After incubation at 37°C for 24 h, the cells that invaded through matrix membrane were fixed with 4% paraform-aldehyde and stained with crystal violet. Then, stained cells were photographed and counted from five random fields per filter for analysis. This assay was performed at least three times.
RNA isolation and qRT-PCR
Total RNA was extracted from cell samples using Trizol reagents (Thermo Fisher Scientific, Waltham, MA, USA) from cell lines, and cDNA was synthesized using PrimeScriptTM cDNA Kit (Takara, Dalian, China) according to the manufacture’s protocols. qRT-PCR was determined using an ABI 7000 Prism Step One plus detection system (Life Technologies, USA). The relative expression was normalized using GAPDH as an internal reference gene, and U6 was used as the endogenous control of miR-140-5p. Fold changes were calculated using the formula 2-ΔΔCt. All qRT-PCR reactions were performed three times independently. The primer sequences used for qRT-PCR as follow.
Western blotting
Cell samples were harvested and lysed using cell lysis buffer on ice. Total proteins were extracted and concentrations were detected using a BCA protein assay kit (Beyotime, China). Same amounts of protein samples were isolated by 12% SDS-PAGE gels and then transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were then blocked with TBS-T containing 5% nonfat milk powder for 2 h at RT and incubated with primary antibodies at 4°C overnight. The membranes were then incubated with secondary antibody for 1 h at RT. The images were visualized using an enhanced chemiluminescence system. Signals were quantified and analyzed using the Image-Pro Plus 6.0 software (Media Cybernetics, Sarasota, USA). The following primary antibodies were applied: anti-Smad3 (1:1000; #9513), anti-E-cadherin (1:1000; #14472), anti-vimentin (1:1000; #3932), anti-N-cadherin (1:1000; #4061) anti-Snail (1:1000; #3879) (All purchased from Cell signaling Technology). The corresponding HRP-conjugated antibody (1: 5000; Abcam, UK) was used as the secondary antibody.
Statistical analysis
Statistical analysis was performed using SPSS 22.0 software (IBM Corporation, NY, USA). All data were expressed as the mean ± standard deviation (SD). All experiments were performed triplicate independently. Comparisons were determined using one-way analysis of variance (ANOVA) or Student’s t-test. P < 0.05 was considered significantly different.
PVT1 forward 5’-AAAACGGCAGCAGGAAATGT-3’ and reverse 5’-GGAGTCATGGGTGTCAGACA-3’.
miR-140-5p forward 5’-GGGCCAGTGGTTTTACCCTA-3’
and reverse 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC CTACCA-3’.
Smad3 forward 5’-CTCCAAACCTATCCCCGAAT-3’
and reverse 5’-CCTGTTGACATTGGAGAGCA-3’. U6 forward 5′-AAAGCAAATCATCGGACGACC-3′
and reverse 5′- GTACAACACATTGTTTCCTCGGA-3′.
GAPDH forward 5′-AGAAGGCTGGGGCTCATTTG -3′
and reverse 5′-AGGGGCCATCCACAGTCTTC-3’.
Results
The expressions of lncRNA PVT1 and Smad3 were increased, while miR-140-5p was reduced in cervical cancer cell lines
To determine the roles of lncRNA PVT1, miR-140-5p and Smad3 in cervical cancer metastasis, expressions of lncRNA PVT1, miR-140-5p and Smad3 in cervical cancer cell lines (HeLa and SiHa) and human normal cervical cell line (End1/E6E7) were determined by qRT-PCR and western blotting. The results in Figure 1A and 1B show that lncRNA PVT1 expression was significantly upregulated in cervical cancer cell lines compared with that in the normal cervical cell line by over 1.6-fold (P < 0.01) and 1.3-fold (P < 0.05) respectively. While, the expression of miR-140-5p was remarkably downregulated in cervical cancer cell lines compared with the normal cervical epithelial cells as their respective reductions were 51.8% and 36.7% (P < 0.01). Furthermore, the mRNA and protein expressions of Smad3 were significantly increased in the cervical cancer cell lines compared with that in the normal cervical epithelial cells with 1.5-fold, 1.3-fold increase in mRNA levels and 3.8-fold, 3.3-fold increase in protein levels, respectively (Figure 1C and 1D, P < 0.01).
Figure 1.
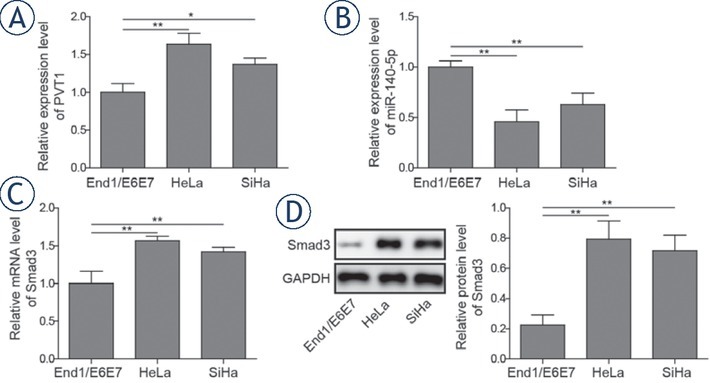
Expressions of PVT1, miR-140-5p and Smad3 in normal cervical epithelial cells and cervical cancer cell lines. Expression levels of PVT1 (A), miR-140-5p (B) and Smad3 (C) were detected by qRT-PCR. (D) Protein expressions of Smad3 were detected by western blotting. T he data are presented as means ± SD of three independent experiments. Statistical significance compared with the normal cervical epithelial cells is indicated by *P < 0.05 and **P < 0.01.
PVT1 directly sponged miR-140-5p and miR-140-5p directly targeted Smad3 in cervical cancer cells
Bioinformatics analysis showed that miR-140-5p might be a target of PVT1 (Figure 2A). Dual luciferase assay was further conducted to determine whether PVT1 could regulate miR-140-5p expression by acting as a molecular sponge, showing that miR-140-5p mimics could significantly inhibit the luciferase activity of PVT1-WT in both cancer cell lines, but had no significant effect on that of PVT1-MUT (Figure 2C and 2D). Furthermore, bio-informatics analysis revealed miR-140-5p can also bind directly to Smad3 (Figure 2B). The results of Figure 2E and 2F showed that miR-140-5p mimics could remarkably suppress the luciferase activity of Smad3-WT in both cervical cancer cell lines, and had no effect on that of Smad3-MUT.
Figure 2.
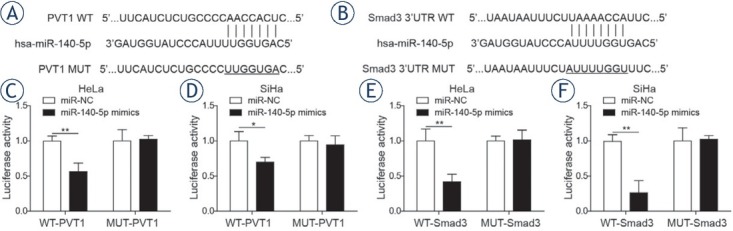
MiR-140-5p was a target of lncRNA PVT1 and Smad3 was a downstream target of miR-140-5p. (A) Binding sites between lncRNA PVT1 and miR-140-5p. (B) Binding sites between miR-140-5p and Smad3. Dual-luciferase assay was applied to explore the interaction between PVT1 and miR-140-5p in HeLa cells (C) and SiHa cells (D). Dual-luciferase assay was applied to explore the interaction between miR-140-5p and Smad3 in HeLa cells (E) and SiHa cells (F). The data are presented as means ± SD of three independent experiments. Statistical significance compared with miR-NC is indicated by *P < 0.05 and **P < 0.01.
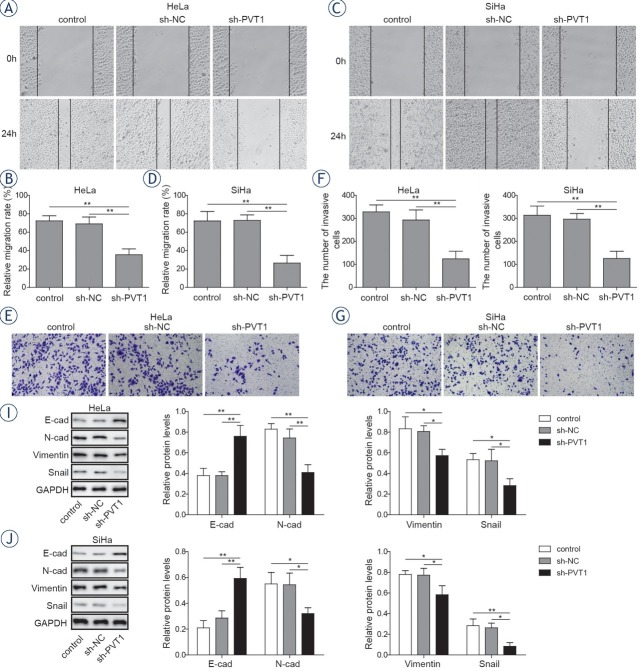
Downregulation of lncRNA PVT1 inhibits proliferation, migration and invasion of cervical cancer cells
Since PVT1 was highly expressed in cervical cancer cells, revealing that lncRNA PVT1 could be a potential oncogene during the progression of cervical cancer. To figure out the function of lncRNA PVT1 during the progression of cervical cancer cells, PVT1 specific sh-RNA was applied to knockdown the expression of PVT1. As shown in Figure 3A and 3B, the expressions of PVT1 were significantly downregulated in sh-PVT1 transfected cervical cancer cells compared with that in cells transfected with sh-NC and the control group as their respective reductions were 39.8% and 43.7% (P < 0.01). While, the expressions of miR-140-5p in sh-PVT1 transfected cervical cancer cells were significantly upregulated compared with cells transfected with sh-NC and the control group by over 1.4-fold (P < 0.05, Figure 3C) and 1.9-fold respectively (P < 0.01, Figure 3D). Meantime, the mRNA levels (Figure 3E and 3F) and protein levels (Figure 3G and 3H) of Smad3 were significantly decreased after transfecting with sh-PVT1 in cervical cancer cells with 25.3%, 38.5% reductions in mRNA levels and 57.2%, 62.3%
Figure 3.
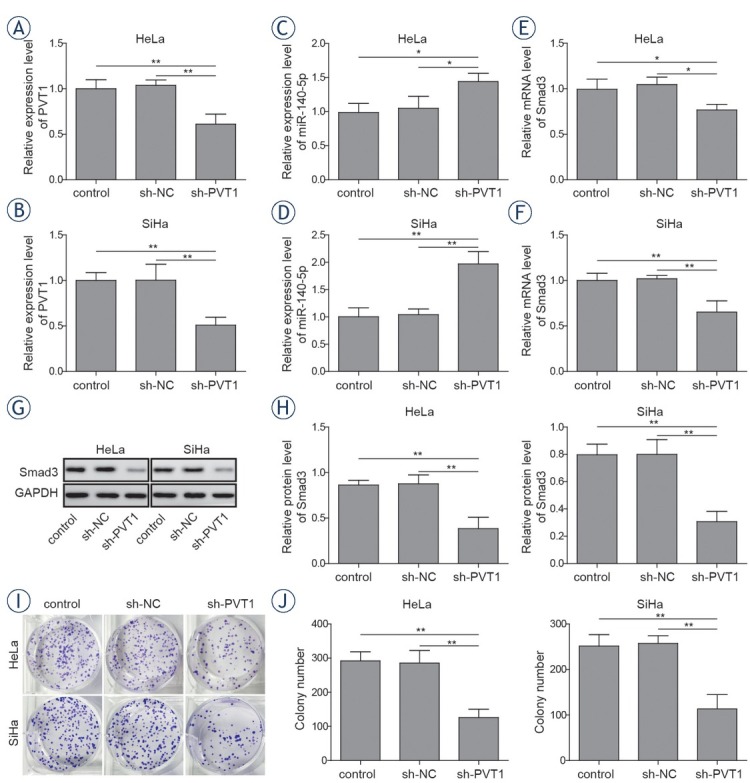
Silencing of lncRNA PVT1 inhibited proliferation of cervical cancer cells. Both HeLa and SiHa cells were transfected with sh-NC or sh-PVT1. Expressions of PVT1 (A, B) and miR-140-5p (C, D) were determined by qRT-PCR. (E-F) mRNA levels and (G, H) proteins levels of Smad3 were determined by qRT-PCR and western blotting respectively. (I, J) Colony formation assays were performed to evaluate the proliferation of both HeLa and SiHa cells. The data are presented as means ± SD of three independent experiments. Statistical significance compared with sh-NC is indicated by *P < 0.05 and **P < 0.01.
reductions in protein levels, respectively (P < 0.01). To explore the role of PVT1 in the proliferation of cervical cancer cells, colony formation assay was performed. As shown in Figure 3I and 3J, PVT1 knockdown significantly inhibit the proliferation of cervical cancer cells compared with the sh-NC group. Next, the migration activities of cervical cancer cells were detected by wound healing assay.
The results showed that PVT1 knockdown could significantly decrease the migration activities of cervical cancer cells compared with the sh-NC groups, with an inhibition rate of 51.2% and 61.3% in both cells lines respectively. (Figure 4A-4D, P < 0.01). Moreover, invasion capacity of cervical cancer cells was further determined by transwell assays. The results showed that PVT1 suppression could remarkably inhibit the invasion capacities of cervical cancer cells compared with the sh-NC groups, as their respective reductions were 59.2% and 61.2%, respectively (Figure 4E-4H, P < 0.01). Besides, expressions of E-cadherin, N-cadherin, vimentin and Snail were determined. The results showed that downregulation of PVT1 lead to significant increase in the expressions of E-cadherin (1.9-fold and 2.1-fold) and significant decrease in the expressions of N-cadherin (41.2% and 37.1%), vimentin (30.1% and 13.6%) and Snail (39.2% and 45%) compared with the sh-NC group (Figure 4I and 4J, P < 0.05). These data indicated that PVT1 could serve as an oncogenic lncRNA in the progression of cervical cancer.
Figure 4.
Silencing of lncRNA PVT1 suppressed the metastasis of cervical cancer cells. Both HeLa and SiHa cells were transfected with sh-NC or sh-PVT1. (A-D) Wound healing assay was conducted to evaluate the migration activities of HeLa and SiHa cells. (E-H) Transwell assay was conducted to evaluate the invasion activities of HeLa and SiHa cells. (I, J) Expressions of E-cadherin, N-cadherin, vimentin and Snail were determined by western blotting in both HeLa and SiHa cells. The data are presented as means ± SD of three independent experiments. Statistical significance compared with sh-NC is indicated by *P < 0.05 and **P < 0.01.
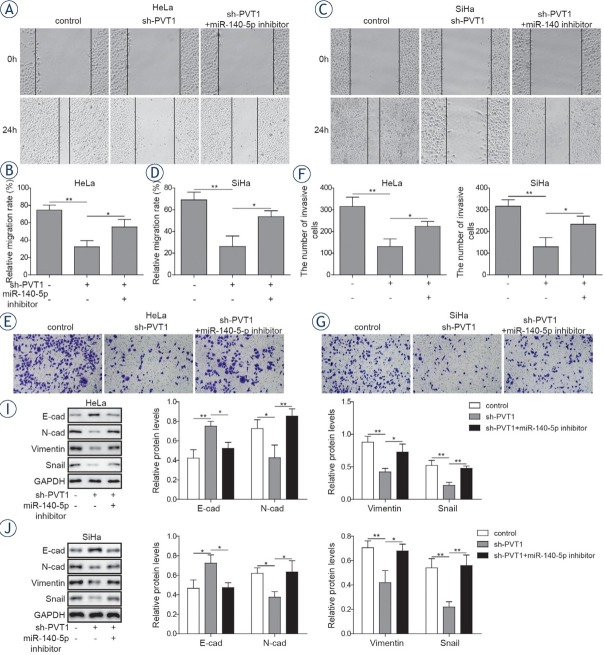
LncRNA PVT1 promoted cervical cancer cells progression via modulating miR-140-5p
To determine whether miR-140-5p was implicated in the effect of lncRNA PVT1 on the progression of cervical cancer cells, sh-PVT1 transfected cervical cancer cells were transfected with or without miR-140-5p inhibitor. The results showed that miR-140-5p inhibitor could decline the PVT1 inhibition-mediated decreasing effect on the Smad3 expression in cervical cancer cells (Figure 5A-5D). Colony formation assay indicated that inhibition of miR-140-5p could reserve the inhibition effects of PVT1
Figure 5.
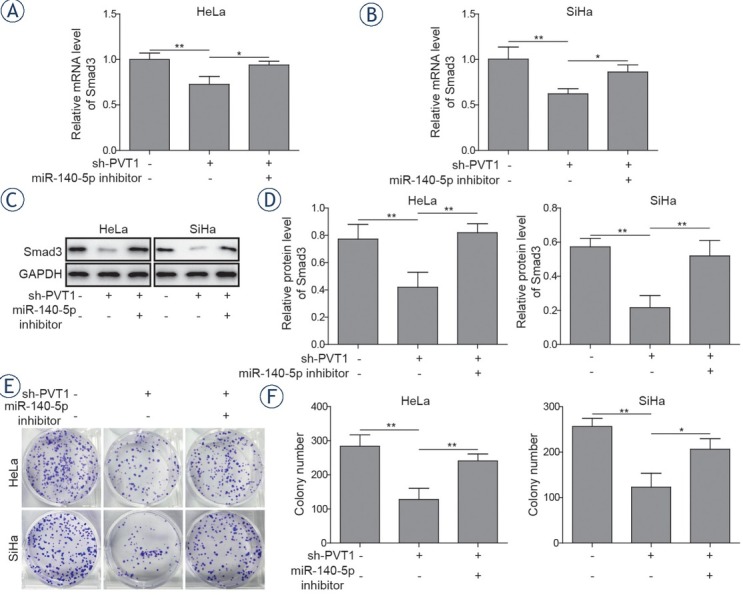
Silencing of miR-140-5p reversed the effects of PVT1 inhibition on the proliferation of cervical cancer cells. Both HeLa and SiHa cells were transfected with or without miR-140-5p inhibitor in the presence of sh-PVT1. (A, B) mRNA expressions and (C, D) protein expressions of Smad3 were determined by qRT-PCR and western blotting in both HeLa and SiHa cells. (E, F) Colony formation assays were performed to evaluate the proliferation of both HeLa and SiHa cells. The data are presented as means ± SD of three independent experiments. Statistical significance compared with sh-PVT1 is indicated by *P < 0.05 and **P < 0.01.
downregulation on proliferation of cervical cancer cells (Figure 5E and 5F). Similar results could also be observed in wound healing and transwell assays, that decreased migration and invasion abilities induced by PVT1 downregulation could be rescued by miR-140-5p inhibitor (Figure 6A-6H). Besides, treatment with miR-140-5p inhibitor could suppress sh-PVT1-induced upregulation of E-cadherin expression and reverse sh-PVT1-induced down-regulation of N-cadherin, vimentin and Snail expressions (Figure 6I-6J). These data indicated that PVT1 knockdown could suppress the progression of cervical cancer cells via miR-140-5p.
Figure 6.
Silencing of miR-140-5p reversed the effects of PVT1 inhibition on the metastasis of cervical cancer cells. (A-D) Wound healing assay was conducted to evaluate the migration activities of HeLa and SiHa cells. (E-H) Transwell assay was conducted to evaluate the invasion activities of HeLa and SiHa cells. (I, J) Expressions of E-cadherin, N-cadherin, vimentin and Snail were determined by western blotting in both HeLa and SiHa cells. The data are presented as means ± SD of three independent experiments. Statistical significance compared with sh-PVT1 is indicated by *P < 0.05 and **P < 0.01.
Discussion
Accumulating evidence has revealed that aberrantly expressed lncRNAs plays crucial role in tumor occurrence, invasion and metastasis including cervical cancer.23 By far, PVT1 is recognized as an oncogene in many cancers, however, the role of PVT1 in the progression of cervical cancer and the potential underlying mechanism by which PVT1 participate in remains uncertain.
Recent studies have shown that PVT1 is upregulated in many kinds of cancers and serves as an oncogenic lncRNA. Zhang et al. have demonstrated that PVT1 could promote the proliferation and invasion by sponging miR-200a in glioma cells.24 Zou et al.25 showed that expressions of PVT1 were significantly elevated in ovarian cancer tissues and cells, and the progression of ovarian cancer cells promoted by PVT1 was mediated by the upregulation of SOX2. Our results also showed that PVT1 expressions were significantly upregulated in cervical cancer cells. Furthermore, knockdown of PVT1 could significantly decrease the proliferation, migration and invasion of cervical cancer cells. These findings showed that PVT1 might act as an oncogene in the progression of cervical cancer, which is in consistent with the previous researchers.26, 27
Despite much progress have been made in the research of lncRNAs, the underlying mechanism remains unclear. Recently, interactions between lncRNAs and miRNAs have proposed that lncR-NAs can serve as a sponge to bind miRNA and regulate the functions of miRNAs, which play a crucial role in the proliferation and tumorigenesis of many cancers.28, 29 So, we further focused on miR-140-5p, due to its critical role in the progression of cervical cancer.12, 13 Our data revealed that expressions of miR-140-5p were significantly downregulated in cervical cancer cells. Inhibition of PVT1 significantly increased the expressions of miR-140-5p in cervical cancer cells. In addition, luciferase assay was further applied to determine the correlation between PVT1 and miR-140-5p. Our results revealed that miR-140-5p upregulation could significantly inhibit the luciferase activities of PVT1-WT but did not affect that of PVT1-MUT, indicating that PVT1 binds to miR-140-5p and regulate its function. To further, clarify the interaction of PVT1 and miR-140-5p during the progression of cervical cancer cells. MiR-140-5p inhibitor were transfected into PVT1 knockdown cervical cancer cells then proliferation, migration and invasion assays of cervical cancer cells were performed. Zhang et al. showed that PVT1 could promote metastasis and proliferation of colon cancer via the inhibiting of miR-26.30 Tian et al. reported that PVT1 could downregulate miR-31 to enhance CDK1 expression thus facilitating the progression of bladder cancer cells.10 Our results illustrate the interaction between PVT1 and miR-140-5p in the progression of cervical cancer cells.
Normally, miRNAs could regulate tumorigenesis by targeting a variety of protein-coding genes.31 Smad3 has been demonstrated increased and could act as an oncogenic role in the progression of many type of cancers. For instance, Yamazaki et al. showed that Smad3 could promote the invasion and migration of pancreatic cancer cells.32 Fan et al. showed that activation of TGFA/Smad3 signal could induce the migration and invasion of cervical cancer cell lines, suggesting that Smad3 plays a vital role in cervical cancer metastasis.33 Our present results also revealed that Smad3 expressions were significantly increased in cervical cancer cells. Besides, bioinformatics analysis showed that Smad3 might be downstream target of miR-140-5p. In addition, the luciferase assay displayed that miR-140-5p could target Smad3 directly. More importantly, our results corroborated that transfection of miR-140-5p inhibitor could partially reverse PVT1 knockdown-induced changes of Smad3 expressions in cervical cancer cells, indicating that miR-140-5p act as a tumor suppressor through the regulation of Smad3.
Collectively, we found that PVT1 is upregulated in cervical cancer cells, and PVT1 might serve as a prognostic indicator for cervical cancer patients.34, 35, 36 Furthermore, PVT1 sponges directly to miR-140-5p and regulates its expression and function. In addition, Smad3 is also upregulated in cervical cancer cells and it is the downstream target of miR-140-5p. Moreover, PVT1 promotes the proliferation, migration and invasion of cervical cancer cells via regulating miR-140-5p and Smad3. Our findings provide new insights into the therapeutic strategy against cervical cancer.
Disclosure
No potential conflicts of interest were disclosed.
Footnotes
Author Contributions
QQC conceived and designed the experiments. CYC guaranteed the integrity of the whole research and made the clinical studies. ZC defined intellectual content, SC did literature research, QQC made data analysis and CYC contributed statistical analysis. All authors were responsible for the editing of the manuscript. All authors read and approved the final manuscript.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel R L, Torre L A, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Pang T, Wang S, Gao M, Kang H, Zhao Y, Yao Y. HPV18 E7 induces the over-transcription of eIF4E gene in cervical cancer. Iran J Basic Med Sci. 2015;18:684–90. et al. PMID: 26351560. [PMC free article] [PubMed] [Google Scholar]
- 3.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–60. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 4.Santin A D, Zhan F, Bignotti E, Siegel E R, Cane S, Bellone S. Gene expression profiles of primary HPV16- and HPV18-infected early stage cervical cancers and normal cervical epithelium: identification of novel candidate molecular markers for cervical cancer diagnosis and therapy. Virology. 2005;331:269–91. doi: 10.1016/j.virol.2004.09.045. et al. [DOI] [PubMed] [Google Scholar]
- 5.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5:10976–96. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun L, Luo H, Liao Q, Bu D, Zhao G, Liu C. Systematic study of human long intergenic non-coding RNAs and their impact on cancer. Sci China Life Sci. 2013;56:324–34. doi: 10.1007/s11427-013-4460-x. et al. [DOI] [PubMed] [Google Scholar]
- 7.Ching T, Peplowska K, Huang S, Zhu X, Shen Y, Molnar J. Pan-cancer analyses reveal long intergenic non-coding RNAs relevant to tumor diagnosis, subtyping and prognosis. EBioMedicine. 2016;7:62–72. doi: 10.1016/j.ebiom.2016.03.023. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang M, Zhai X, Xia B, Wang Y, Lou G. Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumour Biol. 2015;36:7615–22. doi: 10.1007/s13277-015-3465-4. [DOI] [PubMed] [Google Scholar]
- 9.Han D, Wang J, Cheng G. LncRNA NEAT1 enhances the radio-resistance of cervical cancer via miR-193b-3p/CCND1 axis. Oncotarget. 2018;9:2395–409. doi: 10.18632/oncotarget.23416. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Tian Z, Cao S, Li C, Xu M, Wei H, Yang H. LncRNA PVT1 regulates growth, migration, and invasion of bladder cancer by miR-31/ CDK1. J Cell Physiol. 2019;234:4799–4811. doi: 10.1002/jcp.27279. et al. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med. 2018;15:238–250. doi: 10.20892/j.issn.2095-3941.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding Y, Fang Q, Li Y, Wang Y. Amplification of lncRNA PVT1 promotes ovarian cancer proliferation by binding to miR-140. Mamm Genome. 2019 doi: 10.1007/s00335-019-09808-1. [DOI] [PubMed] [Google Scholar]
- 13.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader J S. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One. 2016;11:e0156274. doi: 10.1371/journal.pone.0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong G, Liang X, Wang D, Gao H, Wang L, Wang L. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014;31:57. doi: 10.1007/s12032-014-0057-x. et al. [DOI] [PubMed] [Google Scholar]
- 15.Muhammad S, Tang Q, Wei L, Zhang Q, Wang G, Muhammad B U. miRNA-30d serves a critical function in colorectal cancer initiation, progression and invasion via directly targeting the GNA13 gene. Exp Ther Med. 2019;17:260–272. doi: 10.3892/etm.2018.6902. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma H P, Kong W X, Li X Y, Li W, Zhang Y, Wu Y. miRNA-223 is an anticancer gene in human non-small cell lung cancer through the PI3K/AKT pathway by targeting EGFR. Oncol Rep. 2019 doi: 10.3892/or.2019.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–91. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Z, Yin S, Sun R, Zhang S, Fu M, Wu Y. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16:139. doi: 10.1186/s12943-017-0708-6. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan H, Chen W, He G, Yang S. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed Pharmacother. 2015;75:117–22. doi: 10.1016/j.biopha.2015.07.035. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Xu J. MiR-140-5p regulates hypoxia-mediated human pulmonary artery smooth muscle cell proliferation, apoptosis and differentiation by targeting Dnmt1 and promoting SOD2 expression. Biochem Biophys Res Commun. 2016;473:342–8. doi: 10.1016/j.bbrc.2016.03.116. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Zou K, Yu L, Zhao W, Lu Y, Mao J. MicroRNA-140 inhibits the epithelial-mesenchymal transition and metastasis in colorectal cancer. Mol Ther Nucleic Acids. 2018;10:426–37. doi: 10.1016/j.omtn.2017.12.022. et al. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Dong P, Xiong Y. Hanley S J B. Yue J, Watari H. Musashi-2, a novel onco-protein promoting cervical cancer cell growth and invasion, is negatively regulated by p53-induced miR-143 and miR-107 activation. J Exp Clin Cancer Res. 2017;36:150. doi: 10.1186/s13046-017-0617-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong J, Su M, Chang W, Zhang K, Wu S, Xu T. Long non-coding RNAs on the stage of cervical cancer. Oncol Rep. 2017;38:1923–31. doi: 10.3892/or.2017.5905. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Yang G, Luo Y. Long non-coding RNA PVT1 promotes glioma cell proliferation and invasion by targeting miR-200a. Exp Ther Med. 2019;17:1337–45. doi: 10.3892/etm.2018.7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou M F, Ling J, Wu Q Y, Zhang C X. Long non-coding RNA PVT1 functions as an oncogene in ovarian cancer via upregulating SOX2. Eur Rev Med Pharmacol Sci. 2018;22:7183–8. doi: 10.26355/eurrev_201811_16251. [DOI] [PubMed] [Google Scholar]
- 26.Wan L, Sun M, Liu G J, Wei C C, Zhang EB, Kong R. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15:1082–94. doi: 10.1158/1535-7163.MCT-15-0707. et al. [DOI] [PubMed] [Google Scholar]
- 27.Ping G, Xiong W, Zhang L, Li Y, Zhang Y, Zhao Y. Silencing long noncoding RNA PVT1 inhibits tumorigenesis and cisplatin resistance of colorectal cancer. Am J Transl Res. 2018;10:138–49. PMID: 29423000. [PMC free article] [PubMed] [Google Scholar]
- 28.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Piao H L, Kim B J, Yao F, Han Z, Wang Y. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50:1705–15. doi: 10.1038/s41588-018-0252-3. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R, Li J, Yan X, Jin K, Li W, Liu X. Long noncoding RNA plasmacytoma variant translocation 1 (PVT1) promotes colon cancer progression via endogenous sponging miR-26b. Med Sci Monit. 2018;24:8685–92. doi: 10.12659/MSM.910955. et al. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Huang V. Endogenous miRNAa: miRNA-mediated gene upregulation. Adv Exp Med Biol. 2017;983:65–79. doi: 10.1007/978-981-10-4310-9_5. [DOI] [PubMed] [Google Scholar]
- 32.Yamazaki K, Masugi Y, Effendi K, Tsujikawa H, Hiraoka N, Kitago M. Upregulated SMAD3 promotes epithelial-mesenchymal transition and predicts poor prognosis in pancreatic ductal adenocarcinoma. Lab Invest. 2014;94:683–91. doi: 10.1038/labinvest.2014.53. et al. [DOI] [PubMed] [Google Scholar]
- 33.Fan Q, Qiu M T, Zhu Z, Zhou J H, Chen L, Zhou Y. Twist induces epithelial-mesenchymal transition in cervical carcinogenesis by regulating the TGF-beta/Smad3 signaling pathway. Oncol Rep. 2015;34:1787–94. doi: 10.3892/or.2015.4143. et al. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Xiong D, Ye L, Wang K, Huang L, Mei S. Up-regulated lncRNA XIST contributes to progression of cervical cancer via regulating miR-140-5p and ORC1. Cancer Cell Int. 2019;19:45. doi: 10.1186/s12935-019-0744-y. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su Y, Xiong J, Hu J, Wei X, Zhang X, Rao L. MicroRNA-140-5p targets insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) to suppress cervical cancer growth and metastasis. Oncotarget. 2016;7:68397–411. doi: 10.18632/oncotarget.11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J P, Yang X J, Xiao L, Wang Y. Long noncoding RNA PVT1 as a novel serum biomarker for detection of cervical cancer. Eur Rev Med Pharmacol Sci. 2016;20:3980–6. [PubMed] [Google Scholar]